Abstract
Since the stomach is a first line of defense for the host against ingested microorganisms, an ex vivo swine stomach contents (SSC) assay was developed to search for genes important for Salmonella enterica serovar Typhimurium survival in the hostile gastric environment. Initial characterization of the SSC assay (pH 3.87) using previously identified, acid-sensitive serovar Typhimurium mutants revealed a 10-fold decrease in survival for a phoP mutant following 20 min of challenge and no survival for mutants of rpoS or fur. To identify additional genes, a signature-tagged mutagenesis bank was constructed and screened in the SSC assay. Nineteen mutants were identified and individually analyzed in the SSC and acid tolerance response assays; 13 mutants exhibited a 10-fold or greater sensitivity in the SSC assay compared to the wild-type strain, but only 3 mutants displayed a 10-fold or greater decrease in survival following pH 3.0 acidic challenge. Further examination determined that the lethal effects of the SSC are pH dependent but that low pH is not the sole killing mechanism(s). Gas chromatography analysis of the SSC revealed lactic acid levels of 126 mM. Upon investigating the effects of lactic acid on serovar Typhimurium survival in a synthetic gastric fluid, not only was a concentration- and time-dependent lethal effect observed, but the phoP, rpoS, fur, and pnp genes were identified as involved in protection against lactic acid exposure. These studies indicate a role in gastric survival for several serovar Typhimurium genes and imply that the stomach environment is defined by more than low pH.
The food-borne pathogen Salmonella is a leading cause of bacterial gastroenteritis and food-related death in the United States (30). Salmonella is often transmitted to humans by consumption of contaminated foods of animal origin (dairy, beef, pork, poultry, or eggs) or fruits and vegetables contaminated with animal feces. On swine farms, the prevalence of Salmonella in fecal samples ranges from 38 to 75 percent (5, 13, 36). Since these Salmonella-positive animals often do not present with clinical symptoms, the ubiquitous distribution of Salmonella is perpetuated with carrier pigs shedding Salmonella in their feces and into their surroundings. Moreover, the rooting behavior of swine predisposes them to ingestion of Salmonella bacteria that have been shed into the environment by carrier animals.
Gastric acidity is a major barrier to colonization and infection of the gastrointestinal tract by microorganisms and is a first line of defense against microbial pathogens that infect their host via the oral route. The gastric acidity can be as low as pH 1.5 (42). Furthermore, several organic acids can be present in the stomach, including lactic, acetic, propionic, and butyric acids (31, 32, 41). Therefore, a successful food-borne pathogen must be able to protect itself from the deleterious conditions in the stomach environment. Many microorganisms, including Salmonella enterica serovar Typhimurium and Escherichia coli O157:H7, have been shown to possess adaptive systems to protect against acid stress (3, 19). Several gene products have been shown to be important for the adaptative response of Salmonella to acid stress, including RpoS, an alternate sigma factor involved in stationary-phase physiology and stress responses; PhoP, a response regulator important for macrophage survival and resistance to antimicrobial peptides; and Fur, a regulator of iron metabolism (3). The acid tolerance response (ATR) of serovar Typhimurium is able to protect against two types of acid stress, organic (weak acids) and inorganic (low pH) (4, 6). Regulators of the acid tolerance response that are involved in protection against these two types of acid stress have been identified in serovar Typhimurium, with RpoS and Fur protecting against organic acid stress and PhoP and RpoS protecting against inorganic acid stress (6). The target genes that are controlled by each of these regulators for the protection against acid stress remain to be identified.
To date, most studies that have analyzed the responses of microorganisms to acid stress have been performed in vitro using various minimal or complex media. The acid protection observed for bacteria during acid challenge can be medium dependent. A synthetic gastric fluid has been assembled containing pepsin, lysozyme, and bile to mimic the stomach environment (9). The synthetic gastric fluid has been used by multiple investigators to determine the acid tolerance/resistance of bacterial pathogens in this simulated stomach environment (2, 14, 21). Cotter et al. demonstrated that Listeria monocytogenes is more sensitive to challenge in an ex vivo porcine gastric fluid (pH 2.5) than in the synthetic gastric fluid (pH 2.5), suggesting that the porcine stomach contains additional components that result in stress which are not present in the synthetic fluid (14).
Signature-tagged mutagenesis (STM) is a functional genomics technique that utilizes transposons containing unique sequences (signatures) to identify individual mutants within a complex pool of mutants (25). STM analysis evaluates the transposon tags that are not recovered from an experimental challenge model to identify transposon mutants that are unable to survive the chosen environmental condition. The gene into which the transposon has inserted is identified by nucleotide sequencing of the transposon/bacterial DNA junction. Since its development a decade ago, STM has been performed with 31 bacterial species and has identified more than 1,700 bacterial genes with a role in virulence (39). Most STM studies have utilized in vivo systems for screening transposon banks; however, in principle, any environmental condition that exerts a negative selection on microorganisms can be used for STM analysis.
We developed an ex vivo swine stomach contents (SSC) assay to screen serovar Typhimurium signature-tagged transposon mutants to identify genes involved in protection in the gastric environment. The acid tolerance response was determined for the individual mutants and compared to the phenotype in the SSC assay. Characterization of the SSC indicates that the lethal effect is pH dependent but that low pH is not the only important component for the damaging effects towards Salmonella.
MATERIALS AND METHODS
Bacterial strains, media, and buffers.
The Salmonella enterica serovar Typhimurium strains used in this study are listed in Table 1, and the STM strains constructed herein are listed in Table 2. Bacteria were grown in Luria-Bertani (LB) broth or E salts minimal medium, pH 7.0 (45), containing 0.4% glucose (EG medium). All media and buffers with an acidic pH were lowered using HCl. Antibiotics were used at the concentrations of 100 μg/ml for ampicillin, 30 μg/ml for nalidixic acid, and 50 μg/ml for kanamycin. Unless otherwise indicated, all cultures were grown and assays performed at 37°C.
TABLE 1.
Salmonella enterica serovar Typhimurium strains
| Strain | Genotype | Source |
|---|---|---|
| SX 2 (SF530, χ3761) | UK1 wild type | J. Foster (15) |
| SX 21 (JF2690) | UK1 rpoS::Ap | J. Foster (27) |
| SX 46 (JF3203) | UK1 phoP::Tn10 | J. Foster (6) |
| BSX 30 (JF2043) | LT2 iroA::MudJ fur-1 zbf-5123::Tn10 | J. Foster (24) |
| SB 26 | UK1 Nalr | This study |
| SB 27 | UK1 rpoS::Ap Nalr | SB 26 × SX 21 |
| SB 29 | UK1 rpoS::Ap Nalr STMA1-miniTn5Km2 | This study |
| SB 270 | UK1 fur-1 zbf-5123::Tn10 | SX 2 × BSX 30 |
TABLE 2.
Characterization of signature-tagged mutants
| Function class | Mutant namea | Gene | STM no.b | Insertion site/no. of bpc | Description | Survival ratiof
|
|
|---|---|---|---|---|---|---|---|
| SSCd | ATRe | ||||||
| Protein fate | SB 110/1F1 | dnaK | STM0012 | −127/1,917 | Chaperone | 0.032 | 0.10 |
| Protein fate | SB 156/18G1 | STM2691 | +275/2,181 | Putative ABC transmembrane transporter | 0.37 | 0.44 | |
| Amino acid biosynthesis | SB 200/31G4 | usg | STM2369 | +175/1,014 | Putative aspartate-semialdehyde dehydrogenase | 0.25 | 0.10 |
| Regulatory functions | SB 213/24F5 | barA | STM2958 | +313/2,757 | Sensor kinase for virulence gene expression | 0.050 | 1.13 |
| Energy metabolism | SB 135/16H3 | STM1382 | +616/1,227 | Putative regulatory protein | 0.066 | 0.27 | |
| Central intermediary metabolism | SB 104/2G3 | dgt | STM0208 | +780/1,518 | dGTP triphosphohydrolase | 0.00052 | 1.30 |
| Cellular processes | SB 117/8B6 | sopB/sigD | STM1091 | +709/1,686 | Inositol phosphate phosphatase | 0.015 | 0.35 |
| Cellular processes | SB 162/18F6 | traL | PSLT078 | +218/312 | Conjugative transfer | 0.017 | 0.20 |
| Unclassified | SB 157/18D2 | STM2585 | +468/546 | Transposase-like protein | 0.086 | 0.50 | |
| Unclassified | SB 115/8G3 | pefA | PSLT018 | +108/519 | Plasmid-encoded fimbriae | 0.61 | 1.13 |
| Unclassified | SB 114/6C6 | pefC | pSLT017 | +175/2,409 | Plasmid-encoded fimbriae | 0.031 | 0.27 |
| Unclassified | SB 116/8E6 | rfaL | STM3713 | +211/1,215 | O-antigen ligase | 0.45 | 0.92 |
| Cell envelope | SB 113/7B2 | asmA | STM2120 | +718/1,866 | Outer membrane protein assembly | 0.0035 | 1.11 |
| Cell envelope | SB 126/10B1 | rfbM/manC | STM2084 | +291/1,440 | Mannose-1-phosphate guanylyltransferase | 2.94 | 57.73 |
| Cell envelope | SB 105/11B1 | ynaI | STM1663 | +430/1,032 | Putative integral membrane protein | 0.023 | 48.67 |
| Cell envelope | SB 209/35C6 | ynaI | STM1663 | +94/1,032 | Putative integral membrane protein | 0.012 | 84.25 |
| Transcription | SB 130/12B6 | pnp | STM3282 | +1,584/2,136 | Polynucleotide phosphorylase | 0.00062 | 0.39 |
| Mobile and extrachromosomal element functions | SB 166/18E4 | mig-3 | STM1868 | +532/882 | Macrophage-inducible protein | 0.11 | 0.86 |
| Unknown function | SB 134/16H1 | yjeA/poxR | STM4344 | +713/978 | Lysyl-tRNA synthetase | 0.053 | 0.06 |
Strain name/STM pool.
The STM and pSLT prefixes indicate the gene numbers on the chromosome and virulence plasmid, respectively, as assigned by the S. enterica serovar Typhimurium LT2 complete genome sequencing project.
Location of transposon insertion site/number of base pairs within the gene.
Following 1 h of pH 4.4 adaptation, a 10-min challenge of each mutant was performed independently in the SSC assay.
Following 1 h of pH 4.4 adaptation, a 90-min challenge of each mutant was performed independently in the ATR assay at pH 3.0.
Data are presented as the ratio of the percent survival of the mutant divided by the percent survival of the wild-type strain; therefore, mutants with a ratio below 1 did not survive as well as the wild-type strain, and mutants with a ratio above 1 survived better than the wild-type strain.
Collection and preparation of the SSC.
Without removing the pigs from feed prior to necropsy, the swine stomach contents of 18 pigs at 6 weeks of age were obtained aseptically immediately following euthanasia and placed on ice. The stomach contents were centrifuged at 3,000 rpm at 4°C, the supernatants were filter sterilized (0.22 mm), and the pH of each was determined (pH range of 1.42 to 4.44). Equal volumes of four of the stomach content samples, ranging in pH from 3.80 to 4.02, were combined to produce the challenging agent in the swine stomach contents assay at pH 3.87. An additional SSC pool was created using two different collected samples with pHs of 3.47 and 4.15. All procedures involving animals were approved by the U.S. Department of Agriculture, Agricultural Research Service, National Animal Disease Center Animal Care and Use Committee and complied with federal guidelines.
Generating the signature tag sequences.
Following a similar design described by Hensel et al. (25), a degenerate 118-base oligonucleotide (oSBM 53) containing a 60-base variable region was constructed with restriction enzyme (BamHI, KpnI, and HindIII) sites incorporated into the flanking constant regions for cloning purposes: 5′-CTAGGATCCTGGTACCGTCTGATAAGCTT[MN]30AAGCTTGGTTAGAGGATCCGGGTACCATG-3′ (M = A or C; N = A, G, C, or T; restriction sites are underlined) (Oligos Etc., Wilsonville, OR). This degenerate oligonucleotide has the ability to produce ∼1 × 1027 different sequence molecules. A double-stranded fragment was produced by 30 rounds of PCR using oSBM 44 (5′-CTAGGATCCTGGTACCG-3′) and oSBM 45 (5′-CATGGTACCCGGATCCTC-3′). The STM fragments were digested with KpnI, cloned into the suicide vector pUTminiTn5Km2 (16), and transformed into CC118λpir (26). To select for optimal tag sequences, the following procedures were performed on 125 of the resulting plasmids to screen for signature tags that exhibited strong signals in Southern hybridization and did not cross-hybridize: the signature tag sequences were PCR amplified with oSBM 46 (5′-GGTACCGTCTGATAAGC-3′) and oSBM 47 (5′-GGATCCTCTAACCAAGC-3′), digested with HindIII to remove the constant regions, and blotted to Zeta probe membranes (Bio-Rad Laboratories), and the nylon membranes were probed with various combinations of the resulting fragments by Southern hybridization using the AlkPhos direct labeling and detection system with CDP-Star (Amersham Biosciences, Piscataway, NJ). Forty-eight of the signature tag sequences were selected to create the 47 mutants of each STM pool and the internal control present in each STM pool.
STM bank construction.
The plasmids containing the 47 signature tags were separately transformed into SM10λpir (33) and then independently conjugated into a nalidixic acid-resistant strain of virulent serovar Typhimurium UK1 for random DNA mutagenesis. Serovar Typhimurium mutants (1,598) were stocked individually in microtiter dishes (LB containing 20% glycerol) to create 34 STM pools, each containing 47 signature-tagged mutants. To serve as an internal control for each STM pool, a signature-tagged rpoS mutant (SB 29) was constructed to occupy the “A1” position of each STM pool.
Screening the STM banks in the SSC assay.
To prepare the serovar Typhimurium STM pools for challenge in the swine stomach contents assay, 2 μl of stock culture from each of the 48 mutants in a pool was transferred to 9.9 ml of LB medium containing nalidixic acid and kanamycin for overnight growth. A 1:100 dilution of the overnight culture was prepared in 3 ml of EG minimal medium with nalidixic acid and kanamycin and grown at 37°C, 180 rpm. At an optical density at 600 nm (OD600) of 0.4 (∼2 × 108 CFU/ml), a 2-ml sample of the culture was pelleted and frozen to represent the input pool. Additionally, 200 μl of the culture was pelleted and resuspended in an equal volume of EG, pH 4.4, spent medium for 1 h at 37°C for acid adaptation. The adapted cells were pelleted and resuspended in 200 μl of swine stomach contents for the first SSC challenge. Immediately, a 10-μl aliquot was serially diluted and plated on selective LB media to determine the CFU/ml at T0. After 10 min of challenge in the SSC at 37°C, a 10-μl aliquot of the sample was serially diluted and plated on selective LB media to determine viable counts at T10 min. Simultaneously, 100 μl of the challenged sample was added to 1 ml of EG medium, pH 7.0. Following brief centrifugation, the cell pellet was resuspended in 10 ml of EG medium containing nalidixic acid and kanamycin for overnight growth. A second round of challenge was performed on the surviving mutants of the STM pool as described above. Following overnight growth of the surviving STM mutants from the second challenge, a 1-ml culture was pelleted and frozen as the output pool.
PCR amplification of the signature tags and dot blot analysis.
The cell pellets from the input and output STM pools were each resuspended in 1 ml of H2O, and 2 μl of each served as a template for PCR amplification of the signature tag sequences using oSBM 46 and 47. Cycling conditions were as follows: 95°C for 2 min, 40 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 5 s, followed by a final extension for 10 min at 72°C. The PCR products were digested with HindIII to remove the constant regions, and the 62-bp signature tags were agarose gel extracted (Quantum Prep Freeze ′N Squeeze DNA gel extraction spin columns; Bio-Rad Laboratories, Hercules, CA). One hundred nanograms of DNA was labeled to produce the input and output pool probes for Southern hybridization using the AlkPhos direct labeling and detection systems with CDP-Star (Amersham Biosciences). To create the nylon membranes for Southern hybridization, the 48 signature tag DNA sequences represented in each of the 34 STM pools were prepared by PCR amplification and purification. Following quantitation (GeneQuant pro; Amersham Pharmacia Biosciences), 10 ng of each of the 62-bp PCR-amplified tags was transferred to Zeta probe membranes using a Bio-Dot apparatus (Bio-Rad Laboratories). Signal detection was performed using the AlkPhos direct labeling and detection systems with CDP-Star (Amersham Biosciences) and a ChemiDoc XRS chemiluminescence detection system with Quantity One software (Bio-Rad Laboratories).
Identification of the transposon insertion sites and DNA sequence analysis.
Chromosomal DNA from the 19 identified STM mutants was extracted (DNeasy tissue kit; QIAGEN, Valencia, CA), digested with EcoRI (Invitrogen, Carlsbad, CA), and electrophoresed on a 1% agarose gel. DNA fragments 1.8 kb and larger were gel extracted (Quantum Prep Freeze ′N Squeeze DNA gel extraction spin column; Bio-Rad Laboratories) and ligated into the pBluescript II SK vector (Stratagene, La Jolla, CA) treated with EcoRI and alkaline phosphatase. The ligations were electroporated into DH5α (Invitrogen), and transformants were selected on LB containing ampicillin and kanamycin. Plasmids were isolated using a QIAprep spin miniprep kit (QIAGEN, Valencia, CA) and sequenced by dideoxy chain termination using a primer specific to the O end of the mini-Tn5Km2 transposon sequence (oSBI 116; 5′-GCATGCAAGCTTCGGCC-3′) and an ABI 3700 DNA analyzer (Applied Biosystems Inc., Foster City, CA) at the Iowa State University DNA Sequencing and Synthesis Facility (Ames, IA). DNA sequence similarity searches were conducted using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (1).
SSC assay and ATR analysis of the identified signature-tagged mutants.
Prior to testing the signature-tagged mutants individually in the SSC and ATR assays, each mutation was moved to a clean serovar Typhimurium UK1 background by P22 phage transduction and tested for kanamycin resistance (and nalidixic acid sensitivity). For both assays, the mutants and the wild-type strain were grown overnight in EG medium with the appropriate antibiotics at 37°C. A 1:100 dilution in EG medium was prepared from the overnight culture and grown to an OD600 of 0.4. The cultures were adapted for 1 h at pH 4.4, briefly centrifuged, and resuspended in an equal volume of either swine stomach contents for a 10-min challenge (SSC assay) or EG, pH 3.0, spent medium for a 90-min challenge (ATR assay). The percent survival for each strain was calculated by dividing the number of CFU at the challenge time point (10 or 90 min) by the number of CFU immediately after resuspension of the bacteria in the challenging agents and then multiplying by 100. The data are presented as the ratio of the percent survival of the mutant divided by the percent survival of the wild-type strain; therefore, mutants with a ratio below 1 did not survive as well as the wild-type strain, and mutants with a ratio above 1 survived better than the wild-type strain. For all assays, the data represent the means of experiments performed in triplicate and the error bars represent the standard deviations.
Survival assays.
The following challenging agents were created prior to performing additional survival assays on serovar Typhimurium.
The pH of the SSC was raised to the indicated pHs using a small volume of NaOH (less than a 0.01 dilution). Following 2 h of incubation at pH 7.0, a portion of the SSC, pH 7.0, was lowered to pH 3.87 by HCl (for irreversible alkaline denaturation).
Heat inactivation of the SSC was performed at 75°C for 30 min and by autoclaving at 121°C, 15 lb/in2, 15 min.
The SSC was subjected to filtration using consecutive Centricon centrifugal filter units with 30-, 10-, and 3-kDa cutoffs (Millipore, Billerica, MA). With little volume loss of sample in the retentate, an aliquot of each filtrate from the 30-, 10-, and 3-kDa-cutoff filter units was used to challenge STM pool A containing SB 110, SB 104, SB 156, SB 114, SB 113, SB 117, SB 116, SB 126, SB 134, and SB 135 as well as the rpoS signature-tagged mutant (SB 29) and five mutants that were not sensitive to challenge in the SSC assay (SB 271 [17C1], SB 272 [21H5], SB 273 [24A5], SB 274 [27F3], and SB 275 [35A3]).
Synthetic gastric fluid was prepared as previously described (9), containing proteose peptone (8.3 g/liter), d-glucose (3.5 g/liter), NaCl (2.05 g/liter), KH2PO4 (0.6 g/liter), CaCl2 (0.11 g/liter), KCl (0.37 g/liter), bile (0.05 g/liter), lysozyme (0.1 g/liter), and pepsin (13.3 mg/liter) at pH 3.87. Lactic acid was added to a final concentration of 5, 10, or 20 mM, lactoferrin at 20 μM, and lactoferricin at 2 μM, and the assay was performed with or without pepsin. All chemical compounds listed above were purchased from Sigma-Aldrich (St. Louis, MO).
Gas chromatography for volatile fatty acid analysis.
Gas chromatography of butyl esters was used to determine the concentration of volatile fatty acids, according to the basic concepts of Salanitro and Muirhead (40). For each sample, 500 μl of swine stomach contents was combined with 200 μl of 200 mM heptanoic acid internal standard and 50 μl of 8% NaOH. Sample and standard tubes were shell frozen in dry ice-cooled acetone and then dried overnight using a Virtis Freezemobile lyophilizer (Gardiner, NY). Butylation was accomplished by adding 300 μl of butylation fluid (100 ml butanol plus 25 ml fresh sulfuric acid) and 800 μl chloroform. Samples were immediately capped, vortexed, and placed in an 80°C heating block for 2 h with occasional mixing. Following cooling to room temperature, trifluoroacetic anhydride (300 μl) was added and the mixture incubated at room temperature for 1 h. Samples were washed in triplicate with 1 ml of water (to remove residual salts) by removing the aqueous layer with vacuum after each wash. A 1-μl aliquot of the chloroform layer was analyzed by gas chromatography on an Agilent 6890N gas chromatograph fitted with an Agilent 19095J-121 capillary column (10 m by 530 μm) HP-5, 5% phenylmethyl siloxane (Palo Alto, CA). Nitrogen served as the carrier gas, with a flow rate of 6.0 ml/min. The oven temperature program was initiated at 50°C for 3.5 min followed by a rise to 180°C at 8°C/min. Oven temperature was then held at 180°C for 0.5 min followed by an increase to 280°C at a rate of 30°C/min. The oven temperature was held at 280°C for 10 min for a total run time of 33.6 min. Front inlet and flame ionization detector temperatures were 50°C and 270°C, respectively.
RESULTS
Survival of the phoP, rpoS, and fur mutants in the SSC assay.
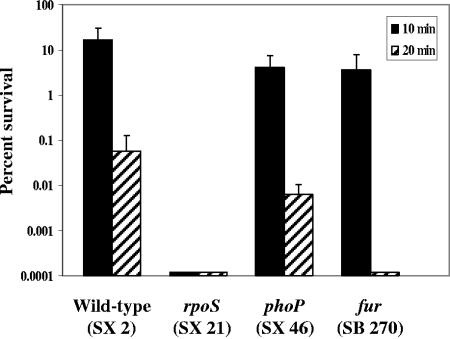
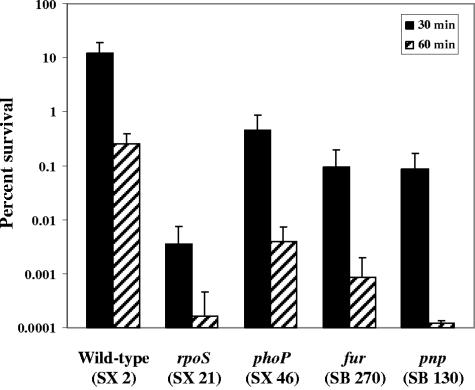
To investigate the initial survival mechanisms that Salmonella may rely upon during its introduction into the host's gastrointestinal environment, an ex vivo challenge assay using the stomach contents of swine was developed. The challenging agent of the swine stomach contents assay was produced by equally combining the filter-sterilized stomach contents of four pigs (final pH of 3.87). The assay was characterized with several strains known to be sensitive to acidic challenge, since a major obstacle for bacteria in the gastric environment is acidic pH. Serovar Typhimurium UK1 (SX 2) and strains containing mutations in rpoS (SX 21), phoP (SX 46), and fur (SB 270) were grown in minimal EG medium to mid-log phase, acid adapted at pH 4.4 for 1 h, and challenged in the SSC for 10 and 20 min (Fig. 1). The phoP mutant was 10-fold more sensitive in the SSC assay than the parent strain following 20 min of challenge. No survival was observed for the rpoS mutant following 10 min of SSC exposure, nor for the fur mutant after 20 min of challenge in the SSC assay.
FIG. 1.
Survival of the rpoS, phoP, and fur mutants in the SSC assay. SX 2 (wild type), SX 21 (rpoS), SX 46 (phoP), and SB 270 (fur) strains were grown in minimal EG medium to a mid-logarithmic phase of an OD600 of 0.4 (∼2 × 108 CFU/ml). The cultures were adapted at pH 4.4 for 1 h. Acid-adapted cells were washed and resuspended in an equal volume of filter-sterilized SSC. Viable counts were determined at 0, 10, and 20 min for determination of percent survival.
Screening the signature-tagged mutants in the SSC assay.
To identify additional genes of serovar Typhimurium that are important for survival during exposure to the contents of the swine stomach, an STM bank of ∼1,600 mutants (34 separate pools of 47 mutants) was constructed. Each pool of mutants endured two rounds of challenge in the SSC assay for 10 min. The input and output pools were screened by Southern hybridization to identify mutants sensitive to the SSC exposure (present in the input pool but absent in the output pool). The Southern hybridization patterns revealed 18 mutants (1.13% of the mutants screened) that were either absent or greatly diminished in the output pools, thereby identifying susceptibility to SSC exposure (Table 2). In addition, one mutant (SB 126) that repeatedly exhibited a stronger hybridization signal in the output pool than the input pool was also selected for further analysis. To confirm the phenotype of the 19 mutants, each was independently challenged in the SSC assay, and a ratio of the percent survival of the mutant compared to the percent survival of the parent strain was determined (Table 2). Five of the mutants exhibited less than a 10-fold decrease in survival following SSC challenge compared to the wild-type strain (SB 116, SB 115, SB 166, SB 156, and SB 200), whereas 10 of the mutants revealed a 10- to 100-fold decrease in survival (SB 110, SB 114, SB 117, SB 105, SB 134, SB 135, SB 157, SB 162, SB 213, and SB 209). Three mutants that displayed a considerable decrease in survival in the SSC assay compared to the parent strain were SB 104 (1,912-fold), SB 113 (285-fold), and SB 130 (1,613-fold). The signature-tagged mutant that repeatedly demonstrated an obvious increase in Southern hybridization signal (SB 126) survived as well as the wild-type strain in the SSC assay. The transposon insertion sites in the 19 mutants were determined by cloning the EcoRI DNA restriction fragments (>1.8 kb) from each mutant into the pBluescript II SK vector, and the resulting plasmids from kanamycin-resistant clones were sequenced. DNA sequence similarity searches using BLAST identified 18 different genes from the 19 mutants (Table 2).
ATR of signature-tagged mutants.
Since acidic pH is a lethal component of the stomach environment, each of the signature-tagged mutants was assayed to determine its acid tolerance response. As demonstrated in Table 2, most of the mutants (16 out of the 19) displayed less than a 10-fold decrease in survival compared to the wild-type strain. SB 110, SB 134, and SB 200 were 10-fold, 17-fold, and 10-fold more sensitive to challenge in the ATR assay than the parent strain, respectively.
Serovar Typhimurium is sensitive to low pH as well as other components in the SSC.
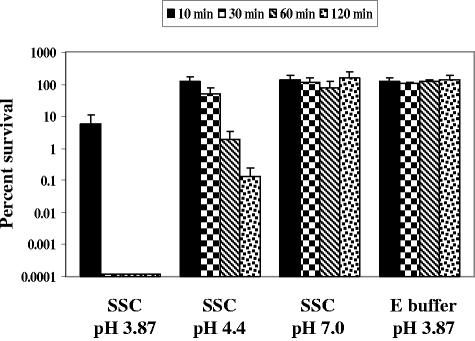
Since a discrepancy was revealed for the importance of the genes identified by signature-tagged mutagenesis during exposure to the SSC compared to the acidic pH in the ATR assay, the role of pH in the SSC assay was examined. The wild-type serovar Typhimurium strain (SX 2) was assayed in the SSC assay at the initial pH of 3.87 as well as pHs 4.4 and 7.0 (Fig. 2). At 30 min of SSC challenge, no survival was observed for the serovar Typhimurium strain at pH 3.87. However, at 2 h postchallenge, 0.11% of the organisms survived the pH 4.4 SSC challenge and 100% survived at pH 7.0 (as well as pH 5.8; data not shown). Therefore, as the pH of the SSC increased, the survival rate of the serovar Typhimurium culture also increased, indicating a pH-dependent lethal mechanism(s) in the SSC. Bacteria challenged in E buffer at pH 3.87 (or synthetic gastric fluid at pH 3.87; see Fig. 4), where only inorganic acid stress (low pH) is present, also survived 100% at 2 h. The large difference in serovar Typhimurium survival observed between the SSC pH 3.87 and E buffer pH 3.87 challenges suggests that low pH is not the only lethal component in the swine stomach. Thus, low pH is an important component of the porcine gastric environment, but additional lethal factors also exist and appear to be low pH dependent.
FIG. 2.
Effect of pH on the survival of serovar Typhimurium in the SSC assay. Wild-type serovar Typhimurium UK1 (SX 2) was grown to a mid-log phase of an OD600 of 0.4. Following acid adaptation at pH 4.4 for 1 h, the cells were pelleted and resuspended in an equal volume of SSC at pHs 3.87, 4.4, and 7.0 (adjusted with NaOH) as well as E buffer at pH 3.87. Viable counts were determined at 0, 10, 30, 60, and 120 min for determination of percent survival.
FIG. 4.
Survival of serovar Typhimurium in synthetic gastric fluid containing lactic acid. Wild-type serovar Typhimurium UK1 (SX 2) was grown to mid-log phase, adapted at pH 4.4 for 1 h, pelleted, and resuspended in an equal volume of synthetic gastric fluid at pH 3.87 containing lactic acid at 0, 5, 10, or 20 mM. Viable counts were determined at 0, 15, 30, 60 and 90 min for determination of percent survival.
Neither heat inactivation (75°C, 30 min, as well as autoclaving at 121°C, 15 lb/in2, 15 min) nor irreversible inactivation at neutral pH (pH 3.87 to pH 7.0 for 2 h and then back to pH 3.87) of susceptible proteins (such as pepsin) affected the ability of the SSC to kill wild-type serovar Typhimurium (data not shown). Also, to determine if the challenging components in the SSC were specific for a particular pig in the SSC pool, an additional challenging agent of swine stomach contents was created using the stomach contents of two different pigs that resulted in a pH similar to that of the original SSC challenging agent. Similar results were observed for the two SSC challenging agents, both in survival studies of the wild-type strain and in Southern hybridization patterns of the identified signature-tagged mutants (data not shown).
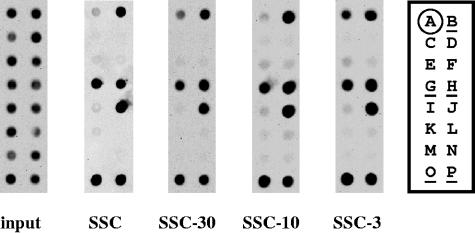
To further characterize the SSC, successive filtration of the SSC using centricon filter units with molecular mass cutoffs of 30, 10, and 3 kDa was performed. Filtrates were generated from which proteins/peptides greater than the molecular mass cutoff of the given filter unit were excluded. The filtrate from each filter unit was used as a challenging agent in comparison to the unfiltered SSC, using a pool of several identified signature-tagged mutants (STM pool A). STM pool A exhibited the same percent survival, following challenge with all three of the SSC filtrates, as the unfiltered SSC (data not shown). Furthermore, the Southern hybridization profiles of the various signature-tagged mutants were the same following challenges in the filtered and unfiltered SSC (Fig. 3). These results suggest that a component(s) capable of passing through a 3-kDa filter unit is responsible for the pH-dependent killing effect of the SSC and that all of the mutants represented in STM pool A are sensitive to the small component(s).
FIG. 3.
Effects of SSC filtration on survival of signature-tagged mutants. The SSC was filtered consecutively through centricon centrifugal filter units with molecular mass cutoffs of 30, 10, and 3 kDa. The filtrate from each was employed to challenge mid-log-phase, pH 4.4-adapted STM pool A for 0 or 10 min. The Southern hybridization patterns of STM pool A were determined following challenge in the filtered and unfiltered SSC. STM pool A input represents the 16 mutants present prior to challenge in the SSC assay, whereas the output pools indicate the absence or presence of signature-tagged mutants following challenge. The signature-tagged mutants represented in STM pool A are as follows: A = SB 29; B = SB 271; C = SB 110; D = SB 117; E = SB 104; F = SB 116; G = SB 272; H = SB 273; I = SB 156; J = SB 126; K = SB 114; L = SB 134; M = SB 113; N = SB 135; O = SB 274; P = SB 275. The controls are indicated in the key by circling (negative) or underlining (positive) the strain designation.
Serovar Typhimurium survival in synthetic gastric fluid with lactic acid.
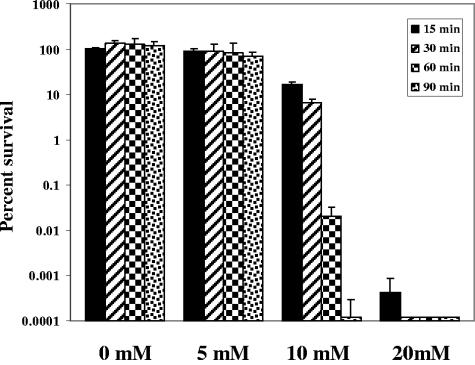
The volatile fatty acid composition of the SSC was determined using gas chromatography. The volatile fatty acid profile indicated that lactic acid, by far, had the highest concentration of all organic acids at 126 mM in the SSC challenging agent (data not shown); therefore, we tested the effects of various lactic acid concentrations on the survival of serovar Typhimurium in a synthetic gastric fluid (9) at pH 3.87 (the same pH as the SSC). The data presented in Fig. 4 demonstrate a concentration-dependent decrease in survival for the wild-type strain of serovar Typhimurium upon exposure to lactic acid in the synthetic gastric fluid. At a lactic acid concentration of 5 mM, the wild-type strain showed a 69% survival rate following 90 min of challenge. The percentages of survival at 10 mM were 16.7%, 6.4%, 0.020%, and 0.00012% at 15, 30, 60, and 90 min of challenge, respectively. A greater than 5-log decrease in bacterial survival was observed using 20 mM lactic acid at 15 min of exposure, and no survival was detected at 30 min. Pepsin, lactoferrin, and lactoferricin were also tested in the synthetic gastric fluid, but no observable difference in bacterial survival was observed (data not shown).
Each of the mutants presented in this study was individually assayed in the synthetic gastric fluid containing 10 mM lactic acid. Of the 19 STM mutants, only the pnp mutant (SB 130) exhibited a decrease in survival compared to the wild-type serovar Typhimurium strain (Fig. 5); a 100-fold decrease in survival was observed following 30 min of exposure and a 1,000-fold decrease at 60 min. An elevated sensitivity to lactic acid was also observed for the phoP mutant (greater than 10-fold at 30 min and 64-fold at 60 min), the rpoS mutant (greater than 1,000-fold at 30 and 60 min), and the fur mutant (greater than 100-fold at 30 and 60 min).
FIG. 5.
Survival of serovar Typhimurium mutants in synthetic gastric fluid containing 10 mM lactic acid. SX 2 (wild type), SX 21 (rpoS), SX 46 (phoP), SB 270 (fur)m and SB 130 (pnp) were grown in minimal EG medium to an OD600 of 0.4. Following acid adaptation at pH 4.4 for 1 h, the cells were washed and resuspended in an equal volume of synthetic gastric fluid containing 10 mM lactic acid. Viable counts were determined at 0, 30, and 60 min for determination of percent survival.
DISCUSSION
For ingested bacteria, including food-borne pathogens, the environment of the stomach can be considered one of the host's first lines of defense. For swine, the oral route of infection is common, since pigs root in their surroundings, an environment that is potentially fecally contaminated with Salmonella, given that 38 to 75% of U.S. swine herds are reported to be infected with Salmonella (5, 13, 36). Thus, the present study was performed to identify genes of Salmonella enterica serovar Typhimurium that are important for survival in the gastric environment. An ex vivo SSC assay was developed to screen for mutants of serovar Typhimurium with a decrease in survival upon challenge in the SSC assay. The pH of the SSC challenging agent was 3.87, a pH that correlates with previous reports (31, 37). Interestingly, we noticed that the more feed present in the porcine stomach at necropsy, the higher the pH of the stomach contents, suggesting a buffering effect provided by foodstuff as has been described by others (29, 34). Initial characterization of the SSC assay involved analysis of mutations in the rpoS, fur, and phoP genes of serovar Typhimurium, since these genes have been shown to be important for survival in acidic conditions. As demonstrated by Foster and colleagues (3, 7), serovar Typhimurium utilizes various acid protection systems during acidic encounters, including an adaptive system termed the acid tolerance response. Furthermore, acid stress is composed of two components, organic (weak acids) and inorganic (low pH) acid stress, whereas the perceived level of acid stress is a combination of both factors (4, 6). The rpoS and fur genes have been shown to be important for survival against organic acid stress, whereas phoP is involved in protection against inorganic acid stress (6). The results of the SSC assay indicate a role for rpoS, fur, and to a lesser extent phoP in survival during exposure to the SSC. As implied by the sensitivities of the rpoS and fur mutants in the SSC assay, organic acids could be an important component of the SSC. Still, both RpoS and Fur are global regulators of gene expression, and mutations in these two genes affect multiple bacterial systems that could be important for protection in the SSC assay. The 10-fold decline in survival for the phoP mutant indicates a slight sensitivity to SSC exposure. The phoP mutant could also be susceptible to weak acids present in the SSC, since a decrease in survival in the presence of 10 mM lactic acid was observed. On the other hand, another SSC component may be responsible for the decrease in survival, for example, an antimicrobial peptide, since a role for PhoP in protection against antimicrobial peptides has been demonstrated (23). The susceptibility of the phoP mutant to lactic acid is contradictory to the findings of Bearson et al. (6), which suggest that a phoP mutant is sensitive to inorganic acid stress, not organic acid stress. However, there are several differences between the two assays, including the weak acid used in the challenge assay, the concentration of the weak acid, the challenging pH, and the challenging media. A mutant of phoP may be vulnerable to weak acids, or at least lactic acid, at higher organic acid concentrations.
To identify additional genes required for survival in the stomach, STM was employed. Upon screening of ∼1,600 mutants in the SSC assay, 19 were selected for further analysis, with 18 demonstrating a reduction in Southern hybridization signal in the output pools compared to the input pools and 1 mutant exhibiting an increase in signal. STM transposon insertion sites were identified in genes involved in a variety of functions, including cellular regulation and processes (barA, sopB, and traL), molecular chaperoning (dnaK), energy metabolism (dgt), transcription (pnp), and translation (usg and poxR), as well as components and enzymes associated with the cellular envelope (ynaI, manC, rfaL, and asmA) and synthesis of fimbriae (pefA and pefC), with many of these genes also involved in bacterial virulence. Furthermore, when each of the identified signature-tagged mutants was individually analyzed in the SSC and ATR assays, obvious differences in the levels of sensitivity between the assays were observed for each mutant. Thirteen of the mutants exhibited a 10-fold or greater decrease in survival during exposure to the SSC compared to the parent strain, but only three mutants showed 10-fold or more sensitivity to the pH 3.0 acidic challenge. These results suggest that, in addition to low pH, other components are present in the SSC that are harmful to Salmonella. Further evidence for this was observed when the survival of serovar Typhimurium was compared in the SSC at pH 3.87 to that in E buffer or synthetic gastric fluid at pH 3.87 (inorganic acid stress only). Within 30 min of challenge, no bacterial survival was observed in the SSC at pH 3.87, yet no decrease in survival was detected in E buffer at pH 3.87 (or synthetic gastric fluid at pH 3.87) following 2 h of challenge. On the other hand, as the pH of the swine stomach contents increased, serovar Typhimurium was able to survive over a longer period of time, indicating that low pH is an important component of the swine stomach environment, since the lethal components of the SSC are low pH dependent.
Previously identified as encoding an acid shock and macrophage-inducible protein (10, 18, 20), a signature-tagged transposon in the promoter region of dnaK resulted in elevated sensitivity in the SSC assay (31-fold) and the ATR (10-fold). As a gene encoding a molecular chaperone involved in protein refolding during stress and directing abnormal proteins to degradation (28), the identification of dnaK is not unexpected and provides validation for the STM screen. This is the first report to describe an acid-sensitive phenotype for a dnaK mutant in serovar Typhimurium and demonstrates the involvement of DnaK in the acid stress response. To our knowledge, this is also the first report to describe usg and poxR mutants having increased sensitivity to acid stress in serovar Typhimurium and rfbM/manC and ynaI mutants having elevated acid tolerance. Mutations in pnp, dgt, and asmA resulted in dramatic decreases in bacterial survival in the SSC assay but not in the ATR assay. Dgt hydrolyzes dGTP to deoxyguanosine and tripolyphosphate and can bind strongly to single-stranded DNA (46), whereas AsmA is involved in the assembly of outer membrane proteins. Mutations in asmA reduce lipopolysaccharide (LPS) levels, thereby altering membrane fluidity and allowing the assembly of mutated outer membrane proteins (17). Reduced amounts of LPS have been shown to affect both the stress tolerance and virulence of S. enterica serovar Dublin (44). The role of Pnp as a polynucleotide phosphorylase and component of the mRNA degradosome has identified it as a potential porcine vaccine candidate for Actinobacillus pleuropneumoniae (22). In E. coli, a decrease in RpoS-regulated transcripts was observed in a pnp mutant (8), including the dps gene involved in acid resistance (12). Another gene associated with RpoS that was identified in the STM screen is barA. The barA mutant exhibited a 20-fold decrease in survival during SSC challenge. In E. coli, barA mutants have decreased rpoS expression, associated with hypersensitivity to hydrogen peroxide (35). BarA is a sensor kinase that phosphorylates the response regulator SirA; this regulatory system directly increases the expression of virulence genes through regulation of the HilA regulon (containing 40 genes associated with type III secretion systems) while indirectly decreasing the expression of motility/chemotaxis genes through the FlhDC regulon (55 genes) (43). Additional genes identified in the STM analysis have previously been shown to be involved in various stages of colonization and virulence (pefA, pefC, sopB, and poxR) or LPS O-antigen synthesis (manC and rfaL), as well as encoding a macrophage-inducible protein regulated by PhoP (mig-3).
The composition of the SSC is affected by both host and environmental factors, with the possibility of multiple elements contributing to the hostile environment of the stomach, including low pH, microbial by-products, antimicrobial peptides, bacteriophage, ammonia, alpha-amylase, lipase, surfactant, pepsin, bile, weak acids, etc. Attempts to determine the component(s) responsible for the lethality of the SSC involved unsuccessfully searching for a proteinaceous factor that might be susceptible to irreversible alkaline denaturation (i.e., pepsin), heat denaturation, or filtration. Challenge of the identified signature-tagged mutants with filtrates produced from consecutive filtration of the SSC through 30-, 10-, and 3-kDa-cutoff filter units revealed similar killing effects for all three filtrates compared to the unfiltered SSC, both in overall survival of the signature-tagged mutant pool and in individual survival of each mutant (Southern hybridization patterns). These results indicate that the lethal component(s) of the SSC, which each of the mutants represented in STM pool A is sensitive to, is smaller than 3 kDa.
Further investigation of the SSC challenging agent by gas chromatography revealed a slightly elevated lactic acid concentration of 126 mM. The lactic acid concentration in the swine stomach has previously been reported to be diet dependent, associated with lactic acid-producing microorganisms in the stomach, and can vary 50-fold or more, as shown with reported gastric levels ranging from 1.6 to 77 mM (11, 31, 32, 41). Since the lactic acid concentration of the swine stomach contents from several individual pigs in this study ranged from 1 to 163 mM, survival analysis was determined for the wild-type strain of serovar Typhimurium with various concentrations of lactic acid in a synthetic gastric fluid. Bacterial survival was both concentration dependent (with little decrease in survival at 5 mM but basically no survival at 20 mM) and exposure time dependent (with bacterial survival decreasing from 16.7% at 15 min postchallenge in 10 mM lactic acid to 0.00012% at 90 min). Recently, Mikkelson et al. (32) demonstrated a correlation between the concentration of undissociated lactic acid and the death rate of serovar Typhimurium in the gastric content of swine. Since the pKa of lactic acid is approximately 3.9, the filterable, pH-dependent killing observed in the SSC assay could be a result of anion accumulation dependent on the ΔpH, whereby undissociated (protonated) fermentation acids can pass across the cell membrane and dissociate in the more alkaline interior of the cell, resulting in an accumulation of anionic species that can affect protein synthesis and function (38). Furthermore, dissociated weak acids cause the intracellular pH of bacteria to drop, resulting in disruption of enzymatic processes and collapse of the proton motive force.
The 19 identified signature-tagged mutants as well as mutants of rpoS, phoP, and fur were assayed in the presence of 10 mM lactic acid in the synthetic gastric fluid. Of the signature-tagged mutants, only the pnp mutant exhibited a decrease in survival compared to the parent serovar Typhimurium strain. Mutations in the rpoS, phoP, and fur genes also rendered serovar Typhimurium sensitive to lactic acid exposure. The role of these genes, whose products are involved in global mRNA synthesis (RpoS, PhoP, and Fur) and degradation (Pnp), in protection against lactic acid exposure may be multifaceted. Furthermore, since only one of the signature-tagged mutants was sensitive to the lactic acid challenge, multiple lethal components are probably present in the SSC. Further characterization of the swine stomach contents is warranted and may identify components that could serve as targets for Salmonella control.
Acknowledgments
We thank Ann Marie Jensen, Holly Dolphin-Rooney, Amanda Toot, and Deborah Lebo for excellent technical assistance and Thomas Casey and Zoe McCuddin for critical review of the manuscript.
This research was supported by USDA ARS CRIS funds.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, K. W., and C. W. Kaspar. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 4.Baik, H. S., S. Bearson, S. Dunbar, and J. W. Foster. 1996. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology 142:3195-3200. [DOI] [PubMed] [Google Scholar]
- 5.Barber, D. A., P. B. Bahnson, R. Isaacson, C. J. Jones, and R. M. Weigel. 2002. Distribution of Salmonella in swine production ecosystems. J. Food Prot. 65:1861-1868. [DOI] [PubMed] [Google Scholar]
- 6.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein, J. A., P. H. Lin, S. N. Cohen, and S. Lin-Chao. 2004. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl. Acad. Sci. USA 101:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beumer, R. R., J. de Vries, and F. M. Rombouts. 1992. Campylobacter jejuni non-culturable coccoid cells. Int. J. Food Microbiol. 15:153-163. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 11.Canibe, N., S. H. Steien, M. Overland, and B. B. Jensen. 2001. Effect of K-diformate in starter diets on acidity, microbiota, and the amount of organic acids in the digestive tract of piglets, and on gastric alterations. J. Anim. Sci. 79:2123-2133. [DOI] [PubMed] [Google Scholar]
- 12.Choi, S. H., D. J. Baumler, and C. W. Kaspar. 2000. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaboration in Animal Health and Food Safety Epidemiology. November 2004, posting date. CAHFSE annual report for July 1, 2003-June 30, 2004. Animal and Plant Health Inspection Service, Riverdale, Md. [Online.] http://www.aphis.usda.gov/cahfse/results/index.htm.
- 14.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 15.Curtiss, R., III, S. B. Porter, M. Munson, S. A. Tinge, J. O. Hassan, C. Gentry-Weeks, and S. M. Kelly. 1991. Nonrecombinant and recombinant avirulent Salmonella live vaccines for poultry, p. 169-198. In L. C. Blankenship, J. S. Bailey, N. A. Cox, N. J. Stern, and R. J. Meinersmann (ed.), >Colonization control of human bacterial enteropathogens in poultry. Academic Press, New York, N.Y.
- 16.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 17.Deng, M., and R. Misra. 1996. Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol. Microbiol. 21:605-612. [DOI] [PubMed] [Google Scholar]
- 18.Foster, J. W. 1993. The acid tolerance response of Salmonella typhimurium involves transient synthesis of key acid shock proteins. J. Bacteriol. 175:1981-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 20.Foster, J. W. 1991. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J. Bacteriol. 173:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fratamico, P. M. 2003. Tolerance to stress and ability of acid-adapted and non-acid-adapted Salmonella enterica serovar Typhimurium DT104 to invade and survive in mammalian cells in vitro. J. Food Prot. 66:1115-1125. [DOI] [PubMed] [Google Scholar]
- 22.Fuller, T. E., S. Martin, J. F. Teel, G. R. Alaniz, M. J. Kennedy, and D. E. Lowery. 2000. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb. Pathog. 29:39-51. [DOI] [PubMed] [Google Scholar]
- 23.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 28.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 29.Mainville, I., Y. Arcand, and E. R. Farnworth. 2005. A dynamic model that simulates the human upper gastrointestinal tract for the study of probiotics. Int. J. Food Microbiol. 99:287-296. [DOI] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen, L. L., and B. B. Jensen. 2003. The stomach as a barrier that reduces the occurrence of pathogenic bacteria in pigs. Presented at the 9th International Symposium on Digestive Physiology in Pigs, Banff, Alberta, Canada.
- 32.Mikkelsen, L. L., P. J. Naughton, M. S. Hedemann, and B. B. Jensen. 2004. Effects of physical properties of feed on microbial ecology and survival of Salmonella enterica serovar Typhimurium in the pig gastrointestinal tract. Appl. Environ. Microbiol. 70:3485-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morelli, L. 2000. In vitro selection of probiotic lactobacilli: a critical appraisal. Curr. Issues Intest. Microbiol. 1:59-67. [PubMed] [Google Scholar]
- 35.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 36.National Animal Health Monitoring System. 1997. Shedding of Salmonella by finisher hogs in the US, swine 1995: grower/finishers. Animal and Plant Health Inspection Service N223.197. Animal and Plant Health Inspection Service, Riverdale, Md. [Online.] http://www.aphis.usda.gov/vs/ceah/ncahs/nahms/swine/swine95/sw95salm.pdf.
- 37.Radcliffe, J. S., Z. Zhang, and E. T. Kornegay. 1998. The effects of microbial phytase, citric acid, and their interaction in a corn-soybean meal-based diet for weanling pigs. J. Anim. Sci. 76:1880-1886. [DOI] [PubMed] [Google Scholar]
- 38.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 39.Saenz, H. L., and C. Dehio. 2005. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 8:612-619. [DOI] [PubMed] [Google Scholar]
- 40.Salanitro, J. P., and P. A. Muirhead. 1975. Quantitative method for the gas chromatographic analysis of short-chain monocarboxylic and dicarboxylic acids in fermentation media. Appl. Microbiol. 29:374-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholten, R. H., C. M. van der Peet-Schwering, L. A. den Hartog, M. Balk, J. W. Schrama, and M. W. Verstegen. 2002. Fermented wheat in liquid diets: effects on gastrointestinal characteristics in weanling piglets. J. Anim. Sci. 80:1179-1186. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J. L. 2003. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 66:1292-1303. [DOI] [PubMed] [Google Scholar]
- 43.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomsen, L. E., M. S. Chadfield, J. Bispham, T. S. Wallis, J. E. Olsen, and H. Ingmer. 2003. Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 228:225-231. [DOI] [PubMed] [Google Scholar]
- 45.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 46.Wurgler, S. M., and C. C. Richardson. 1993. DNA binding properties of the deoxyguanosine triphosphate triphosphohydrolase of Escherichia coli. J. Biol. Chem. 268:20046-20054. [PubMed] [Google Scholar]