Abstract
The nysL gene, encoding a putative P450 monooxygenase, was identified in the nystatin biosynthetic gene cluster of Streptomyces noursei. Although it has been proposed that NysL is responsible for hydroxylation of the nystatin precursor, experimental evidence for this activity was lacking. The nysL gene was inactivated in S. noursei by gene replacement, and the resulting mutant was shown to produce 10-deoxynystatin. Purification and an in vitro activity assay for 10-deoxynystatin demonstrated its antifungal activity being equal to that of nystatin. The NysL protein was expressed heterologously in Escherichia coli as a His-tagged protein and used in an enzyme assay with 10-deoxynystatin as a substrate. The results obtained clearly demonstrated that NysL is a hydroxylase responsible for the post-polyketide synthase modification of 10-deoxynystatin at position C-10. Kinetic studies with the purified recombinant enzyme allowed determination of Km and kcat and revealed no inhibition of recombinant NysL by either the substrate or the product. These studies open the possibility for in vitro evolution of NysL aimed at changing its specificity, thereby providing new opportunities for engineered biosynthesis of novel nystatin analogues hydroxylated at alternative positions of the macrolactone ring.
The genetics and biochemistry of antibiotic biosynthesis in bacteria attract much attention due to the possibility of using this information to design and produce new pharmaceuticals (26). Filamentous gram-positive bacteria of the genus Streptomyces are known to produce a wide range of chemically diverse secondary metabolites with different biological activities. Over 200 gene clusters governing antibiotic biosynthesis in Streptomyces have been isolated and analyzed, giving rise to a wealth of information regarding the enzymology of antibiotic formation. Biosynthetic pathways for many antibiotics produced by Streptomyces include steps catalyzed by P450 monooxygenase enzymes, which mostly perform hydroxylation of pathway intermediates (19).
P450 monooxygenases are widely distributed in nature and are mostly known to perform monooxygenation, although other catalytic functions, such as dehydrogenation, C—C and C=N bond cleavage, and dehydration, have also been reported for P450 enzymes (16). The “classical” reaction catalyzed by these enzymes is the transfer of one oxygen atom from O2 to a variety of substrates, using electrons supplied by NAD(P)H through ferredoxin (Fdx) and ferredoxin oxidoreductase (Fdr). Although in some bacteria the genes for P450 monooxygenases are usually clustered with the Fdx and Fdr genes, such a gene arrangement is rare in Streptomyces, suggesting that in streptomycete bacteria these enzymes are rather nonselective in their choice of electron transfer partners (19).
A considerable number of streptomycete P450 monooxygenases involved in antibiotic biosynthesis have been unraveled, including those performing monooxygenation of intermediates in the biosynthetic pathways for macrolides (5, 15, 17), anthracyclines (27), glycopeptides (6), coumarins (12) etc. Interestingly, some bacterial P450 monooxygenases exhibit substantial flexibility regarding the structural features of their substrates (13, 28), suggesting the possibility for using these enzymes in combinatorial biosynthesis. Recently, characterizations of P450 monooxygenase genes involved in the biosynthesis of two polyene macrolide antibiotics, pimaricin and amphotericin B, have been published (10, 11, 17). Deletion of pimD in the pimaricin producer Streptomyces natalensis resulted in the accumulation of 4,5-deepoxy pimaricin, suggesting a role for PimD as a 4,5-epoxydase. The latter was recently confirmed via an enzymatic assay with heterologously expressed protein (18). Inactivation of the amphL gene in the amphotericin producer S. nodosus led to the production of 8-deoxy amphotericins A and B, implying that AmphL is a C-8 hydroxylase in amphotericin biosynthesis. Another P450 enzyme from the amphotericin biosynthetic pathway was shown to be responsible for the oxidation of the methyl group, resulting in the appearance of the exocyclic carboxyl moiety. Most interestingly, the amphotericin B analogue lacking the latter group was purified and shown to have considerably less hemolytic activity than amphotericin B (11). A similar result was obtained upon inactivation of RimG, the P450 monooxygenase responsible for oxidation of the exocyclic methyl group on the rimocidin/C-108 precursor in S. diastaticus (22). Considering the importance of polyene macrolides as antifungal agents used in human therapy, a better understanding of the reactions catalyzed by these P450 monooxygenases and their substrate range might help in combinatorial biosynthesis of novel polyene macrolides aimed at the development of new antibiotics.
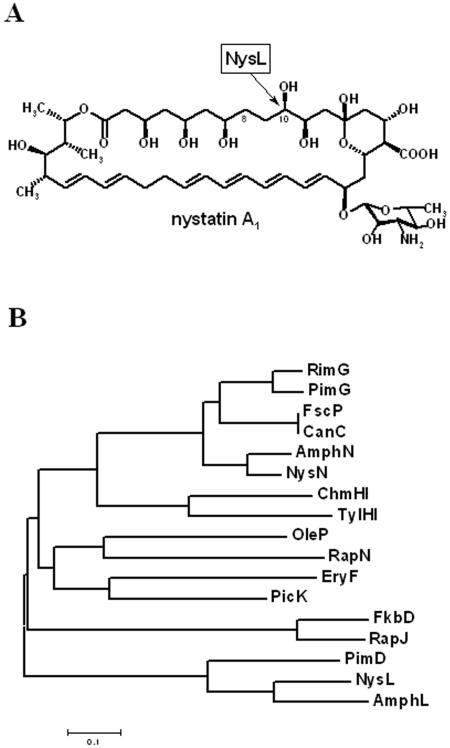
Streptomyces noursei ATCC 11455 produces a complex mixture of polyene macrolides with antifungal activity which are generally referred to as nystatins (9). The major component of the nystatin complex, nystatin A1, is composed of a 38-membered macrolactone ring synthesized by six large polyketide synthase (PKS) proteins (7). The macrolactone ring of nystatin A1 is further modified in the course of biosynthesis via the addition of a mycosamine deoxysugar moiety, oxidation of the methyl group at C-16, and hydroxylation at C-10 (14). The last two modifications in the biosynthesis of nystatin were suggested to be performed by P450 monooxygenase enzymes encoded by the genes nysL and nysN in the nystatin biosynthetic gene cluster, although experimental evidence for the enzymatic activities of these monooxygenases was not available. In the present work, we have proven the role of NysL as a C-10 hydroxylase (Fig. 1A) involved in nystatin biosynthesis via gene inactivation in S. noursei, heterologous expression of NysL, and in vitro enzyme assays for the recombinant protein.
FIG. 1.
(A) Chemical structure of nystatin A1. The site of NysL-catalyzed hydroxylation is indicated by an arrow. (B) Phylogenetic tree of actinomycete P450 monooxygenases known to be responsible for post-PKS modification of macrolide antibiotic precursors in the biosynthetic pathways of amphotericin (AmphL and AmphN), chalcomycin (ChmHI), candicidin (CanC), erythromycin (EryF), FK520 (FkbD), FR-008 (FscP), nystatin (NysL and NysN), oleandomycin (OleP), picromycin (PicK), pimaricin (PimD and PimG), rapamycin (RapH and RapJ), rimocidin (RimG), and tylosin (TylHI).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Some of the plasmids are described below. S. noursei strains were maintained on ISP2 agar medium (Difco) and grown in tryptic soy broth (Oxoid) for DNA isolation. Escherichia coli strains were handled using standard techniques (21). Conjugation from E. coli ET12567(pUZ8002) to S. noursei and the gene replacement procedure were performed as described previously (23). Assessments of nystatin and 10-deoxynystatin production and accurate mass determinations were done by liquid chromatography-mass spectroscopy (LC-MS) of dimethyl sulfoxide extracts of 5-day-old cultures from well plate cultivations (9). Well plate cultivations were performed in semidefined 0.5× SAO-23 medium according to a previously described protocol (25).
TABLE 1.
Bacterial strains, plasmids, and phage used for this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | General cloning host | BRL |
| ET12567(pUZ8002) | Strain for intergeneric conjugation; Kmr Cmr | 23 |
| Streptomyces noursei strains | ||
| ATCC 11455 | Wild type, nystatin producer | ATCC |
| NDA59 | Mutant with in-frame nysA deletion, nystatin nonproducer | 8 |
| NLD101 | NDA59 with in-frame deletion of the nysL gene | This work |
| Phage | ||
| N95 | Recombinant λ phage containing the nysL gene and flanking regions | 7 |
| Plasmids | ||
| pGEM3Zf(−) | ColE1 replicon; Apr; 3.2 kb | Promega |
| pQE2 | N-terminal six-His tag expression vector | QIAGEN |
| pQNL2 | Vector for heterologous expression of the six-His-NysL protein | This work |
| pSOK20 | pSG5 and ColE1 replicons; oriT; Amr; E. coli-Streptomyces conjugative vector | 29 |
| pSOK804 | E. coli-Streptomyces conjugative and integrative vector; VWBint; ColE1 replicon; oriT; Amr | 24 |
| pNLD1 | nysL replacement vector | This work |
Abbreviations: Am, apramycin; Ap, ampicillin; Km, kanamycin; Tc, tetracycline; Cm, chloramphenicol.
DNA manipulation.
General techniques for DNA manipulation were used as described elsewhere (21). Isolation of the DNA fragments from agarose gel was done with a QIAEX kit (QIAGEN, Germany). Southern blot analysis was performed with a DIG High Prime labeling kit (Roche Biochemicals, Germany) according to the manufacturer's manual. Oligonucleotide primers were purchased from MWG-Biotech AG. The DNA sequence of the nystatin biosynthetic gene cluster has been reported previously (7).
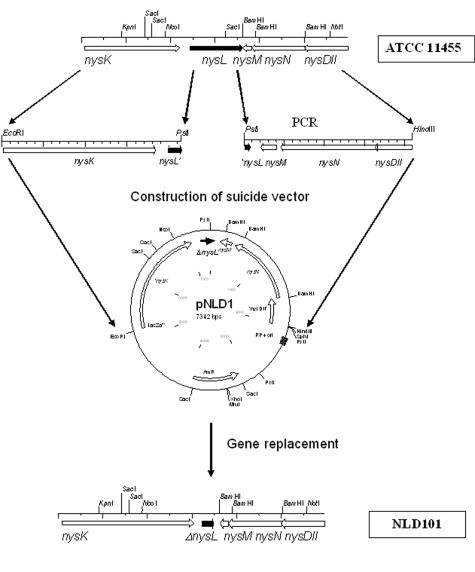
Construction of nysL deletion mutant.
A 2,148-kb DNA fragment designated Nldel12, encompassing part of nysL and the downstream region, was amplified from the pL95X template using primers NLD1 (5′-GACGAATTCAACTGGTGGCGGAGCTGA-3′) and NLD2 (5′-GACCTGCAGCTGCTTGAGTTCGGTG-3′). A 2,089-kb DNA fragment designated NLdel34, containing the upstream region and some of the coding region of nysL, was amplified from the plasmid pL95X template using primers NLD3 (5′-GACCTGCAGTCGAGGAACTGCGGGTGCT-3′) and NLD4 (5′-GCAAAGCTTTGCGGGCGATGGCGTTCAC-3′). The NLdel12 and NLdel34 PCR products were digested with the EcoRI/PstI and PstI/HindIII endonucleases, respectively, and ligated together with the 3.0-kb EcoRI/HindIII fragment from pSOK201, yielding the nysL replacement vector pNLD1. Using this vector, we obtained the nysL in-frame deletion mutant NLD101 by double homologous recombination in the previously constructed non-nystatin-producing mutant NDA59, which can be complemented with the nysA gene to restore nystatin production (8).
Purification of 10-deoxynystatin and test for antifungal activity.
The purification of 10-deoxynystatin was performed according to a previously described procedure (25). The antifungal activities of nystatin and 10-deoxynystatin were determined using Candida albicans as a test organism according to a method described previously (9).
Heterologous expression of NysL.
The nysL gene was amplified for insertion into the N-terminal His tag expression vector pQE-2 (QIAGEN) by PCR. The forward primer NLQE21 (5′-GGACCATATGAGCACACCGACCGCAC-3′) introduced a unique NdeI site at the 5′ end of the gene, while the reverse primer NLQE22 (5′-CCAGAAGCTTCATCACGTCACCAGGTGAC-3′) carried a HindIII site downstream from translational stop codons. The amplified DNA fragment was digested with NdeI and HindIII and cloned into the same sites of pQE2 to generate pQNL2, the vector for heterologous expression of the N-terminal six-His-NysL protein. The cloned fragment was sequenced in order to check its integrity after PCR amplification. Expression and purification of six-His-NysL were performed essentially as described elsewhere (20), but after induction of the culture with 1 mM IPTG (isopropyl-ß-d-thiogalactopyranoside) and incubation for 4 h at 30°C.
NysL enzyme assay and studies of enzyme kinetics.
NysL activity was measured in a mixture containing 0.3 to 105 μM 10-deoxynystatin, 16 to 48 nM six-His-NysL, 100 μg/ml spinach ferredoxin, 0.2 U/ml spinach ferredoxin-NADP+ reductase, 1.4 mM NADPH, 10 mM glucose-6-phosphate, and 8 U/ml glucose-6-phosphate dehydrogenase in 50 mM Tris-HCl, pH 8.0 (16). The reaction mixture was incubated at 30°C in the temperature-controlled autoinjector of the Agilent 1100 LC-MS equipment used in the assay. The reaction was started by adding the enzymes to the preheated substrate mixture. Every fifth minute after the reaction was started, an aliquot from the reaction mixture was injected into the LC-MS instrument for determination of the nystatin and 10-deoxynystatin concentrations. The LC-MS analyses were performed on an Agilent 1100 high-performance liquid chromatography (HPLC) system connected to a diode array detector and an Agilent single-quadrupole mass spectrometer using electrospray ionization in negative mode. A Zorbax Bonus-RP (2.1 × 50 mm) chromatographic column operated at 0.3 ml/min was used for analyte separation. The mobile phase consisted of 10 mM ammonium acetate, pH 4.0, and 30% acetonitrile.
RESULTS
Inactivation of nysL gene in S. noursei leads to production of 10-deoxynystatin.
The nysL gene in the S. noursei nystatin biosynthetic gene cluster is located downstream of nysK, the gene encoding the last module of the nystatin PKS and a terminal thioesterase (7). Amino acid sequence analysis of the deduced nysL product revealed a high degree of similarity of this polypeptide to bacterial P450 monooxygenases, including those involved in antibiotic biosynthesis (data not shown). Further phylogenetic analysis (Fig. 1B) revealed clustering of the NysL amino acid sequence with those of PimD and AmphL, proven to act as an epoxidase and a hydroxylase in the biosynthesis of the polyene macrolides pimaricin and amphotericin, respectively. Although we have suggested earlier that NysL is most probably a C-10 hydroxylase, the experimental evidence for this assumption was lacking. Repeated attempts to inactivate nysL by in-frame deletion via double homologous recombination in an S. noursei wild-type strain failed. We then used a previously constructed mutant that does not produce nystatin, NDA59, with an inactivated nysA gene encoding the nystatin PKS loading module, which can be complemented with the wild-type copy of nysA to restore nystatin production (8). The use of the latter mutant, along with the extension of flanking regions for the gene replacement construct up to 2 kb, allowed isolation of the ΔnysL mutant designated NLD101. The scheme for NDL101 mutant construction is presented in Fig. 2. The deletion within the nysL gene in NLD101 was confirmed by Southern blot analysis (data not shown).
FIG. 2.
Schematic representation of ΔnysL mutant construction in S. noursei.
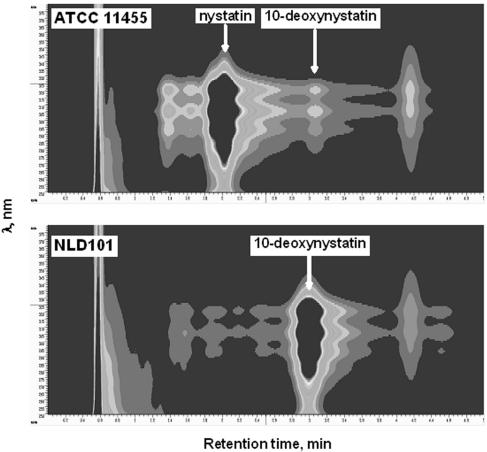
To analyze polyene macrolide production by the ΔnysL mutant, the integrative plasmid pNA0 (8) carrying the nysA gene was introduced into the NLD101 mutant. The resulting recombinant strain, NLD101(pNA0), was incubated for 5 days in defined production medium, and culture extracts were analyzed for the presence of nystatin-related polyene macrolides by using HPLC and LC-MS. The latter analysis (Fig. 3) confirmed the production of a 910-Da nystatin analogue by the NLD101(pNA0) strain at a volumetric yield of 1.06 ± 0.04 g/liter, which was somewhat lower than the nystatin yield in the wild-type strain (1.51 ± 0.03 g/liter). The accurate atomic mass of this analogue, its retention time during HPLC, and the UV spectrum exactly matched those for 10-deoxynystatin, which was identified during studies of the NysH/NysG transporter system in S. noursei (25). The 10-deoxynystatin produced was then purified from the NLD101 culture extract for further testing.
FIG. 3.
Diode array detector-HPLC profiles of culture extracts from S. noursei ATCC 11455 and the NLD101 mutant.
To test whether the hydroxy group at C-10 has significance for the antifungal activity of nystatin, the MIC50 was determined for 10-deoxynystatin, using Candida albicans as a test organism and nystatin as a reference. The MIC50s of 10-deoxynystatin and nystatin were found to be identical (0.45 μg/ml under the conditions tested), implying that C-10 hydroxylation has little significance for the activity of nystatin, at least against C. albicans.
Recombinant NysL protein efficiently converts 10-deoxynystatin to nystatin.
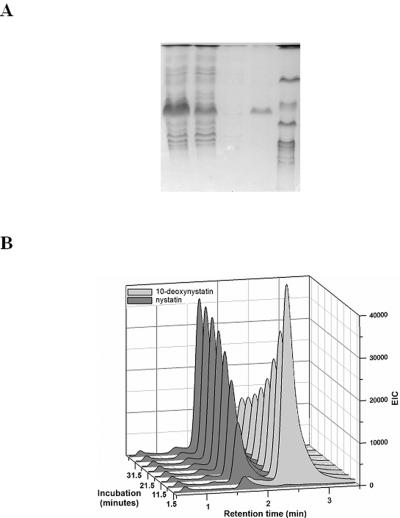
To allow for enzymatic studies of the NysL monooxygenase, we heterologously expressed this protein in Escherichia coli with an N-terminal six-His affinity tag and purified the recombinant protein. The PCR-amplified product containing the coding region of the nysL gene was cloned into the pQE2 expression vector immediately after the six-His tag sequence (pQNL2) (see Materials and Methods). Induction of the T5 promoter in the culture of E. coli DH5α carrying the pQNL2 expression plasmid resulted in high-level production of the partially soluble 43-kDa protein, whose molecular mass corresponded to that of NysL. The recombinant six-His-NysL protein was purified on a Ni-nitrilotriacetic acid agarose column, and its purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 4A).
FIG. 4.
(A) Heterologous expression of NysL in Escherichia coli as a His-tagged protein. Lanes: 1, crude cell extract; 2, flowthrough; 3, wash; 4, eluted His-NysL; 5, molecular weight marker. (B) Enzymatic assay monitored by HPLC and LC-MS showing conversion of 10-deoxynystatin to nystatin by His-NysL and the redox partners.
The ability of the purified six-His-NysL protein to convert its putative substrate 10-deoxynystatin to nystatin was analyzed in vitro. The reaction mixture (see Materials and Methods) contained an NADPH-regenerating system based on glucose-6-phosphate and glucose-6-phosphate dehydrogenase in addition to the buffer solution, spinach ferredoxin, spinach ferredoxin-NADP+ reductase, NADPH, and 10-deoxynystatin. Only in the presence of such an electron-donating system was six-His-NysL able to convert 10-deoxynystatin to nystatin, and no conversion was observed without the participation of the NADPH-regenerating system. The progress of the reaction in terms of the conversion of 10-deoxynystatin to nystatin was monitored over 40 min (Fig. 4B). The conversion of >75% of the 10-deoxynystatin to nystatin was observed under these conditions.
Kinetic studies of recombinant NysL protein.
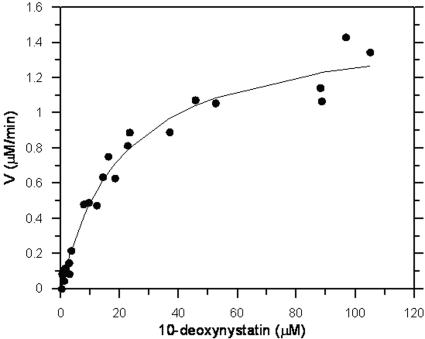
The kinetics of the recombinant NysL-catalyzed reaction was studied next. With a substrate concentration of 45 μM, the initial rate of hydroxylation was found to increase linearly with the enzyme concentration in the range of 16 to 45 nM six-His-NysL. At 30°C, the phase of the reaction where the enzyme concentration and the rate of hydroxylation correlated linearly lasted ca. 15 to 20 min. An enzyme concentration of 16.1 nM and a reaction time of 5 min were chosen for kinetic studies. Enzymatic assays were performed with increasing concentrations of 10-deoxynystatin and nystatin. Data from the assays, monitored by LC-MS, did not reveal inhibition of the recombinant NysL enzyme by either the substrate or the product. At substrate concentrations in the range of 0.3 to 105 μM, the data could be fitted well to the Michaelis-Menten equation (Fig. 5). Further increases in the substrate concentration resulted in precipitation of 10-deoxynystatin and were therefore not considered. The Km and Vmax values were determined (based on three independent assays) to be 24 ± 3 μM and 1.5 ± 0.2 μM/min, respectively. Provided that all of the enzyme is active, this corresponds to a kcat value of 1.6 ± 0.2 s−1 and a kcat/Km value of 0.07 ± 0.01 μM−1 s−1.
FIG. 5.
Kinetics of six-His-NysL-catalyzed reaction. The presented data are based on three independent enzyme assays.
DISCUSSION
P450 monooxygenases play an important role in the biosynthesis of many secondary metabolites produced by Streptomyces bacteria. Recent advances in engineered antibiotic biosynthesis in streptomycetes make P450 monooxygenases attractive targets for genetic manipulation that can lead to the production of novel antibiotic analogues (6, 10, 11, 17, 22). The biosynthesis of all polyene macrolide antibiotics known to possess potent antifungal activity involves oxidative steps apparently catalyzed by the P450 monooxygenases (1). Inactivation of the monooxygenase genes in the producers of the polyene macrolides pimaricin and amphotericin yielded new analogues of these antibiotics with properties different from those of the final products of the respective biosynthetic pathways (10, 11, 17).
In the present study, we have characterized the P450 monooxygenase NysL encoded by the nystatin biosynthetic gene cluster in S. noursei ATCC 11455. An alignment of the NysL amino acid sequence with those for P450 enzymes known to be involved in antibiotic biosynthesis, followed by phylogenetic analysis, clearly revealed that NysL is related to PimD and AmphL, an epoxidase and a hydroxylase in the pimaricin and amphotericin biosynthetic pathways, respectively. PimD, AmphL, and NysL represent a separate clade on the phylogenetic tree which is distant from that containing P450 monooxygenases from the polyene macrolide biosynthetic pathways known or assumed to catalyze the oxidation of an exocyclic methyl group (1, 11, 22).
Inactivation of the nysL gene in S. noursei yielded a recombinant strain producing 10-deoxynystatin as a major polyene macrolide (Fig. 3). This metabolite is present in trace amounts in wild-type strain extracts and was assumed to be accumulated due to the failure of NysL to hydroxylate this precursor at C-10 (9). Sletta et al. have demonstrated that the accumulation of 10-deoxynystatin by S. noursei also occurs upon inactivation of the presumed nystatin efflux system NysH-NysG (25). This observation suggested a link between the process of NysL-catalyzed hydroxylation of 10-deoxynystatin and active transport of the nystatin, and we hypothesized that the NysH-NysG transporter provides conditions favorable for C-10 hydroxylation by NysL.
Based on the current model for the polyene macrolide mode of action, which involves the formation of ion-permeable channels in fungal membranes (2), it could be assumed that removal of the C-10 hydroxyl group from the nystatin polyol region will decrease channel permeability. Indeed, according to the model, hydroxyl groups in the polyol region of the molecule are located on the inner part of the channel, creating a hydrophilic environment allowing ions and other small molecules to leak out of the cell. However, we have demonstrated that the antifungal activity of 10-deoxynystatin is equal to that of nystatin, at least for C. albicans. This observation was in contrast to the data for 4,5-deepoxypimaricin and 8-deoxyamphotericin B, which were obtained upon inactivation of the genes encoding P450 monooxygenases performing epoxidation and hydroxylation of pimaricin and amphotericin precursors, respectively, in polyol regions of the molecules. In both cases, a notable decrease in antifungal activity was observed (10, 17). One possible explanation for this phenomenon might be that the conformation of the polyol region of the nystatin molecule is different from those of pimaricin and amphotericin. This difference might result in limited participation of the nystatin C-10 hydroxyl group in channel formation and, thus, its low significance for antifungal activity.
Heterologous expression and purification of His-tagged NysL allowed for preliminary characterization of this enzyme in vitro. Recombinant NysL was shown to efficiently convert 10-deoxynystatin to nystatin A1, and no inhibition was observed by either the substrate or the product. This is in contrast with the apparent substrate inhibition shown for the epoxidase PimD, the close homologue of NysL (18). The Km and kcat values determined for the recombinant NysL protein are similar to those reported for other macrolide-specific P450 monooxygenases and presented by Mendes et al. (18). According to the kcat/Km criterion, His-tagged NysL has a somewhat higher substrate specificity toward 10-deoxynystatin than recombinant PimD toward its substrate, 4,5-deepoxypimaricin. It could be interesting to test whether recombinant NysL can use other polyene macrolides as substrates, since this could open possibilities for combinatorial biosynthesis. The latter can be achieved through expression of the nysL gene in polyene macrolide-producing hosts in order to obtain novel analogues with different post-PKS hydroxylation patterns. For example, the expression of NysL in the amphotericin producer devoid of AmphL activity (10) could yield an amphotericin analogue hydroxylated at C-10 instead of C-8 (Fig. 1A). Alternatively, NysL can be engineered via either site-specific mutagenesis (4) or directed evolution (3) in order to change its specificity towards 10-deoxynystatin. This approach might provide novel nystatin analogues hydroxylated at alternative macrolactone ring positions in hope of obtaining antifungal compounds with improved pharmacologic properties.
Acknowledgments
We are grateful to Per Bruheim and Randi Aune for help with analytical methods.
This work was supported by the Research Council of Norway.
REFERENCES
- 1.Aparicio, J. F., P. Caffrey, J. A. Gil, and S. B. Zotchev. 2003. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 61:179-188. [DOI] [PubMed] [Google Scholar]
- 2.Baginski, M., H. Resat, and E. Borowski. 2002. Comparative molecular dynamics simulations of amphotericin B-cholesterol/ergosterol membrane channels. Biochim. Biophys. Acta 1567:63-78. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. G., C. F. Harford-Cross, and L. L. Wong. 2001. Engineering the CYP101 system for in vivo oxidation of unnatural substrates. Protein Eng. 14:797-802. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. G., X. Chen, F. Xu, Z. Rao, and L. L. Wong. 2003. Engineering substrate recognition in catalysis by cytochrome P450cam. Biochem. Soc. Trans. 31:558-562. [DOI] [PubMed] [Google Scholar]
- 5.Betlach, M. C., J. T. Kealey, G. W. Ashley, and R. McDaniel. 1998. Characterization of the macrolide P-450 hydroxylase from Streptomyces venezuelae which converts narbomycin to picromycin. Biochemistry 37:14937-14942. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, D., S. Pelzer, A. Holtzel, G. J. Nicholson, S. Stockert, W. Wohlleben, G. Jung, and R. D. Sussmuth. 2001. The biosynthesis of vancomycin-type glycopeptide antibiotics—new insights into the cyclization steps. Angew Chem. Int. Ed. Engl. 40:1693-1696. [DOI] [PubMed] [Google Scholar]
- 7.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Strøm, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 8.Brautaset, T., S. E. Borgos, H. Sletta, T. E. Ellingsen, and S. B. Zotchev. 2003. Site-specific mutagenesis and domain substitutions in the loading module of the nystatin polyketide synthase, and their effects on nystatin biosynthesis in Streptomyces noursei. J. Biol. Chem. 278:14913-14919. [DOI] [PubMed] [Google Scholar]
- 9.Bruheim, P., S. E. Borgos, P. Tsan, H. Sletta, T. E. Ellingsen, J.-M. Lancelin, and S. B. Zotchev. 2004. Chemical diversity of polyene macrolides produced by Streptomyces noursei ATCC 11455 and recombinant strain ERD44 with genetically altered polyketide synthase NysC. Antimicrob. Agents Chemother. 48:4120-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne, B., M. Carmody, E. Gibson, B. Rawlings, and P. Caffrey. 2003. Biosynthesis of deoxyamphotericins and deoxyamphoteronolides by engineered strains of Streptomyces nodosus. Chem. Biol. 10:1215-1224. [DOI] [PubMed] [Google Scholar]
- 11.Carmody, M., B. Murphy, B. Byrne, P. Power, D. Rai, B. Rawlings, and P. Caffrey. 2005. Biosynthesis of amphotericin derivatives lacking exocyclic carboxyl groups. J. Biol. Chem. 280:34420-34426. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., and C. T. Walsh. 2001. Coumarin formation in novobiocin biosynthesis: beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 8:301-312. [DOI] [PubMed] [Google Scholar]
- 13.Cupp-Vickery, J. R., C. Garcia, A. Hofacre, and K. McGee-Estrada. 2001. Ketoconazole-induced conformational changes in the active site of cytochrome P450eryF. J. Mol. Biol. 311:101-110. [DOI] [PubMed] [Google Scholar]
- 14.Fjaervik, E., and S. B. Zotchev. 2005. Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl. Microbiol. Biotechnol. 67:436-443. [DOI] [PubMed] [Google Scholar]
- 15.Gaisser, S., R. Lill, J. Staunton, C. Mendez, J. Salas, and P. F. Leadlay. 2002. Parallel pathways for oxidation of 14-membered polyketide macrolactones in Saccharopolyspora erythraea. Mol. Microbiol. 44:771-781. [DOI] [PubMed] [Google Scholar]
- 16.Mansuy, D. 1998. The great diversity of reactions catalyzed by cytochromes P450. Comp. Biochem. Physiol. C 121:5-14. [DOI] [PubMed] [Google Scholar]
- 17.Mendes, M. V., E. Recio, R. Fouces, R. Luiten, J. F. Martin, and J. F. Aparicio. 2001. Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis. Chem. Biol. 8:635-644. [DOI] [PubMed] [Google Scholar]
- 18.Mendes, M. V., N. Anton, J. F. Martin, and J. F. Aparicio. 2005. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 386:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Keefe, D. P., and P. A. Harder. 1991. Occurrence and biological function of cytochrome P450 monooxygenases in the actinomycetes. Mol. Microbiol. 5:2099-2105. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz de Orue Lucana, D., and H. Schrempf. 2000. The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol. Gen. Genet. 264:341-353. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Seco, E. M., S. Fotso, H. Laatsch, and F. Malpartida. 2005. A tailoring activity is responsible for generating polyene amide derivatives in Streptomyces diastaticus var. 108. Chem. Biol. 12:1093-1101. [DOI] [PubMed] [Google Scholar]
- 23.Sekurova, O., H. Sletta, T. E. Ellingsen, S. Valla, and S. Zotchev. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC 11455. FEMS Microbiol. Lett. 177:297-304. [DOI] [PubMed] [Google Scholar]
- 24.Sekurova, O. N., T. Brautaset, H. Sletta, S. E. Borgos, O. M. Jakobsen, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186:1345-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sletta, H., S. E. Borgos, P. Bruheim, O. N. Sekurova, H. Grasdalen, R. Aune, T. E. Ellingsen, and S. B. Zotchev. 2005. Nystatin biosynthesis and transport: nysH and nysG genes encoding a putative ABC transporter system in Streptomyces noursei ATCC 11455 are required for efficient conversion of 10-deoxynystatin to nystatin. Antimicrob. Agents Chemother. 49:4576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosburg, D. A., and C. T. Walsh. 2005. Natural product biosynthetic assembly lines: prospects and challenges for reprogramming. Ernst Schering Res. Found. Workshop 51:261-284. [DOI] [PubMed] [Google Scholar]
- 27.Walczak, R. J., M. L. Dickens, N. D. Priestley, and W. R. Strohl. 1999. Purification, properties, and characterization of recombinant Streptomyces sp. strain C5 DoxA, a cytochrome P-450 catalyzing multiple steps in doxorubicin biosynthesis. J. Bacteriol. 181:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue, Y., D. Wilson, L. Zhao, H. Liu, and D. H. Sherman. 1998. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem. Biol. 5:661-667. [DOI] [PubMed] [Google Scholar]
- 29.Zotchev, S., K. Haugan, O. Sekurova, H. Sletta, T. E. Ellingsen, and S. Valla. 2000. Identification of a gene cluster for antibacterial polyketide-derived antibiotic biosynthesis in the nystatin producer Streptomyces noursei ATCC 11455. Microbiology 146:611-619. [DOI] [PubMed] [Google Scholar]