Abstract
We previously discovered that yliH and yceP are induced in Escherichia coli biofilms (D. Ren, L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood, Appl. Microbiol. Biotechnol. 64:515-524, 2004). Here, it is shown that deletion of yceP (b1060) and yliH (b0836) increases biofilm formation in continuous-flow chambers with minimal glucose medium by increasing biofilm mass (240- to 290-fold), surface coverage (16- to 31-fold), and mean thickness (2,800-fold). To determine the genetic basis of the increase in biofilm formation, we examined the differential gene expression profile in biofilms for both the mutants relative to the wild-type strain in rich medium with glucose and found that 372 to 882 genes were induced and that 76 to 337 were repressed consistently >2-fold (P ≤ 0.05). The increase in biofilm formation was related to differential expression of genes related to stress response (8 to 64 genes) for both mutants, including rpoS and sdiA. More importantly, 42 to 130 genes related to autoinducer 2 cell signaling were also differentially expressed, including gadAB and flgBCEGHIJLMN, as well as signaling through indole, since 17 to 26 indole-related genes were differentially expressed, including phoAER, gltBD, mtr (encodes protein for indole import), and acrEF (encodes proteins for indole export). Increased biofilm formation in the yliH and yceP mutants in LB supplemented with 0.2% glucose (LB glu) occurred through a reduction in extracellular and intracellular indole concentrations in both mutants (50- to 140-fold), and the addition of indole to the culture restored the wild-type biofilm phenotype; hence, indole represses biofilms. Additionally, both mutants regulate biofilms through quorum sensing, since deletion of either yliH or yceP increased extracellular autoinducer 2 concentrations 50-fold when grown in complex medium (most notably in the stationary phase). Both proteins are involved in motility regulation, since YliH (127 amino acids) and YceP (84 amino acids) repressed motility two to sevenfold (P ≤ 0.05) in LB, and YceP repressed motility sevenfold (P ≤ 0.05) in LB glu. Heightened motility in the yceP mutant occurred, due to increased transcription of the flagella and motility loci, including fliC, motA, and qseB (3- to 86-fold). We propose new names for these two loci: bssR for yliH and bssS for yceP, based on the phrase “regulator of biofilm through signal secretion.”
Bacteria spend the majority of their existence in biofilm communities (9) attached to a solid or a liquid (6, 33) in an extracellular matrix (10). The study of temporal biofilm development has shown that biofilm formation involves many regulatory mechanisms (15, 45), and DNA microarrays have been used to determine global gene expression patterns in Escherichia coli biofilms (4, 34, 42) and to determine how to control biofilms with plant-derived inhibitors (37). It has been discerned that sulfur metabolism impacts E. coli biofilm formation through CysB and CysE (37, 49), that stress responses (e.g., RpoS, RpoE, SoxS, RecA, and DnaK) influence biofilm formation (4, 8, 34, 42), that carbon flux plays an important part in biofilms (25, 29), and that glucose represses biofilm formation in six E. coli K-12 strains (24). Carbon catabolism genes were differentially expressed in all E. coli biofilm DNA microarray studies (4, 34, 42). Specifically, Schembri et al. (42) reported that 124 carbon compound catabolism genes were differentially expressed, including slp; Beloin et al. (4) found that 16 carbohydrate transport and metabolism genes were differentially expressed, including lamB and rbsB; and Ren et al. (34) noted that 4 carbon metabolism genes were differentially expressed (lacZ, hyaA, furK, and gatD).
Autoinducer 2 (AI-2) and the stationary-phase signals affect gene expression in Escherichia coli (13, 35, 36), and cell signaling is important for biofilm formation (11, 14, 17). Specifically, AI-2 directly stimulates biofilms for cells that lack a conjugation plasmid in E. coli (17), and AI-1 is important for biofilm differentiation in Pseudomonas aeruginosa (11). LuxS synthase forms AI-2 (41), LsrACDB imports AI-2 (59), YdgG (TqsA) exports AI-2 (21), and LsrK phosphorylates AI-2 (59). AI-2 concentrations in suspension are highest during the exponential phase but decrease in the stationary phase (59), and a stationary-phase signal has been shown to repress AI-2 concentrations (36).
Indole is another putative extracellular signal (55) which is also involved in biofilm formation in E. coli (14) and is prevalent in the stationary phase (36). Indole is formed from tryptophan by tryptophanase (TnaA) (55), is exported by AcrEF (27), and is imported by Mtr (60).
Catabolite repression is an important regulatory mechanism involved in both the synthesis and uptake of AI-2 (59), as well as the synthesis of indole (55). Cyclic AMP (cAMP) and the cAMP receptor protein (CRP) (56) activate AI-2 uptake via lsrACDB (56) and indole synthesis via tnaAB (55) and inhibit AI-2 synthesis via luxS (56). Further, cAMP-independent catabolite repression through RpoS (σS; stationary-phase regulator) (20) negatively regulates AI-2 uptake (56).
In E. coli, catabolite repression is mediated in part through the cAMP-CRP complex (5). The presence of glucose interferes with the activation of membrane-bound adenylate cyclase (Cya), decreases cAMP levels in the cell, and represses the expression of CRP (23). In the absence of glucose, cAMP binds to CRP and forms a complex that regulates transcription by binding to the promoter regions of cAMP-regulated genes (24).
Alternatively, catabolite repression can also be regulated through cAMP-independent pathways; CsrA, the carbon storage regulator, represses metabolic pathways including glycogen biosynthesis and gluconeogenesis (39). It also represses biofilm formation and activates biofilm dispersal in E. coli (25). Similarly, the global regulator Crc protein (catabolite repression control) (57) of P. aeruginosa activates biofilm formation in the same strain (29).
We have found that differential gene expression in biofilms allows the identification of genes that are involved in biofilm formation (17, 18, 21, 34), although altered expression does not prove that a gene will in turn regulate biofilms. In all three E. coli biofilm reports (4, 34, 42), yceP was induced in biofilms (3- to 12-fold). Two of the three reports observed that yliH was induced (3- to 28-fold) relative to exponential-growth planktonic cultures (34, 42). However, both YliH and YceP are uncharacterized proteins; YliH (127 amino acids; B0836) is a putative receptor that is induced in the stationary phase (43), and YceP (84 amino acids; B1060) is a conserved hypothetical protein (44). The goals of this study were to confirm that YliH and YceP control biofilm formation in E. coli, to investigate their impact on biofilm architecture, and to investigate their mechanism. By studying the differential gene expression in biofilms of the two isogenic mutants relative to the wild-type strain, we determined that these two proteins play a role in catabolite repression, through which synthesis of the putative stationary-phase signal indole, quorum sensing, and stress response are regulated.
MATERIALS AND METHODS
Bacterial strains and growth media.
The E. coli strains and plasmids are listed in Table 1. Wild-type E. coli K-12 BW25113 (Yale University Coli Genetic Stock Center) was used to compare the isogenic E. coli K-12 BW25113 ΔyliH, ΔyceP, and ΔcreC deletion mutants (Genome Analysis Project in Japan) (1). The kanamycin (Kan) gene insertion and the deletions were corroborated with five primers each in PCRs (three primers homologous to sequences internal to the Kan gene and two primers homologous to the sequence upstream and downstream of each deletion) (1). Biofilm flow chamber experiments were performed using strains harboring the green fluorescent protein plasmid pCM18 (19) to visualize the biofilm in the flow cell experiments. To ensure that the deletion of yliH was responsible for both the motility and biofilm phenotypes, plasmid pCA24N yliH+ (1) was used, which expresses yliH+ under tight control of the lacIq repressor; hence, it was induced by isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma, St. Louis, Mo.) (0 to 0.50 mM in Luria-Bertani medium [LB] for the motility complementation experiment and 0. to 1.25 mM in LB supplemented with 0.2% glucose [LB glu] for the biofilm complementation experiment).
TABLE 1.
E. coli strains and plasmids used
| Strain or plasmid | Genotypea | Source or reference |
|---|---|---|
| Strains | ||
| K-12 BW25113 | lacIq ΔlacZWJ16 ΔaraBADAH33 ΔrhaBADLD78hsdR514 rrnBT14 | Yale CGSG Stock Center |
| K-12 BW25113 ΔyliH | Kmr; K-12 ΔyliH Ω | 1 |
| K-12 BW25113 ΔyceP | Kmr; K-12 ΔyceP Ω | 1 |
| K-12 BW25113 ΔcreC | Kmr; K-12 ΔcreC Ω | 1 |
| V. harveyi BB170 | Lacking AI-1 sensor and with AI-2 sensor | 50 |
| Plasmids | ||
| pCA24N | Cmr; lacIq | 1 |
| pCA24N yliH+ | Cmr; pCA24N pT5/lac::yliH+ | 1 |
| pCM18 | Emr; pTRKL2-PCP25RBSII-gfp3*-To-T1 | 19 |
| pVS159 | Ampr; qseB::lacZ in pRS551 | 47 |
| pVS176 | Ampr; motA::lacZ in pRS551 | 47 |
| pVS175 | Ampr; fliC::lacZ in pRS551 | 47 |
| pVS183 | Ampr; fliAehK-12::lacZ in pRS551 | 47 |
| pVS182 | Ampr; flhD::lacZ in pRS551 | 47 |
| pLW11 | Ampr; lsrACDBFG::lacZ in pFZY1 | 56 |
Ampr, Kmr, Cmr, and Emr are ampicillin, kanamycin, chloramphenicol, and erythromycin resistance, respectively.
LB (40), M9 minimal medium supplemented with 0.4% Casamino Acids (M9C) (38), LB glu, and M9C supplemented with 0.4% glucose (M9C glu) were used to study biofilm formation in the 96-well biofilm assay. M9C glu was used in the biofilm flow chamber experiment. For plasmid selection, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 30 μg/ml chloramphenicol, or 300 μg/ml erythromycin was used.
Ninety-six-well biofilm assay.
Biofilm formation was quantified in 96-well polystyrene plates as reported previously (37). Biofilm formation of E. coli wild-type and mutant strains yceP and yliH was measured in LB, LB glu, M9C, and M9C glu. Ten to 20 replicate wells were averaged to obtain each data point. Three independent cultures were used.
Flow cell biofilm experiments and image analysis.
M9C glu medium supplemented with 300 μg/ml erythromycin to maintain plasmid pCM18 (19) was used to form biofilms at 37°C in a continuous flow cell (BST model FC81; Biosurface Technologies Corp., Bozeman, MT). The flow cell contains a standard glass microscope slide on one side and a plastic coverslip on the other side, and the flow channel has dimensions of 47.5 mm by 12.7 mm with a 1.6-mm gap between the glass plates. The constitutive green fluorescent protein plasmid pCM18 allowed visualization of the biofilm with a TCS SP2 scanning confocal laser microscope with a 40× N PLAN L dry objective with correction collar and a numerical aperture of 0.55 (Leica Microsystems, Heidelberg, Germany). Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 and used to inoculate the flow chamber for 2 h at 10 ml/h; this provided laminar flow (Reynolds number of 0.526) to establish the biofilm (5.7 × 107 cells/ml for E. coli ΔyceP/pCM18, 4.55 × 107 cells/ml for E. coli ΔyliH/pCM18, and 2.5 × 107 cells/ml for the wild-type strain were used to inoculate each of the flow chamber experiments). Fresh medium was then introduced at the same flow rate and circulated for 24 h.
Color confocal flow cell images were converted to gray scale with an Image Converter (Neomesh Microsystems, Wainuiomata, Wellington, New Zealand). Biomass (measured as cubic micrometers per square micrometer), substratum coverage (measured as a percentage), surface roughness, and mean thickness (measured in micrometers) were determined using COMSTAT image-processing software (22), written as a script in Matlab 5.1 (The MathWorks) equipped with the Image Processing Toolbox. Thresholding was fixed for all image stacks, and 25 to 50 planar images were processed for each of nine positions. Nine positions with 25 images each were obtained for the E. coli K-12 wild-type strain biofilm, but only three positions were analyzed for this strain, since only scattered biofilm was observed, and 6 of the image stacks obtained did not show enough biofilm to be analyzed by the COMSTAT software at the set threshold value. Values are means of data from the different positions at the same time point, and standard deviations were calculated based on these mean values for each position. Simulated three-dimensional images were obtained using IMARIS (Bitplane, Zurich, Switzerland).
Motility assay and growth rate measurement.
LB and LB glu overnight cultures were used to assay motility as described previously (18, 47) (motility plates with and without 0.2% glucose were used). Between five and eight plates were used to evaluate motility for each independent culture. Three independent cultures were tested for each strain.
The specific growth rates of E. coli K-12 and its mutants were determined in LB medium using turbidity (OD600) and calculated using the linear portion of the natural logarithm of OD600 versus time (OD600, from 0.05 to 0.7). Three independent cultures were used to measure the growth rates for each strain.
Promoter transcriptional assays.
E. coli strains with the flhD::lacZ, fliA::lacZ, fliC::lacZ, motA::lacZ, and qseB::lacZ fusions (via plasmids pVS182, pVS183, pVS175, pVS176, and pVS159, respectively) (Table 1) (47), as well as the lsrACDFG::lacZ fusion via plasmid pLW11 (56), were cultured overnight in LB and LB glu with ampicillin (100 μg/ml) and kanamycin (50 μg/ml, for the deletion strains), diluted 1:100 in LB and LB glu media (both supplemented with 100 μg/ml ampicillin) to create exponentially growing cells that were harvested at an OD600 value of ∼1 (exponential phase) and an OD600 value of ∼6 (stationary phase), and assayed as reported previously (58). The β-galactosidase activities were calculated, based on a protein concentration of 0.24 mg protein/ml/OD600 unit (53). Each experiment was repeated with a second independent culture.
AI-2 activity Vibrio harveyi assay.
E. coli cell-free culture fluids were tested for the presence of extracellular AI-2 by recording luminescence in the Vibrio harveyi BB170 reporter strain, as described previously (36). Luminescence readings were taken using a Turner Design 20/20 luminometer (Sunnyvale, Calif.) and were reported as relative light units per cell.
Indole assay.
Extracellular and intracellular indole was measured in the E. coli wild-type strain and its yliH and yceP mutants, based on a modified protocol by Kawamura-Sato et al. (27). The strains were first cultured overnight in LB and LB glu with kanamycin (50 μg/ml) for the deletion strains. The cultures were then diluted to a starting OD600 value of ∼0.1 in fresh medium, and indole concentrations were measured at 4 and 24 h. To measure extracellular indole, 5 ml of cell-free culture fluid was mixed for 2 min with 2 ml of Kovac's reagent (10 g of p-dimethylaminobenzaldehyde, 50 ml of HCl, and 150 ml of amyl alcohol), 100 μl of the reaction mixture was diluted in 900 μl of HCl-amyl alcohol solution (50 ml of HCl and 150 ml of amyl alcohol), and the absorbance at 540 nm was measured; concentrations were calculated based on the calibration curve. To measure intracellular indole, cells were harvested from 20 ml of culture, washed, resuspended in 5 ml of cold LB medium, and sonicated at a power level of 15 W for 1 min (four intervals of 15 s each; Fisher Scientific model 60 Sonic Dismembrator); the cellular debris was then removed by centrifugation (10,000 × g) for 1 min, and the concentration of indole in supernatant was determined as described above.
Microarray analysis.
To isolate RNA from the biofilm cells, E. coli wild-type K-12 BW25113 and the isogenic strains with the yceP and yliH mutations were cultured overnight in LB glu medium (with kanamycin added to the two knockout mutants). The overnight cultures were then used to inoculate 250 ml fresh LB glu medium with 10 g untreated glass wool (Corning Glass Works, Corning, NY). The cells were incubated for 24 h at 37°C and 250 rpm, forming a biofilm on the surface of the glass wool. The glass wool was then quickly removed from the reactor (<30 s) and gently washed two times in 100 ml of 0.85% NaCl buffer at 0°C. Biofilm cells were removed from glass wool by sonication for 2 min in 200 ml of 0.85% NaCl buffer at 0°C, and RNA was isolated as described previously (34) with the RNeasy kit (QIAGEN). To inhibit RNase and ensure high-quality RNA, two agents were used: (i) beta-mercaptoethanol, which acts as a reducing agent to irreversibly denature RNases, and (ii) guanidinium isothiocyanate contained in the RLT buffer (RNA Lysis Tissue, RNeasy mini kit; QIAGEN), which is a strong but temporary RNase-denaturing agent. In addition, to determine the impact of glucose on gene expression in the biofilms and suspension cells, the wild-type strain was cultured in both LB and LB glu with glass wool, and RNA from the wild-type suspension cells biofilm cells was isolated (suspension and biofilm cells were harvested from the same reactor).
The E. coli GeneChip antisense genome array (part no. 900381; Affymetrix), which contains probe sets for all 4,290 open reading frames, rRNA, tRNA, and 1,350 intergenic regions, was used. The procedures are described in the Gene Expression Technical Manual (June 2004 update). Using the GeneChip operating software (Affymetrix), individual strain reports for both the wild-type strain and mutant cDNA samples were obtained, as well as comparison reports of the yliH and yceP mutants to wild-type E. coli. Total cell intensity was scaled automatically in the software to an average value of 500. To ensure the reliability of the induced and repressed gene list, genes were identified as differentially expressed if the P value was <0.05 and if the expression ratio was >2-fold for all genes, since the standard deviation for the expression ratio for the genes in the two data sets was 2 and 4. Additionally, since comparisons were made with two separate wild-type biofilm samples, only those genes which were induced or repressed consistently with respect to both samples were chosen for further study.
The data quality was assessed based on the manufacturer's guidelines (GeneChip Expression Analysis: Data Analysis Fundamentals; Affymetrix). In addition, the expected signals based on the E. coli K-12 BW25113 genotype (Table 1) were obtained (e.g., a low signal for deleted genes araA and rhaA and no signals for yliH and yceP in their respective experiments).
Statistical analysis.
The differences observed between the wild-type strain and yliH and yceP mutants in biofilm formation, motility, and transcription of the motility genes were determined to be significant by using analysis of variance. The wild-type strain and yliH and yceP mutants were fixed factors, and the independent cultures for each bacterial strain were the random factors nested within the bacterial strain. Furthermore, the number of repetitions varied from culture to culture, which led to an unbalanced data set. In summary, the model is nested, unbalanced, and mixed with the following form:
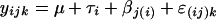 |
where yijk is the observed multivariate value (biofilm, motility, etc.) for bacterium i, culture j, and observation k. The experimental factors were as follows: bacterial strain, τi (treatment effect of bacterial strain i, with i = 1, 2, and 3 where 1 is the wild type, 2 is the ΔyliH mutant, and 3 is the ΔyceP mutant), culture βj(i) (random culture j nested within the bacterial strain i, with j = 1, 2, and 3), overall mean μ (mean value of the experiment), and random error ɛ(ij)k (error term for observation k in bacterium i and culture j). The assumptions for the parameters are Στi = 0, βj(i) ≈ N(0,Σβ), and ɛ(ij)k ≈ N(0, Σɛ), where Σβ and Σɛ are the covariance matrix with nonzero diagonal elements and zero elsewhere. The P values were calculated based on this model with a critical P value of ≤0.05 considered significant. Statistical analysis was performed using SAS software.
Microarray accession numbers.
The expression data for biofilm samples of the E. coli wild type and yliH and yceP mutants have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession number GSE3937 (2, 16).
RESULTS
YceP and YliH and biofilm formation.
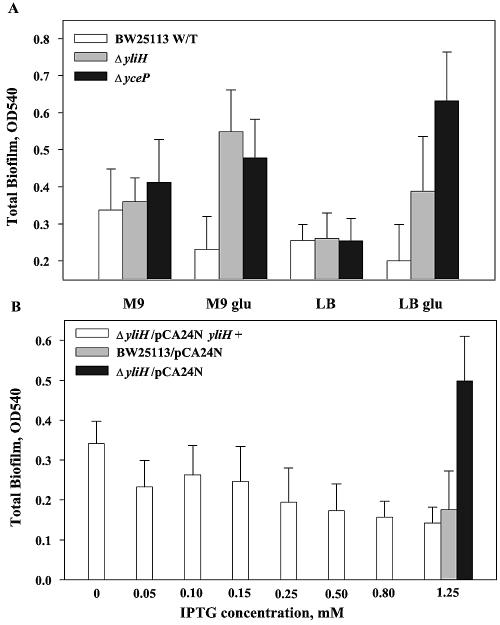
We focused on E. coli biofilm formation, since this is the most thoroughly studied bacterium; hence, many pathways in this organism have been described, and many isogenic mutants are available (26). To confirm the microarray results from previous studies which indicated that YliH and YceP influence biofilm formation (4, 34, 42), 96-well plates were used to quantify biofilm formation in LB, LB glu, M9C, and M9C glu media. Culturing of the mutants and wild-type strain in LB did not lead to a variation in observed biofilm formation (Fig. 1A), while twofold (P = 0.052) and threefold (P = 0.001) increases in biofilm occurred in LB glu upon deletion of yliH and yceP, respectively (Fig. 1A); hence, these mutations increased biofilm formation when glucose was present in the medium and this behavior made the mutations interesting. Similarly, adding glucose to minimal medium (M9C glu) and deleting yliH or yceP resulted in a roughly twofold (P ≤ 0.0003) increase in biofilm, while no significant changes in biofilm formation were observed in experiments conducted with M9C only (Fig. 1A); therefore, these two loci influence E. coli biofilm formation as predicted by the DNA microarrays. The deletions of genes yceP and yliH did not affect the growth of the strains, as shown by the specific growth rates of the strains measured in inverse hours (Table 2).
FIG. 1.
Effects of deleting yliH and yceP on wild-type (W/T) E. coli biofilm formation in microtiter plates in LB, LB glu, M9C, and M9C glu media (A); complementation of yliH using pCA24N yliH+ in LB glu medium (B). The biomass was measured after 24 h. Experiments were performed in triplicate.
TABLE 2.
Specific growth rates of strains in LB media and COMSTAT analysis from the flow cell using M9C glu (after 24 h)
| Strain | Growth rate in LB (h−1) | Biomass (μm3/μm2) | Substratum coverage (%) | Mean thickness (μm) | Roughness coefficient |
|---|---|---|---|---|---|
| K-12 BW25113 | 1.52 ± 0.02 | 0.09 ± 0.04 | 3.1 ± 0.2 | 0.01 ± 0.01 | 2.0 ± 0.0 |
| K-12 BW25113 ΔyceP | 1.42 ± 0.06 | 21 ± 16 | 49 ± 24 | 33 ± 27 | 0.7 ± 0.4 |
| K-12 BW25113 ΔyliH | 1.63 ± 0.03 | 25 ± 8 | 94 ± 8 | 32 ± 21 | 0.24 ± 0.1 |
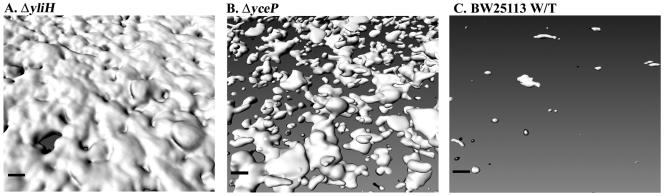
To study the influence of these deletions on biofilm architecture, a continuous-flow system was used with M9C glu medium (Fig. 2). Both mutants showed significant increases in total biofilm relative to the wild-type strain, with the most dramatic change seen in the yliH mutant, which formed a particularly thick biofilm, covering the whole surface of the slide (Fig. 2A). For E. coli ΔyliH, the biomass was 290-fold larger than that of the wild-type strain, thickness increased 2,700-fold, and surface coverage increased 31-fold (Table 2). E. coli ΔyceP had 240-fold-greater biomass, 2,800-fold-greater average thickness, and 16-fold-more surface coverage than wild-type BW25113 (Fig. 2B). Hence, the wild-type strain formed a very poor biofilm in this medium (characterized by a few isolated cells that covered <4% of the total observable surface) (Fig. 2C), while the mutations yliH and yceP caused the cells to colonize more than half of the entire surface provided. The yliH mutation caused the cells to form a wall of biofilm covering over 90% of the viewable area, which was comprised of dense, smooth masses with an undulating texture (Fig. 2A). The yceP deletion caused the cells to form a more random, scattered, and globular biofilm with far less surface coverage (Fig. 2B).
FIG. 2.
Effects of deleting yliH (A) and yceP (B) on wild-type E. coli biofilm formation (C) in flow chambers with M9C glu medium after 24 h. Images were analyzed with IMARIS, and the scale bar is 10 μm.
YceP and YliH and motility.
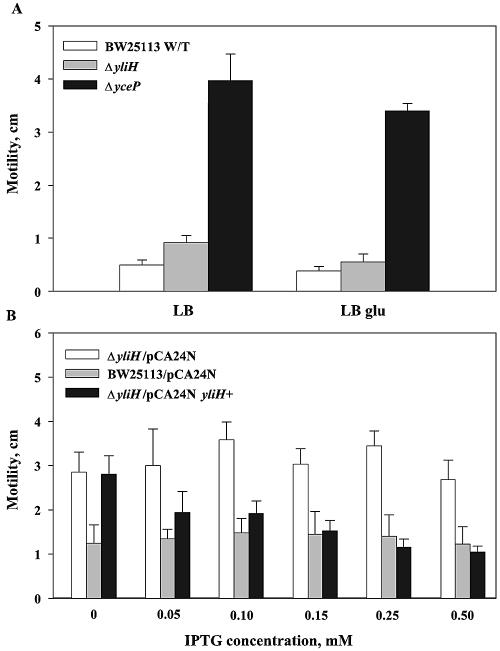
To investigate whether motility was involved in the dramatically altered biofilms, we measured swimming for the two mutant strains and determined that deleting yliH and yceP led to a two- and sevenfold (P ≤ 0.04) increase in motility, respectively, relative to that for the wild-type strain in LB medium (Fig. 3A). When cultured in LB glu, the yceP mutant retained this phenotype (ninefold increase relative to the wild-type strain) (P = 0.001), while the motility of the ΔyliH mutant was inhibited by the addition of glucose. The two mutations affected cell motility, but this phenotype was not directly linked with biofilms, since increased motility but not increased biofilm formation was observed when LB cultures were used.
FIG. 3.
Effects of deleting yliH and yceP on the motility (after 8 h) of wild-type E. coli in LB and LB glu (A) and motility (after 24 h) while complementing yliH in trans using pCA24N yliH+ in LB (B). IPTG had no affect on the negative controls E. coli K-12 BW25113 ΔyliH/pCA24N and E. coli K-12 BW25113/pCA24N. Each experiment was done in duplicate.
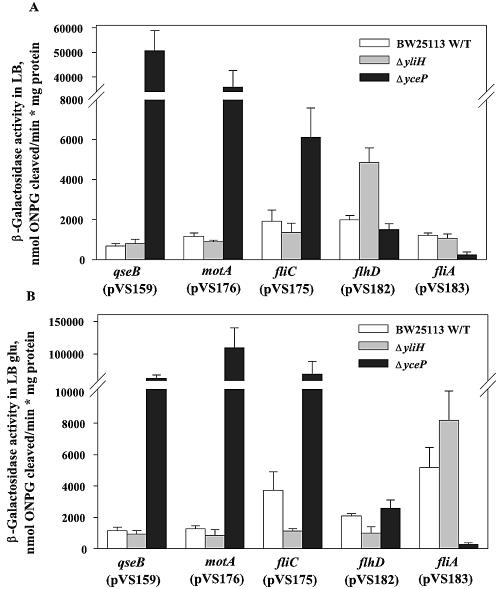
To determine the genetic mechanism of the altered motility, lacZ transcriptional fusions were used to measure transcription of the promoters of the motility and flagellar genes flhD, fliA, fliC, motA, and qseB (47); these genes encode proteins for the synthesis and motion of flagella in E. coli (7). The assay was performed with both LB and LB glu using suspension cells (Fig. 4). For the yceP mutant, there was a dramatic increase in the transcription of qseB (75-fold in LB and 54-fold in LB glu) (P ≤ 0.001), motA (31-fold in LB and 86-fold in LB glu (P ≤ 0.004), and fliC (3-fold in LB and 19-fold LB glu) (P ≤ 0.01) relative to the wild-type strain in suspension cells, which was consistent with the observed increase in motility. These results were also confirmed by the microarray experiment for the biofilms, which showed that 16 flagellum- and motility-related genes were induced >2-fold (e.g., flhDC, motAB, cheRZ, and fliACDSJMNOQR).
FIG. 4.
Effects of deleting yliH and yceP on the transcription of qseB::lacZ, motA::lacZ, fliC::lacZ, flhD::lacZ, and fliA::lacZ in LB (A) and LB glu (B). The assay was performed with exponential-phase cells (OD600, ∼0.5), and each experiment was repeated in triplicate.
For the yliH mutant suspension cells, only flhD was induced twofold (P ≤ 0.011) relative to the wild-type strain in LB medium (Fig. 4). In LB glu medium, flhD and fliC were repressed two- and threefold (P ≤ 0.02), which is consistent with the observed reduction in the motility of the yliH mutant when it was cultured in LB glu. The microarray data for biofilm cells in LB glu showed an opposite result, with 26 flagellum- and motility-related genes induced >2-fold (P ≤ 0.05), including qseAB, flhD, fliA, and motAB, as well as cheABZ, fliDSTGHIJKMNOPQRE, and flgMG. This result is not surprising, since it was previously shown that significant differences exist between gene expression patterns observed with biofilm cells (used here for the microarray experiments) and suspension cells (used in the transcription assay). Additionally, yliH was reported to be induced 1,000-fold in the stationary phase versus the exponential phase (43); therefore, its deletion is more likely to play a role in the biofilm rather than suspension cells.
Complementation of motility and biofilm formation.
To show conclusively that the yliH gene is responsible for the altered motility phenotype in LB, YliH was added in trans using the complementation plasmid pCA24N yliH+. Increasing transcription of yliH+ by varying the IPTG concentration from 0 to 0.50 mM led to a gradual decrease in motility relative to that of the wild-type strain (Fig. 3B); hence, YliH represses motility in LB. The motility of the negative control strain E. coli ΔyliH/pCA24N was not altered by the addition of IPTG.
Biofilm formation was also complemented in LB glu medium by increasing the IPTG concentration from 0 to 1.25 mM (Fig. 1B). This led to a gradual decrease in total biofilm, reducing it to that of the wild-type strain; hence, YliH represses biofilm formation in LB glu. The biofilm of the control strains was measured at the highest IPTG concentration to ensure that the plasmid was not affected by IPTG addition.
Genes regulated by YliH and YceP.
Comparing the differential gene expression in the biofilms of the yliH and yceP mutant strains relative to the biofilm of the wild-type strain revealed that YliH and YceP controlled a large number of genes. Two different microarray data sets were compared, and only the genes consistently induced or repressed in both sets are reported. The results showed that 882 genes were induced and 337 genes were repressed consistently >2-fold (P ≤ 0.05) in the yliH mutant relative to the wild-type strain (see Table S1 in the supplemental material). Similarly, 372 genes were induced and 76 genes were repressed consistently >2-fold (P ≤ 0.05) in the yceP mutant relative to the wild-type strain (see Table S2 in the supplemental material).
The genes affected by the mutations were categorized into 19 main groups based on the EcoCyc Database (28) and the Clusters of Orthologous Groups database (54); the main groups affected for the yliH and yceP mutants, based on changes of ≥2-fold, are translation, ribosomal structure, biogenesis (99 and 35 genes, respectively, including many ribosomal proteins), transcription (84 and 35 genes, respectively, including induced alpA and sdiA and repressed rpoAS), cell envelope biogenesis and outer membrane (41 and 21 genes, respectively, including induced acrE, agaS, and wcaAE), cell motility (51 and 16 genes were induced, respectively, including motB, fimD, and yhcA in both data sets), energy production and conversion (85 and 31 genes, respectively, with succinate- and formate-related genes repressed and fumarase- and hydrogenase-related genes induced in both sets), carbohydrate transport and metabolism (99 and 30 genes, respectively, with agaBCDIWZ genes and yihN induced in both sets and gatYZ repressed in the yliH mutant), amino acid transport and metabolism (71 and 26 genes, respectively, including astAB and gabABPT genes repressed for both), inorganic ion transport and metabolism (43 and 11 genes, respectively, with b1995 and ygaP most induced in both sets), and genes with predicted or unknown functions (404 and 148 genes, respectively).
In addition, several operons previously identified to be important to biofilm formation were induced in both of the mutants relative to the wild-type strain, including eight fimbria-related operons (e.g., yadCKM and yehABCD), two curli production loci (csgGFED and csgAB), five polysaccharide metabolism operons (e.g., yipS and wcaFEDCBA), and seven flagellum- and motility-related operons.
YceP and YliH regulate indole transport.
Genes related to the stationary-phase signal indole (3, 36) were also altered in the biofilms. The yliH mutation affected 26 of these signaling genes (16 induced and 10 repressed), and the yceP mutation altered expression of 17 of these genes (11 induced and 6 repressed). These genes include the stationary-phase signal-related phoAE related to phosphate (induced two- to sixfold in both mutants) (36).
Genes encoding proteins involved in the export of indole were induced in both the yliH and yceP mutant biofilms, since acrE was induced about 20-fold, and acrF was induced 4- to 7- fold. Additionally, the gene encoding a protein involved in the uptake of indole, mtr, was repressed 2.5-fold in both mutants. Hence, the enhanced biofilms by both mutants seem related to the reduced levels of intracellular indole. The genes for proteins involved in indole synthesis, tnaA and tnaB, were only slightly induced (<3-fold), and genes previously reported as induced by indole (3) were repressed by the deletion of yliH and yceP, including astD (2- to 4-fold) and gabT (4- to 8-fold), again supporting lower intracellular indole levels. These expression ratios indicate that both YliH and YceP may repress indole export and induce indole import and that yliH and yceP mutations in these genes may lead to decreased intracellular indole concentrations when cultured in LB glu.
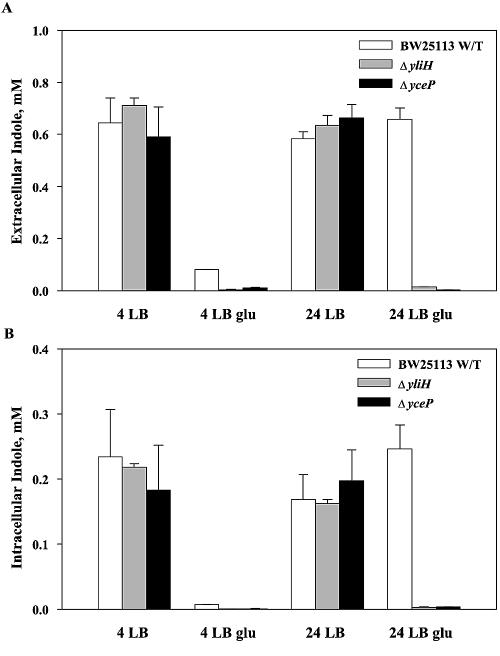
To test this hypothesis, we measured the extracellular (see Fig. 6A) and intracellular (see Fig. 6B) indole concentrations of the E. coli wild-type strain and its yliH and yceP mutants in both LB and LB glu medium. Our results showed that both mutant strains were deficient in intracellular and extracellular indole when grown in LB glu (see Fig. 6) but retained the same indole concentrations as the wild-type strain when grown in LB. Hence, the increased biofilm formation with the two mutants corresponded to reduced intracellular and extracellular indole concentrations.
FIG. 6.
Extracellular (A) and intracellular (B) indole concentrations, measured 4 h and 24 h after inoculation. Experiments were performed in duplicate.
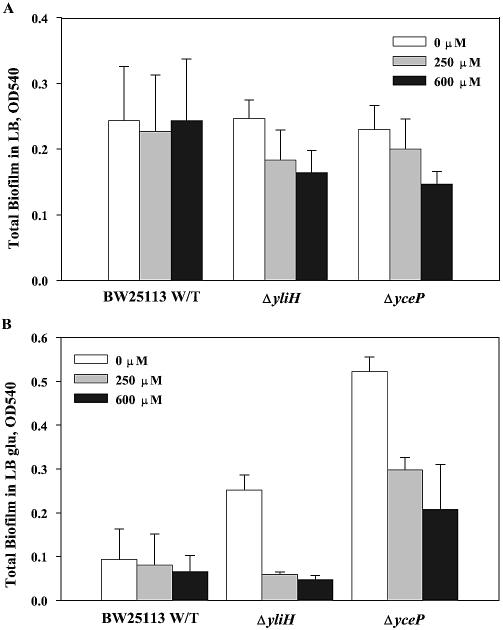
In addition, we tested the biofilm formation of these strains in 96-well polystyrene plates with the addition of exogenous indole in LB glu and found that the wild-type biofilm phenotype was restored by the addition of indole to the culture medium (see Fig. 7). Indole was not toxic to the strains at these concentrations (data not shown) and had no effect on the biofilm formation of the wild-type strain. This confirms that indole represses biofilms and that yliH and yceP mutants increase biofilm formation by repressing indole concentrations through a catabolite repression-related process. It should be noted that adding exogenous indole has previously been shown to induce biofilm formation in E. coli (14), and inactivation of tnaA was associated with the absence of indole production and decreased biofilm formation (14), so our results disagree with this initial result.
FIG. 7.
Effects of indole on biofilm formation of yliH and yceP mutants and wild-type E. coli in LB (A) and LB glu (B). The biomass was measured after 24 h. Experiments were performed in triplicate.
YceP and YliH regulate AI-2 uptake and processing.
Based on the microarray data from 24-h biofilms, the yliH mutation affected the expression of 130 genes (84 genes induced and 46 genes repressed) by >2-fold (P ≤ 0.05), which are affected by quorum sensing via AI-2 (13, 35, 46); deletion of yceP affected 42 genes (32 induced and 10 repressed) by >2-fold (P ≤ 0.05) (see Tables S1 and S2 in the supplemental material). For example, the AI-2-regulated central intermediary metabolism genes gadAB previously identified by our group (35) were repressed 24- and 8-fold in the yliH mutant, and the AI-2-regulated flagellum-related genes flgB, flgH, flgM, and flgN were induced up to 13-fold in both mutants (35). Additionally 99% of the genes known to be induced by AI-2 were also induced in the yceP mutant, and 77% of genes known to be repressed by AI-2 were also repressed in this mutant. Similarly, 77% of AI-2-induced genes were induced in the yliH mutant, and 93% of genes repressed by AI-2 were repressed in this mutant. It is interesting that 22 of the genes known to be AI-2 regulated were stress response related, which means there is a strong correlation between the genes affected by the yliH and yceP mutations, quorum sensing, and stress response.
We further analyzed the microarray data (LB glu) for genes encoding proteins involved with AI-2 synthesis (luxS) and uptake (lsrACDB), as well as lsrK encoding LsrK kinase, which acts to phosphorylate AI-2 once inside the cell (52, 59). The microarray results with LB glu indicated that lsrA was induced 11- and 6-fold and lsrD was induced 5- and 4-fold for the yliH and yceP mutants relative to the wild type in the biofilm cells, leading to the hypothesis that 24-h biofilm cells internalize AI-2. Additionally, lsrK was induced 2- to 2.6-fold more, supporting our hypothesis of increased intracellular AI-2 levels in the yliH and yceP mutant biofilm cells. The mutations did not impact the export of AI-2 via YdgG (21). Since we have shown that addition of exogenous AI-2 directly stimulates E. coli biofilm formation as much as 30-fold (17), we hypothesize that the increase in biofilm formation for yliH and yceP mutants, when cultured in LB glu (Fig. 1A and Fig. 2), occurs due to the increased internalization of AI-2 in the biofilm cells based on the induction of AI-2-stimulated genes in the biofilm cells.
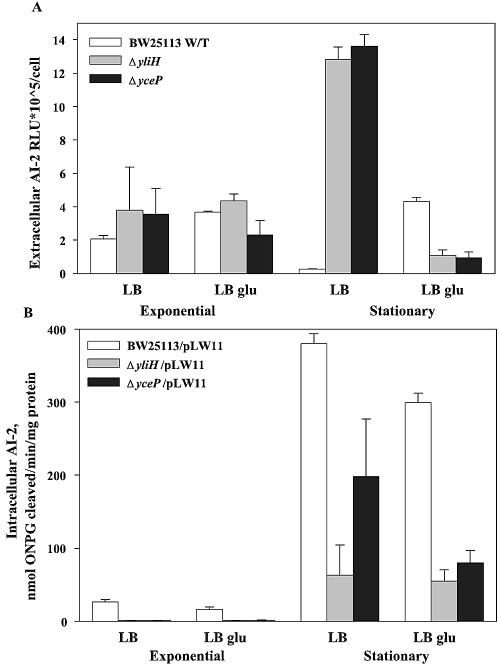
Because of the large number of AI-2-related genes affected, the V. harveyi bioluminescence assay was used to determine extracellular AI-2 concentrations in LB and LB glu medium with the yliH and yceP mutants. E. coli culture fluids were collected at two time points, one at the exponential phase (OD600, ∼0.5) and another one in the stationary phase 6 h after inoculation (OD600, ∼7). As previously reported, the AI-2 concentration for the wild-type strain in LB medium was higher in the exponential phase and declined substantially in the stationary phase (59). In LB glu, the extracellular AI-2 concentrations remained high throughout the exponential and stationary phases (Fig. 5A), which is consistent with previous reports (56).
FIG. 5.
Effects of deleting yliH and yceP on extracellular (A) and intracellular (B) AI-2 activity in LB and LB glu media. Data are shown for exponential-phase culture fluids (OD600, ∼0.6) and stationary-phase culture fluids (6 h). The experiment was done in duplicate.
For the yceP and yliH mutants, the extracellular concentrations of AI-2 were only slightly higher than those of the wild-type strain in the exponential phase but increased dramatically (50-fold) as cells entered the stationary phase of growth in LB (Fig. 5A). This suggests that yceP and yliH mutations may induce AI-2 synthesis, induce AI-2 export, or repress AI-2 import in this medium.
To investigate this hypothesis further, the effect of the yliH and yceP mutations on the intracellular AI-2 concentration was analyzed using plasmid pLW11 (56) with a lacZ fusion containing the lsrACDBFG promoter region; transcription of this operon increases due to internal AI-2 (51). Intracellular AI-2 was repressed 2- to 6-fold in stationary-phase suspension cells in LB medium for the yliH and yceP mutants (Fig. 5B), while extracellular AI-2 was increased 50-fold (Fig. 5A), indicating that the mutations acted to repress internalization of AI-2 when cultured in LB. Since extracellular AI-2 is known to induce biofilm formation and this phenotype was not observed in our mutants when grown in LB, this indicates that AI-2 uptake is necessary for biofilm formation. When cultured in LB glu, intra- and extracellular AI-2 was repressed four- to fivefold in stationary-phase suspension cells (Fig. 5A and B) in the yliH and yceP mutants, respectively. This result was surprising, since we expected intracellular levels of AI-2 in suspension cells to be higher than those of the wild-type strain, based on our microarray data (biofilm cells). It may therefore be possible that the expression patterns observed in our microarrays are unique to biofilm cells only and are not observed in suspension cells; hence, we can hypothesize that AI-2 is internalized in biofilm but not suspension cells. This line of evidence is suggested by our microarray data, which showed 77 to 99% of AI-2-repressed genes were repressed in our mutants and that 77 to 98% of AI-2-induced genes were induced in our mutants, a correlation which is unlikely to be a coincidence.
YliH and YceP and stress response.
The microarray data indicate a significant impact of the yliH and yceP mutations on the expression of stress response genes in biofilms. The yliH mutation altered the expression of 64 genes (2% of the genome) previously reported to be involved in the stress response (12, 30), with 13 genes induced >2-fold (e.g., sdiA, ydaD, and ydaK) and 51 genes repressed >2-fold (P ≤ 0.05) (e.g., yodC, yjbJ, and rpoS). The yceP mutation influenced eight stress-related genes, of which four were induced >2-fold (e.g., sdiA and fliI), and four were repressed >2-fold (P ≤ 0.05) (e.g., tktB and rpoS).
Stress response is largely regulated by the stationary-phase sigma S factor (σs) encoded by gene rpoS, which plays a critical role in adaptation to nutrient deprivation and other stresses (30). The rpoS gene was repressed 8.6- and 3.7-fold for the yliH and yceP mutations, respectively. RpoS has been reported to repress biofilm formation (8) and to act as a positive regulator of >100 stationary-phase genes (30). Additionally, RpoS negatively regulates yliH and many of the flagella and motility genes in stationary phase when grown in LB (30). This is consistent with other reports where yliH expression was induced in late stationary phase >1,000-fold (43), since rpoS transcript is known to peak in the late exponential phase and decrease thereafter, which would allow for increased transcription of motility genes observed here. The effects observed here are probably mediated through RpoS, since most of the genes induced in our experiment are repressed by RpoS and vice versa.
YliH and YceP and catabolite repression.
The observation that the deletion of yliH and yceP led to a substantial increase of biofilm when grown in LB glu led us to believe that these mutations are involved in catabolite repression in E. coli. In the wild-type strain, the addition of glucose inhibited biofilm formation 4.2-fold in LB glu relative to LB and 1.5-fold in M9C glu relative to M9C (data not shown), consistent with other reports (25, 48).
The transport of all carbohydrates occurs via the carbohydrate phosphotransferase system (PTS) through permease enzyme II (EII) with specific complexes for each carbohydrate such as PtsG (EIICBGlc) and Crr (EIIAGlc), which transport glucose (31). The phosphorylation of PTS carbohydrates occurs through the carbon-nonspecific enzyme I (PtsI) and histidine protein (PtsH).
The microarrays showed that genes encoding proteins involved in glucose phosphorylation and transport were repressed for both the yliH and yceP mutant biofilms, with ptsI repressed 4- and 2-fold, ptsH repressed 3- and 1.6-fold, and crr (encoding part of the glucose-specific transport EIIGlc) repressed 3- and 2-fold. Conventional catabolite repression occurs by lowering levels of cAMP. When glucose is absent from the medium, cAMP is present at high concentrations and binds with CRP to influence gene transcription (24). For the yliH and yceP mutants, no difference was observed in the expression of crp in the biofilm, but a twofold induction was observed in the transcription ratio of cya relative to that of wild-type strain, which indicates that the mutations may act to block catabolite repression.
In addition, genes involved in cAMP-independent catabolite repression were also observed to have altered transcription levels in both of our mutant biofilms. This type of catabolite repression is mediated by proteins from creC (20), rpoS (20), and csrA (25). Deletion of yliH represses csrA (6.1-fold) and induces creC (4.5-fold) in the biofilm. Deletion of yceP represses csrA slightly (1.5-fold) and induces creC (3.5-fold). CsrA represses biofilm formation through its role in central carbon flux (25), and CreC is a presumed regulator of gene expression in response to environmental catabolites and appears here to induce biofilms; to our knowledge this is the first link of this regulator to biofilm formation. To confirm the hypothesis that this gene is indeed involved in biofilm formation of E. coli, we measured total biofilm formed by mutant creC relative to that of the wild-type E. coli strain and found that biofilm formation was repressed about 30% when grown in LB medium but was substantially increased (sixfold) when grown in LB glu (data not shown). These results indicate that CreC represses biofilm formation in LB glu, similar to our mutants.
Catabolite repression of genes in biofilm and suspension cells.
To further investigate the effects of catabolite repression on the expression of csrA, creC, rpoS, crp, and cya, we conducted two additional sets of microarray experiments where gene expression was compared for biofilm and suspension cells in both LB glu and LB medium. The results showed that the genes related to catabolite repression are inversely regulated in biofilm cells compared to suspension cells; for example, csrA and rpoS were repressed 1.5- and 3-fold in suspension cells in LB glu relative to LB but were induced 2- and 4-fold in biofilm cells under the same conditions. Similarly, creC was induced eightfold in suspension cells due to the addition of glucose but was repressed threefold in biofilm cells. The expression of crp was repressed 2.5-fold in biofilm cells due to glucose, but no change in the expression of cya was observed. These results indicate that the regulation of catabolite repression was substantially different in biofilm cells than in suspension cells and further confirms that this regulation plays a big role in the formation of biofilms in E. coli. These results also corroborate our results showing that biofilm formation increases in LB glu for both of our mutants (Fig. 1A). Additionally, these results show that the gene expression patterns between biofilm and suspension cells can have great variability; hence, they may explain the discrepancies in our yceP and yliH mutant microarray data for biofilm cells and the assays carried out with suspension cells.
DISCUSSION
In this study, we show clearly that YliH and YceP repress biofilm formation in M9C glu and LB glu media (Fig. 1A) but not in M9C and LB media. This difference in biofilm formation was correlated to the vastly reduced extracellular and intracellular indole concentrations in LB glu with the yliH and yceP strains (Fig. 6), and we found that the addition of exogenous indole repressed biofilm formation in both of our mutants and restored the wild-type phenotype in LB glu (Fig. 7). In further agreement with this, we found that mutations in trpE and tnaL eliminated indole intracellular concentrations (as expected) and induced biofilm formation in E. coli, which provides further evidence that indole represses biofilms (unpublished data). These experiments were performed after noting that YliH and YceP repressed indole export genes in biofilm cells (acrEF was induced 6- to 11-fold in the mutants), induced indole import (mtr repressed 2.5-fold in the mutants), and induced other indole-related genes (e.g., astD and gabT), showing that microarrays are a good tool for investigating the role of specific genes and their proteins.
Based on the microarray data, we also hypothesize that YliH and YceP reduce biofilm formation by reducing intracellular AI-2 in biofilm cells; the AI-2 uptake gene lsrA was induced 6- to 11-fold in the yliH and yceP mutants; 77 to 99% of AI-2-repressed genes were repressed in the mutants, while 77 to 98% of the AI-2-induced genes were induced in our mutants. Therefore, these two proteins reduce biofilm by presumably reducing intracellular AI-2 concentrations in biofilm cells (based on the microarray data). As expected, the intra- and extracellular AI-2 concentrations in stationary-phase suspension cells were changed dramatically for the yliH and yceP mutants (Fig. 5A and B); however, the AI-2 concentration was unexpectedly decreased in LB glu medium. This indicates the expression patterns observed in our microarrays are unique to biofilm cells and are not the same for suspension cells; hence, we hypothesize that AI-2 is internalized in biofilm cells but not in suspension cells.
The induction of biofilm formation and the repression of indole in these mutants upon the addition of glucose to the culture medium led us to believe that these mutations are involved in catabolite repression. Considering the microarray data, we hypothesize that these mutants repress glucose phosphorylation and transport, leading to a reduction of catabolite repression, and increased AI-2 internalization (based on microarray data), as shown by an increase in the transcription of cya. Increased cAMP levels serve to induce transcription of the lsr operon (56). Additionally, the repression shown in our microarray data of rpoS, which negatively regulates the lsr operon, increases induction of the lsr operon transcription with subsequent AI-2 uptake (56).
The expression of genes involved with cAMP-independent catabolite repression was also altered significantly; the mutations repressed the expression of csrA and led to an increase in transcription of creC. This is the first record of CreC affecting biofilm formation for any strain. The overexpression of csrA has previously been shown to inhibit biofilm formation in E. coli (25); therefore, the inhibition of this gene in our mutants was consistent with the observed phenotype and previously published results. As expected, the expression of creC and csrA was found to be regulated by the addition of glucose to the growth medium in both suspension and biofilm cells, as shown by our wild-type strain microarray data for LB glu relative to that for LB. Interestingly, the addition of glucose had substantially different effects on the suspension and biofilm cells, indicating further that biofilm formation is strongly dependent on carbon flux and that its regulation differs greatly between suspension and biofilm cells.
Previously, in the absence of a conjugation plasmid, an increase in biofilm formation has been linked to increases in transcription of motility and flagellum genes (32). YliH represses motility in LB, and YceP represses motility in both LB and LB glu (i.e., deleting yliH and yceP increases motility) (Fig. 3A), which was corroborated by increased transcription of flagellum and motility genes fliC, motA, and qseB (Fig. 4), as well as by the DNA microarray results. However, the increased motility does not correlate directly to the increased biofilm phenotype in LB glu, since it was observed with both LB and LB glu, whereas the biofilm was not altered in LB (Fig. 1A).
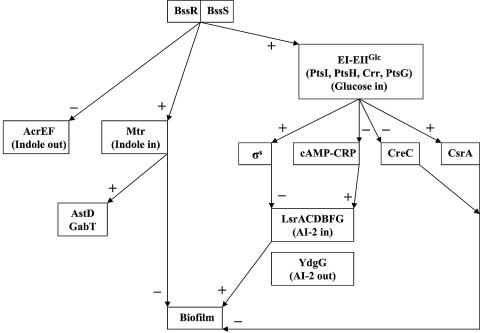
YliH and YceP appear to be global regulators of several genes involved in catabolite repression and stress response and regulation of the uptake and export of signaling pathways, including quorum sensing and the putative stationary-phase signal. We propose here that YliH and YceP are involved in the regulation of indole, as well as uptake and export of AI-2 through a cAMP-dependent pathway, as discussed above. Therefore, we propose new names for these two loci (bssR for yliH and bssS for yceP), based on the phrase “regulator of biofilm through signal secretion.” Additionally, a conceptual model for their regulation, which is consistent with our observed results for gene expression in biofilms with LB glu, is shown in Fig. 8.
FIG. 8.
Conceptual biofilm model including YliH and YceP in LB glu medium. +, positive regulation; −, negative regulation.
Supplementary Material
Acknowledgments
We thank A. Heydorn from the Technical University of Denmark for kindly providing COMSTAT; S. Molin from the Technical University of Denmark for plasmid pCM18; H. Mori of the Mara Institute of Science in Japan for the yliH, yceP, and creC mutants and plasmids pCA24N and pCA23N yliH+; B. Bassler from Princeton University for Vibrio harveyi BB170; J. Kaper from the University of Maryland for plasmids pVS159, pVS176, pVS175, pVS182, and pVS183; and Ming-Hui Chen and Feng Guo from the University of Connecticut for providing help with statistical analysis of the data.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. Wanner, and H. Mori. 21 February 2006, posting date. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Systems Biol. [Online.] doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed]
- 2.Barrett, T., T. O. Suzek, D. B. Troup, S. E. Wilhite, W.-C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash, W. Fujibuchi, and R. Edgar. 2005. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 33:D562-D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beca-DeLancey, R. R., M. M. T. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J.-M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 5.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagella gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corona-Izquierdo, F. P., and J. Membrillo-Hernández. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Let. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., L. Montanaro, and C. R. Arciola. 2005. Biofilm in implant infections: its production and regulation. Int. J. Artif. Organs 11:1062-1068. [DOI] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Biotechnol. 183:2918-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa, M. P., C.-F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino, P., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 15.Domka, J., J. Lee, and T. K. Wood. Submitted for publication.
- 16.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González Barios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González Barios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K-12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, M. C., R. J. Palmer, C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 20.Hardie, K. R., C. Cooksley, A. D. Green, and K. Winzer. 2003. Autoinducer 2 activity in Escherichia coli culture supernatants can be actively reduced despite maintenance of an active synthase, LuxS. Microbiology 149:715-728. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Ishizuka, H., A. Hanamura, T. Inada, and H. Aiba. 1994. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 13:3077-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura-Sato, K., K. Shibayama, T. Horii, Y. Iimuma, Y. Arakawa, and M. Ohta. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Let. 179:345-352. [DOI] [PubMed] [Google Scholar]
- 28.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole, G. A., K. A. Gibbs, P. W. Hager, J. P. V. Phibbs, and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 31.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 33.Ren, D., L. A. Bedzyk, P. Setlow, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Bacillus subtilis surface biofilms with and without sporulation and the importance of yveR for biofilm maintenance. Biotechnol. Bioeng. 86:344-364. [DOI] [PubMed] [Google Scholar]
- 34.Ren, D., L. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 35.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 88:630-642. [DOI] [PubMed] [Google Scholar]
- 36.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ. Microbiol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren, D., R. Zuo, A. F. González Barios, L. A. Bedzyk, G. R. Eldridge, M. E. Pasmore, and T. K. Wood. 2005. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 71:4022-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez, R. L., and R. C. Tait. 1983. Recombinant DNA techniques: an introduction. Benjamin/Cummings Publishing, Menlo Park, Calif.
- 39.Sabnis, N. A., J. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270:29096-29104. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 42.Schembri, M. A., K. Kjærgaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 43.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 44.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southey-Pilling, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilm. J. Bacteriol. 187:8114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 48.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturgill, G., C. M. Toutain, J. Komperda, G. A. O'Toole, and P. N. Rather. 2004. Role of CysE in production of an extracellular signaling molecule in Providencia stuartii and Escherichia coli: loss of cysE enhances biofilm formation in Escherichia coli. J. Bacteriol. 186:7610-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 52.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 53.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. 2004. Altering toluene 4-monooxygenase by active-site engineering for the synthesis of 3-methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J. Bacteriol. 186:4705-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., Y. Hashimoto, C.-Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and J. P. V. Phibbs. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 173:4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood, T. K., and S. W. Peretti. 1991. Effect of chemically induced, cloned-gene expression on protein synthesis in E. coli. Biotechnol. Bioeng. 38:397-412. [DOI] [PubMed] [Google Scholar]
- 59.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanofsky, C., V. Horn, and P. Gollnick. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 173:6009-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.