Abstract
The Florida Everglades is one of the largest freshwater marshes in North America and has been subject to eutrophication for decades. A gradient in P concentrations extends for several kilometers into the interior of the northern regions of the marsh, and the structure and function of soil microbial communities vary along the gradient. In this study, stable isotope probing was employed to investigate the fate of carbon from the fermentation products propionate and butyrate in soils from three sites along the nutrient gradient. For propionate microcosms, 16S rRNA gene clone libraries from eutrophic and transition sites were dominated by sequences related to previously described propionate oxidizers, such as Pelotomaculum spp. and Syntrophobacter spp. Significant representation was also observed for sequences related to Smithella propionica, which dismutates propionate to butyrate. Sequences of dominant phylotypes from oligotrophic samples did not cluster with known syntrophs but with sulfate-reducing prokaryotes (SRP) and Pelobacter spp. In butyrate microcosms, sequences clustering with Syntrophospora spp. and Syntrophomonas spp. dominated eutrophic microcosms, and sequences related to Pelospora dominated the transition microcosm. Sequences related to Pelospora spp. and SRP dominated clone libraries from oligotrophic microcosms. Sequences from diverse bacterial phyla and primary fermenters were also present in most libraries. Archaeal sequences from eutrophic microcosms included sequences characteristic of Methanomicrobiaceae, Methanospirillaceae, and Methanosaetaceae. Oligotrophic microcosms were dominated by acetotrophs, including sequences related to Methanosarcina, suggesting accumulation of acetate.
The Florida Everglades is one of the largest freshwater marshes in North America (16). The northern regions of the marsh, particularly Water Conservation Area 2A (WCA-2A), have been subject to eutrophication in recent decades, due to nutrient runoff from the adjacent Everglades Agricultural Area. WCA-2A is characterized by a gradient in P concentrations in surface soils ranging from approximately 1,500 mg/kg in eutrophic regions to approximately 400 mg kg−1 in oligotrophic areas (18, 27). A large body of research has shown that eutrophication resulted in a number of ecosystem level changes in the marsh (8, 13, 14, 25, 27), including a shift in the dominant plant species from sparse stands of sawgrass (Cladium jamaicense Crantz) in ridges and spike rush (Eleocharis sp.) in sloughs to dense stands of cattail (Typha domingensis Pers.). Significantly greater primary productivity in eutrophic regions resulted in greater carbon input to the soils, leading to greater peat accumulation (2, 27, 37) and higher rates of methanogenesis and anaerobic respiration (4-7). Concomitant with larger amounts of available carbon in eutrophic regions are changes in the composition and increases in the sizes of specific microbial groups responsible for carbon cycling, such as methanogens, fermenters, and sulfate-reducing prokaryotes (SRP) (4-7, 14, 37; I. Uz and A. Ogram, submitted for publication).
The general pathways for carbon mineralization in anoxic soils are well known (10, 30), but the relationship between eutrophication and specific pathways through which simple fermentation products such as volatile fatty acids (VFA) are channeled by anaerobic microorganisms is not clear at this time. In addition, relatively little is known of the specific prokaryotes responsible for these transformations in natural systems. In methanogenic soils, VFA are converted to methane (CH4) through consortia of hydrogen-producing, fatty-acid-consuming bacteria (syntrophs) and methanogens, although these consortia may compete with SRP for VFA in soils with sufficient sulfate to support SRP (14, 30).
Our objective in this study was to probe soil communities with substrates labeled with stable isotopes to identify prokaryotes responsible for metabolism of carbon from selected VFA (butyrate and propionate) in eutrophic, transition, and oligotrophic soils of WCA-2A. This work extends previous studies from this laboratory that document changes in the structure and function of microbial communities as a result of eutrophication in the Everglades (4-7).
MATERIALS AND METHODS
Site description and sampling procedures.
Soil samples were collected from three sites in December 2004 along the nutrient gradient in WCA-2A of the northern Florida Everglades (4, 13, 39, 41), under flooded (water depth, 0.2 to 0.29 m) conditions. A map showing sampling locations was previously published (4). Samples were collected from sites F1 (eutrophic), F4 (transition region), and U3 (oligotrophic). At each sampling station, three soil cores were obtained within an area of approximately 25 m2 and to a depth of 20 cm. Cores were stored on ice and transported to the laboratory in Gainesville, Fla., where approximately 8 cm of floc was discarded. This was followed by sectioning of the cores from 0 to 10 cm of substrata of soil, which were used in this study. This section of soil represents nutrient impacts for longer than the past 3 years, as indicated by peat accretion rates calculated from 137Cs distribution (27). Three replicate cores were collected from each site and composited to form a single composite sample for each site. Large root and plant materials were removed from the composite samples. Samples were kept at 4°C until analysis, which was within 2 days of sampling.
Microcosms for stable isotope probing.
Preliminary studies indicated that modified BCYT medium (basal carbonate yeast extract trypticase-peptone) (35), BCYT-R (containing 0.01-g/liter trypticase-peptone), yielded minimal background fermentation and propionate- and butyrate-induced methanogenesis, similar to that observed with BCYT (data not shown). BCYT-R was therefore chosen for these studies.
Each 55-ml serum bottle contained 10 ml of BCYT-R medium with 0.1% resazurin. Soil samples (10 g [wet weight]) were introduced into the bottle containing BCYT-R medium under a constant stream of N2 to prevent exposure to oxygen and immediately crimped using butyl rubber septa and aluminum seals (Bellco Glass, Inc., Vineland, NJ). Before addition of the 13C-labeled substrate, microcosms were preincubated for 10 days after reduction with cysteine (2%) to a final redox potential of approximately −110 to −200 mV (18). All incubations were carried out at 28°C in the dark. Following preincubation, 10 mM (each) [13C]propionate or [13C]butyrate (Isotec, Miamisburg, OH) from anaerobic stock solutions was added to the vials. Methanogenesis was quantified in weekly intervals for 4 weeks, at which point methanogenesis stabilized, indicating complete oxidation of the added fatty acids. A parallel set contained unlabeled propionate and butyrate to compare methanogenesis with labeled fatty acids. Another 10 mM (each) [13C]propionate or [13C]butyrate was anaerobically added to the microcosms, and CH4 was followed until the seventh week. Endogenous CH4 production obtained from control microcosms (without the addition of exogenous fatty acids) was subtracted from the gross CH4 produced from experimental microcosms.
Analytical methods.
CH4 in the headspace was measured by gas chromatography with a Shimadzu 8A GC as described previously (4). All determinations were carried out in triplicate, and average values with 1 standard deviation are reported.
Statistical analyses.
All statistical analyses in this study were performed by a general linear model to test for the main effects and their interactions on all response variables with PROC GLM (SAS Institute). Population normality was tested for each variable before parametric statistics for comparisons and testing were used.
Nucleic acid extraction, purification of DNA, and PCR amplifications.
The entire 20-ml volume of soil in BCYT-R was centrifuged in a 50-ml tube, and the biomass was pelleted. Total DNA was extracted with the Ultra Clean Soil DNA mega kit (Mo Bio Laboratories, Solana Beach, CA) after 7 weeks of incubation. The quality of the DNA was evaluated by electrophoresis through a 0.7% agarose gel with Tris-acetate-EDTA buffer, and concentrations of total DNA were estimated by UV absorbance at 260 nm (29).
Separation of [13C]DNA from [12C]DNA.
To evaluate the possibility of [12C]DNA carryover in [13C]DNA following ultracentrifugation, 150 to 200 ng of unlabeled Escherichia coli DNA (TOP10F′ cells grown in Luria-Bertani medium) was mixed with experimental DNA prior to ultracentrifugation as described previously (32). Total DNA was subjected to CsCl-ethidium bromide density gradient centrifugation in a VTI 65.2 rotor at 55,000 rpm for 18 h at 20°C, as previously described (26). DNA bands were visualized with a handheld long-wavelength (365-nm) UV lamp. Separation was observed between “lighter” and “denser” DNA bands, although smearing of DNA was observed between bands. The lower band was extracted and recentrifuged as above for additional purification. Following the second ultracentrifugation, CsCl and ethidium bromide were removed by standard methods (29). DNA was concentrated by passage through a Centricon YM-100 filter assembly (Millipore Corporation, Bedford, MA), and the concentrated DNA was resuspended in sterile double-distilled water for molecular analyses. The purity of labeled DNA was confirmed by screening dense and light bands for the presence of the added E. coli DNA by E. coli-specific PCR primers as previously described (28). E. coli DNA was not detected in the denser [13C]DNA fractions, but it was detected in all the lighter [12C]DNA fractions (data not shown).
PCR amplification.
PCR primers for amplification of bacterial 16S rRNA gene sequences were 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (19). Archaeal 16S rRNA genes were amplified with the universal primer 1492R and Archaea-specific primer 23F (5′-TGCAGAYCTGGTYGATYCTGCC-3′) (3). PCR amplification was performed with a 96-well format iCycler PCR system (Bio-Rad, Hercules, CA) using HotStarTaq Master Mix (QIAGEN, Valencia, CA), as reported previously (36).
Cloning and RFLP analyses.
Fresh PCR amplicons obtained with universal bacterial and archaeal primers from labeled DNA were ligated into a pCRII-TOPO cloning vector and transformed into Escherichia coli TOP10F′ cells according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Colonies with inserts were screened by direct PCR amplification in a 96-well PCR plate, with promoter-specific primers SP6 and T7 (Invitrogen, Carlsbad, CA) for screening bacterial inserts and 23F in combination with 1492R for screening archaeal inserts with the previously described PCR program (36). Restriction fragment length polymorphism (RFLP) analyses were conducted using the restriction enzymes HhaI and AluI in separate reactions and electrophoresed through a 2% agarose gel. RFLP groups were assigned to operational taxonomic units (OTUs), and OTUs were subjected to analysis by the analytic rarefaction software, aRarefactWin (version 1.3; S. Holland, Stratigraphy Lab, University of Georgia, Athens; http://www.uga.edu/∼strata/software/), to confirm that sufficient numbers of RFLP groups were selected for sequencing to represent the entire microbial diversity in the libraries.
DNA sequencing and phylogenetic analysis.
At least two to three representatives of each RFLP group were sequenced at the DNA Sequencing Core Laboratory of the University of Florida with 27F and 23F primers. Chimera evaluation was performed online via Bellerophon, available at the Ribosomal Database Project II website (9). Sequences generated from this study were compared with sequences previously submitted to the National Center for Biotechnology Information database using the Basic Local Alignment Search Tool (1). Sequences were aligned with ClustalX, version 1.8 (34), and phylogenetic trees were generated with PAUP, version 4.0b8, using the maximum parsimony algorithm with default settings (D. L. Swofford, Sinauer Associates, Sunderland, Mass.). Bootstrap resampling analysis for 100 replicates was performed to estimate the confidence of tree topologies.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences obtained in this study were deposited in GenBank under accession numbers DQ173775 to DQ173822 for bacterial sequences from [13C]propionate microcosms, DQ173867 to DQ173919 for bacterial sequences from [13C]butyrate microcosms, and DQ173823 to DQ173866 for archaeal sequences from both substrates.
RESULTS
Methanogenesis rates from [13C]propionate and [13C]butyrate.
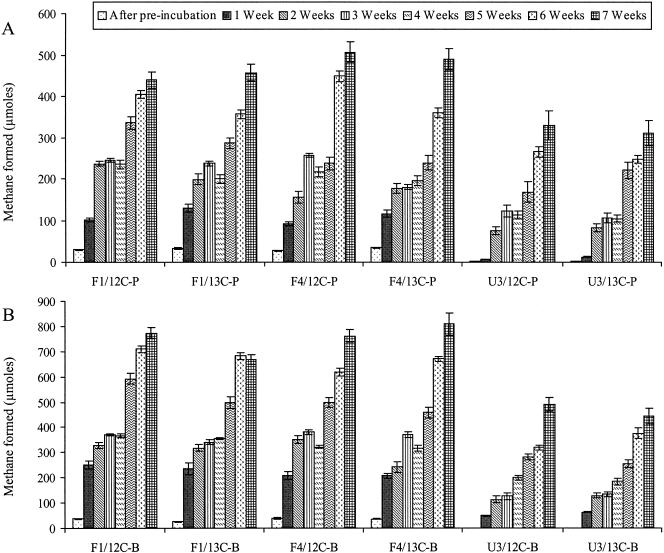
The average methanogenesis rates from 7 weeks of incubation with [13C]propionate were similar in F1 (6.7 μmol g−1 week−1) and F4 (6.1 μmol g−1 week−1). Methanogenesis rates were significantly lower in U3 microcosms (3 μmol g−1 week−1) (Fig. 1A). Methanogenesis rates with [13C]butyrate were also higher in F1 and F4 (11 to 12 μmol g−1 week−1) than in U3 microcosms (5.2 μmol g−1 week−1) (Fig. 1B). Similar rates were observed between microcosms incubated with [12C]- and [13C]-labeled substrates. A total of 400 μmol (each) of [13C]propionate and [13C]butyrate was consumed in the microcosms at the time the experiments were halted.
FIG. 1.
VFA-induced methane formation in microcosms with soil from F1 (eutrophic), F4 (transition), and U3 (oligotrophic) regions of the Florida Everglades. Propionate (A) and butyrate (B) results are shown. 12C labels indicate the addition of unlabeled carbon donor; 13C labels indicate the labeled isotope. Analyses were conducted in duplicate, and mean values are presented; error bars represent ±1 standard deviation. A second addition of [13C]butyrate or [13C]propionate was performed at 4 weeks.
Significant differences were observed in methanogenesis rates between microcosms with soils from the different regions. Propionate-induced methanogenesis in U3 was significantly lower than that observed in F1 (P ≤ 0.0001) and F4 (P ≤ 0.01) microcosms. Butyrate-induced methanogenesis in U3 was significantly lower than that observed for F1 (P ≤ 0.01) and F4 (P ≤ 0.003). Methanogenesis in F1 microcosms did not differ significantly from that observed for F4 for either substrate.
Comparisons of predicted versus observed methane production indicated significant differences between the soils with regard to the fate of the added substrates. In theory, 1 mol of propionate oxidation yields 1.75 mol of CH4, and 1 mol of butyrate oxidation yields 2.5 mol of CH4 (24, 30). Assuming that all of the fatty acids added resulted in the formation of CH4, the [13C]propionate and [13C]butyrate microcosms should yield approximately 700 and 1,000 μmol of CH4, respectively. F1 microcosms formed 65% of the predicted CH4 propionate, F4 formed 69%, and U3 yielded only 44% of the predicted CH4. For butyrate microcosms, F1 produced 67% of the predicted methane, F4 formed 81%, and U3 yielded only 44% of the predicted CH4. U3 microcosms yielded less CH4 than predicted, compared to other microcosms, suggesting a more complex fate of fatty acids in U3 soils. Previous studies indicated that terminal electron acceptors such as O2, NO3−, Fe(III), and Mn(IV) are rapidly depleted in Everglades soils, such that they likely do not play a significant role in the mineralization of organic matter (13, 27). Sulfate concentrations at the time of sampling ranged from 0.13 to 0.46 mM in F1 and F4 and 0.05 to 0.2 mM in U3 (A. Chauhan, H. F. Castro, K. Sand, K. R. Park, K. R. Reddy, and A. Ogram, Abstr. 105th Gen. Meet. Am. Soc. Microbiol., poster N-150, 2005), suggesting that SRP may be active but limited by sulfate in these sites. The relatively lower amounts of naturally available carbon than of sulfate in U3 than in F1 and F4 may have resulted in a greater importance of SRP in propionate and butyrate metabolism in U3 than in F1 and F4.
Phylogeny of bacterial sequences in [13C]propionate microcosms.
Of a total of 96 clones screened from each labeled DNA library, 14 OTUs were observed from F1, 8 were observed from F4, and 9 were observed from U3 microcosms. Most sequences were associated with either δ-Proteobacteria (see Fig. S2A in the supplemental material) or low-G+C gram-positive bacteria (see Fig. S2B in the supplemental material), and most of these sequences clustered closely with those of species previously implicated in syntrophy. A few sequences clustering with other divisions were also observed and were not included in the phylogenetic trees presented due to their low numbers. These sequences are presented as part of the total distributions within the clone libraries in Table 1.
TABLE 1.
Relative bacterial phylotype abundances from F1 (eutrophic), F4 (transition), and U3 (oligotrophic) soil microcosmsa
| Closest phylogenetic relative | Abundance (%) from:
|
|||||
|---|---|---|---|---|---|---|
| F1
|
F4
|
U3
|
||||
| Propionate | Butyrate | Propionate | Butyrate | Propionate | Butyrate | |
| Pelotomaculum sp. | 20 | 0 | 35 | 0 | 0 | 0 |
| Smithella sp. | 15 | 0 | 21 | 0 | 0 | 0 |
| Syntrophobacter sp. | 10 | 0 | 0 | 0 | 0 | 0 |
| SRP (δ-Proteobacteria) | 18 | 12 | 19 | 33 | 31 | 0 |
| Clostridium sp. | 0 | 10 | 8 | 26 | 0 | 0 |
| Pelobacter sp. | 0 | 0 | 0 | 0 | 19 | 0 |
| Syntrophospora sp. | 0 | 35 | 0 | 0 | 0 | 0 |
| Syntrophomonas sp. | 2 | 18 | 0 | 0 | 0 | 0 |
| Pelospora sp. | 0 | 0 | 0 | 10 | 0 | 60 |
| Blastocholoris sp. | 4 | 0 | 0 | 0 | 9 | 0 |
| Acidobacteria | 3 | 0 | 4 | 0 | 0 | 0 |
| Cytophaga sp. | 3 | 0 | 0 | 0 | 0 | 0 |
| Spirochaeta sp. | 6 | 0 | 4 | 8 | 0 | 0 |
| Chlorobi | 4 | 0 | 0 | 0 | 8 | 3 |
| Streptococcus sp. | 4 | 0 | 0 | 0 | 8 | 0 |
| Fusobacterium sp. | 2 | 0 | 0 | 0 | 0 | 0 |
| SBR1039 group | 4 | 0 | 0 | 0 | 0 | 0 |
| Magnetospirillum sp. | 2 | 0 | 4 | 0 | 0 | 0 |
| Eubacterium sp. | 3 | 4 | 2 | 6 | 5 | 5 |
| Planctomycetes | 0 | 0 | 3 | 0 | 0 | 5 |
| Anaerobaculum sp. | 0 | 0 | 0 | 6 | 6 | 5 |
| Haloanerobacter sp. | 0 | 5 | 0 | 0 | 0 | 0 |
| Aminobacterium sp. | 0 | 6 | 0 | 0 | 0 | 0 |
| Acidobacterium sp. | 0 | 0 | 0 | 0 | 0 | 4 |
| Bacteroidetes sp. | 0 | 4 | 0 | 5 | 0 | 5 |
| Holophaga sp. | 0 | 6 | 0 | 0 | 0 | 8 |
| Verrucomicrobia | 0 | 0 | 0 | 0 | 0 | 5 |
| Bacterium, Ellin6067 group | 0 | 0 | 0 | 6 | 8 | 0 |
| Uncultured pLW-42 group | 0 | 0 | 0 | 0 | 6 | 0 |
A total of 96 clones were compared from each library. F1, eutrophic; F4, transition; and U3, oligotrophic. Two or more representatives from each phylotype were sequenced, and 16S rRNA gene sequences were compared to their closest cultured phylogenetic relative from the National Center for Biotechnology Information database.
In F1 microcosms, 45% of the clones were closely related with known syntrophic propionate oxidizers (Table 1). Sequences associated with Pelotomaculum spp., recently identified as propionate oxidizers (12), formed the dominant group, and sequences aligning with Smithella propionicus and Syntrophobacter spp. comprised the remaining sequences associated with known propionate utilizers. Further, 2% of the sequences clustered with Syntrophomonas spp., a genus previously shown to oxidize butyrate and not propionate (30). Sequences associated with SRP constituted 18% of this guild. In F4, 56% of the sequences clustered with known propionate oxidizers, novel Pelotomaculum-like sequences, and Smithella spp. Sequences clustering with Syntrophobacter spp. were not found in this system. SRP accounted for 19% of the clones. In U3, no clones were associated with known syntrophs but were associated primarily with SRP and with Pelobacter spp.
Of particular interest in the F1 and F4 libraries is the strong representation by sequences clustering with Smithella spp. Smithella propionica has been shown to dismutate propionate to acetate and butyrate and to further metabolize butyrate to acetate (11). Relatively high proportions of Pelotomaculum and Smithella spp. were also recently observed in a stable-isotope-probing (SIP) study of propionate oxidizers in anoxic paddy soils (22). A relatively low proportion (2%) of sequences clustering with Syntrophomonas spp., a genus which includes known butyrate oxidizers, were also identified in F1 propionate microcosms; these may have fed on labeled butyrate formed by Smithella spp. or may represent a novel lineage of propionate and butyrate oxidizing bacteria, as was recently suggested by Lueders and coauthors (22).
Phylogeny of butyrate utilizers in [13C]butyrate microcosms.
Of a total of 96 clones screened from each library, 9 OTUs were observed for F1, 9 were observed for F4, and 8 OTUs were observed for U3 microcosms. As with the butyrate libraries, most sequences clustered with either the δ-Proteobacteria or the low-G+C gram-positive division, and many were associated with known syntrophs (see Fig. S3A in the supplemental material). A few sequences clustering with other divisions were also observed and were not included in the phylogenetic tree, due to their low numbers.
General trends similar to those observed in the propionate microcosms were observed in the butyrate microcosms; most sequences in F1 and F4 libraries clustered with known butyrate syntrophs (Table 1). In F1 libraries, most sequences clustered with Syntrophospora and Syntrophomonas; both genera included strains known to oxidize butyrate in anoxic environments (30). In F4, however, most clones clustered with Pelospora, a genus that, along with Syntrophospora and Syntrophomonas, belongs to the family Syntrophomonadaceae (23).
As with the propionate library, the dominant cluster in the U3 [13C]butyrate library was associated with SRP. In contrast to the U3 propionate library, sequences clustering with Clostridium sp. comprised a significant portion (26%). Clostridium species are typically considered to be primary fermenters, but some may also function syntrophically (42). Sequences clustering with Pelospora also comprised a significant proportion of this library (10%), although at a smaller percentage than in the F4 library (Table 1).
A minor proportion of propionate and butyrate clone libraries from F1, F4, and U3 clustered in lineages not identified earlier as syntrophs, including Planctomycetes, Verrucomicrobia, and Bacteroidetes (Table 1). Some clones clustered with genera known to include homoacetogens, such as Eubacterium, Clostridium, Spirochaeta, and Holophaga (20, 21). Some of these species may also function syntrophically, or they may have been labeled by cross-feeding on by-products of syntrophs.
Phylogeny of methanogens in [13C]DNA fractions.
Propionate and butyrate [13C]DNAs were screened for archaea that may have been labeled by consumption of metabolites of fatty acid oxidation, such as acetate or CO2. A total of 32 clones each from F1, F4, and U3 microcosms were screened. In propionate microcosms, four OTUs were observed from F1, seven OTUs were observed from F4, and five OTUs were observed from U3. Microcosms incubated with labeled butyrate yielded five OTUs from F1, five OTUs from F4, and seven OTUs from U3. The phylogenetic tree of these sequences is presented in Fig. S4 in the supplemental material. Many of the sequences obtained in this study clustered closely with well-known cultivated representatives of both acetotrophic and hydrogenotrophic methanogens. Of interest is the relatively deep branch within the hydrogenotrophic branch, branching with strong bootstrap support (97%) between sequences associated with Methanospirillum and Methanoculleus. It is not known at this time if this clade represents a novel genus or is merely a branch of either Methanospirillum or Methanoculleus.
Archaeal sequences from F1, F4, and U3 microcosms incubated with [13C]propionate and [13C]butyrate included sequences clustering with hydrogenotrophs, such as Methanospirillium spp., Methanoculleus sp., and Methanosaeta spp. (see Fig. S4 in the supplemental material). U3 sequences were more diverse and included sequences clustering with the acetotroph Methanosarcina sp. and 3% clustered within the Crenarchaeota (Table 2). Crenarchaeota have been recognized as a metabolically versatile group that may gain energy and carbon by either chemoorganotrophy or chemolithoautotrophy (40). Several species use CO2 as a sole carbon source and gain energy by oxidation of inorganic forms of sulfur and H2, reducing sulfur or nitrate (38). Their role in U3 soils is not known at this time.
TABLE 2.
Relative archaeal phylotype abundances from F1 (eutrophic), F4 (transition), and U3 (oligotrophic) soil microcosmsa
| Closest phylogenetic relative | Abundance (%) from:
|
|||||
|---|---|---|---|---|---|---|
| F1
|
F4
|
U3
|
||||
| Propionate | Butyrate | Propionate | Butyrate | Propionate | Butyrate | |
| Methanospirillum sp. | 40 | 43 | 43 | 43 | 20 | 20 |
| Methanoculleus sp. | 25 | 27 | 27 | 25 | 15 | 15 |
| Methanosaeta sp. | 35 | 30 | 30 | 32 | 34 | 35 |
| Methanosarcina sp. | 0 | 0 | 0 | 0 | 28 | 30 |
| Crenarchaeota | 0 | 0 | 0 | 0 | 3 | 0 |
A total of 32 clones were compared from each library. F1, eutrophic; F4, transition; and U3, oligotrophic. Two representatives from each phylotype were sequenced, and 16S rRNA gene sequences were compared to their closest cultured phylogenetic relative from the NCBI database.
DISCUSSION
Eutrophication of the northern Florida Everglades resulted in a number of significant ecosystem level changes, many of which were due to changes in the species and density of the dominant plant communities. Increases in carbon input to soils from increased plant biomass resulted in increases in the rates of various biogeochemical processes, particularly those associated with carbon metabolism, such as respiration and methanogenesis (4-7, 27, 37). Little is known of the impacts of eutrophication on specific pathways for carbon mineralization in these soils; in this study, we investigated the pathways of prokaryotes through which VFA are funneled in eutrophic and oligotrophic soils of the marsh. A range of VFA are produced via fermentation in these soils (Uz and Ogram, submitted), and they serve as important precursors for methane production.
Methanogenesis from VFA requires cooperation between secondary fermenters (syntrophs) and methanogens. Syntrophs are responsible for conversion of VFA to acetate, CO2, and H2. Fermentation of VFA yields very low levels of energy, and the products must be maintained at low concentrations for the fermentation to proceed (30). A variety of environmental factors may affect the composition and activities of syntrophic consortia, including the availability of carbon.
Investigation of syntrophic consortia in soils is limited by available methods. Limitations of standard enrichment approaches are well documented (7), and PCR-based methods are of little value at this time because no molecular marker for syntrophs, either specific or general, has been reported. SIP provides a very good alternative to studying the composition of syntrophic consortia in soils, although data obtained from SIP must be interpreted carefully. Cross-feeding by nonsyntrophs on labeled metabolic by-products of the consortia or feeding on dead labeled biomass could result in labeling of nonsyntrophs. Incubations in this study were conducted for 7 weeks, such that methanogens were sufficiently labeled to be separated from [12C]DNA by density gradient ultracentrifugation. This length of time may also have been sufficient for some labeling of nonsyntrophs. The majority of strains identified as syntrophs by these experiments were likely syntrophs, however; clone libraries constructed from [13C]DNA isolated from the eutrophic F1 and F4 microcosms incubated with either [13C]propionate or [13C]butyrate were dominated by sequences clustering with those of known syntrophs, whose primary known physiological attribute is syntrophy.
Dominance by members of the family Syntrophomonadaceae in F1 and F4 libraries is in contrast with those libraries derived from the oligotrophic U3 microcosms. Both [13C]propionate and [13C]butyrate libraries from U3 were dominated by sequences clustering with δ-Proteobacteria SRP, with significant representation by sequences associated with Clostridium sp. (26%) in the butyrate library. Sulfate concentrations in these soils typically range from 0.13 to 0.46 mM in F1 and F4 and from 0.05 to 0.2 mM in U3 soils (Chauhan et al., Abstr. 105th Gen. Meet. Am. Soc. Microbiol.). This sulfate was further diluted 1:1 (vol/vol) in BCYT-R medium, and microcosms were preincubated 1 week prior to addition of the labeled substrate. It is unlikely that sufficient SO42− was present in these microcosms to account for the oxidation of propionate; it is more likely that SRP syntrophically fermented propionate in U3 microcosms.
Clostridium spp. are typically considered to be primary fermenters, but some may also participate in syntrophic associations. For example, Clostridium bryantii has been shown to oxidize fatty acids or acetate in consortia with hydrogen-scavenging bacteria (31, 33). Clostridium bryantii was recently reassigned to Syntrophospora bryantii gen. nov., comb. nov., after careful evaluation established that it was not related to previously cultured Clostridium spp. (42). Since Syntrophospora spp. are known butyrate oxidizers, it may be that the Clostridium sequences identified in the U3 library represent a novel lineage of butyrate-oxidizing bacteria.
The composition of archaeal libraries from F1 and F4 [13C]propionate or [13C]butyrate microcosms was similar, with approximately similar proportions of clones clustering with the hydrogenotrophic methanogenic genera Methanospirillum and Methanoculleus and the acetotrophic genus Methanosaeta. In contrast, the U3 archaeal library was dominated by acetotrophic methanogens and included significant representation by the acetotrophic genus Methanosarcina. Methanosarcina species are typically capable of utilizing both H2 and acetate as electron donors (15) and are more efficient at utilizing high concentrations of acetate than is Methanosaeta (17). A previous study of methanogen phylotypes in these soils reported similar distributions of acetotrophs between F1 and U3 for samples collected during the spring (5), as for the samples used in this study. The significant representation by sequences clustering with Methanosaeta and Methanosarcina suggests that acetate accumulated with time in both propionate and butyrate U3 microcosms but did not similarly accumulate in F1 and F4 microcosms. It is not known at this time if alternative, nonmethanogenic groups in F1 and F4 are responsible for acetate metabolism or if total numbers of Methanosaeta are sufficiently high in F1 and F4 to accommodate any excess acetate. Previous studies on these soils found that potential methanogenesis rates from acetate were significantly higher in F1 soils than in U3 soils (5), although the most probable numbers of acetotrophs were almost similar in F1 and U3 soils (7).
Significantly, potential CH4 yields from propionate and butyrate were higher in F1 and F4 (65% and 69% for propionate, respectively, and 67% and 81% for butyrate, respectively) than for U3 (44% for both substrates), suggesting the possibility of a different route for mineralization of these substrates in U3 soils. Sulfate-reducing guilds in U3 are dominated by incomplete acetate oxidizers (4, 6), such that SRP may be limited in their ability to metabolize acetate in these soils. In addition, low sulfate concentrations in these microcosms likely prevented enrichment of SRP in U3 microcosms, possibly resulting in increased acetate accumulation. Stable-isotope probing of acetate metabolizing guilds in these soils would likely aid in elucidation of the fate of acetate in these soils.
Supplementary Material
Acknowledgments
This study was supported by grant DEB-0078368 from the National Science Foundation.
We thank K. Sand and P. Jasrotia for technical assistance, Sue Newman for sample collection, and Madeline Rasche for use of the ultracentrifuge.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bachoon, D., and R. D. Jones. 1992. Potential rates of methanogenesis in sawgrass marshes with peat and marl soils in the Everglades. Soil Biol. Biochem. 24:21-27. [Google Scholar]
- 3.Burggraf, S., K. O. Stetter, P. Rouviere, and C. R. Woese. 1991. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 4.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro, H. F., A. Ogram, and K. R. Reddy. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida Everglades. Appl. Environ. Microbiol. 70:6559-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro, H. F., S. Newman, K. R. Reddy, and A. Ogram. 2005. Distribution and stability of sulfate reducing prokaryotic and hydrogenotrophic methanogenic assemblages in nutrient-impacted regions of the Florida Everglades. Appl. Environ. Microbiol. 71:2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan A., A. Ogram, and K. R. Reddy. 2004. Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 70:3475-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chimney, M. J., and G. Goforth. 2001. Environmental impacts to the Everglades ecosystem: a historical perspective and restoration strategies. Water Sci. Technol. 44:93-100. [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 11.de Bok, F. A., A. J. Stams, C. Dijkema, and D. R. Boone. 2001. Pathway of propionate oxidation by a syntrophic culture of Smithella propionica and Methanospirillum hungatei. Appl. Environ. Microbiol. 67:1800-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bok, F. A., H. J. Harmsen, C. M. Plugge, M. C. de Vries, A. D. Akkermans, W. M. de Vos, and A. J. Stams. 2005. The first true obligately syntrophic propionate-oxidizing bacterium, Pelotomaculum schinkii sp. nov., co-cultured with Methanospirillum hungatei, and emended description of the genus Pelotomaculum. Int. J. Syst. Evol. Microbiol. 55:1697-1703. [DOI] [PubMed] [Google Scholar]
- 13.DeBusk, W. F., and K. R. Reddy. 1998. Turnover of detrital organic carbon in a nutrient-impacted Everglades marsh. J. Environ. Qual. 30:1438-1446. [Google Scholar]
- 14.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grieβhammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediments: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleason, P. J., and P. Stone. 1994. Age, origin and landscape evolution of the Everglades peatland, 149-197. In S. M. Davis and J. C. Ogden (ed.), Everglades: the ecosystem and its restoration. St. Lucie Press, Delray Beach, Fla.
- 17.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Ecol. 88:181-198. [Google Scholar]
- 18.Koch-Rose, M. S., K. R. Reddy, and J. P. Chanton. 1994. Factors controlling nutrient profiles in a subtropical peatland of the Everglades. J. Environ. Qual. 23:526-533. [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 20.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 21.Leaphart, A. B., and C. R. Lovell. 2001. Recovery and analysis of formyltetrahydrofolate synthetase gene sequences from natural populations of acetogenic bacteria. Appl. Environ. Microbiol. 67:1392-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lueders, T., B. Pommerenke, and M. W. Friedrich. 2004. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70:5778-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthies, C., N. Springer, W. Ludwig, and B. Schink. 2000. Pelospora glutarica gen. nov., sp. nov., a glutarate-fermenting, strictly anaerobic, spore-forming bacterium. Int. J. Syst. Evol. Microbiol. 50:645-648. [DOI] [PubMed] [Google Scholar]
- 24.Mormile, M. R., K. R. Gurijala, J. A. Robinson, M. J. McInerney, and J. M. Suflita. 1996. The importance of hydrogen in landfill fermentations. Appl. Environ. Microbiol. 62:1583-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry, W. 2004. Elements of south Florida's comprehensive Everglades restoration plan. Ecotoxicology 13:185-193. [DOI] [PubMed] [Google Scholar]
- 26.Radajewski S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 27.Reddy, K. R., J. R. White, A. Wright, and T. Chua. 1999. Influence of phosphorus loading on microbial processes in the soil and water column of wetlands, p. 249-273. In K. R. Reddy, G. A. O'Connor, and C. L. Schelske (ed.), Phosphorus biogeochemistry in subtropical ecosystems. Lewis Publishers, New York, N.Y.
- 28.Sabat, G., P. Rose, W. J. Hickey, and J. M. Harkin. 2000. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 66:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnurer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 32.Singleton, D. R., S. N. Powell, R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken. 2005. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil 71:1202-1209. [DOI] [PMC free article] [PubMed]
- 33.Stieb, M., and B. Schink. 1985. Anaerobic oxidation of fatty acids by Clostridium bryantii sp. nov., a sporeforming, obligately syntrophic bacterium. Arch. Microbiol. 140:387-390. [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Touzel, J. P., and G. Albagnac. 1983. Isolation and characterization of Methanococcus mazei strain MC3. FEMS Microbiol. Lett. 16:241-245. [Google Scholar]
- 36.Uz, I., M. E. Rasche, T. Townsend, A. V. Ogram, and A. S. Lindner. 2003. Characterization of methanogenic and methanotrophic assemblages in landfill samples. Proc. Biol. Sci. 270:S202-S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaithiyanathan, P., and C. J. Richardson. 1997. Nutrient profiles in the Everglades: examination along the eutrophication gradient. Sci. Total Environ. 205:81-95. [DOI] [PubMed] [Google Scholar]
- 38.Vorholt, J. A., D. Hafenbradl, K. O. Stetter, and R. K. Thauer. 1997. Pathways of autotrophic CO2 fixation and of dissimilatory nitrate reduction to N2O in Ferroglobus placidus. Arch. Microbiol. 167:19-23. [DOI] [PubMed] [Google Scholar]
- 39.White, J. R., and K. R. Reddy. 1999. Influence of nitrate and phosphorus loading on denitrification enzymes activity in Everglades wetland soils. Soil Sci. Soc. Am. J. 63:1945-1954. [Google Scholar]
- 40.Woese, C. R. 1993. The archaea: their history and significance, p. vii-xxviii. In M. Kates et al. (ed.), The biochemistry of archaea (Archaebacteria). Elsevier Science Publishers, Amsterdam, The Netherlands.
- 41.Wright, A. L., and K. R. Reddy. 2001. Heterotrophic microbial activities in northern Everglades wetland. Soil Sci. Soc. Am. J. 65:1856-1864. [Google Scholar]
- 42.Zhao, H. X., D. C. Yang, C. R. Woese, and M. P. Bryant. 1990. Assignment of Clostridium bryantii to Syntrophospora bryantii gen. nov., comb. nov. on the basis of a 16S rRNA sequence analysis of its crotonate-grown pure culture. Int. J. Syst. Bacteriol. 40:40-44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.