Abstract
Toxigenic Vibrio cholerae, rarely isolated from the aquatic environment between cholera epidemics, can be detected in what is now understood to be a dormant stage, i.e., viable but nonculturable when standard bacteriological methods are used. In the research reported here, biofilms have proved to be a source of culturable V. cholerae, even in nonepidemic periods. Biweekly environmental surveillance for V. cholerae was carried out in Mathbaria, an area of cholera endemicity adjacent to the Bay of Bengal, with the focus on V. cholerae O1 and O139 Bengal. A total of 297 samples of water, phytoplankton, and zooplankton were collected between March and December 2004, yielding eight V. cholerae O1 and four O139 Bengal isolates. A combination of culture methods, multiplex-PCR, and direct fluorescent antibody (DFA) counting revealed the Mathbaria aquatic environment to be a reservoir for V. cholerae O1 and O139 Bengal. DFA results showed significant clumping of the bacteria during the interepidemic period for cholera, and the fluorescent micrographs revealed large numbers of V. cholerae O1 in thin films of exopolysaccharides (biofilm). A similar clumping of V. cholerae O1 was also observed in samples collected from Matlab, Bangladesh, where cholera also is endemic. Thus, the results of the study provided in situ evidence for V. cholerae O1 and O139 in the aquatic environment, predominantly as viable but nonculturable cells and culturable cells in biofilm consortia. The biofilm community is concluded to be an additional reservoir of cholera bacteria in the aquatic environment between seasonal epidemics of cholera in Bangladesh.
Toxigenic Vibrio cholerae O1 and O139 are causative agents of cholera (38), an acute dehydrating diarrhea, which occurs in epidemic (13, 31) and pandemic (23) forms. Since the first pandemic was recorded in 1817, as many as seven cholera pandemics have occurred (35). The most recent, the seventh pandemic, began in Indonesia (14), but cholera pandemics have usually begun in the Gangetic delta of the Indian subcontinent and then in other continents (10, 40). Of the 206 O serogroups of V. cholerae, serovar O1 was the only recognized cause of cholera until late 1992. At that time an outbreak of acute watery diarrhea clinically resembling cholera erupted in India and southern Bangladesh (1, 10). The bacterium causing cholera-like diarrhea failed to agglutinate with any of the then existing 138 V. cholerae O antisera (1) and was thus designated O139 with the synonym “Bengal” to commemorate its emergence in the coast of the Bay of Bengal. Since then, O1 and O139 remain the two recognized serogroups causing epidemics of cholera.
Epidemiological studies of V. cholerae O139, including its emergence, prevalence, and coexistence with O1 El Tor V. cholerae, have been conducted primarily in Bangladesh and India via systematic surveillance (19). In the Ganges delta region, cholera outbreaks occur seasonally (13, 14), but variations in prevalence of the two epidemic serogroups O1 and O139 of V. cholerae are distinct (11). Despite being autochthonous to the aquatic environment (5, 21, 30), toxigenic V. cholerae O1 is isolated only infrequently from surface water by culturing methods during epidemic periods (20) and in interepidemic periods it is rarely isolated in culture (18). However, fluorescent monoclonal antibody combined with molecular genetics-based methods have clearly demonstrated the presence of V. cholerae O1 in the aquatic environment during the interepidemic periods, which is now known as the viable-but-nonculturable state (6, 16, 18, 34, 37). The same is assumed to be true for V. cholerae O139 (19), since reports of detection and isolation of V. cholerae O139 from water samples are few (20).
After an initial appearance in the coastal areas of southern Bangladesh in 1992 (1), recurrent cholera caused by V. cholerae O139 indicates a ubiquitous and continued presence in the region (11). While correlations of sea surface temperature and sea surface height in the Bay of Bengal with onset of cholera epidemics have been established in Bangladesh (7, 28, 30), field studies in the Bay of Bengal have not been done. Hence, little is known about the geographic distribution of toxigenic strains of V. cholerae O1 and O139. The present study was undertaken to determine where and how V. cholerae O1 and O139 live in the aquatic environment immediately adjacent to the Bay of Bengal. Data showing the natural occurrence of V. cholerae in the region adjacent to the Bay of Bengal, with the bacteria predominantly in the nonculturable state between epidemics and as aggregates of structured biofilms, are provided here.
MATERIALS AND METHODS
Sample collection and processing.
Water and plankton samples were collected every 2 weeks between March and December 2004 from six ponds (natural bodies of water used by resident people for washing utensils and drinking) located in Mathbaria (Fig. 1), which is geographically adjacent to the coast of the Bay of Bengal and approximately 400 km southwest of Dhaka, the capital city of Bangladesh. For purposes of comparison, water and the plankton samples were also collected in July 2004 from a flowing river in Matlab, a site recognized to be endemic for cholera and located approximately 50 km southeast of Dhaka. All samples were collected by using aseptic techniques in sterile dark Nalgene bottles (Nalgene Nunc International, St. Louis, Mo.), placed in an insulated plastic box, and transported at ambient air temperature from the site of collection to the central laboratory of the International Center for Diarrheal Disease Research, Bangladesh (ICDDR,B), in Dhaka. All samples were processed the following day, with approximately 20 h elapsing between sample collection in the field and processing in the laboratory.
FIG. 1.
Map of Bangladesh showing areas where environmental samples were collected. Mathbaria and Matlab are two of the important foci of cholera endemicity in this region.
For sample collection, 100 liters of water was filtered successively through 64- and 20-μm-pore-size nylon nets (Millipore Corp., Bedford, Mass.) (with a 64-μm-pore-size [64-μm] net placed sequentially in front of the 20-μm nylon net, with each having a collecting bucket at the base), and 50-ml portions of the concentrates were collected initially to determine crude estimations of the zooplankton and phytoplankton, respectively. During this process of filtration, 200 ml of filtrate water from the 20-μm mesh net was collected as representative of water, to be analyzed for planktonic (unattached, free-living) bacteria. Both 64- and 20-μm plankton samples were further concentrated in laboratory using respective size plankton nets (specially devised, netted plastic beakers) to a final volume of 5 ml each. For bacteriological analysis, the plankton samples were crushed by using a glass homogenizer (Elberbach Corp., Ann Arbor, Mich.) to release attached bacteria. Such homogenates were used for multiplex-PCR (M-PCR), direct fluorescent antibody (DFA), and direct plating analyses, as well as for enrichment of V. cholerae in alkaline peptone water (APW; Difco Laboratories, Detroit, Mich.). Water samples were concentrated by filtration through a 0.22-μm-pore-size bacteriological membrane filter (Millipore), and the retained contents on the membrane filter were washed into phosphate-buffered saline (pH 8.0) for the reasons described above. The DFA was also carried out on whole plankton samples when homogenates tested positive for V. cholerae O1 and/or O139.
Enrichment and plating.
Samples were enriched in APW at 37°C for 6 to 8 h before plating as described previously (18, 20). Approximately 5 μl of enriched APW broth was streaked by using an inoculating loop onto both thiosulfate-citrate-bile salts-sucrose (Eiken, Tokyo, Japan) and taurocholate-tellurite-gelatin agar (Difco) and incubated at 37°C for 18 to 24 h. Colonies with the characteristic appearance of V. cholerae were confirmed by biochemical and serological tests and, in the case of the latter, by using polyvalent and monoclonal antibodies specific for V. cholerae O1 or O139 produced by the ICDDR,B and, finally, by molecular methods (33).
DFA.
DFA counting was done according to a method described elsewhere (2). Briefly, samples were preincubated overnight, in the dark, with 0.025% yeast extract (Difco) and 0.002% nalidixic acid (Sigma). The samples were then centrifuged, and the pellet was stained with fluorescein isothiocyanate-labeled antiserum specific for O1 or O139 obtained from New Horizon Diagnostic Corp. (Columbia, Md.). Stained samples were observed under UV light by using an epifluorescence microscope (Olympus BX51) connected to a digital camera (Olympus DP20).
PCR.
A single-primer-pair PCR for the amplification of V. cholerae species-specific gene ompW, encoding outer membrane protein OmpW, was carried out as described elsewhere (33). The genes responsible for O-antigen biosynthesis and for generation of serotype-specific determinants are located in the rfb region on the V. cholerae chromosome. The rfb genes specific for V. cholerae O1 and O139, and the ctxA gene, encoding subunit A of cholera toxin, were amplified by using M-PCR. Samples preincubated overnight with 0.025% yeast extract and 0.002% nalidixic acid were subjected to PCR amplification by using a method described previously (15). Briefly, broth was centrifuged at 10,000 rpm to collect cell pellets and to extract DNA. The cell pellets were diluted 10-fold in 10 mM Tris-HCl (pH 8.0) buffer containing 1 mM EDTA disodium salt (EDTA) and boiled for 10 min. After centrifugation at 13,000 rpm for 10 min at 4°C, the supernatant was used as a template for PCR (15). Primer sequences for genes, e.g., V. cholerae O1 rfb, V. cholerae O139 rfb, and ctxA, are listed in Table 1. Amplification with the three primer pair (O1 rfb, O139 rfb, and ctxA [forward and reverse for each pair]) genes was performed simultaneously in 0.2-ml microcentrifuge tubes. Samples (3 μl) were added to the PCR mixture to achieve a 30-μl final volume containing a 0.21 mM concentration of each deoxynucleoside triphosphate mixture, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 0.17 μM ctxA primer pair, 0.27 μM each of the O1 and O139 rfb primer pairs, and 0.75 U of Taq polymerase (Takara, Kyoto, Japan). Amplification conditions used were 5 min at 94°C for initial denaturation of DNA and 35 cycles, each consisting of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final round of extension for 7 min at 72°C in a DNA RoboCycler gradient temperature cycler (Stratagene, La Jolla, Calif.). After amplification, 6 μl of each reaction mixture was subjected to electrophoresis on a 3% agarose gel (11 by 14 cm) using a horizontal electrophoresis apparatus (Horizon 11.14; Life Technologies/Gibco-BRL). The gel containing the amplified DNA was stained with ethidium bromide and visualized with a UV transilluminator, and images of the transilluminator were digitized with a one-dimensional gel documentation system (Bio-Rad).
TABLE 1.
PCR primers used in this study
| Primer no. | Primer name | Primer sequence | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| 1 | ctxAF | 5′-CTCAGACGGGATTTGTTAGGCACG-3′ | 302 | 15 |
| 2 | ctxAR | 5′-TCTATCTCTGTAGCCCCTATTACG-3′ | 302 | 15 |
| 3 | O1 rfbF | 5′-GTTTCACTGAACAGATGGG-3′ | 192 | 15 |
| 4 | O1 rfbR | 5′-GGTCATCTGTAAGTACAAC-3′ | 192 | 15 |
| 5 | O139 rfbF | 5′-AGCCTCTTTATTACGGGTGG-3′ | 449 | 15 |
| 6 | O139 rfbR | 5′-GTCAAACCCGATCGTAAAGG-3′ | 449 | 15 |
| 7 | ompW F | 5′-CACCAAGAAGGTGACTTTATTGTG-3′ | 304 | 32 |
| 8 | ompW R | 5′-GGTTTGTCGAATTAGCTTCACC-3′ | 304 | 32 |
RESULTS AND DISCUSSION
Of the total 297 samples (99 each of the water and two size fractions of plankton) analyzed between March and December 2004 for V. cholerae, only 8 (3%) yielded V. cholerae O1 and 4 (1%) yielded V. cholerae O139 Bengal in culture (Table 2), using APW enrichment followed by plate culture. Analysis by a month of sampling, i.e., for culture, showed that the eight strains of V. cholerae O1 were isolated in December and, of the four strains of V. cholerae O139, one was isolated in March and three were isolated in September. In contrast, V. cholerae non-O1/non-O139 strains were isolated in culture from nearly every sample collected during the 18 rounds of sampling between March and December 2004 (Table 2), with the frequency of isolation varying significantly, lowest numbers of isolates being obtained between August and October.
TABLE 2.
Detection and isolation of V. cholerae O1, O139, and non-O1/non-O139 from water and two size fraction plankton samples collected from six pond sites in Mathbaria, adjacent to the Bay of Bengal
| Site | Samplea | No. of samplesb | No. of samples positive
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Culturec
|
DFA
|
Multiplex-PCR
|
||||||||
| O1 | O139 | Non-O1/O139 | O1 | O139 | O1 rfb | O139 rfb | ctxA | |||
| 1 | Water | 10 | 0 | 0 | 6 | 6 | 0 | 0 | 0 | 1 |
| 20-μm fraction (plankton) | 10 | 1 | 0 | 0 | 5 | 1 | 0 | 1 | 1 | |
| 64-μm fraction (plankton) | 10 | 1 | 0 | 4 | 4 | 0 | 0 | 0 | 3 | |
| 2 | Water | 17 | 0 | 0 | 10 | 7 | 2 | 0 | 0 | 0 |
| 20-μm fraction (plankton) | 17 | 0 | 0 | 10 | 3 | 3 | 1 | 1 | 3 | |
| 64-μm fraction (plankton) | 17 | 0 | 0 | 7 | 5 | 5 | 0 | 1 | 0 | |
| 3 | Water | 18 | 0 | 1 | 8 | 5 | 4 | 0 | 2 | 3 |
| 20-μm fraction (plankton) | 18 | 0 | 2 | 9 | 8 | 3 | 1 | 3 | 2 | |
| 64-μm fraction (plankton) | 18 | 0 | 0 | 11 | 5 | 3 | 0 | 3 | 3 | |
| 4 | Water | 18 | 0 | 0 | 8 | 4 | 1 | 0 | 1 | 0 |
| 20-μm fraction (plankton) | 18 | 0 | 0 | 10 | 3 | 4 | 0 | 0 | 1 | |
| 64-μm fraction (plankton) | 18 | 0 | 0 | 9 | 4 | 5 | 0 | 0 | 2 | |
| 5 | Water | 18 | 1 | 0 | 5 | 2 | 1 | 2 | 2 | 4 |
| 20-μm fraction (plankton) | 18 | 1 | 0 | 6 | 6 | 2 | 1 | 1 | 1 | |
| 64-μm fraction (plankton) | 18 | 1 | 0 | 7 | 3 | 2 | 1 | 0 | 4 | |
| 6 | Water | 18 | 1 | 0 | 4 | 3 | 1 | 1 | 2 | 3 |
| 20-μm fraction (plankton) | 18 | 1 | 0 | 6 | 6 | 5 | 1 | 1 | 3 | |
| 64-μm fraction (plankton) | 18 | 1 | 1 | 5 | 3 | 2 | 0 | 0 | 0 | |
Six each of water and two size fractions of plankton samples were collected per round every fortnight.
Sites 1 and 2 could not be sampled for all 18 rounds.
One representative colony from each culture-positive sample was isolated and stored for further study.
M-PCR for amplification of the genes ctxA (302 bp) and rfb for V. cholerae serovars O1 (192 bp) and O139 (449 bp) was carried out with 297 environmental samples that comprised 99 each of the water and two size fractions of plankton. As shown in Table 2, 34 (11%) samples were determined to be positive for ctxA, 21 (7%) of which included ctxA but not rfb for V. cholerae O1 or O139, 8 (3%) V. cholerae O1 rfb, and 5 (2%) V. cholerae O139 rfb. In addition, 13 (4%) samples were positive for V. cholerae O139 rfb alone, and 2 (0.7%) were positive for V. cholerae O1 rfb alone. The overall incidence of toxigenic V. cholerae as measured by M-PCR did not show significant difference between the two size fractions of plankton (data not shown), that is, the two size groups obtained by filtration through 64- and 20-μm mesh size plankton nets.
Using the DFA detection method, 82 (28%) of the 297 samples tested were positive for V. cholerae O1 and 45 (15%) were positive for V. cholerae O139 Bengal (Table 2). Of the 99 samples each of water and the two size fractions of plankton, 27 (27%) water, 31 (31%) 20-μm plankton, and 24 (24%) 64-μm plankton samples were positive for V. cholerae O1, whereas 9 (9%) water, 19 (19%) 20-μm plankton, and 17 (17%) 64-μm plankton samples were positive for V. cholerae O139 as determined by DFA. As shown in Table 3, DFA counts, which varied significantly between months, were higher in the water samples for both V. cholerae O1 and O139 than in the plankton.
TABLE 3.
DFA counts of V. cholerae O1 and O139 in the samples collected from six pond sites in Mathbaria, adjacent to the Bay of Bengala
| Mo | Sample type | DFA count (cells/liter) of indicated V. cholerae serovar
|
|||
|---|---|---|---|---|---|
| O1
|
O139
|
||||
| Round 1 | Round 2 | Round 1 | Round 2 | ||
| March | 64-μm fraction (plankton) | <10 | ND | <10 | ND |
| 20-μm fraction (plankton) | 1.3 × 101 | ND | 1.9 × 101 | ND | |
| Water | <10 | ND | <10 | ND | |
| April | 64-μm fraction (plankton) | ND | ND | ND | ND |
| 20-μm fraction (plankton) | ND | ND | ND | ND | |
| Water | ND | ND | ND | ND | |
| May | 64-μm fraction (plankton) | 7.5 × 101 | <10 | <10 | 3.8 × 101 |
| 20-μm fraction (plankton) | <10 | <10 | <10 | <10 | |
| Water | 2.3 × 104 | <10 | 5.0 × 101 | <10 | |
| June | 64-μm fraction (plankton) | 2.5 × 101 | 5.3 × 102 | 3.0 × 101 | 3.2 × 101 |
| 20-μm fraction (plankton) | 1.7 × 101 | 3.8 × 101 | 5.4 × 101 | 2.6 × 101 | |
| Water | <10 | 1.8 × 105 | <10 | <10 | |
| July | 64-μm fraction (plankton) | 8.8 × 101 | <10 | 1.0 × 102 | 1.3 × 101 |
| 20-μm fraction (plankton) | 1.9 × 101 | 1.3 × 101 | 3.4 × 101 | 1.3 × 101 | |
| Water | 2.3 × 104 | 5.0 × 103 | 5.0 × 103 | 1.3 × 101 | |
| August | 64-μm fraction (plankton) | 1.6 × 103 | <10 | 2.0 × 102 | 2.6 × 101 |
| 20-μm fraction (plankton) | 3.8 × 101 | <10 | 3.4 × 101 | 6.0 × 101 | |
| Water | 2.5 × 105 | 5.0 × 103 | 3.0 × 104 | <10 | |
| September | 64-μm fraction (plankton) | 3.4 × 102 | 2.6 × 102 | <10 | <10 |
| 20-μm fraction (plankton) | 3.6 × 102 | 3.7 × 102 | <10 | 5.1 × 101 | |
| Water | 3.9 × 104 | 2.0 × 104 | <10 | 3.0 × 104 | |
| October | 64-μm fraction (plankton) | <10 | <10 | <10 | <10 |
| 20-μm fraction (plankton) | <10 | <10 | <10 | 5.0 × 102 | |
| Water | <10 | <10 | <10 | <10 | |
| November | 64-μm fraction (plankton) | <10 | <10 | 1.0 × 102 | <10 |
| 20-μm fraction (plankton) | 2.5 × 102 | <10 | <10 | <10 | |
| Water | <10 | 5.0 × 103 | <10 | <10 | |
| December | 64-μm fraction (plankton) | 3.5 × 103 | <10 | <10 | <10 |
| 20-μm fraction (plankton) | 4.5 × 102 | 4.4 × 101 | <10 | <10 | |
| Water | 3.4 × 107 | 3.3 × 104 | <10 | <10 | |
Values are the geometric means of six replicates of each sample. Samples were collected twice each month. Rounds 1 and 2 represent biweekly counts in Mathbaria. ND, not done.
Noteworthy in the DFA results was the observation that cells of both V. cholerae O1 and O139 were solitary or dispersed (Fig. 2A and C) but also could be seen in clusters, notably in the case of V. cholerae O1 (Fig. 2B). The clusters of cells were observed predominantly in the water samples and more frequently during the noncholera season. Fluorescent stained micrographs of such clusters showed V. cholerae O1 cells residing in thin biofilms. The biofilm clusters harbored cells that varied in size and shape and, in most cases, were atypical of V. cholerae, i.e., did not appear as curved rods (Fig. 3A and B). However, in spite of the diverse physical appearances, whether as single cells or in clusters, as visualized by DFA (Fig. 3C and D), the first isolation by direct culture was achieved only in early December, when three of the six pond sites were simultaneously positive for V. cholerae O1 (Table 2). As shown in Fig. 4, large clusters of V. cholerae O1 in biofilms, either as a free-floating biofilm or attached to detritus, were observed after the first isolation in culture from these pond sites in Mathbaria. The successful isolation of V. cholerae O1 and the sharp rise in the number of V. cholerae cells observed in the water samples (Table 3) by DFA in this round of surveillance suggested recent significant multiplication of the bacterium in the water. The appearance of these intact and robust cells in clusters within biofilms suggests that biofilms may be significant in the persistence and proliferation of V. cholerae O1 (3, 8, 9, 12). Notably, pond sites from which V. cholerae O1 had been isolated in a follow-up sampling were again determined to be negative by culture (Table 2).
FIG. 2.
Epifluorescent micrographs of V. cholerae O1 and O139 detected in environmental samples collected between March and December 2004. Fluorescent monoclonal antibodies (DFA) specific for V. cholerae serovar O1 or O139, obtained from New Horizon Diagnostic, were used, and the stains were visualized (18, 19) using an epifluorescence microscope (Olympus model BX51, BX2 series). Microscopic images were captured digitally (Olympus DP20) and processed by using Adobe Photoshop (version 5). (A and C) Typical single cells of V. cholerae O1 and O139, respectively; (B and D) cell clusters of V. cholerae O1 in water samples collected from Mathbaria and Matlab, respectively, during July 2004.
FIG. 3.
Epifluorescent micrographs of V. cholerae O1 detected in environmental samples collected in September and November 2004. Samples were stained by using DFA reagents specific for V. cholerae serovar O1, obtained from New Horizon Diagnostic, and visualized (18, 19) by using an epifluorescence microscope (Olympus model BX51, BX2 series). Microscopic images were captured digitally (Olympus DP20) and processed by using Adobe Photoshop version 5. (A and B) Different stages of coccoid V. cholerae O1 cells attached to thin film (biofilm) on surfaces, either free-floating (A) or attached to plankton or other aggregates (B) in samples collected in September from pond sites 1 and 3, respectively; (C and D) short rods of V. cholerae O1 cells in in situ biofilms in samples collected in November from ponds at sites 1 and 3, respectively.
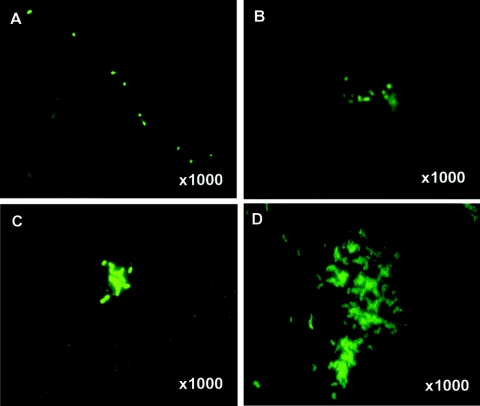
FIG. 4.
Epifluorescent micrographs of V. cholerae O1 detected in environmental samples collected in December 2004. Samples were stained with direct fluorescent monoclonal antibody (DFA) specific for V. cholerae serovar O1 (obtained from New Horizon Diagnostic) and visualized (18, 19) by using an epifluorescence microscope (Olympus model BX51, BX2 series). Microscopic images were captured digitally (Olympus DP20) and processed by using Adobe Photoshop version 5. (A and B) V. cholerae O1 in the initial stage of in situ biofilm formation, either free-swimming (A) or attached to plankton or other aggregates (B) in samples collected from pond site 1; (C and D) V. cholerae O1 in the initial stage of in situ biofilm formation in a sample collected from ponds at sites 5 and 6, respectively.
To determine whether the occurrence of the biofilm aggregates of V. cholerae O1 observed in the aquatic environment represented a general phenomenon or was limited to Mathbaria, water and plankton samples collected from Matlab, a historic site of cholera epidemics in Bangladesh (Fig. 1), were tested for the same. As shown in Fig. 2D, fluorescence microscopy analysis of V. cholerae O1 revealed cell clusters, as had been observed in the Mathbaria pond samples (Fig. 2B). Interestingly, the DFA data indicated V. cholerae O139 also to be present in the Matlab samples. However, cellular aggregates of V. cholerae O139 were not observed in either sampling location.
During epidemics, cholera usually strikes villages in the coastal regions before cases occur inland (26, 32). As was shown many years ago, V. cholerae is native to the aquatic environment (7, 16, 24, 30, 46), thriving and multiplying seasonally (5). However, the phenomenon of no recovery of V. cholerae O1/O139 (18, 23) by standard culture methods during interepidemic periods has posed a lingering question as to where V. cholerae O1/O139 resides between epidemics and what triggers its annual bloom. From the many studies carried out previously, it has been proposed that a nongrowth or slow-growth state for V. cholerae is a mechanism of persistence (4, 6, 18, 32, 42), when the presence of V. cholerae O1/O139 can be detected only by molecular (29, 36) and immunochemical methods and not by culture (18, 21). In addition, laboratory-based studies have implicated biofilms as a mechanism for responding to unfavorable environmental conditions (12, 25, 26, 43), but environmental data supporting this hypothesis are lacking (44). The present study is the first to provide data on the detection and isolation of V. cholerae O1 and O139 in the remote areas of cholera endemicity adjacent to the Bay of Bengal, which has been historically known as the birthplace of Asiatic cholera (1, 31, 32). The recovery of V. cholerae O139 in culture from only 1% of the 297 environmental samples collected in the present study between March and December 2004 was typical, although an earlier study reported as many as 12% of the environmental samples yielded culturable V. cholerae O139 (20). The recovery of V. cholerae O1 in only 3% of the samples collected in the present study is similarly typical, i.e., <1% of the environmental samples were reported in earlier studies (18, 24). V. cholerae O139 was recovered in culture in the present study in December during and immediately after the two usual peaks of cholera. During the time period of the present study, cholera cases recorded in Mathbaria were attributed exclusively to V. cholerae O1 (A. K. Siddique et al., unpublished data).
PCR is useful for rapid detection of toxigenic V. cholerae O1 and O139 (15, 29, 36), and whereas culture yielded toxigenic strains in only 2 to 3% of environmental samples, the direct detection of ctxA and rfb for both V. cholerae O1 and O139 in the same water and plankton samples by using M-PCR provided a powerful clue to the presence of toxigenic V. cholerae O1/O139. Earlier studies using direct PCR-based detection of toxigenic V. cholerae O1 and O139 in water and plankton samples provided evidence for an environmental reservoir (36). Since <10% of clinical and <1% of environmental isolates of V. cholerae non-O1/non-O139 possess ctx (20, 27, 45), our finding that amplification of a 302-bp ctxA, irrespective of the rfb gene of either V. cholerae O1 or O139, indicated an alternative reservoir(s) for ctx in the environment in the Bay of Bengal. Furthermore, amplification of either V. cholerae O1 or O139 rfb but not ctx in M-PCR analyses suggests the presence of a progenitor of the pandemic strains, since surface water isolates of V. cholerae O1 are rarely positive for cholera toxin (45). Most noteworthy may be the conclusion that, whereas culture methods failed to isolate toxigenic V. cholerae directly from the environment (4, 18, 24), results of M-PCR supported the hypothesis that pandemic strains, or their progenitors, nevertheless exist in the aquatic environment (15, 29, 36).
Whereas conventional culture methods reflect metabolically active and dividing cells under standard bacteriological laboratory conditions, the DFA-direct counting method provides enumeration of both culturable and viable-but-nonculturable cells of V. cholerae (2, 46). DFA results obtained in the present study specifically support the conclusion that V. cholerae O1 and O139 are present year-round in the aquatic environment of Bangladesh, and this was further substantiated by the results of M-PCR, as discussed above. DFA also is efficient in detecting both V. cholerae O1 and O139 in zooplankton in Bangladesh (18, 19) and attached in and on zooplankton (16, 17, 39, 41) and phytoplankton (22), explaining the seasonal distribution of V. cholerae in the aquatic environment. Except for the observation of the specific attachment of V. cholerae to planktonic copepods, also observed in the present study, the overall occurrences of V. cholerae O1 and O139 in both the 20- and 64-μm plankton size fractions were not significantly different. Furthermore, DFA counting results for both V. cholerae O1 and O139 in Mathbaria were higher for water than for the two size classes of plankton. The Mathbaria area of Bangladesh is rural and adjacent to heavily forested mangroves, very likely providing nutrients and thereby contributing to these results. In contrast, culture yielded little or no recovery of V. cholerae O1/O139 isolates, even during cholera epidemics, whereas DFA detection of both culturable and nonculturable V. cholerae O1 and O139 serovars in the aquatic environment of Mathbaria was much more consistent and, when done in combination with M-PCR, supported earlier reports of year-round detection of V. cholerae serovar O1 in the aquatic environment of Bangladesh (5, 18, 21, 30, 41).
Acknowledgments
This research was funded by the National Institutes of Health research grant 1R01A139129-01 under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health, the International Center for Diarrhoeal Disease Research, Bangladesh, and the University of Maryland. The ICDDR,B is supported by donor countries and agencies, which provide unrestricted support to the Center for its operations and research.
We gratefully acknowledge Sirajul Islam Khan and Ziaur Rahman of the Department of Microbiology, Dhaka University, for the excellent fluorescence microscopic support. Finally, we acknowledge the contribution of the NIH environmental surveillance team for their support and commitment to the research.
REFERENCES
- 1.Albert, M. J., A. K. Siddique, M. S. Islam, A. S. G. Faruque, M. Ansaruzzamam, S. M. Faruque, and R. B. Sack. 1993. A large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. [DOI] [PubMed] [Google Scholar]
- 2.Brayton, P. R., and R. R. Colwell. 1987. Fluorescent antibody staining method for enumeration of viable environmental Vibrio cholerae O1. J. Microbiol. Methods 6:309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldwell, D. E. 1995. Cultivation and study of biofilm communities, p. 64-79. In H. M. L. Scott and J. W. Costerton (ed.), Microbial biofilms. University Press, Cambridge, United Kingdom.
- 4.Colwell, R. R., P. R. Brayton, D. B. Roszak, A. Huq, and L. M. Palmer. 1985. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Bio/Technology 3:817-820. [Google Scholar]
- 5.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. I. Greenouh (ed.), Cholera. Plenum Press, Inc., New York, N.Y.
- 6.Colwell, R. R., and A. Huq. 1994. Vibrios in the environment: viable but non-culturable Vibrio cholerae, p. 117-133. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 7.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 9.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., Asadulghani, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, Q. S. Ahmed, A. S. G. Faruque, M. A. Salam, T. Ramamurthy, G. B. Nair, A. Weintraub, and D. A. Sack. 2003. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 9:1116-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flemming, H. C. 1993. Biofilms and environmental protection. Water Sci. Technol. 27:1-10. [Google Scholar]
- 13.Glass, R. I., M. I. Huq, B. J. Stoll, M. U. Khan, M. H. Merson, J. V. Lee, and R. E. Black. 1982. Endemic cholera in rural Bangladesh, 1966-1980. Am. J. Epidemiol. 116:959-970. [DOI] [PubMed] [Google Scholar]
- 14.Glass, R. I., and R. E. Black. 1992. The epidemiology of cholera, p. 129-154. In D. Barua and W. B. I. Greenough (ed.), Cholera. Plenum Press, Inc., New York, N.Y.
- 15.Hoshino, K., S. Yamasaki, A. K. Mukhopadhyay, S. Chakraborty, A. Basu, S. K. Bhattacharya, G. B. Nair, T. Shimada, and Y. Takeda. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20:201-207. [DOI] [PubMed] [Google Scholar]
- 16.Huq, A., E. B. Small, P. A. West, and R. R. Colwell. 1984. The role of planktonic copepods in the survival and multiplication of Vibrio cholerae in the aquatic environment, p. 521-534. In R. R. Colwell (ed.), Vibrios in the environment. John Wiley & Sons, Inc., New York, N.Y.
- 17.Huq, A., S. A. Huq, D. J. Grimes, M. O'Brien, K. H. Chu, J. McDowell Capuzzo, and R. R. Colwell. 1986. Colonization of the gut of the blue crab (Callinectes sapidus) by Vibrio cholerae. Appl. Environ. Microbiol. 52:586-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huq, A., R. R. Colwell, R. Rahaman, A. Ali, M. A. R. Chowdhury, S. Parveen, D. A. Sack, and E. R. Cohen. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huq, A., R. R. Colwell, M. A. Chowdhury, B. Xu, S. M. Moniruzzaman, M. S. Islam, M. Yunus, and M. J. Albert. 1995. Coexistence of Vibrio cholerae O1 and O139 Bengal in plankton in Bangladesh. Lancet 345:1249. [DOI] [PubMed] [Google Scholar]
- 20.Islam, M. S., M. K. Hasan, M. A. Miah, M. Yunus, K. Zaman, and M. J. Albert. 1994. Isolation of Vibrio cholerae O139 synonym Bengal from the aquatic environment in Bangladesh: implications for disease transmission. Appl. Environ. Microbiol. 60:1684-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam, M. S., B. S. Drasar, and R. B. Sack. 1994. The aquatic environment as reservoir of Vibrio cholerae: a review. J. Diarrhoeal Dis. Res. 11:197-206. [PubMed] [Google Scholar]
- 22.Islam, M. S., Z. Rahim, M. J. Alam, S. Begum, S. M. Moniruzzaman, A. Maeda, K. Amako, M. J. Albert, R. B. Sack, A. Huq, and R. R. Colwell. 1999. Association of Vibrio cholerae O1 with the cyanobacterium, Anabaena sp., elucidated by polymerase chain reaction and transmission electron microscopy. Trans. R. Soc. Trop. Med. Hyg. 93:36-40. [DOI] [PubMed] [Google Scholar]
- 23.Kaper, J. B., J. G. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan, M. U., M. Shahidullah, M. S. Haque, and W. U. Ahmed. 1984. Presence of vibrios in surface water and their relation with cholera in a community. Trop. Geogr. Med. 36:335-340. [PubMed] [Google Scholar]
- 25.Kierec, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kierec, K., and P. I. Watnick. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. USA 100:14357-14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodama, H., Y. Gyobu, N. Tokuman, I. Okada, H. Uetake, T. Shimada, and R. Sakazaki. 1984. Ecology of non-O1 Vibrio cholerae in Toyama prefecture. Microbiol. Immunol. 28:311-328. [DOI] [PubMed] [Google Scholar]
- 28.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipp, E. K., I. N. G. Rivera, A. I. Gil, E. M. Espeland, N. Choopun, V. R. Louis, E. Russek-Cohen, A. Huq, and R. R. Colwell. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69:3676-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobitz, B., L. Beck, A. Huq, B. Wood, G. Fuchs, A. S. G. Faruque, and R. R. Colwell. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. USA 97:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longini, I. M., M. Yunus, K. Zaman, A. K. Siddique, R. B. Sack, and A. Nizam. 2002. Epidemic and endemic cholera trends over a 33-year period in Bangladesh. J. Infect. Dis. 186:246-251. [DOI] [PubMed] [Google Scholar]
- 32.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, A. K. Mukhopadhyay, S. Garg, M. K. Bhattacharya, T. Takeda, T. Shimada, Y. Takeda, and B. C. Deb. 1994. Spread of Vibrio cholerae O139 Bengal in India. J. Infect. Dis. 169:1029-1034. [DOI] [PubMed] [Google Scholar]
- 33.Nandi, B., R. K. Nandi, S. Mukhopadhyay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver, J. D. 1993. Formation of viable but non-culturable cells, p. 239-271. In S. Kielleberg (ed.), Starvation in bacteria. Plenum Press, Inc., New York, N.Y.
- 35.Politzer, R. 1959. Cholera, monograph series, no. 43. World Health Organization, Geneva, Switzerland.
- 36.Rivera, I. N. G., E. K. Lipp, N. Choopun, A. Gil, A. Huq, and R. R. Colwell. 2003. Method for DNA extraction and application of multiplex PCR to detect toxigenic Vibrio cholerae O1 and O139 in aquatic ecosystems. Appl. Environ. Microbiol. 5:599-606. [DOI] [PubMed] [Google Scholar]
- 37.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sack, R. B., A. K. Siddique, I. M. Longini, Jr., A. Nizam, M. Yunus, M. S. Islam, J. G. Morris, Jr., A. Ali, A. Huq, G. B. Nair, F. Qadri, S. M. Faruque, D. A. Sack, and R. R. Colwell. 2003. A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J. Infect. Dis. 187:96-101. [DOI] [PubMed] [Google Scholar]
- 39.Shukla, B. N., D. V. Singh, and S. C. Sanyal. 1995. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the river Ganga, Varanasi. FEMS Immunol. Med. Microbiol. 12:113-120. [DOI] [PubMed] [Google Scholar]
- 40.Siddique, A. K., K. Zaman, A. H. Baqui, K. Akram, P. Mutsuddy, A. Eusof, K. Haider, M. S. Islam, and R. B. Sack. 1992. Cholera epidemics in Bangladesh: 1985-1991. J. Diarrhoeal. Dis. Res. 10:79-86. [PubMed] [Google Scholar]
- 41.Tamplin, M. L., A. L. Gauzens, A. Huq, D. A. Sack, and R. R. Colwell. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56:1977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wai, S. N., Y. Mizunoe, and S. Yoshida. 1999. How Vibrio cholerae survive during starvation. FEMS Microbiol. Lett. 180:123-131. [DOI] [PubMed] [Google Scholar]
- 43.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of flagellum leads to altered colony morphology, biofilm development, and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.World Health Organization. 1984. Report of the Third Meeting of the Scientific Working Group on Bacterial Enteric Infections: microbiology, epidemiology, immunology, and vaccine development, p. 1-17. World Health Organization, Geneva, Switzerland.
- 46.Xu, H. S., N. C. Roberts, F. L. Singleton, R. W. Attwell, D. J. Grimes, and R. R. Colwell. 1983. Survival and viability of non-culturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microbiol. Ecol. 8:213-223. [DOI] [PubMed] [Google Scholar]