Abstract
The increased incidence of bacterial antibiotic resistance has led to a renewed search for novel antimicrobials. Avoiding the use of broad-range antimicrobials through the use of specific peptidoglycan hydrolases (endolysins) might reduce the incidence of antibiotic-resistant pathogens worldwide. Staphylococcus aureus and Streptococcus agalactiae are human pathogens and also cause mastitis in dairy cattle. The ultimate goal of this work is to create transgenic cattle that are resistant to mastitis through the expression of an antimicrobial protein(s) in their milk. Toward this end, two novel antimicrobials were produced. The (i) full-length and (ii) 182-amino-acid, C-terminally truncated S. agalactiae bacteriophage B30 endolysins were fused to the mature lysostaphin protein of Staphylococcus simulans. Both fusions display lytic specificity for streptococcal pathogens and S. aureus. The full lytic ability of the truncated B30 protein also suggests that the SH3b domain at the C terminus is dispensable. The fusions are active in a milk-like environment. They are also active against some lactic acid bacteria used to make cheese and yogurt, but their lytic activity is destroyed by pasteurization (63°C for 30 min). Immunohistochemical studies indicated that the fusion proteins can be expressed in cultured mammalian cells with no obvious deleterious effects on the cells, making it a strong candidate for use in future transgenic mice and cattle. Since the fusion peptidoglycan hydrolase also kills multiple human pathogens, it also may prove useful as a highly selective, multipathogen-targeting antimicrobial agent that could potentially reduce the use of broad-range antibiotics in fighting clinical infections.
Mastitis in dairy cattle is caused by environmental pathogens that enter the mammary gland through the teat canal when the animal is exposed to contaminated soil or bedding or during the milking process. In the United States alone, losses due to mastitis are estimated to be between $1.7 billion and $2 billon annually (46). Commercial losses from mastitis are due to poor milk quality, veterinary and antibiotic costs, and, in the most extreme cases, premature culling of infected animals. In a regional survey of Pennsylvania and New York, Staphylococcus aureus and Streptococcus agalactiae (group B streptococcus [GBS]) were shown to be responsible for up to 20% each of the mastitis cases in the region (53). Although implementation of a five-point mastitis control plan (8) has reduced the incidence of mastitis caused by S. agalactiae in some countries, the streptococcal pathogens Streptococcus dysgalactiae (group C streptococcus [GCS]) and Streptococcus uberis are still significant mastitis-causing pathogens in the United States. S. uberis is also responsible for 20% of mastitis cases in the United Kingdom (50).
Treatment of mastitis has historically been limited to the use of broad-range antibiotics such as tetracycline, penicillin, and pirlimycin, which are often <50% successful (12). The use of broad-range antibiotics, while still common, is now discouraged due to the fear that antibiotic resistance will develop in both the target organism and commensal bacteria. In addition to causing mastitis infections of dairy cattle, GBS, GCS, and S. aureus are also known human pathogens (19, 45, 54), with multidrug-resistant S. aureus being a major concern in modern health care. There is much debate and little consensus about whether antibiotic resistance can be transferred from the farm to the clinic (15, 16). However, one hyperinvasive human GBS strain has been shown to have a bovine-associated ancestor (5), providing even more impetus to the search for more pathogen-specific antimastitis agents.
In the quest for more specific agents, alternative antimicrobials such as bacteriophages (23) and phage endolysins (17, 24) are receiving more interest. Phage endolysins are produced inside the bacterial cell during phage development and participate in cell lysis by degrading the peptidoglycan of the host (3). Peptidoglycan is a major structural component of both gram-positive and gram-negative bacterial cell walls. It is composed of a complex molecule with a sugar backbone of alternating N-acetylglucosamine and N-acetyl muramic acid residues cross-linked with peptide bridges. There are three major peptidoglycan hydrolase activities, namely, (i) glycosidase, (ii) amidase, and (iii) endopeptidase activities (32). The peptidoglycan hydrolase activities of both bacterial autolysins and phage endolysins are generally localized to short modular domains (<200 amino acids [aa]) and share homology across species and genera (2, 18, 43). Bacteriophage endolysins are attracting increasing attention in the field of antimicrobials due to their high specificities. In addition, some workers have suggested that resistance to them is unlikely to develop (17, 29).
An alternative to the use of antibiotics for mastitis prevention was recently reported by Wall and colleagues (13, 51), who showed that the expression of a staphylococcal peptidoglycan hydrolase, lysostaphin, as a transgene in the mammary glands of cloned cattle resulted in cows that were resistant to a mastitis challenge with live S. aureus injected directly into the teat canal. Lysostaphin is a peptidoglycan hydrolase that is secreted as part of the natural defense of Staphylococcus simulans, presumably to give it a competitive advantage over S. aureus when both organisms are present. Lysostaphin specifically cleaves the pentaglycine bridge of the staphylococcal peptidoglycan. Significantly, it is active against multidrug-resistant strains of S. aureus (4), and it also exhibits limited activity against members of a related group of mastitis-causing bacteria, the coagulase-negative staphylococci (10). Lysostaphin, however, has no lytic activity against streptococci.
The GBS bacteriophage B30 endolysin was recently cloned and reported to be lytic for the mastitis-causing species S. agalactiae and S. dysgalactiae as well as most hemolytic streptococci (40). This 443-aa peptidoglycan hydrolase contains an endopeptidase domain (CHAP) and a lysozyme domain (Acm), each of which functions independently of the other (40). A 99% identical variant of this protein was subsequently described, with similar properties (9). The GBS B30 endolysin has no activity against S. aureus.
It is not uncommon for peptidoglycan hydrolases to possess multiple hydrolytic domains or modules (38, 43) or for these domains to act independently of each other and even maintain their enzymatic activities in the presence of a nonfunctional second domain (40). It has been shown that some pneumococcal phage lysins possess an interchangeable, C-terminal choline-binding domain that, when deleted, results in the loss of peptidoglycan hydrolase activity. However, the domain can be exchanged with other pneumococcal choline-binding domains to recreate a functional chimeric endolysin (33).
The long-term goal of this project is to create transgenic cattle expressing antimicrobials that are effective against multiple mastitis-causing bacteria. We reasoned that since the hydrolase activity modules of phage endolysins appear to act independently while maintaining their specificities, fusions of different domains might create a novel antimicrobial that could specifically target multiple mastitis-causing pathogens. We report here the construction of fusion proteins between lysostaphin and the endolysin of phage B30. These fusion proteins possess enzymatic domains that degrade both streptococcal and staphylococcal peptidoglycans and can kill living cells from either species when the cells are exposed to the enzyme externally. This is the first report of chimeric peptidoglycan hydrolase fusion proteins that maintain unique peptidoglycan hydrolase activities against both parent organisms. The antimicrobial activities of these fusion proteins were also tested in a milk environment as well as against several lactic acid bacteria used in fermented dairy products. Finally, we demonstrate that the B30 endolysin-lysostaphin fusion can be expressed in cultured mammalian cells as a prelude to the future goal of creating transgenic animals.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Gram-positive bacteria used in this study are listed in Table 1. Lactobacillus delbrueckii was grown at 37°C in MRS medium (Difco). Lactococcus lactis strains were grown at 30°C in M17 medium supplemented with 0.5% glucose. Leuconostoc and Pediococccus strains were grown at 30°C in MRS medium. Staphylococcus aureus, Streptococcus agalactiae, Streptococcus dysgalactiae, Streptococcus uberis, and Streptococcus suis were grown at 37°C in either brain heart infusion or Todd-Hewitt broth (Difco). Streptococcus salivarius was grown at 37°C in brain heart infusion broth (Difco). Streptococcus thermophilus strains were grown at 42°C in M17 medium supplemented with 0.5% lactose. Cloning and vector constructs were maintained in Escherichia coli DH5α (Invitrogen, Carlsbad, CA) or BL21(DE3) (Novagen, Madison, WI).
TABLE 1.
Gram-positive bacterial strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| Lactobacillus delbrueckii subsp. bulgaricus SMQ-97 | Industrial strain used in yogurt manufacture | QIa |
| Lactococcus lactis IL1403 | Laboratory strain | 7 |
| Lactococcus lactis SMQ-86 | Laboratory strain | 14 |
| Leuconostoc cremoris HER1286 | Mesophilic starter for viili fermentation | 44 |
| Leuconostoc mesenteroides HER1273 | Commercial cheese starter Pro 2 | 47 |
| Pediococcus pentosaceus FBB63C | Industrial strain used in sausage manufacture | 20 |
| Pediococcus acidilactici PAC 1.17 | Industrial strain used in sausage manufacture | 20 |
| Staphylococcus aureus ATCC29740 | Mastitis pathogen | ATCC |
| Streptococcus agalactiae ATCC13813 | Type strain, nonhemolytic | 35 |
| Streptococcus dysgalactiae | Mastitis pathogen isolate | USDA |
| Streptococcus salivarius ATCC25975 | Oral isolate | 21 |
| Streptococcus suis 89-9999 | Serotype 2 strain causing septicemia in pigs | 41 |
| Streptococcus thermophilus LMG18311 | Industrial strain used in yogurt manufacture | 6 |
| Streptococcus thermophilus RD534 | Industrial strain used in yogurt manufacture | 28 |
| Streptococcus thermophilus SMQ119 | Industrial strain used in yogurt manufacture | 36 |
| Streptococcus thermophilus SMQ-301 | Industrial strain used in cheese manufacture | 49 |
| Streptococcus uberis | Mastitis pathogen | USDA |
QI, Quest International, Chicago, IL.
Plasmid constructs and DNA manipulation.
The 1,329-bp B30 endolysin gene was cloned previously by Pritchard and coworkers (40). They inserted the gene between the unique NdeI and XhoI sites of pET21a (Novagen) to create pSD101. The vector pET21a has a lacZ-T7 promoter fusion that is strongly induced by IPTG (isopropyl-β-d-thiogalactopyranoside). Deleted versions of the B30 endolysin gene were constructed by introducing, with the use of PCR primers, XhoI sites (underlined) at aa codons 90 (90R [5′-TGC TAC ATG CTC GAG AGG CGT-3′]) and 182 (182R [5′-TCG GAT ACC TCG AGA ATC GT-3′]). The same forward primer (BLGF [5′-TCC GGC GTA GAG GAT CGA GAT-3′]), located at the unique BglII site (partial site is underlined) in the T7 promoter region of pSD101, was used for each amplification. PCR products were gel purified with a Qiaex kit (QIAGEN), digested with endonucleases (BglII and XhoI), desalted via Micro Bio-Spin 30 columns (Bio-Rad), and ligated to similarly restricted, gel-purified, dephosphorylated (shrimp alkaline phosphatase; Roche) pET21a vector using standard techniques. The final constructs permit the expression of the lysin truncated after aa 90 or 182 but maintain the XhoI site and His6 tag in frame.
The truncated (182 aa) and full-length (443 aa) phage endolysins were fused in frame with a previously reported lysostaphin construct (pBLG-lys) (25). In pBLG-lys, the lysostaphin coding region has been truncated to remove the 143-aa bacterial signal peptide, and two asparagine residues (N125 and N232) are mutated into glutamine residues (to remove potential eukaryotic oligosaccharide attachment sites). This lysostaphin lacks the signal sequence and is the same size as the mature form, with 246 aa. The pBLG-lys construct was digested with NotI and XbaI to obtain a 4-kb fragment containing the 246-aa lysostaphin protein coding region, a 0.6-kb bovine growth hormone poly(A) addition region, and 2.5 kb of ovine beta-lactoglobulin 3′-proximal gene sequence (25). The fragment was gel purified (Qiaex; QIAGEN), made blunt with T4 polymerase and deoxynucleoside triphosphates (Invitrogen), and ligated into the XhoI site of pSD101 or the truncated B30-182 construct, which had been blunt ended and dephosphorylated with shrimp alkaline phosphatase (Roche). Fusion of the blunt-ended, NotI- and XbaI-cut lysostaphin gene to the blunt-ended, XhoI-cut 443-aa-endolysin gene (or B30-182 gene truncation) generates an in-frame protein fusion at aa 443 (or 182) of the phage endolysin to aa 1 of the mature lysostaphin sequence.
Introduction of the 182-lysostaphin (182-Lyso) fusion into the eukaryotic expression vector pCDNA3.1 (Invitrogen) was a two-step process. First, the 182-Lyso coding region was introduced into pET21a to reconstitute the His6 tag at the C-terminal end of the fusion. Amplification of the 182-Lyso coding sequence was performed with the forward primer BLGF (described above) and a reverse primer (LYSOR [5′-GCC CTC AGT CGA CAC TAG TCC ACA GAA CAC-3′]) with a unique SalI site (underlined) inserted immediately before the translational stop signal of the lysostaphin open reading frame. The PCR product was gel purified, digested with BglII and SalI, and desalted as described above. This BglII-SalI blunt-ended fragment was ligated into pET21a that was doubly digested with BglII and XhoI and dephosphorylated as described above. Transformations were done as described by the supplier of the DH5α cells (Invitrogen, Carlsbad, CA), with the final construct verified by restriction enzyme mapping and peptidoglycan hydrolase activity measurements. Constructs containing the T7-lacZ promoter fusion were first transformed into E. coli DH5α (Invitrogen), and positive clones (by restriction enzyme mapping) were then introduced into E. coli BL21(DE3) (Novagen) cells. After induction and sonication of the cells, extracts were tested for enzyme activity. In the second step, the 182-Lyso His-tagged construct was moved into pCDNA3.1. Amplification was performed using a forward primer (KOZF [5′-TTC GAT ATC GCG GCC GCA ACC ATG GCA ACT TAT CAA-3′]) harboring an EcoRV (underlined) site immediately 5′ to a consensus eukaryotic translational start site (italics) (27) and a reverse primer (182HR [5′-CTT TCG GAA TTC GTT AGC AGC CGG AAT C-3′]) harboring an EcoRI site (underlined) following the His6 tag and translational stop signal. The amplification product was digested with EcoRI and EcoRV and subcloned into similarly cut and dephosphorylated pCDNA3.1 as described above. The inactive construct (B30-90) was DNA sequence verified. All other constructs were enzymatically active or produced an immunohistochemically verifiable product.
Peptidoglycan hydrolase expression.
All enzymatic assays were performed with extracts of E. coli BL21(DE3) cells harboring either pSD101 or the constructs described above. Cell extracts were prepared by growth of bacteria in Superbroth (Becton Dickinson) supplemented with 100 μg/ml ampicillin incubated at 37°C with shaking. At mid-log phase (optical density at 600 nm [OD600] of 0.3 to 0.5), the culture was either placed on ice for 30 min, after which induction of the T7-lacZ hybrid promoter was achieved with a final concentration of 1 mM IPTG followed by incubation at 19°C with shaking overnight (∼18 h) (26), or induced with 1 mM IPTG and incubated for 4 h with shaking at 37°C. Cells were then pelleted and washed once with lysin buffer A (50 mM ammonium acetate, 10 mM CaCl2, 1 mM dithiothreitol, pH 6.2), and pellets were frozen at −80°C. Cells were thawed, resuspended (up to 20 days later) in 1 to 3 ml lysin buffer A, and disrupted with six 5-second pulses of sonication on ice, with 5-second rest periods between pulses. Lysates were clarified with a 30-min spin in a microcentrifuge at 4°C, filter sterilized through a Millex 0.22-μm-pore-size sterile filter, and, if not used immediately, stored at −80°C.
Protein purification.
Fusion proteins were purified from E. coli cell extracts by nickel-affinity chromatography using gravity-flow QIAGEN Ni-nitrilotriacetic acid (Ni-NTA) agarose as described by the manufacturer. Briefly, cells were harvested and washed with lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8), and pellets were frozen at −80°C. Frozen cell pellets were thawed (up to 20 days later), resuspended in 10 ml lysis buffer, and disrupted with 15 10-second pulses of sonication on ice, with 10-second rest periods between pulses. Lysates were clarified with a 30-min spin at 6,800 rpm in an HS4 Sorvall rotor (8,500 × g) at 4°C and batch incubated with 5 ml of Ni-NTA slurry (QIAGEN) for 1 h at 4°C with gentle rolling. The slurry was immediately washed, and protein was eluted in elution buffer with 250 mM imidazole as described by the manufacturer. Protein eluates were desalted in Micro Bio-Spin 30 columns (Bio-Rad) prior to protein determination with the BCA protein assay (Pierce) and subsequent use in turbidity assays.
SDS-PAGE and zymogram analysis.
Zymograms were run in parallel with an identical 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, with the same gel mix used for both. Cells were added to the gel mix in lysin buffer A. These target cells were obtained from 100 ml of mid-log-phase culture, washed once with lysin buffer A, and resuspended in 1 ml of lysin buffer A. Four hundred microliters of cells in lysin buffer A and 4 μl 10% SDS were added to the zymogram gel mix prior to the addition of ammonium persulfate and TEMED (N,N,N′,N"-tetramethylethylenediamine). Four hundred microliters of lysin buffer A was substituted for the cell suspension in the control PAGE lane. Gels were run at 131 V for 1.5 h in a Bio-Rad Mini-PROTEAN 3 gel apparatus. Zymograms were soaked for 1 h in distilled water to remove SDS and then incubated at room temperature in lysin buffer A overnight. Control SDS gels were put directly into Bio-Rad BioSafe Coomassie stain (Bio-Rad) for 1 hour and then rinsed in distilled water overnight.
Plate lysis assay.
Cell lysates of the constructs were prepared, clarified, and sterilized as described above and were spotted onto 10 ml tryptic soy broth-0.7% agar plates that contained 500 μl of either heat-killed (60°C for 30 s) or live (mid-log phase) target bacteria (S. agalactiae, S. dysgalactiae, S. uberis, or S. aureus) that had been pelleted and resuspended at a 50× concentration in lysin buffer A and added to the 50°C liquid agar. Plates were allowed to incubate overnight at 4°C.
Turbidity assay.
Target cells were grown to mid-log phase (A600 of 0.4 to 0.6). Cells were pelleted and resuspended in lysin buffer A to an A600 of ∼2.0. Preincubation of 25 to 100 μl of endolysin-containing extracts at either 63°C or 37°C for 30 min was performed in 450 μl lysin buffer A with 500 μl of either whey or water added (replacing the water with lysin buffer A had no effect on the assay results). Whey was isolated from whole milk by low-speed (5,000 rpm) centrifugation for 5 min at room temperature to remove fat, followed by two other centrifugation steps at 30,000 × g for 30 min at 4°C. Whey was frozen and thawed when needed. The turbidity assay was initiated by the addition of target cells to a final OD600 of approximately 1.0 to 1.2, and changes in OD600 were recorded for 1 h. The protein concentrations of the extracts were determined by means of the BCA protein assay (Pierce). Activity was expressed as the change in OD600 per minute per mg total protein after correcting for a control value (cells alone). All activities were calculated from the linear portion of the turbidity assay, obtained from plots of time versus OD600. Changes in OD600 of <0.1 after subtracting the cell-only control value following 1 hour of incubation were considered background. All activities were derived from extracts of E. coli cells harboring the constructs.
Cell culture and immunohistochemistry.
CHO cells were cultured in standard Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (GIBCO). Transient transfection was performed with Lipofectamine (Invitrogen) per the manufacturer's instructions. Immunohistochemistry was performed as reported previously (48), using a lysostaphin primary antibody provided by David Kerr and described previously (25).
DNA sequence analysis and homology searches.
FASTA sequence comparison at the University of Virginia website (http://fasta.bioch.virginia.edu) and the NCI-Frederick Cancer Research Facility Seqweb (http://seqweb.ncifcrf.gov) GCG program were utilized for DNA sequence manipulations.
RESULTS
Lytic activities of constructs.
The B30 endolysin gene (443 codons) was previously cloned into pET21a with the addition of eight codons (the XhoI site [LE] and the His6 tag) at the end encoding the C terminus of the protein (40). This construct (B30-443) possesses both endopeptidase and lysozyme-like Acm peptidoglycan hydrolase activity domains, as illustrated in Fig. 1. A series of C-terminal deletion mutants of the GBS B30 endolysin gene were created in an E. coli vector to identify short protein fragments that maintain lytic activity. The native enzyme is fully active against S. agalactiae, S. dysgalactiae, and S. uberis but has no observable activity against S. aureus in the plate lysis assay (Fig. 1). Mutant B30-90, containing only the first 90 aa of the endolysin, interrupts the CHAP endopeptidase domain and has a deletion of the entire Acm muramidase domain. This truncated construct has no lytic activity against the streptococcal test species or S. aureus (Fig. 1). On the other hand, the truncated protein B30-182, which includes the entire CHAP endopeptidase region but bisects the Acm glycosidase domain between two essential aa (D158 and E185), is highly active (Fig. 1).
FIG. 1.
Plate lysis assays of B30 endolysin and lysostaphin constructs. Crude E. coli extracts were prepared from cells harboring pET21a-derived expression vectors. Spots of cleared lawn represent lysis of the target organisms. Lysostaphin (Sigma bact lysostaphin) in lysin buffer A was spotted as a control. B30-90 is an inactive truncation and served as a negative control. S. agal, S. agalactiae; S. aur, S. aureus; S. uber, S. uberis; S. dys, S. dysgalactiae.
To test the hypothesis that a fusion between the B30 endolysin and lysostaphin would be effective against both streptococcal and staphylococcal pathogens, two different fusion proteins were constructed. First, the 445-aa version of the B30 endolysin (minus the His6 tag) was fused to the mature form of secreted lysostaphin (aa 235 to 480) (22), which had been modified (Gln125,232) to remove potential oligosaccharide attachment sites (25), to create the final fusion, 443-Lyso. The truncated protein B30-182 was then fused to the same lysostaphin sequence to create 182-Lyso. Both fusion constructs actively lyse S. aureus and the same streptococcal test species as the parent B30 endolysin (Fig. 1).
Zymograms were run with either S. aureus or S. agalactiae cells embedded in the SDS-PAGE gel matrix to verify that the fusion proteins were the only proteins in the crude E. coli extracts that were responsible for lysis of both bacterial species (Fig. 2). In all cases, the locations of the cleared zones were similar for the crude extracts and the purified proteins, confirming that the lysis activities in the crude extracts were due to the endolysin or fusion protein constructs (Fig. 2). Moreover, the locations of the cleared zones in the zymograms are consistent with the predicted sizes of the truncation and fusion constructs (B30-443, 50,742 kDa; B30-182, 20,958 kDa; 443-L, 76,778 kDa; 182-L, 47,820 kDa) (Fig. 2). Nearly all activity was lost in the fractions containing the purified B30-182 protein after 1 week of storage at either 4°C or −80°C. On the other hand, purified B30-443 was stable for months (data not shown). There was no obvious change in size or apparent degradation of the purified B30-182 protein, as determined by SDS-PAGE, following this loss of activity. Thus, in order to obtain a high sensitivity and maintain consistency in forthcoming turbidity assays, all measurements were performed with crude E. coli extracts, as they were more active and maintained activity longer than the nickel-column-purified proteins stored at either −80°C or 4°C.
FIG. 2.
SDS-PAGE and zymogram analysis of lysin constructs. (A) Analysis of endolysin B30-443 and the fusion protein 443-Lyso. Lane M, markers; lanes 1 and 2, SDS-PAGE of E. coli crude extract and purified B30-443, respectively; lane 3, SDS-PAGE of E. coli crude extract of 443-Lyso; lanes 4 and 5, zymogram with S. agalactiae of E. coli crude extract and purified B30-443, respectively; lane 6, zymogram with S. agalactiae and E. coli crude extract of 443-Lyso; lane 7, zymogram with S. aureus and E. coli crude extract of 443-Lyso. The 443-Lyso fusion was not constructed with a His tag and thus was not purified. (B) Analysis of truncated endolysin B30-182 and the fusion protein 182-Lyso. Lane M, markers; lanes 1 and 2, SDS-PAGE of E. coli crude extract and purified B30-182, respectively; lanes 3 and 4, SDS-PAGE of E. coli crude extract and purified 182-Lyso, respectively; lanes 5 and 6, zymograms with S. agalactiae of E. coli crude extract and purified B30-182, respectively; lanes 7 and 8, zymograms with S. agalactiae of E. coli crude extract and purified 182-Lyso, respectively; lanes 9 and 10, zymograms with S. aureus of E. coli crude extract and purified 182-Lyso, respectively.
Lysis activity in the presence of whey.
With the ultimate goal of expressing antimicrobials in transgenic cows, it is important to determine if they are effective in a milk-like environment. E. coli extracts containing fusion and nonfusion proteins were preincubated at 37°C for 30 min with or without whey proteins and then tested in a turbidity reduction assay. (Whole milk [containing fat] and higher concentrations of whey were tested but were found to interfere with the OD600 measurements.) All extracts that gave clearing zones in the plate assays (Fig. 1) also showed reductions in the turbidity assay following incubation at 37°C (Table 2). As expected, full-length lysostaphin was active against S. aureus, while the full-length phage B30 endolysin had no effect (Table 2). Similarly, the B30-182 and full-length B30-443 endolysins were active against S. agalactiae and had no effect on S. aureus. The B30 endolysin-lysostaphin fusions were active against both species. However, the presence of whey generally reduced the lytic activities of all constructs by half or more, except for that of purified lysostaphin. B30-90 is an inactive B30 truncation (Fig. 1), as it showed no lysis in the turbidity assay (data not shown), and B30-90 extracts did not interfere with or enhance the activity of lysostaphin in the turbidity assay (Table 2). It is important to note that the exact levels of expression of the B30-derived proteins in E. coli extracts are unknown, and although zymogram analysis suggests similar levels of expression, a comparison of the specific activities of all constructs will only be possible when known amounts of stable and purified B30-derived proteins are assayed in turbidity reduction assays.
TABLE 2.
Effects of whey proteins on lytic activity of lysin constructsa
| Construct |
S. agalactiae growth
|
S. aureus growth
|
||
|---|---|---|---|---|
| No whey | Whey | No whey | Whey | |
| B30-443 | 0.030 | 0.017 | ||
| B30-443-Lyso | 0.058 | 0.025 | 0.127 | 0.026 |
| B30-182 | 0.020 | 0.015 | ||
| B30-182-Lyso | 0.027 | 0.014 | 0.059 | 0.011 |
| Lysostaphin | 1.772 (1.929) | 1.654 (1.103) | ||
| Lyso + B30-90 | 2.044 (ND) | 1.992 (1.428) | ||
All values represent changes in OD600/min/mg protein (E. coli extract) in the turbidity assay. Values without parentheses reflect values after a 37°C 30-minute preincubation in the presence or absence of whey. Values in parentheses are the results of a 63°C 30-minute preincubation. ND, not done. All proteins containing B30 sequences were completely inactivated by a 63°C 30-minute incubation, and these values are not shown. For the lysostaphin and “Lyso + B30-90” samples, 50 ug of purified bacterial lysostaphin (Sigma) was added as a control. B30-90 is a negative control extract from E. coli harboring the truncated B30-90 construct that does not contribute to lysis of the target cells.
Lysis spectra of lysostaphin, B30 endolysin, and chimeric enzymes.
For the purpose of expressing proteins in transgenic cow milk, it is important to know whether the antimicrobials will have a deleterious effect on the lactic acid bacteria used in the manufacture of various fermented dairy products. Turbidity assays indicated that strains of Streptococcus thermophilus used in cheese and yogurt manufacture as well as a Leuconostoc cremoris strain used in sour cream and some specialty cheeses were lysed by the fusion and nonfusion constructs (Table 3). Moreover, the oral bacterium S. salivarius, which is phylogenetically closely related to S. thermophilus, was also lysed by these enzymes (Table 3). Similar results were reported for lysis of S. salivarius ATCC 9222 with a GBS phage lysin 99% identical to the B30 endolysin (9). Other dairy lactic acid bacteria, such as Lactobacillus delbrueckii, Lactococcus lactis, and Leuconostoc mesenteroides, as well as Pediococcus acidilactici and Pediococcus pentosaceus, used in meat starter cultures, were not lysed by these antimicrobial proteins. The absence of lysis with Lactobacillus is consistent with a previous report for another Lactobacillus delbrueckii subspecies (40). Fifty micrograms of purified bacterial lysostaphin did not lyse any of the milk-processing bacteria tested (data not shown).
TABLE 3.
Effect of B30 endolysin and fusion constructs on down stream milk processing bacteria
| Strain | Effect of constructa
|
|||
|---|---|---|---|---|
| B30-443 | B30-443-Lyso | B30-182 | B30-182-Lyso | |
| Streptococcus agalactiae ATCC 13813 | 0.025 | 0.056 | 0.012 | 0.035 |
| Streptococcus thermophilus SMQ-301 | 0.079 | 0.107 | 0.031 | 0.085 |
| Streptococcus thermophilus SMQ-119 | 0.114 | 0.213 | 0.014 | 0.129 |
| Streptococcus thermophilus RD534 | 0.148 | 0.159 | 0.096 | 0.125 |
| Streptococcus thermophilus LMG18311 | 0.057 | 0.078 | 0.015 | 0.050 |
| Streptococcus suis 89-9999 | 0.001 | 0.003 | 0.003 | 0.003 |
| Streptococcus salivarius ATCC25975 | 0.004 | 0.035 | 0.005 | 0.007 |
| Leuconostoc cremoris HER1286 | 0.011 | 0.029 | 0.006 | 0.008 |
Values represent changes in OD600/min/mg protein (E. coli extract) in the turbidity assay.
Effect of pasteurization on antimicrobial activity.
The presence of endolysins in milk with bactericidal activities against cheese-making bacteria could potentially reduce the usefulness of milk from transgenic cows expressing these constructs. However, in most countries, milk is pasteurized before reaching the consumer or being used in cheese making. In order to test the effect of pasteurization on these lysins, E. coli extracts containing the fusion and nonfusion proteins were tested for activity after a preincubation for 30 min at 63°C with or without the addition of whey. The pasteurization treatment completely inactivated the B30 native endolysin and all B30-derived constructs, with or without lysostaphin, in the presence or absence of whey. These data suggest that the presence of B30 endolysin (or its derivatives) in pasteurized milk should not affect dairy bacteria and milk processing. Interestingly, 50 μg of pure lysostaphin was not inactivated by the heat treatment (Table 2).
Expression of 182-Lyso in mammalian cells.
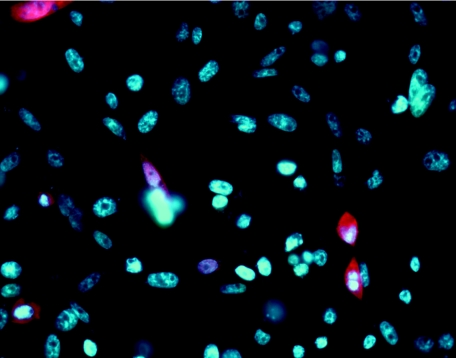
Because of its lower molecular mass, its lytic activity against all of the mastitis pathogens tested in this work (S. agalactiae, S. aureus, S. dysgalactiae, and S. uberis), and its inactivation by pasteurization, the 182-Lyso fusion is a strong candidate for further study. As a first step toward making a bovine transgene construct, the 182-Lyso construct was introduced into the pCDNA3.1 eukaryotic expression vector. A Kozak translational start sequence (27) was added to the vector, which utilizes the cytomegalovirus promoter for high expression. Chinese hamster ovary (CHO) cells were transiently transfected with this construct, and immunohistochemistry with lysostaphin antibodies demonstrated intracellular expression of the fusion protein at 48 h posttransfection (Fig. 3).
FIG. 3.
Immunohistochemistry of 182-Lyso expression in CHO cells with lysostaphin antibody. Transiently transfected CHO cells are shown. Blue, DAPI (4′,6′-diamidino-2-phenylindole)-stained nuclei; red, Alexa 594 chromophore.
DISCUSSION
Two peptidoglycan hydrolases, B30 endolysin and lysostaphin, with bactericidal activity against two different mastitis-causing pathogens have been fused. The fusion protein maintains the lytic properties of both parent proteins against hemolytic streptococci and S. aureus. The B30 endolysin was known to lyse mastitis-causing pathogens such as S. agalactiae (GBS) and S. dysgalactiae (GCS) (40). Here we show that Streptococcus uberis can now be added to this list. Lysostaphin is known to target a fourth major mastitis-causing organism, i.e., S. aureus. When the B30 endolysin is fused to lysostaphin, the product is bactericidal for all four of these mastitis-causing pathogens, which cumulatively account for most of the mastitis cases in the world. Both lysostaphin and the B30 phage endolysin are able to lyse intact bacterial cells when exposed externally. There are some examples of functional pneumococcal chimeric phage endolysins in which the cell wall (choline) binding domains were exchanged (33). However, this is the first report of a chimeric fusion endolysin that maintains both parental lytic activities. The species specificity of each parental lysin makes them strong candidates for creating a mastitis-resistant transgenic cow.
The B30 endolysin contains two hydrolytic domains, a CHAP endopeptidase domain and an Acm muramidase domain. Site-directed inactivation of these catalytic domains has shown that each domain is sufficient to cause lysis of S. agalactiae (40). In order to determine if a shortened B30 endolysin harboring just one complete domain would be sufficient to lyse S. agalactiae, the B30 endolysin was truncated at aa 182. This truncated protein, B30-182, lyses not only S. agalactiae, but also S. dysgalactiae, S. uberis, and even S. thermophilus. The >80% removal of the Acm domain, including the highly conserved Glu 185 residue that is essential for glycosidase function (40), indicates that the observed lytic activity is undoubtedly due to the endopeptidase activity (the endopeptidase domain homology resides between aa 6 and 107; Fig. 1). We suspect that the additional amino acids after the CHAP domain may help to ensure proper folding of the enzyme. This strong lytic activity of the truncated protein is particularly interesting because the truncation not only removes the muramidase domain but also deletes the C-terminal SH3b binding domain (40, 52). Specific binding of peptidoglycan hydrolases to bacterial cell walls is often conferred by the bacterial SH3 (SH3b) cell wall binding domain (30). The B30 endolysin has an SH3b domain located between aa 354 and 443 (http://www.ebi.uniprot.org/entry/Q8HA43). The absence of the SH3b domain in the B30-182 construct indicates that it is not essential for activity, consistent with a recent study of Bacillus (34). The C-terminal 92 aa of lysostaphin constitute an SH3b domain that directs the protein to the cell wall of S. aureus and is essential for its specificity (1). The presence of two SH3b binding domains in the 443-Lyso construct (one from B30 endolysin and one from lysostaphin) clearly does not interfere with the multiple hydrolase specificities of the fusion protein (Tables 2 and 3; Fig. 1).
It is interesting that the full-length B30 endolysin (B30-443) can be readily reconstituted in a zymogram, as can both of the B30-lysostaphin fusion constructs (443-Lyso and 182-Lyso) (Fig. 2). However, for the B30-182 construct (Fig. 2B, lanes 5 and 6), a clear zone was more difficult to obtain using either the purified protein or the crude extracts, which was likely due to an incorrect or partial refolding of the protein following the removal of SDS.
It has been reported that the removal of choline binding domains from the C termini of some pneumococcal lysins greatly reduces the peptidoglycan hydrolase activity (33). However, there are other cases in which truncated lysins maintain their lytic activity (34, 37). One possible explanation for how our truncated lysin recognizes peptidoglycan is that the CHAP domain also plays a role in substrate recognition, in addition to its catalytic role.
As is true for most domains, the SH3b domain aa sequence is not identically conserved between proteins, but rather shows conservation at key positions and functional side chains, allowing much variance in the primary aa sequence (39). When comparing the lysostaphin SH3b sequence to the B30 endolysin SH3b sequence, there is only weak homology at the C termini of both domains (Fig. 4A). These weakly conserved lysostaphin sequences might contribute to streptococcal cell wall binding of the 182-Lyso fusion, helping to explain the range of streptococci lysed by this construct. However, there are no predicted SH3b sequences present in the B30-182 construct, which shows an identical lytic range of streptococci. A similar sequence comparison between the B30-182 sequence and the full-length B30 SH3b domain (aa 354 to 443) (Fig. 4B) also revealed weak homology. Experiments are in progress to determine if either of these weakly conserved sequences is sufficient to bind to either S. aureus or streptococcal cell walls.
FIG. 4.
SH3b sequences in B30 endolysin and lysostaphin proteins. (A) Conserved sequences between SH3b domains of lysostaphin and the B30 endolysin. (B) Conserved sequences between the B30 SH3b domain and the intrahydrolytic region of the B30 endolysin. Solid underlining shows the Acm lysozyme domain (aa 145 to 344); dashed underlining shows the B30 SH3b domain (aa 356 to 443).
It is of interest to understand how the truncated B30-182 and 182-Lyso proteins recognize the streptococcal peptidoglycan in the absence of the B30 endolysin SH3b domain. This concern might prove important in the bovine mammary gland, where both pathogens could be present simultaneously. A preference of the 182-Lyso fusion protein for S. aureus due to the presence of the full-length lysostaphin SH3b domain could be suboptimal for control of streptococcal pathogens. Future experiments are required to address this concern, to determine if the interhydrolytic domain of B30 endolysin can bind to streptococcal cell walls, and to determine if the B30-182 construct cleaves the correct peptide bond with the same high specificity as the full-length protein.
Needless to say, the mammary gland and milk are very complex environments, and these initial studies do not address many factors that will become important when expressing the fusion protein in the mammary gland. We do not know if the fusion protein will be inactivated or bind to other components (fat and proteins) found in whole milk, effectively removing the lysin from action against the target pathogens. Other concerns include the unknown effect of a secreted fusion protein on mammary gland physiology. Many of these concerns can only be addressed in live animals. We have expressed the 182-Lyso protein in CHO cells to show that it can be expressed in a mammalian cell and was not lethal. This expression gives us confidence that the introduction of the Kozak translational start site and the codon usage of the fusion lysin are acceptable for animal studies. We hope to create a transgenic mouse with a secreted fusion protein using the same mammary expression vector, as previously reported (25).
Despite these concerns, it is encouraging that the 182-Lyso fusion protein was effective at 37°C in a milk-like environment and was inactivated during pasteurization conditions. Inactivation during pasteurization is critical due to the effect that the fusion can have on some lactic acid bacteria. Interestingly, bacterial lysostaphin itself was not inactivated by pasteurization, but the fusion proteins lost all activity during heat treatment. The lysostaphin protein used in this study contains two aa changes. The Glu 125 and 232 residues were converted to Lys (Gln125,232Lys) to remove putative glycosylation sites that interfered with eukaryotic expressed protein activity (25). It is possible that one or both of these residues are critical for the heat protection observed for the wild-type protein, but we suspect that it is more likely that the attached B30 sequences (which are inactivated by high heat exposure) are deleterious to the conformational requirements necessary for the lysostaphin endopeptidase to function while it is a part of the fusion protein(s). Although it is highly unlikely that the lysin would survive in the human gut, it is still desirable to inactivate it prior to human consumption. The presence of antimicrobials in foodstuffs is a worldwide concern, due in part to the high rate of resistant strains that are becoming apparent in hospitals worldwide. Lysostaphin-resistant strains of S. aureus have been reported and characterized (11). Many of the mastitis-causing bacteria that would potentially be destroyed by this B30-lysostaphin fusion protein are also causative agents of human diseases, further augmenting concerns about human pathogens developing antibiotic resistance. However, there is no evidence that resistance to a bacteriophage endolysin can develop (17, 29).
A concern that needs to be addressed in the use of bacteriophage endolysins as antimicrobials is the possibility of a host immune response to a foreign protein. Antibiotics avoid the immune response due to their small size. This is also not an issue for transgenic animals because the transgene product is recognized as self in the developing embryo. Antigenic responses in people are unlikely to be induced by external exposure to or ingestion of bacterial endolysins. There are many infections, such as acute pharyngitis in children caused by group C streptococci (S. dysgalactiae) (54), potentially fatal neonatal infections caused by GBS (42), and nocosomial infections in hospitals caused by staphylococcus, that might be reduced or cured by such an antimicrobial. Several animal models have been reported to test the use of bacteriophage endolysins in external or mucosal membrane applications (9, 17; reviewed in reference 31).
This designer antimicrobial approach demonstrates the facility of constructing multifunctional but highly specific protein antimicrobials appropriate for the treatment of any nonsystemic infectious diseases in animals and humans that can be caused by multiple unrelated pathogens. Currently, only broad-range antibiotics have this facility. Bacteriophage endolysins have the added advantage of being refractory to resistance development in their target organism despite repeated in vitro efforts to identify resistant strains (17, 29). Considering the high diversity within bacterial and bacteriophage populations, opportunities for novel antimicrobial design appear prodigious and might help reduce the ever-increasing incidence of antibiotic resistance that plagues therapeutic efforts worldwide.
Acknowledgments
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
We thank Julie Samson, Juli Foster-Frey, and Denise Tremblay for technical assistance with enzymatic activity, Max Paape, USDA, for bacterial strains, and David Kerr, University of Vermont, for the lysostaphin antibody.
This work was funded in part by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (S.M.) and by funds from Public Health Service grant AI054897 (D.G.P.).
REFERENCES
- 1.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2002. Breaking free: “protein antibiotics” and phage lysis. Res. Microbiol. 153:493-501. [DOI] [PubMed] [Google Scholar]
- 4.Bhakta, M., S. Arora, and M. Bal. 2003. Intraspecies transfer of a chloramphenicol-resistance plasmid of staphylococcal origin. Indian J. Med. Res. 117:146-151. [PubMed] [Google Scholar]
- 5.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B Streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramley, A. J., and F. H. Dodd. 1984. Reviews of the progress of dairy science: mastitis control—progress and prospects. J. Dairy Res. 51:481-512. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Q., D. Nelson, S. Zhu, and V. A. Fischetti. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 49:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisani, G., P. E. Varaldo, G. Grazi, and O. Soro. 1982. High-level potentiation of lysostaphin anti-staphylococcal activity by lysozyme. Antimicrob. Agents Chemother. 21:531-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeHart, H. P., H. E. Heath, L. S. Heath, P. A. LeBlanc, and G. L. Sloan. 1995. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 61:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deluyker, H. A., S. N. Van Oye, and J. F. Boucher. 2005. Factors affecting cure and somatic cell count after pirlimycin treatment of subclinical mastitis in lactating cows. J. Dairy Sci. 88:604-614. [DOI] [PubMed] [Google Scholar]
- 13.Donovan, D. M., D. E. Kerr, and R. J. Wall. 2005. Engineering disease resistant cattle. Transgenic Res. 14:563-567. [DOI] [PubMed] [Google Scholar]
- 14.Emond, E., B. J. Holler, I. Boucher, P. A. Vandenbergh, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1997. Phenotypic and genetic characterization of the phage abortive infection AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferber, D. 2002. Antibiotic resistance. Livestock feed ban preserves drugs' power. Science 295:27-28. [DOI] [PubMed] [Google Scholar]
- 16.Ferber, D. 2003. Antibiotic resistance. WHO advises kicking the livestock antibiotic habit. Science 301:1027. [DOI] [PubMed] [Google Scholar]
- 17.Fischetti, V. A. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Ann. N. Y. Acad. Sci. 987:207-214. [DOI] [PubMed] [Google Scholar]
- 18.Garcia, P., J. L. Garcia, E. Garcia, J. M. Sanchez-Puelles, and R. Lopez. 1990. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene 86:81-88. [DOI] [PubMed] [Google Scholar]
- 19.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez, C. F., and B. S. Kunka. 1987. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl. Environ. Microbiol. 53:2534-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton, I. R. 1968. Synthesis and degradation of intracellular polyglucose in Streptococcus salivarius. Can. J. Microbiol. 14:65-77. [DOI] [PubMed] [Google Scholar]
- 22.Heinrich, P., R. Rosenstein, M. Bohmer, P. Sonner, and F. Gotz. 1987. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol. Gen. Genet. 209:563-569. [DOI] [PubMed] [Google Scholar]
- 23.Huff, W. E., G. R. Huff, N. C. Rath, J. M. Balog, and A. M. Donoghue. 2005. Alternatives to antibiotics: utilization of bacteriophage to treat colibacillosis and prevent foodborne pathogens. Poult. Sci. 84:655-659. [DOI] [PubMed] [Google Scholar]
- 24.Jado, I., R. Lopez, E. Garcia, A. Fenoll, J. Casal, and P. Garcia. 2003. Spanish pneumococcal infection study network. Phage lytic enzymes as therapy for antibiotic-resistant Streptococcus pneumoniae infection in a murine sepsis model. J. Antimicrob. Chemother. 52:967-973. [DOI] [PubMed] [Google Scholar]
- 25.Kerr, D. E., K. Plaut, A. J. Bramley, C. M. Williamson, A. J. Lax, K. Moore, K. D. Wells, and R. J. Wall. 2001. Lysostaphin expression in mammary glands confers protection against staphylococcal infection in transgenic mice. Nat. Biotechnol. 19:66-70. [DOI] [PubMed] [Google Scholar]
- 26.Korf, U., T. Kohl, H. van der Zandt, R. Zahn, S. Schleeger, B. Ueberle, S. Wandschneider, S. Bechtel, M. Schnolzer, H. Ottleben, S. Wiemann, and A. Poustka. 2005. Large-scale protein expression for proteome research. Proteomics 5:3571-3580. [DOI] [PubMed] [Google Scholar]
- 27.Kozak, M. 1984. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12:857-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lévesque, C., M. Duplessis, J. Labonté, S. Labrie, C. Fremaux, D. Tremblay, and S. Moineau. 2005. Genomic organization and molecular analysis of the virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 71:4057-4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 30.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 31.Loessner, M. J. 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8:480-487. [DOI] [PubMed] [Google Scholar]
- 32.Lopez, R., and E. Garcia. 2004. Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 28:553-580. [DOI] [PubMed] [Google Scholar]
- 33.Lopez, R., E. Garcia, P. Garcia, and J. L. Garcia. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3:199-211. [DOI] [PubMed] [Google Scholar]
- 34.Low, L. Y., C. Yang, M. Perego, A. Osterman, and R. C. Liddington. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433-35439. [DOI] [PubMed] [Google Scholar]
- 35.Melancon, D., and D. Grenier. 2003. Production and properties of bacteriocin-like inhibitory substances from the swine pathogen Streptococcus suis serotype 2. Appl. Environ. Microbiol. 69:4482-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moineau, S., S. A. Walker, B. J. Holler, E. R. Vedamuthu, and P. A. Vandenbergh. 1995. Expression of a Lactococcus lactis phage resistance mechanism by Streptococcus thermophilus. Appl. Environ. Microbiol. 61:2461-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, M., Y. Tanji, Y. Orito, K. Mizoguchi, A. Soejima, and H. Unno. 2001. Functional analysis of antibacterial activity of Bacillus amyloliquefaciens phage endolysin against gram-negative bacteria. FEBS Lett. 500:56-59. [DOI] [PubMed] [Google Scholar]
- 38.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729-745. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard, D. G., S. Dong, J. R. Baker, and J. A. Engler. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079-2087. [DOI] [PubMed] [Google Scholar]
- 41.Quessy, S., J. D. Dubreuil, M. Caya, and R. Higgins. 1995. Discrimination of virulent and avirulent Streptococcus suis capsular type 2 isolates from different geographical origins. Infect. Immun. 63:1975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regan, J. A., M. A. Klebanoff, R. P. Nugent, D. A. Eschenbach, W. C. Blackwelder, Y. Lou, R. S. Gibbs, P. J. Rettig, D. H. Martin, and R. Edelman. 1996. Colonization with group B streptococci in pregnancy and adverse outcome. VIP Study Group. Am. J. Obstet. Gynecol. 174:1354-1360. [DOI] [PubMed] [Google Scholar]
- 43.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 44.Saxelin, M.-L., E.-L. Nurmiaho-Lassila, V. T. Meriläinen, and R. I. Forsén. 1986. Ultrastructure and host specificity of bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and Leuconostoc cremoris from Finnish fermented milk “Viili.” Appl. Environ. Microbiol. 52:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah, P. M. 2005. The need for new therapeutic agents: what is the pipeline? Clin. Microbiol. Infect. 3:36-42. [DOI] [PubMed] [Google Scholar]
- 46.Sordillo, L. M., and K. L. Streicher. 2002. Mammary gland immunity and mastitis susceptibility. J. Mammary Gland Biol. Neoplasia 7:135-146. [DOI] [PubMed] [Google Scholar]
- 47.Sozzi, T., J. M. Poulin, R. Maret, and R. Pousaz. 1978. Isolation of a bacteriophage of Leuconostoc mesenteroides from dairy products. J. Appl. Bacteriol. 44:159-161. [Google Scholar]
- 48.Talbot, N. C., A. M. Powell, and W. M. Garrett. 2002. Spontaneous differentiation of porcine and bovine embryonic stem cells (epiblast) into astrocytes or neurons. In Vitro Cell Dev. Biol. Anim. 38:191-197. [DOI] [PubMed] [Google Scholar]
- 49.Tremblay, D. M., and S. Moineau. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63-76. [DOI] [PubMed] [Google Scholar]
- 50.Veterinary Laboratories Agency and Scottish Agricultural College Survey. VIDA table 4. [Online.] http://www.defra.gov.uk/corporate/vla/science/documents/science-vida-cattle95-02.pdf.
- 51.Wall, R. J., A. M. Powell, M. J. Paape, D. E. Kerr, D. D. Bannerman, V. G. Pursel, K. D. Wells, N. C. Talbot, and H. W. Hawk. 2005. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat. Biotechnol. 23:445-451. [DOI] [PubMed] [Google Scholar]
- 52.Whisstock, J. C., and A. M. Lesk. 1999. SH3 domains in prokaryotes. Trends Biochem. Sci. 24:132-133. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, D. J., R. N. Gonzalez, and H. H. Das. 1997. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 80:2592-2598. [DOI] [PubMed] [Google Scholar]
- 54.Zaoutis, T., M. Attia, R. Gross, and J. Klein. 2004. The role of group C and group G streptococci in acute pharyngitis in children. Clin. Microbiol. Infect. 10:37-40. [DOI] [PubMed] [Google Scholar]