Abstract
We assessed the effects of different arcA mutations on poly(3-hydroxybutyrate) (PHB) synthesis in recombinant Escherichia coli strains carrying the pha synthesis genes from Azotobacter sp. strain FA8. The arcA mutations used were an internal deletion and the arcA2 allele, a leaky mutation for some of the characteristics of the Arc phenotype which confers high respiratory capacity. PHB synthesis was not detected in the wild-type strain in shaken flask cultures under low-oxygen conditions, while ArcA mutants gave rise to polymer accumulation of up to 24% of their cell dry weight. When grown under microaerobic conditions in a bioreactor, the arcA deletion mutant reached a PHB content of 27% ± 2%. Under the same conditions, higher biomass and PHB concentrations were observed for the strain bearing the arcA2 allele, resulting in a PHB content of 35% ± 3%. This strain grew in a simple medium at a specific growth rate of 0.69 ± 0.07 h−1, whereas the deletion mutant needed several nutritional additives and showed a specific growth rate of 0.56 ± 0.06 h−1. The results presented here suggest that arcA mutations could play a role in heterologous PHB synthesis in microaerobiosis.
Escherichia coli and other enterobacteria can adapt their metabolism to oxygen availability in the environment. Aerobic and anaerobic respiration and fermentation are different metabolic pathways that enable bacteria to optimize energy generation according to the oxygen levels in the surrounding medium (25). Sophisticated and interrelated regulatory networks switch the expression of these pathways on and off as needed. Such adaptive responses are mainly coordinated by two regulators, Fnr and ArcA (11). At the transcriptional level, the two-component signal transduction system ArcAB modulates the expression of many operons according to the redox state of the environment (20). ArcB is a transmembrane sensor kinase which, under anaerobic or microaerobic conditions, undergoes stable phosphorylation and then transphosphorylates the response regulator ArcA (16). The main targets for repression by the phosphorylated regulator are the genes that encode the enzymes involved in aerobic respiration, such as those of the tricarboxylic acid cycle. On the other hand, the cytochrome d oxidase, with high affinity for oxygen, and fermentation enzymes such as pyruvate-formate lyase are activated under microaerobic conditions (20). The effect of ArcA in the transcription of the whole genome has been recently analyzed (18, 29), and it was shown that many other genes in addition to those involved in redox metabolism are the putative targets of ArcA regulation.
Many bacterial species synthesize polyhydroxyalkanoic acids (PHAs), which accumulate in the cytoplasm as hydrophobic granules and function as a carbon reservoir and as an electron sink (2). These thermoplastic polymers have drawn great interest since their discovery, due to their degradability and the potential to produce them from renewable carbon and nitrogen sources (21). Poly(3-hydroxybutyrate) (PHB) is the best-characterized PHA. The PHB biosynthetic pathway includes a 3-ketothiolase, an NADPH- or NADH-dependent reductase, and a PHB synthase (33). By means of the reactions catalyzed by the former enzymes, a highly reduced polymer is accumulated intracellularly.
The use of recombinant E. coli is an alternative to natural PHA producers in fermentation processes. It grows quickly using several carbon sources, and it offers a well-defined physiological environment for the construction and manipulation of various metabolic pathways. Moreover, as it does not synthesize PHAs, it lacks the enzymatic depolymerization machinery; hence, a high PHA content can be attained. For instance, a recombinant E. coli strain carrying the pha genes from Azotobacter sp. strain FA8 (27) was able to accumulate PHB up to 65% of the cell dry weight from agroindustrial by-products (22).
The E. coli arc mutants are unregulated for aerobic respiration under microaerobic conditions. As a consequence, the tricarboxylic acid cycle enzymes are not repressed, and the pool of reducing equivalents (such as NADH or NADPH) is elevated (1). The synthesis of PHAs consumes a great amount of reducing equivalents. In view of this, we hypothesize that the accumulation of PHB under low-oxygen growth conditions will be favored in an arcA genetic background, due to the availability of an excess of reducing equivalents that could be funneled into an electron sink like this polymer.
In this study, we constructed two different arcA mutants from the above-mentioned strain (22). As ArcA is a global regulator with pleiotropic effects, a ΔarcA mutation and the arcA2 mutation, which is leaky for some of the phenotypic characteristics of arcA mutants, were chosen. Both mutants were characterized regarding their growth and respiratory capacity and used to analyze the synthesis of PHB in microaerobiosis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this work are summarized in Table 1. All plasmid constructions were propagated in E. coli DH5α. For maintenance and transfer of pKNG101 and its derivatives, E. coli CC118 λpir and S17-1 λpir were used. P1 transduction was performed according to Sternberg and Maurer (34).
TABLE 1.
Escherichia coli strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| K1060a | F−fadE62 lacI60 tyrT58(AS) fabB5 mel-1 supF58 | 24 |
| DH5α | F− φ80d lacZΔM15 recA endA1 Δ(lacZYA-argF)U169 deoR gyrA96 thi-1 hsdR17 supE44 relA1 | Gibco-BRL |
| S17-1 λpir | StrrrecA thi-1 hsdRM+ RP4::2-Tc::Mu::Kmr Tn7 λpir lysogen | 9 |
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir lysogen | 12 |
| SP314a | Δ(galK-bioD)76 relA1 spoT1 thi-1 deoC7 Δ(deoD-arcA)253 | 28 |
| ECL618 | arcA2 zij::Tn10 | 13 |
| ECL547 | sdh+ φ(sdh-lac) | 14 |
| CT548 | arcA2 zij::Tn10, by P1 transduction of arcA2 to ECL547 | This work |
| CT1061 | arcA2 zij::Tn10, by P1 transduction of arcA2 to K1060 | This work |
| CT1062 | As K1060, ΔarcA gene lacking 290 nt between internal primers obtained by mutagenesis with pKΔarcAc | This work |
| pJP24 | pQE32 derivative, carrying phaBAC genes from Azotobacter sp. strain FA8; Apr | 22 |
| pKNG101b | Mobilizable suicide vector; sacBR; Strr | 15 |
| pGEM-T Easy | A/T cloning vector; Apr | Promega |
| pGΔarcA | pGEM-T Easy derivative, carrying an arcA gene lacking 290 nt between the internal primers used for mutagenesis | This work |
| pKΔarcA | pKNG101 derivative, carrying an arcA gene lacking 290 nt between the internal primers used for mutagenesis | This work |
Obtained through CGSC.
Obtained through BCCM/LMBP.
nt, nucleotides.
Media and growth conditions.
During DNA manipulations and strain construction, cultures were grown at 37°C with reciprocal agitation (130 strokes · min−1) in LB (10 g · liter−1 tryptone, 5 g · liter−1 yeast extract, and 5 g · liter−1 NaCl, pH 7.2). For antibiotic selection, the concentrations of antibiotics were 100 μg · ml−1 (ampicillin), 25 μg · ml−1 (streptomycin), and 30 μg · ml−1 (tetracycline). For P1 transduction, selection was done on LB-tetracycline plates. For selection against sacB, Blomfield medium containing 60 g · liter−1 sucrose was used (3). The arcA mutants were phenotypically identified by plating the cells onto toluidine blue agar medium (10 g · liter−1 tryptone, 8 g · liter−1 NaCl, and 200 μg · ml−1 toluidine blue) (10). When necessary, media were solidified by the addition of agar at 15 g · liter−1.
For the diamide sensitivity test, SP314/pQE32 and SP314/pJP24 cells were grown for 24 h under PHB-synthesizing conditions in medium B (10 g · liter−1 tryptone and 8 g · liter−1 NaCl) supplemented with xylose (20 g · liter−1), washed twice, and resuspended in cold 20 mM HEPES to an optical density at 600 nm of approximately 1.0. A 100-μl aliquot of the bacterial suspension was spread on B medium-xylose plates, and two sterile filter paper disks embedded in either 0.5 M or 0.1 M diamide in dimethyl sulfoxide were applied over the bacterial lawn. After overnight incubation at 37°C, sensitivity was determined by measuring the diameter of the growth inhibition zone. For qualitative detection of PHB inclusion bodies, cells resuspended from the plate were observed by fluorescence microscopy after being stained with the basic oxazin Nile blue A (23).
Characterization of the recombinant strains.
Prior to each cultivation, seed cultures were prepared by transference of a loopful of cells from solid-medium slants to 250-ml Erlenmeyer flasks containing 200 ml of the corresponding fermentation medium and grown at 37°C for 18 h with moderate agitation (50 rpm).
For preliminary characterization in shaken flasks, strains K1060/pJP24 and SP314/pJP24 were grown in a rich medium (MYAG) containing (per liter of deionized water) 6.0 g Na2HPO4, 3.0 g KH2PO4, 1.4 g (NH4)2SO4, 0.5 g NaCl, 10.0 g yeast extract, and 5.0 g casein amino acids. Filter-sterilized (0.22-μm-pore-size filter) glucose and MgSO4 · 7H2O solutions were added to the sterilized medium at a final concentration of 30.0 g · liter−1 and 0.2 g · liter−1, respectively. Strain CT1061/pJP24 was grown in a synthetic medium (SMAG) containing the same salts as MYAG, plus 0.3 g · liter−1 casein amino acids (to prime growth), 5.0 mg · liter−1 thiamine hydrochloride, 5.0 ml · liter−1 trace elements solution, and 30.0 g · liter−1 glucose. Trace elements solution was composed (per liter of 5 M HCl) of 10 g FeSO4 · 7H2O, 2 g CaCl2 · 2H2O, 2.2 g ZnSO4 · 7H2O, 0.5 g MnSO4 · 4H2O, 1.0 g CuSO4 · 5H2O, 0.1 g (NH4)6Mo7O24 · 4H2O, 0.1 g NiCl2, 0.1 g CoCl2 · 4H2O, and 0.02 g Na2B4O7 · 10H2O.
Cultures were grown in 250-ml Erlenmeyer flasks under three different aeration conditions, defined as follows. Standard aerobiosis was achieved using a 1:5 medium volume/flask volume ratio with vigorous rotatory agitation (250 rpm). For maximum aerobiosis, the same agitation conditions were employed but using a 1:20 medium volume/flask volume ratio. For microaerobiosis, flasks were completely filled with growth medium, and cells were kept in suspension with a magnetic stirrer (50 rpm).
Bioreactor cultivation.
Batch cultures were carried out in a 5.6-liter BioFlo 110 fermenter (New Brunswick Scientific Co., Edison, NJ). Strains K1060/pJP24 and CT1062/pJP24 were grown in MYAG medium. Strain CT1061/pJP24 was grown in MYAG and SMAG media. Ampicillin was added at 100 mg · liter−1 to avoid plasmid loss.
To prevent foam formation, 30 μl · liter−1 Antifoam 289 (Sigma Chemicals, St. Louis, Mo.) was added at the onset of cultivation. Inoculum, prepared as described above, was added at 0.01 g (initial cell dry weight [CDW] ) · liter−1.
Cultures were developed in 4.5 liters of either MYAG or SMAG at 37.0 ± 0.2°C. To ensure microaerobic conditions, no air was supplied during the fermentation, and a constant agitation speed of 75 rpm was used to maintain homogeneous conditions and prevent biomass sedimentation. pH was controlled at 7.20 ± 0.03 by the automatic addition of 5 M NaOH or 5 M H2SO4. Dissolved oxygen concentration was measured using an Ag/AgCl2 polarometric oxygen probe (Mettler Toledo, Greifensee, Switzerland). Samples were withdrawn aseptically at selected times until the end of cultivation (48 h).
Construction of K1060 ΔarcA mutant.
Plasmid DNA preparation, DNA ligation, bacterial transformation, agarose gel electrophoresis, and screening followed standard methods (30) and instructions from the manufacturers. Restriction enzymes and DNA modification enzymes were purchased from Promega (Madison, WI).
PCR, crossover PCR deletions, and subcloning were done according to the procedure described by Link et al. (17), modified as follows: PCR mixtures contained 10 mM Tris · HCl (pH 9.0), 50 mM KCl, 0.1% (wt/vol) Triton X-100, 1.25 mM MgCl2, 0.1 mM each deoxynucleoside triphosphate, 6 μM each primer, and 1 U Taq DNA polymerase (GoTaq; Promega). The PCR was denatured at 94°C for 5 min before the amplification was run. The thermal cycle profile was 15 s at 94°C, 15 s at 55°C, and 30 s at 72°C (for a total of 30 cycles) and a final 5-min 72°C extension step.
Crossover PCR deletion products were constructed in two steps. In the first step, two different 50-μl asymmetric PCRs were used to generate fragments to the left and right of the sequence targeted for deletion. The PCR conditions were as described above, except that the primers were used in a 10:1 molar ratio (6 μM outer primer and 0.6 μM inner primer). For the amino-terminal sequence, primers arcANLow (5′-CCC GTC GAC AAA GCC CTT TAC TTA GCT TA-3′) and arcANUp (5′-CCG GAT CCT CCG CGC CAT CTG TCG CTT C-3′) were used. For the carboxy-terminal sequence, primers arcACLow (5′-GAA GCG ACA GAT GGC GCG GAG GAT CCG GAA AGC TAC AAG TTC AAT GGT-3′) and arcACUp (5′-GGG GAG CTC GGT TGA AAA ATA AAA ACG GC-3′) were used. In the second step, the left and right fragments were annealed at their overlapping region and amplified by PCR as a single fragment, using the external primers (1 μl of each of the two asymmetric PCR mixtures and 6 μM concentrations of the two external primers were mixed and amplified).
The 1-kb amplification fragment was ligated in vector pGEM-T Easy, resulting in pGΔarcA. This construction was propagated in E. coli DH5α, purified, and subsequently cloned into pKNG101, a broad-host-range, R6K-based suicide vector, by ligating the fragment cut at the NotI restriction sites situated at both sides of the insert in pGΔarcA with NotI-cut pKNG101 to generate the mutagenic plasmid pKΔarcA. This plasmid was then transferred to E. coli K1060 by competent cell transformation. To obtain allele replacement mutants, the protocol of Kaniga et al. (15) was modified as follows. After the transformants were plated in Blomfield medium, sucrose-resistant revertants were selected from sucrose-sensitive, streptomycin-resistant (Strr) clones in which the complete plasmid had theoretically integrated into the chromosome by a single homologous recombination at the wild-type arcA locus. A proportion of these revertants have the wild-type gene replaced by the mutated gene. The mutation was characterized by growth on toluidine blue agar and by colony PCR using the external primers.
Biomass determination.
Growth was monitored by measuring optical density at 600 nm using a spectrophotometer (UV1203; Shimadzu Scientific Instruments Inc., Columbia, MD). Samples taken from the fermentor (usually 50 ml each) were immediately chilled to 0°C in an ice bath. Cell concentration, defined as grams of CDW per liter, was determined by placing an accurately measured volume of culture broth (approximately 10 ml) into a previously dried and weighed 15-ml polypropylene centrifuge tube. Cells were centrifuged at 10,000 × g for 10 min at 4°C, the supernatant was decanted, and the pellet was washed twice with deionized water by resuspension and centrifugation. After the supernatant was decanted, the cell pellet was dried in an oven at 85°C for 36 h, cooled in a desiccator, and weighed.
Analytical determinations.
Oxygen consumption was measured with a Clark-type polarographic electrode (Biological Oxygen Monitor model 53; Yellow Springs Instruments, Inc., Yellow Springs, Ohio). The assay mixture in the corresponding growth medium was equilibrated for 3 min at 37°C under magnetic stirring and in an air atmosphere. An aliquot of the cellular suspension was added, and oxygen consumption was measured. The total volume in the measurement chamber was 3 ml. Oxygen uptake was expressed as the variation of percentage of oxygen saturation per minute per milligram of protein. Protein concentration was determined by the Folin phenol reagent method of Lowry et al. (19), using bovine serum albumin as a standard.
PHB was determined by gas chromatography using a slight modification of the method described by Braunegg et al. (4). About 10 mg of freeze-dried biomass (0.01 mPa; 8 h) was accurately weighed and placed in Teflon-stoppered vials. Methanolysis was carried out by the addition of 2 ml CH3OH/H2SO4 (85:15 [vol/vol]) and 2 ml CHCl3 to the samples, which were incubated at 100°C for 150 min, cooled to room temperature, and mixed with 2 ml of 1 M NaHCO3. The aqueous phase was discarded, and the organic phase was extracted twice with deionized water. After settling of the layers, the lower phase was dried over anhydrous Na2SO4. The methyl esters of 3-hydroxybutyrate were quantified in a Hewlett Packard HP 5890 gas chromatograph equipped with a Hewlett Packard FFAP column. Pure PHB was used as a standard.
PHB concentration was defined as the grams of polymer per liter. PHB content was defined as the ratio of grams of PHB per gram of CDW.
RESULTS AND DISCUSSION
Preliminary characterization of a strain carrying an arcA deletion mutation for PHB accumulation in shaken flask cultures at different oxygen concentrations.
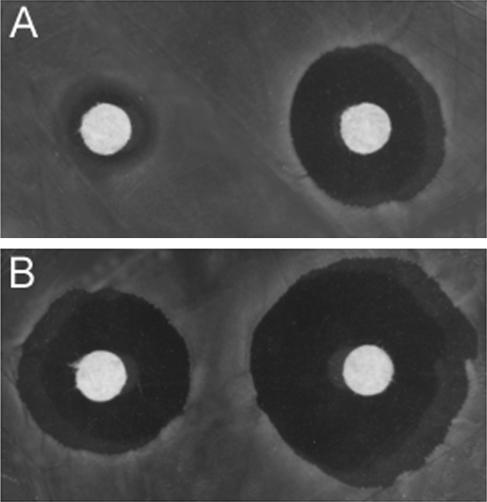
To test the hypothesis that the accumulation of PHB under low-oxygen growth conditions will be favored in an arcA genetic background, we first analyzed the behavior of a well-characterized E. coli ΔarcA mutant at different oxygen concentrations. SP314, which bears a deletion including the arcA gene (28), was the strain of choice into which we introduced plasmid pJP24, harboring the pha structural genes from Azotobacter sp. strain FA8, previously characterized in our laboratory (26, 27). Considering the role of PHB as an electron sink, we reasoned that redox deregulation would have a positive effect on the synthesis of this compound. A simple test was performed with SP314 to verify that the excess of reducing equivalents was directed towards PHB synthesis. Diamide [1,1′-azobis-(N,N-dimethylformamide)] causes the formation of cytoplasmic disulfides in low-molecular-weight thiols and in proteins. In E. coli, such oxidative stress is neutralized by thioredoxin reductase, which uses NADPH to reduce these deleterious disulfide bonds (7). Thus, diamide sensitivity can be used as a rough estimation of NADPH availability, as was previously demonstrated for Streptomyces lividans (5). Diamide sensitivity of strain SP314 containing either vector pQE32 or plasmid pJP24 was analyzed by a plate assay (Fig. 1). The strain bearing the pha genes showed increased sensitivity to diamide compared with the strain carrying the control vector, corresponding to lower levels of NADPH. Intracellular PHB content of SP314/pJP24 was checked by Nile blue A staining (23).
FIG. 1.
Diamide sensitivity test for E. coli SP314 carrying either pQE32 (A) or pJP24 (B) grown on medium B-xylose plates. Filter paper disks containing 0.1 M (left) and 0.5 M (right) diamide were applied over a bacterial lawn, and the sensitivity was scored as the diameter of the growth inhibition zone.
Even when the only theoretical nutritional requirements for the SP314 auxotroph are serine and biotin, minimal medium supplemented with these nutrients did not support SP314/pJP24 growth in batch cultures. The low or null growth of different ΔarcA mutants in minimal medium has been reported in the literature, together with the requirement for the addition of different concentrations of casein amino acids to the medium to reach full growth (10, 18). SP314/pJP24 was then grown in shaken flasks in the complex medium MYAG, as indicated in Materials and Methods. Xylose is commonly used in most of the studies involving ArcA regulation to avoid the repression exerted by glucose on many operons that are controlled by ArcA (14). Preliminary experiments designed to choose the carbon source indicated that there were no significant differences between glucose and xylose regarding polymer yield (data not shown). Therefore, we supplemented the medium with glucose, due to better availability of this carbon source.
The analysis of PHB synthesis by SP314/pJP24 in MYAG during 48 h at different oxygen concentrations is shown in Table 2. A significant increase in PHB accumulation levels was obtained in microaerobiosis, although low biomass was generated. The increase in PHB accumulation correlated with the decrease in dissolved oxygen tension.
TABLE 2.
PHB accumulation in 48-h shaken flasks cultures of SP314/pJP24 under different conditions of oxygen availability
| Culture conditiona | CDW (g · liter−1) | % PHBb |
|---|---|---|
| High aerobiosis | 4.81 ± 0.38 | 13 ± 3 |
| Normal aerobiosis | 4.36 ± 0.25 | 16 ± 2c |
| Microaerobiosis | 1.73 ± 0.15 | 24 ± 4c |
Given by the ratio of medium volume to flask volume and shaking measured in revolutions per minute, as indicated in Materials and Methods.
The amount of PHB is given as a weight percentage (average ± standard deviation) of the CDW for three replicated experiments.
Differences were significant with a P value of <0.05 (analysis of variance [ANOVA]).
In a previous work, we described the production of PHB from agroindustrial by-products by E. coli K1060/pJP24, which accumulates PHB up to 65% of CDW (22). To compare the behavior of this strain with SP314/pJP24 at low oxygen concentrations, both strains were grown in microaerobiosis, and biomass and PHB content were monitored. Table 3 shows the results obtained under these conditions, together with control experiments performed in aerobiosis as described in Materials and Methods. SP314/pJP24 presented the highest PHB yield in microaerobiosis, while K1060/pJP24, the wild-type (ArcA+) strain, did not accumulate detectable amounts of PHB under these conditions. The PHB yield for SP314/pJP24 in microaerobiosis was comparable to the yield obtained for K1060/pJP24 under aerobic growth conditions, although SP314 carries extended deletions that cause poor growth.
TABLE 3.
PHB accumulation in 48-h shaken flasks cultures of SP314/pJP24 and K1060/pJP24 under aerobic and microaerobic conditions
| Strain | Conditiona | CDW (g · liter−1) | PHB (g · liter−1) | % PHBb |
|---|---|---|---|---|
| SP314/pJP24 | Aerobic | 2.95 ± 0.09 | 0.35 ± 0.07 | 12 ± 2c |
| K1060/pJP24 | Aerobic | 3.85 ± 0.14 | 1.04 ± 0.21 | 27 ± 2c |
| SP314/pJP24 | Microaerobic | 2.21 ± 0.12 | 0.51 ± 0.06 | 23 ± 2d |
| K1060/pJP24 | Microaerobic | 1.45 ± 0.05 | <0.05 | <4d |
Given by the ratio of medium volume to flask volume and shaking measured in revolutions per minute, as indicated in Materials and Methods.
The amount of PHB is given as a percentage of weight (average ± standard deviation) of the CDW for three replicated experiments.
Differences were significant with a P value of <0.05 (ANOVA).
The results obtained with the collection strain harboring a ΔarcA mutation validated our hypothesis and simultaneously presented uncertainty about which were the best arcA mutations to pursue the research.
Characterization of the respiratory capacity and growth requirements of E. coli strains carrying different arcA mutations.
ArcA is a global regulator, which controls the expression of 51 operons either by repression or by activation (18). It is possible to assume that the lack of control of such a large number of genes has a negative effect on the growth rate of E. coli at low oxygen concentrations. Another mutation in the arcA gene, named arcA2, has been used in previous work for experiments performed with minimal medium (10). This mutation could eventually result in the deregulation of the genes encoding enzymes involved in respiration without significantly affecting gene functions related to the growth rate.
According to the results obtained with strain SP314 and data from the literature (10, 18), arcA deletion mutants have demanding nutrient requirements for full growth. In view of this, we evaluated the use of strain ECL618, which bears the arcA2 mutation, to study its effect on PHB synthesis. This mutant was first described as having an arcA mutation which suppresses the expression of the tra genes of the F plasmid (13), activates the expression of cytochrome d, and grows in minimal medium primed with 0.3 g · liter−1 casein amino acids (10).
To have a better understanding of the behavior of the arcA2 mutant, we completed the analysis of its pleiotropic phenotype as follows. Mutations in the arcA gene confer sensitivity to dyes such as methylene blue and toluidine blue, and deletion mutants do not grow on media containing these dyes (14). We found that the arcA2 mutant formed small- to medium-sized colonies on toluidine blue agar. Further characterization of this mutant was done, taking into account the respiration capabilities of the arcA mutants. Oxygen consumption rates were measured for 24-h shaken flask cultures of E. coli SP314 and ECL618 in LB supplemented with xylose. Surprisingly, oxygen consumption, determined as indicated in Materials and Methods and expressed as the percentage of oxygen consumed (ΔO2%) per minute per milligram of protein, was twofold higher in the strain bearing the arcA2 mutation (62.45 ± 4.93) than in SP314 (32.72 ± 4.17). We have previously shown that the heterologous expression of pha genes restores the wild-type phenotype in ΔarcA mutants, including their respiratory capacity (J. A. Ruiz, R. O. Fernández, P. I. Nikel, B. S. Méndez, and M. J. Pettinari, unpublished data). Oxygen consumption rates were determined for strains CT548/pQE32 and CT548/pJP24, both carrying the arcA2 mutation, and the values obtained were 42.15 (±10.54) and 45.03 (±8.85), respectively. According to these data, the expression of the pha genes did not decrease the respiratory activity of the strains carrying the arcA2 mutation. These results indicated that strains bearing this mutation could be adequate for further study of PHB accumulation in microaerobiosis.
Synthesis of PHB by recombinant E. coli ΔarcA and arcA2 strains.
The analysis of the behavior of the previously characterized mutations regarding PHB synthesis required their expression in the same genetic background. The strain of choice for such a study was K1060, a prototroph formerly used for PHB production (22). An arcA2 derivative of K1060 was constructed by P1 transduction. Strain SP314 contains a large deletion involving several genes; thus, to have a strain containing only a well-defined arcA mutation, crossover PCR and a gene replacement vector were used to construct an ΔarcA mutation in K1060. Plasmid pJP24 was transformed into the resulting strains, which were named CT1062 (ΔarcA) and CT1061 (arcA2).
Their capacity to accumulate PHB in microaerobiosis and that of the recombinant parental strain were evaluated with a 5.6-liter bioreactor during 48 h using a rich medium (MYAG) and a synthetic medium supplemented with casein amino acids at a concentration of 0.3 g · liter−1 (SMAG) for CT1061/pJP24. Cultures were carried out without air sparging, which corresponds to an initial dissolved oxygen concentration of approximately 20% of air saturation.
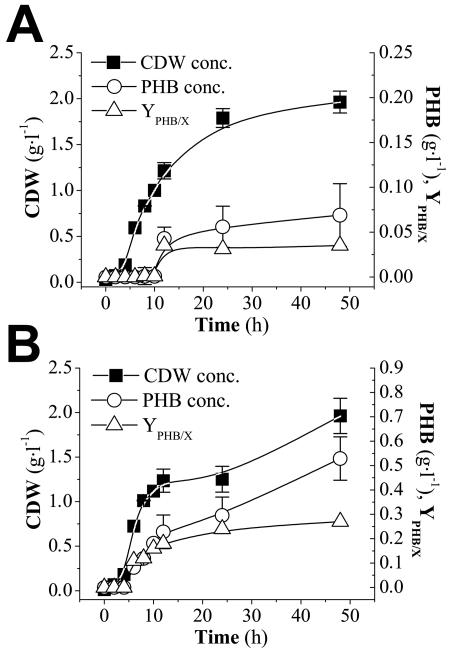
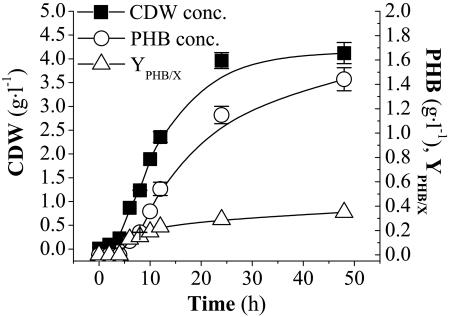
Figure 2 shows the results of the bioreactor cultivations of K1060/pJP24 and CT1062/pJP24, the ΔarcA mutant, in MYAG. Both strains could support growth under microaerobic conditions, the cultures reached the stationary phase after approximately 8 h of exponential growth, and similar biomass concentrations were attained. PHB was not significantly accumulated in K1060/pJP24. The results of the bioreactor cultivation of CT1061/pJP24 in SMAG are indicated in Fig. 3. Biomass and PHB concentrations were higher in this strain, bearing the arcA2 mutation, than in the deletion mutant strain, resulting in a higher PHB content. Besides, this enhanced PHB accumulation was attained in SMAG, a simple salts medium, while CT1062/pJP24 was grown in MYAG medium, as it is unable to grow in SMAG. The maximum specific growth rates for the recombinant strains were 0.55 ± 0.08 h−1 for K1060 and 0.56 ± 0.06 h−1 for CT1062, both cultivated in MYAG, while CT1061 reached a specific growth rate of 0.69 ± 0.07 h−1 in a minimal salts medium. Similar results regarding biomass, PHB concentrations, and specific growth rate were achieved by CT1061/pJP24 grown in MYAG under the same conditions (data not shown).
FIG. 2.
Bioreactor cultures of K1060/pJP24 (ArcA+) (A) and CT1062/pJP24 (ΔarcA) (B) were carried out in MYAG in microaerobiosis. No air was supplied during the cultivation, and a constant agitation speed of 75 rpm was applied to maintain homogeneous conditions.
FIG. 3.
Bioreactor cultures of CT1061/pJP24 (arcA2) were carried out in SMAG in microaerobiosis. No air was supplied during cultivation, and a constant agitation speed of 75 rpm was applied to maintain homogeneous conditions.
Table 4 summarizes the properties of the strains related to the accumulation of PHB in microaerobiosis. Their oxygen consumption is also included, and it corroborates the particular respiratory capacity conferred by the ΔarcA and the arcA2 mutations in strains otherwise isogenic.
TABLE 4.
Summary of the recombinant wild-type strain and the arcA derivative characteristics referred to 48-h batch cultures in a bioreactor
| Strain | Medium | CDW (g · liter−1) | PHB (g · liter−1) | % PHBa | μmaxb (h−1) | O2 consumptionc (ΔO2 % · min−1 · mg protein−1) |
|---|---|---|---|---|---|---|
| K1060/pJP24 | MYAG | 1.95 ± 0.12 | 0.07 ± 0.04 | <4d | 0.55 ± 0.08 | 11.36 ± 1.15 |
| CT1062/pJP24 | MYAG | 1.96 ± 0.05 | 0.53 ± 0.03 | 27 ± 2d,e | 0.56 ± 0.06 | 19.45 ± 3.18 |
| CT1061/pJP24 | SMAG | 4.12 ± 0.21 | 1.44 ± 0.05 | 35 ± 3d,e | 0.69 ± 0.07 | 45.79 ± 4.05 |
The amount of PHB is given as a percentage of weight (average ± standard deviation) of the CDW for at least two replicated experiments.
Maximum specific growth rate.
Oxygen consumption was determined with aliquots taken from the bioreactor, as indicated in Materials and Methods.
Differences were significant with a P value of <0.05 (ANOVA).
Conclusions.
Characterization of strain SP314 showed that it has low biomass yields and high nutritional requirements. As this strain carries two extended deletions, one of them starting at the deo genes and expanding beyond the arcA gene and the other including the gal and bio genes, the results could in principle be the consequence of mutations other than those affecting the arcA gene. On the other hand, strain CT1062 has a defined deletion that affects only the arcA gene, but its growth requirements are similar to those observed for SP314. These results suggest that these phenotypic features are due only to the ΔarcA mutation.
The fact that the recombinant strain CT1061 does not require a rich medium to achieve full growth facilitates the pursuit of physiological studies, as rich medium introduces many variables that have to be considered when such studies are performed. Casein amino acids were added only at very low concentrations to prime growth. Experiments are currently under way to manipulate growth conditions to avoid the need of nutritional additives.
Recent publications demonstrated that the intracellular levels of nicotinamide nucleotides are enhanced in E. coli cultures grown at low oxygen concentrations and in an arcA genetic background (1, 8). There are several studies reporting the manipulation of metabolic reactions to increase NADPH availability to produce PHAs in recombinant E. coli, but all of them relied on highly aerobic processes (35, 36). Microaerobiosis experiments presented in this work are based on the fact that the arcA mutations will cause an increase in the NADPH levels; the experiments were performed without aeration, using only slight agitation (75 rpm) to maintain culture homogeneity. Besides, since aerobic pathways in arcA mutants are active even when the culture reaches oxygen limitation, a greater availability of acetyl coenzyme A is expected to be maintained throughout the growth phases without leading to substrate limitation.
The growth of E. coli under conditions that were not fully aerobic deserves increased interest, as has been shown by recent publications. Salmon et al. (29) described the effect of the ArcA regulator in global gene expression of E. coli under growth conditions of different oxygen availability, thus expanding and refining the repertoire of ArcA-regulated genes. Our results are in good agreement with those of Shalel-Levanon et al. (31, 32). In analyzing E. coli mutants deleted for the arcA gene in glucose-limited chemostat cultures supplied with different oxygen concentrations, the authors found that the most relevant role for the ArcA regulator is in the transition from aerobic to microaerobic conditions. They also found, coincidently with previous results of Alexeeva et al. (1), a high redox potential associated with the ArcA mutant under these conditions, as well as an augmented expression of some of the genes related to glycolysis and the tricarboxylic acid cycle.
Carlson et al. (6) observed that recombinant E. coli DH5α, carrying pha genes from Ralstonia eutropha, can support PHB accumulation in anaerobiosis when growing in rich medium. Our results demonstrated that arcA strains of E. coli are capable of PHB synthesis under similar conditions in microaerobiosis. Residual biomass and PHB concentrations are comparable to those reported by these authors; however, the arcA2 mutant analyzed in this work grew with a higher maximum specific growth rate, even when a poorer medium was used.
In this study, we analyzed the effect of redox deregulation caused by arcA mutations on PHB synthesis. Furthermore, this work considered the difference between some pleiotropic effects caused by two different mutations in the arcA gene and their effects on heterologous PHB accumulation.
Acknowledgments
We thank Dimitris Georgellis for kindly giving us E. coli strains ECL547 and ECL618. We are also indebted to Rubén O. Fernández for assistance with some oxygen consumption measurements.
This work was supported by grants from UBA and CONICET. M.J.P., M.A.G., and B.S.M. are career investigators from CONICET. P.I.N. has a graduate student fellowship from CONICET.
REFERENCES
- 1.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 4.Braunegg, G., B. Sonnleitner, and R. M. Lafferty. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in bacterial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6:29-37. [Google Scholar]
- 5.Butler, M. J., P. Bruheim, S. Jovetic, F. Marinelli, P. W. Postma, and M. J. Bibb. 2002. Engineering of primary carbon metabolism for improved antibiotic production in Streptomyces lividans. Appl. Environ. Microbiol. 68:4731-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson, R., A. Wlaschin, and F. Srienc. 2005. Kinetic studies and biochemical pathway analysis of anaerobic poly-(R)-3-hydroxybutyric acid synthesis in Escherichia coli. Appl. Environ. Microbiol. 71:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 8.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixeira de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 10.Fu, H. A., S. Iuchi, and E. C. C. Lin. 1991. The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol. Gen. Genet. 226:209-213. [DOI] [PubMed] [Google Scholar]
- 11.Gunsalus, R. P. 1992. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J. Bacteriol. 174:7069-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iuchi, S., D. Furlong, and E. C. C. Lin. 1989. Differentiation of arcA, arcB, and cpxA mutant phenotypes of Escherichia coli by sex pilus formation and enzyme regulation. J. Bacteriol. 171:2889-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iuchi, S., and E. C. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 16.Kwon, O., D. Georgellis, and E. C. C. Lin. 2000. Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J. Bacteriol. 182:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 19.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 20.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen. ASM Press, Washington, D.C.
- 21.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikel, P. I., M. J. Pettinari, B. S. Méndez, and M. A. Galvagno. 2005. Statistical optimization of a culture medium for biomass and poly(3-hydroxybutyrate) production by a recombinant Escherichia coli strain using agroindustrial byproducts. Int. Microbiol. 8:243-250. [PubMed] [Google Scholar]
- 23.Ostle, A. G., and J. G. Holt. 1982. Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl. Environ. Microbiol. 44:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overath, P., H. U. Schairer, and W. Stoffel. 1970. Correlation of in vivo and in vitro phase transitions of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 67:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patschkowski, T., D. N. Bates, and P. J. Kiley. 2000. Mechanisms for sensing and responding to oxygen deprivation, p. 61-78. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 26.Pettinari, M. J., L. Chanetón, G. Vazquez, A. Steinbüchel, and B. S. Méndez. 2003. Insertion sequence-like elements associated with putative polyhydroxybutyrate regulatory genes in Azotobacter sp. FA8. Plasmid 50:36-44. [DOI] [PubMed] [Google Scholar]
- 27.Pettinari, M. J., G. J. Vazquez, D. Silberschmidt, B. Rehm, A. Steinbüchel, and B. S. Méndez. 2001. Poly(3-hydroxybutyrate) synthesis genes in Azotobacter sp. strain FA8. Appl. Environ. Microbiol. 67:5331-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roeder, W., and R. Somerville. 1979. Cloning the trpR gene. Mol. Gen. Genet. 176:361-368. [DOI] [PubMed] [Google Scholar]
- 29.Salmon, K. A., S. P. Hung, N. R. Steffen, R. Krupp, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2005. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J. Biol. Chem. 280:15084-15096. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1988. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 92:147-159. [DOI] [PubMed] [Google Scholar]
- 32.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab. Eng. 7:364-374. [DOI] [PubMed] [Google Scholar]
- 33.Steinbüchel, A., and S. Hein. 2001. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71:81-123. [DOI] [PubMed] [Google Scholar]
- 34.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 35.van Wegen, R. J., S. Y. Lee, and A. P. Middelberg. 2001. Metabolic and kinetic analysis of poly(3-hydroxybutyrate) production by recombinant Escherichia coli. Biotechnol. Bioeng. 74:70-80. [DOI] [PubMed] [Google Scholar]
- 36.Wang, F., and S. Y. Lee. 1997. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl. Environ. Microbiol. 63:4765-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]