Abstract
Free-living amoebae (FLA) are ubiquitous organisms that have been isolated from various domestic water systems, such as cooling towers and hospital water networks. In addition to their own pathogenicity, FLA can also act as Trojan horses and be naturally infected with amoeba-resisting bacteria (ARB) that may be involved in human infections, such as pneumonia. We investigated the biodiversity of bacteria and their amoebal hosts in a hospital water network. Using amoebal enrichment on nonnutrient agar, we isolated 15 protist strains from 200 (7.5%) samples. One thermotolerant Hartmannella vermiformis isolate harbored both Legionella pneumophila and Bradyrhizobium japonicum. By using amoebal coculture with axenic Acanthamoeba castellanii as the cellular background, we recovered at least one ARB from 45.5% of the samples. Four new ARB isolates were recovered by culture, and one of these isolates was widely present in the water network. Alphaproteobacteria (such as Rhodoplanes, Methylobacterium, Bradyrhizobium, Afipia, and Bosea) were recovered from 30.5% of the samples, mycobacteria (Mycobacterium gordonae, Mycobacterium kansasii, and Mycobacterium xenopi) were recovered from 20.5% of the samples, and Gammaproteobacteria (Legionella) were recovered from 5.5% of the samples. No Chlamydia or Chlamydia-like organisms were recovered by amoebal coculture or detected by PCR. The observed strong association between the presence of amoebae and the presence of Legionella (P < 0.001) and mycobacteria (P = 0.009) further suggests that FLA are a reservoir for these ARB and underlines the importance of considering amoebae when water control measures are designed.
Pneumonia is an important cause of morbidity and mortality. Identification of the etiological agent is essential to select the appropriate antibiotic therapy. However, the etiology is identified in only about 50% of cases of community-acquired pneumonia (39) and in about 35% of cases of nosocomial pneumonia (50). Intracellular bacteria that grow poorly or not at all on the media used routinely in clinical diagnostic laboratories could be the agents responsible for pneumonia whose etiology is unknown. The candidates for these bacteria include intracellular colonizers of free-living amoebae (FLA), such as Legionella (an established agent of pneumonia [14]) and Parachlamydia (an emerging human pathogen [19]). These so-called amoeba-resisting bacteria (ARB) resist the microbicidal effector mechanisms of amoebae (18) and use the amoebae as a “training ground” for resistance to destruction by macrophages (38). Moreover, amoebae that are a reservoir for ARB are widely spread in the environment and in domestic water systems (44). The internalized bacteria may be protected from adverse conditions, particularly from agents used for water disinfection since amoebae are resistant to most of these disinfectants (53), especially when they are encysted (26). Human infection occurs via inhalation of aerosols containing free bacteria (9). It has also been suggested that infected amoebae could be the infectious particles that bring bacteria to the lungs (46).
Since ARB are probably able to resist human macrophages, amoebal coculture techniques that selectively isolate the ARB may be ideal tools for recovering potentially pathogenic bacteria. In this cell culture system, samples that potentially contain ARB are seeded onto axenic amoebae. Amoebal coculture has the additional advantage of reducing interference from rapidly growing species present in the sample that generally overwhelm traditional agar plates, thereby allowing recovery of fastidious species from a complex microflora. This technique has been successfully used for recovery of Legionella sp. from sputum (33) and feces (47), of new Alphaproteobacteria from hospital water supplies and nasal mucosa (17, 31), of Mycobacterium sp. from sputum (2), and of new Chlamydiales from activated sludge (11).
In this study, we used amoebal coculture to isolate ARB directly from samples from a hospital water network and also from indigenous amoebae recovered from the samples using amoebal enrichment. Even though we focused on Legionella, mycobacteria, and Chlamydia-like organisms, our goal was to isolate all ARB species present, including novel species, since such species may be unknown pathogenic agents that are potentially involved in nosocomial pneumonia whose etiology is unknown.
MATERIALS AND METHODS
Study design.
From May to August 2004, 200 samples were collected from the water network of the University Hospital in Lausanne, Switzerland. A total of 153 tap water swab samples, 26 water samples, and 21 showerhead swab samples were collected. The origins of the samples included the intensive care unit (45 samples), a surgery ward (59 samples), and an internal medicine ward (61 samples). The 35 remaining samples were collected in other wards of the hospital. The water temperature was recorded for the tap water samples. Amoebal coculture and amoebal enrichment on nonnutrient agar were performed for all 200 samples.
Specific broth and media for amoebal culture.
Peptone-yeast extract-glucose medium contained (in 5 liters of distilled water) 100 g proteose peptone (Difco, Sparks, MD), 10 g yeast extract (Difco), 4.9 g MgSO4 · 7H2O, 5 g sodium citrate · 2H2O, 0.1 g Fe(NH4)2(SO4)2 · 6H2O, 1.7 g KH2PO4, 1.97 g Na2HPO4 · 7H2O, 45 g glucose, and 0.295 g CaCl2. Page's modified Neff's amoeba saline (PAS) contained (in 1 liter of distilled water) 120 mg NaCl, 4 mg MgSO4 · 7H2O, 4 mg CaCl2 · 2H2O, 142 mg Na2HPO4, and 136 mg KH2PO4. To prepare nonnutritive agar plates, 1.5 g agar (Research Organics, Cleveland, OH) was diluted in 100 ml of PAS.
Amoebal microplates.
Acanthamoeba castellanii strain ATCC 30010 was grown at 28°C in 75-cm2 cell culture flasks (Corning) with 30 ml peptone-yeast extract-glucose medium (17). When cells formed a homogeneous monolayer, the amoebae were harvested and washed three times in 50 ml of PAS (with centrifugation at 2,000 × g for 10 min to pellet the amoebae). After the last centrifugation, the amoebae were resuspended in PAS, and 1 ml of a suspension containing 5 × 105 A. castellanii cells/ml was distributed into each well of a 24-well Costar microplate (Corning).
Processing of samples.
Each water sample consisted of two 500-ml portions collected at 1- to 2-min intervals in order to recover both proximal and distal microorganisms. The samples were filtered through a 0.2-μm cellulose nitrate membrane. Then the membrane was resuspended in 10 ml of sterile water. To recover ARB (see below), 200-μl portions of water were spread onto amoebal microplates. To isolate amoebae that were potentially present in the samples (see below), 200-μl portions were also spread on nonnutritive agar plates that had previously been seeded with a layer of living Escherichia coli ATCC 25922 (17).
Swabs were vortexed for 30 s in 1 ml of PAS in individual sterile tubes. The suspensions were centrifuged at 800 × g for 10 min. Two hundred-microliter portions of the supernatants were spread onto amoebal microplates. The pellets were resuspended in 100 μl of PAS and spread on nonnutritive agar plates seeded with E. coli.
Amoebal coculture.
Once inoculated, the microplates were centrifuged at 1,500 × g for 30 min and incubated at 32°C. The amoebal cocultures (F0) were subcultured with fresh amoebae on day 6, and the subcultures (F1) were incubated for 14 days at 32°C. Amoebal cocultures were examined daily for amoebal lysis. When amoebal lysis was observed, at the time of subculture, and after 14 days of subculture, the cultures were screened for intra-amoebal bacteria.
This screening was performed by gently shaking the microplates to suspend the amoebae. Then 50-μl portions of the suspensions were deposited on 12-well slides (Erie Scientific, Portsmouth, United Kingdom), dried at 100°C, and stained with Gimenez stain (16). When Gimenez-stained bacteria were observed or when bacterial proliferation or amoebal lysis occurred, 100 μl was seeded on buffered charcoal-yeast extract (BCYE) agar, on charcoal-yeast extract (CYE) agar, and on new amoebal microplates. The BCYE agar plates were incubated for 20 days at 37°C, and the CYE agar plates were incubated for 20 days at 32°C. Seeding on CYE agar plates was also systematically performed from F1 subcultures after 14 days of incubation.
All cocultures, with or without evidence of bacterial growth, were also tested for detection of Chlamydiales and Legionella using previously described specific primers. Primers 16SIGF and 16SIGR and primers Leg225 and Leg858 were used to detect Chlamydiales and Legionella spp., respectively. PCRs were performed with F0 cultures at day 6 and with F1 cultures at day 14. Cocultures were homogenized by scraping, 200 μl of supernatant was collected and inactivated by heating at 90°C for 30 min, and 5 μl was then used undiluted and diluted 10-fold as the DNA template. A total of 40 cycles of amplification were performed, with an annealing temperature of 51°C for primers 16SIGF and 16SIGR and an annealing temperature of 55°C for primers Leg225 and Leg858. The success of the amplification was determined by electrophoresis in a 0.7% agarose gel of PCR products stained with ethidium bromide. PCR products were purified by using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany). Sequencing was performed with the dRhodamine terminator cycle sequencing Ready Reaction with the primers used for PCR and with AmpliTaq DNA (Perkin-Elmer Biosystems, Warrington, England), according to the manufacturer's instructions. Sequences were determined with an ABI Prism 3100 automated sequencer (Applied Biosystems, Rotkreuz, Switzerland).
Ziehl-Neelsen staining was also systematically performed with 50-μl portions of the homogenized initial cocultures (F0) after 20 days of incubation at 32°C. When acid-fast stained bacteria were detected, two 100-μl portions were seeded onto 7H10 agar and incubated for 2 months, one at 32°C and one at 37°C. At the same time, 200 μl of supernatant was collected and inactivated by heating at 90°C for 30 min. DNA was then extracted by using an AquaPure genomic DNA extraction kit and proteinase K (Bio-Rad) according to the manufacturer's recommendations for extraction of DNA of gram-positive bacteria. A specific PCR was then performed using primers 285b and 264 (27). The PCR conditions were as follows: 94°C for 5 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 1.5 min and then a final cycle at 72°C for 10 min. PCR products were purified and sequenced with primer 264 and with internal primer 271 (5′-CTTAACACATGCAAGTCGAAC-3′).
Amoebal enrichment.
Nonnutritive agar plates were incubated at 28°C in a humidified atmosphere and examined daily for the presence of amoebal cells. When plates were positive, the amoebae were subcultured on new nonnutritive agar plates seeded with viable E. coli ATCC 25922. After three or four subcultures, the amoebae were harvested by scraping and resuspended in 1 ml of PAS. To lyse amoebae, the cell suspension was then passed three times through a 27-gauge syringe and vortexed at the maximum speed for 30 s. To identify amoebae, DNA was extracted with an AquaPure genomic DNA extraction kit and proteinase K (Bio-Rad), and an 18S rRNA PCR was performed with primers Ami6F1 (5′-CCAGCTCCAATAGCGTATATT-3′), Ami6F2 (5′-CCAGCTCCAAGAGTGTATATT-3′), and Ami9R (5′-GTTGAGTCGAATTAAGCCGC-3′) at a concentration of 0.5 μM together in the same reaction tube containing 2 mM MgCl2 and 2.5 U of Taq DNA polymerase (GibcoBRL, Life Technologies) (adapted from the method of Serra [49]). After a first step consisting of 94°C for 5 min, 40 cycles of amplification were performed by using denaturation at 94°C for 1 min, annealing at 55°C for 30 s, and elongation at 72°C for 2 min and a final cycle at 72°C for 10 min. Sequencing reactions were then performed with each primer. To recover ARB that were potentially present within the amoebae, 200 μl of suspension was seeded onto A. castellanii in 1 ml of PAS plus 100 μg ampicillin (to kill the E. coli). This amoebal coculture was then handled as described above.
Genotypic and phylogenetic characterization.
To identify the bacteria recovered, PCR amplification and sequencing of the 16S rRNA-encoding gene were performed directly with agar-grown bacteria resuspended in sterile phosphate-buffered saline (PBS) using primers fD1 and rP2 (55). The complete 16S rRNA-encoding gene was sequenced for every new species (homology with the closest neighbor, <99%) and for previously uncultured species, using previously described primers (1). For bacteria belonging to the genera Afipia and Bosea, we also amplified and sequenced the discriminative partial sequence of the rpoB gene (25). All sequences were compared with sequences available in the GenBank database in December 2004 by using the BLAST 2.2.2 program available on the NCBI website (www.ncbi.nlm.nih.gov). When the most similar sequence was that of an unpublished bacterial species, the level of sequence homology with the most similar published bacterial species was also recorded (Table 1). The 16S rRNA and rpoB sequences of the isolates were aligned with the sequences of the best BLAST hits and with the sequences of the closest relative type strains of each isolate. Sequences were edited by removal of the longer 5′ and 3′ ends so that their lengths matched the length of the shortest sequence. rpoB sequences were aligned with the complete rpoB sequence of Afipia felis (accession no. AY242824) in order to select the hypervariable region corresponding to positions 3,380 to 3,800 (25). The homology of the edited sequences was then analyzed by the distance matrix program of the MEGA3 software (29). For identification at the species and genus levels, levels of 16S rRNA sequence similarity with the most similar sequences in the GenBank database of >99% and >97%, respectively, were used as cutoffs (13). For Afipia and Bosea, we considered isolates members of the same species when the level of sequence similarity of the hypervariable region of the rpoB gene was ≥98%, whereas isolates belonged to different species when the level of sequence similarity was ≤96% (25). With these 16S rRNA and rpoB sequences, neighbor-joining (p-distance), minimum-evolution (p-distance), and parsimony (standard parsimony) trees were constructed using the MEGA3 software.
TABLE 1.
ARB isolated from water network samples by amoebal coculture
| Isolate | GenBank accession no.a | No. of isolatesb | % 16S rRNA gene homology with most similar GenBank sequence (accession no.)c | % 16S rRNA gene homology with closest previously described species (accession no.)c | Closest previously described species or subspecies |
|---|---|---|---|---|---|
| Alphaproteobacteria | |||||
| Bradyrhizobium japonicum strain 1 | DQ123628 | 6 (1w, 5t) | 99.7 (AF510592) | 99.6 (AF530468) | Bradyrhizobium japonicum |
| Bradyrhizobium japonicum strain 2 | DQ123629 | 3 (2w, 1t) | 99.8 (AF510592) | 99.7 (AF530468) | Bradyrhizobium japonicum |
| Caulobacter crescentus | DQ123627 | 1 (1s) | 99.6 (AE005930) | 99.6 (AE005930) | Caulobacter crescentus |
| Methylobacterium extorquens strain 1 | 20 (4w, 15t, 1s) | 100 (AF293375) | 100 (AF293375) | Methylobacterium extorquens | |
| Methylobacterium extorquens strain 2 | DQ123631 | 1 (1t) | 99.9 (AF293375) | 99.9 (AF293375) | Methylobacterium extorquens |
| Muricoccus roseus | DQ123632 | 1 (1t) | 99 (AJ488505) | 99 (AJ488505) | Muricoccus roseus |
| “Rhodoplanes sp. strain laus-1” | DQ123619 | 23 (1w, 21t, 1s) | 99.2 (AF407716) | 92.6 (AJ563931) | Beijerinckia indica subsp. lacticogenes |
| “Craurococcus related sp. strain laus-1” | DQ123620 | 1 (1t) | 93.4 (D85828) | 93.4 (D85828) | Craurococcus roseus |
| “Rhodoplanes sp. strain laus-2” | DQ123621 | 1 (1t) | 96.6 (Y12598) | 92.2 (AJ294349) | Chelatococcus asaccharovorans |
| Rasbo bacterium | 1 (1t) | 100 (AF007948) | 100 (AF007948) | Rasbo bacterium | |
| Rhodoplanes elegans | 1 (1t) | 100 (D25311) | 100 (D25311) | Rhodoplanes elegans | |
| Roseomonas gilardii | 1 (1s) | 100 (AY220740) | 100 (AY220740) | Roseomonas gilardii | |
| “Afipia sp. strain laus-1”d | DQ123622 | 1 (1t) | 99.5 (AF288304) | 99.5 (AF288304) | Afipia birgiae |
| Afipia birgiae strain 1d | 2 (1t, 1s) | 100 (AF288304) | 100 (AF288304) | Afipia birgiae | |
| Afipia birgiae strain 2d | 2 (1w, 1t) | 100 (AF288304) | 100 (AF288304) | Afipia birgiae | |
| Bosea eneaed | 3 (2t, 1s) | 100 (AF288305) | 100 (AF288305) | Bosea eneae | |
| “Bosea sp. strain laus-1”d | 1 (1t) | 100 (AF288302) | 100 (AF288302) | Bosea vestrisii | |
| Other bacterial species | |||||
| Mycobacterium gordonae strain 1 | 15 (3w, 10t, 2s) | 100 (AY215258) | 100 (AY215258) | Mycobacterium gordonae | |
| Mycobacterium gordonae strain 2 | DQ123634 | 20 (2w, 14t, 4s) | 99.7 (AY215258) | 99.7 (AY215258) | Mycobacterium gordonae |
| Mycobacterium xenopi | 5 (5t) | 100 (AY215375) | 100 (AY215375) | Mycobacterium xenopi | |
| Mycobacterium kansasii | DQ123633 | 3 (3t) | 99.4 (AY438074) | 99.4 (AY438074) | Mycobacterium kansasii |
| Pseudomonas aeruginosa | 2 (2t) | 100 (AY268175) | 100 (AY268175) | Pseudomonas aeruginosa | |
| Legionella pneumophila | DQ123630 | 2 (2w) | 99.7 (CR628336) | 99.7 (CR628336) | Legionella pneumophila |
| Legionella anisa | 9 (2w, 7t) | 100 (AY744776) | 100 (AY744776) | Legionella anisa |
Sequences exhibiting 100% identity to GenBank sequences for previously described species were not deposited.
The numbers of isolates per sample type are indicated in parentheses. w, water sample; t, tap water swab; s, shower swab.
The accession numbers are GenBank accession numbers. BLAST analysis was used to determine the level of 16S rRNA gene sequence homology with the most similar GenBank sequence.
Species subsequently identified by partial sequencing of the rpoB gene.
Phenotypic tests.
The phenotypes of the strains corresponding to new species were thoroughly studied. Morphological and tinctorial properties were determined by Gram and Gimenez staining. Growth was tested at 32°C on Columbia agar with 5% sheep blood, chocolate agar, and CYE agar. Growth on CYE agar was examined at 32 and 37°C. Oxidase activity was detected using a dimethyl-p-phenylenediamine oxalate disk (Pasteur Diagnostic). Catalase activity was detected by emulsifying a colony in 3% hydrogen peroxide and checking for the presence of microscopic bubbles. Other biochemical tests were performed by inoculation of API 20NE and API 50CH strips (bioMerieux), according to the manufacturer's instructions. These strips were incubated for 7 and 15 days, respectively, at 32°C. The API 20NE strips tested for any reduction of nitrates, indole production, urease activity, glucose acidification, arginine dihydrolase activity, hydrolysis of gelatin and esculin, ß-galactosidase activity, and assimilation of glucose, arabinose, mannose, mannitol, N-acetylglucosamine, maltose, gluconate, caprate, adipate, malate, citrate, and phenylacetate. The API 50CH strips tested for any acidification of glycerol, erythritol, d-arabinose, l-arabinose, ribose, d-xylose, l-xylose, adonitol, methyl-ß-d-xyloside, galactose, glucose, fructose, mannose, sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, methyl-α-d-mannoside, methyl-α-d-glucoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, trehalose, inulin, melezitose, raffinose, starch, glycogen, xylitol, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, gluconate, 2-ketoglutarate, and 5-ketoglutarate.
Growth of new bacterial species within A. castellanii.
To further characterize the new bacterial species, we evaluated their ability to grow within A. castellanii. Briefly, bacteria were grown on CYE agar at 32°C for 10 days. Then they were harvested, washed three times in PAS, and filtered with a 5-μm filter, and the turbidity was adjusted to a McFarland standard of 1. Twenty-microliter portions of the suspensions were used to seed amoebal microplates prepared as described above. The microplates were centrifuged for 30 min at 1,500 × g and incubated for 2 h at 32°C. The amoebal cocultures were washed with PAS to remove extracellular bacteria and then incubated for 1 week at 32°C in a humidified atmosphere. To determine the number of viable bacteria, cocultures were scraped, serially diluted in sterile PBS, and seeded on CYE agar, and the cultures were then incubated for 14 days at 32°C. The numbers of CFU were then compared with the numbers of CFU obtained in the absence of amoebal cells.
Transmission electron microscopy.
Electron microscopy was also used to further characterize the new bacterial species. Briefly, 10-ml portions of 5-day cocultures were harvested, washed in PBS, and fixed in 4% glutaraldehyde. Fixed samples were then treated as described previously (17) and observed with a Philips EM 201 C transmission electron microscope (Philips, Eindhoven, The Netherlands).
Statistical analysis.
The mean numbers of bacterial strains isolated from amoeba-colonized samples were compared to the mean numbers of strains isolated from other samples by using the chi-square test (STATA software, version 7.0; Stata Corporation, College Station, TX).
RESULTS
Amoebal enrichment and temperature tolerance.
Using amoebal enrichment, PCR, and sequencing of the 18S rRNA-encoding gene, we recovered and identified 15 protist isolates from 15 of 200 samples (7.5%). Six identical isolates, corresponding to the same Hartmannella vermiformis strain (strain 1; GenBank accession no. DQ123623), were recovered from water samples (two isolates; water temperature, 42°C) and tap swabs (four isolates) from the emergency ward and intensive care unit. This strain had an intron in the 18S rRNA-encoding gene. Seven other isolates, corresponding to an intronless strain of H. vermiformis (strain 2; sequence identical to GenBank accession no. M95168 sequence), were found at seven locations (one water sample [temperature, 60°C], four tap water swabs, and two shower swabs). We also isolated one strain of Acanthamoeba polyphaga (sequence identical to GenBank accession no. U07415 sequence) and one strain of an unidentified protist (GenBank accession no. DQ123626), each from one tap water swab. The new amoebal species had an intron and exhibited less than 90% 18S rRNA gene similarity in the intronless sequence with other eukaryotic species. It was unable to feed on E. coli ATCC 25922, but it was isolated together with a bacterial species that grew on nonnutritive agar plates and served as food for the protist. This bacterial species was identified by 16S rRNA gene sequencing as Sphingomonas yanoikuyae.
H. vermiformis strain 1 was able to grow on nonnutrient agar seeded with E. coli at 28°C, 37°C, and 44°C and exhibited only limited growth at 47°C. H. vermiformis strain 2 could grow at 28°C, but it exhibited limited growth at 37°C and was inactivated after 3 days of incubation at 44°C (there was no subsequent growth at 28°C). The A. polyphaga strain could grow at 28°C and 37°C, and although it did not grow at 44 and 47°C, it survived at these high temperatures.
Coculture from indigenous amoebae.
Coculture from indigenous amoeba lysates led to the recovery of Legionella pneumophila and Bradyrhizobium japonicum (strain 1) from one isolate of intron-containing H. vermiformis. No Chlamydiales were recovered by coculture from any of the 15 amoebae or were detected by PCR.
Coculture from water and swab samples.
A total of 125 bacterial isolates were recovered by amoebal coculture from 91 of the 200 samples. Thus, 42.3% of water samples, 52.4% of shower swabs, and 45.1% of tap water swabs were found to be colonized with at least one ARB.
Coculture from water and swab samples: mycobacteria.
Mycobacteria were frequently recovered, and they were easily detected using Ziehl-Neelsen staining performed with primary cultures 20 days postinoculation. Forty-three mycobacterial isolates recovered from 41 samples were identified by PCR and sequencing, and two cocultures harbored two different species. Two strains of Mycobacterium gordonae, differing by only one base in a partial 16S rRNA sequence, were recovered from 35 samples. They were present in water, as well as on tap water swabs and shower swabs. Strain 1 was isolated from 15 samples, and strain 2 was isolated from 20 samples. We also isolated a strain of Mycobacterium kansasii (subtype 1) from three samples and a strain of Mycobacterium xenopi from five samples. Mycobacteria were isolated more frequently from samples from which an amoeba was also recovered (7/15 samples, 46.7%) than from samples from which no amoebae were isolated (34/185, 18.4%) (P = 0.009).
Coculture from water and swab samples: various alphaproteobacterial species.
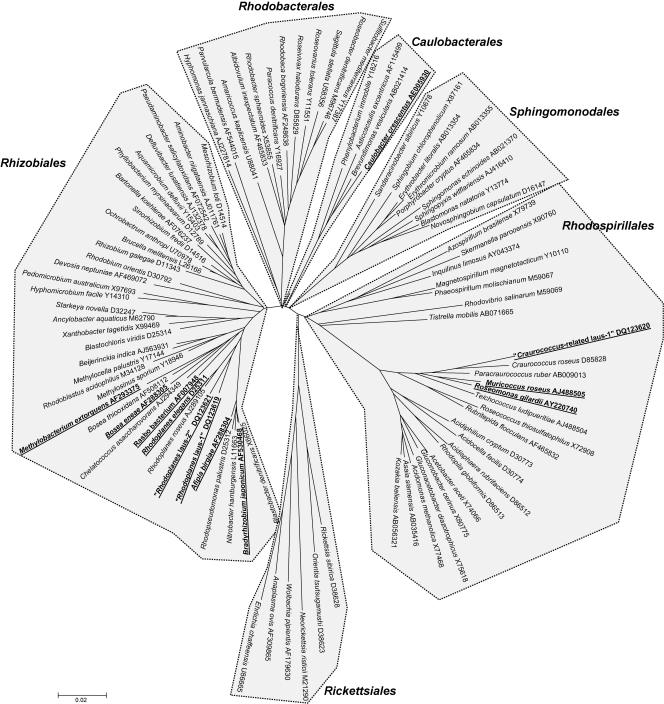
Bacteria belonging to the class Alphaproteobacteria were isolated after the cocultures were plated on CYE and/or BCYE agar (Fig. 1). Two different strains of Methylobacterium extorquens were recovered on CYE and BCYE agar plates, one from 20 samples and one from one sample. Two strains that exhibited >99% 16S rRNA gene similarity with B. japonicum were also recovered, one from six samples and one from three samples. Additional alphaproteobacteria, including the Rasbo bacterium, Muricoccus roseus, Rhodoplanes elegans, Roseomonas gilardii, and Caulobacter crescentus, were isolated from single samples. Sphingomonas sp. (n = 36) and Brevundimonas sp. (n = 4) were also recovered by amoebal coculture, but they could not be considered ARB sensu stricto.
FIG. 1.
Phylogenetic tree showing representative species of alphaproteobacteria. The tree was constructed by using the neighbor-joining method, based on the nearly complete sequence (1,283 nucleotides) of the 16S rRNA gene coding sequence. Only one species per genus was used. Taxonomic names and GenBank accession numbers are indicated at the ends of the branches. Each order is clearly delimited by dashed lines and gray shading and is branched deeply. Best BLAST with known-species isolates recovered in this study, as well as new species recovered in this study, are indicated by boldface type and underlining.
Overall, alphaproteobacteria were not recovered more frequently from samples with amoebae than from samples without amoebae (Table 2).
TABLE 2.
Correlation between the presence of amoebae and the presence of ARB
| Taxon | No. positive/no. tested (%)
|
P value | |
|---|---|---|---|
| Amoebae | No amoeba | ||
| Any ARB | 12/15 (80) | 78/185 (42) | 0.005 |
| Legionella | 5/15 (33) | 6/185 (3) | <0.001 |
| Mycobacterium sp. | 7/15 (47) | 34/185 (18) | 0.009 |
| Alphaproteobacteria | 5/15 (33) | 55/185 (30) | >0.05 |
Coculture from water and swab samples: alphaproteobacterial strains related to Rhodoplanes.
A strain related to the genus Rhodoplanes, “Rhodoplanes sp. strain laus-1,” was recovered from 23 samples. This strain is a fastidious, slow-growing, gram-negative bacterium that was recovered on CYE agar plates incubated at 32°C. Small white colonies were observed after 2 to 3 weeks of incubation. The results of an API 20NE analysis are shown in Table 3. As determined by API 50CH tests, only d-saccharose oxidation and weak potassium 5-ketogluconate oxidation were detected after 14 days of incubation at 32°C. As determined by BLAST analysis, the 16S rRNA gene sequence (1,450 bp) of this isolate exhibited 92.6% similarity with the sequence of Beijerinckia indica subsp. lacticogenes (accession no. AJ563931) and 92.4% similarity with the sequence of Bosea thiooxidans (accession no. AF508112). Genetic analysis predicted that this isolate belongs to the genus Rhodoplanes, as it exhibited 96.9 to 97.5% 16S rRNA gene sequence homology with Rhodoplanes spp.; the most similar sequence was that of Rhodoplanes roseus (accession no. D25313).
TABLE 3.
Phenotypic characteristics of the four new alphaproteobacterial species isolated by coculture of water samples with A. castellaniia
| Test | “Afipia sp. strain laus-1” | “Bosea sp. strain laus-1” | “Craurococcus- related sp. strain laus-1” | “Rhodoplanes sp. strain laus-1” |
|---|---|---|---|---|
| Gram reaction | − | − | − | − |
| Gimenez staining | + | + | + | + |
| Growth at 32°C on: | ||||
| 5% sheep blood agar | + | + | − | + |
| Chocolate agar | − | + | − | + |
| Hemolysis on 5% sheep blood | − | − | ND | − |
| Aerobic growth on charcoal-yeast extract agar at: | ||||
| 32°C | + | + | + | + |
| 37°C | − | − | − | + |
| Anaerobic growth on CYE agar at 32°C | − | − | − | − |
| Oxidase activity | + | +w | + | + |
| Catalase activity | − | − | + | − |
| Arginine dihydrolase activity | − | − | − | − |
| Beta-galactosidase activity | − | − | − | − |
| Urease activity | − | − | + | +w |
| Glucose fermentation | − | − | − | − |
| Hydrolysis of esculin | − | − | − | − |
| Hydrolysis of gelatin | + | + | + | + |
| Indole production | − | − | − | − |
| Reduction of nitrates to nitrites | + | + | − | + |
| Assimilation of: | ||||
| Glucose | − | − | − | − |
| Arabinose | − | − | − | − |
| Mannose | − | +w | + | − |
| Mannitol | − | − | − | − |
| N-Acetylglucosamine | − | − | − | − |
| Maltose | − | − | − | − |
| Potassium gluconate | + | + | − | − |
| Capric acid | − | − | − | − |
| Adipic acid | +w | +w | − | − |
| Malic acid | +w | +w | − | − |
| Trisodium citrate | − | − | − | − |
| Phenylacetic acid | − | − | − | − |
+, positive reaction within 7 days; +w, weak positive reaction within 7 days; −, negative reaction within 7 days; ND, not determined.
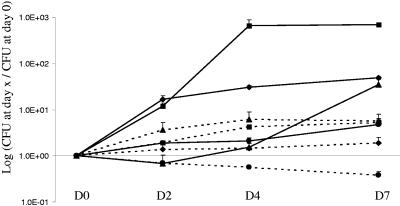
We also isolated another strain affiliated with this genus, “Rhodoplanes sp. strain laus-2.” This strain is a very slowly growing bacterium that was recovered on CYE agar plates incubated at 32°C. Loose and colorless colonies were observed after 3 weeks of incubation. As determined by BLAST analysis, it exhibited only 92.2% 16S rRNA sequence similarity (1,454 bp) with the most closely related previously described species, Chelatococcus asaccharovorans. Interestingly, genetic analysis predicted that this isolate also belongs to the genus Rhodoplanes (97.3 to 97.8% sequence homology), and the most closely related strain in the genus is “Rhodoplanes sp. strain laus-1” (98.4% sequence homology). All phylogenetic analyses confirmed that “Rhodoplanes sp. strain laus-1” and “Rhodoplanes sp. strain laus-2” belong to the Bradyrhizobiaceae family (order Rhizobiales) and are more closely related to species of the genus Rhodoplanes. The two strains clustered together, with bootstrap values of 89%, 96%, and 64% in the neighbor-joining, minimum-evolution, and maximum-parsimony trees, respectively, supporting the fork separating these two strains from their closest relatives, the members of the genus Rhodoplanes (Fig. 1). Although the increase in the number of CFU of “Rhodoplanes sp. strain laus-1” was limited (0.7 log) in the presence of A. castellanii (Fig. 2), these results strongly contrasted with the decrease in the number of CFU in the negative control (without amoebae). This is congruent with the presence of few bacteria inside amoebae after 2 and 5 days of coculture as determined by electron microscopy (see Fig. 4). We were not able to grow sufficient numbers of strain laus-2 to perform additional phenotypic tests and to study its intra-amoebal growth. “Rhodoplanes strain laus-1” has been deposited in the Collection de l’Institut Pasteur as “R. lausannensis strain CIP 108886T.”
FIG. 2.
Growth of new alphaproteobacterial species in coculture with A. castellanii ATCC 30010. “Afipia sp. strain laus-1” (⧫), “Bosea sp. strain laus-1” (▪), “Craurococcus-related sp. strain laus-1” (▴), and “Rhodoplanes sp. strain laus-1” (•) were grown for 7 days in PAS with (solid lines) or without (dashed lines) A. castellanii. The data are means of three independent experiments. D, day.
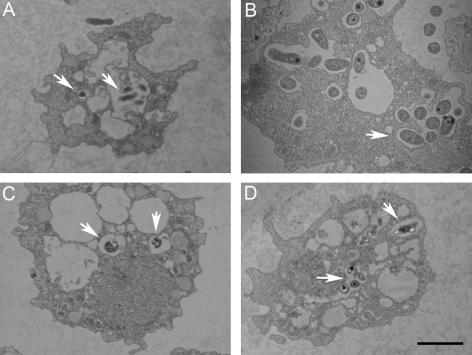
FIG. 4.
New alphaproteobacterial isolates “Afipia sp. strain laus-1” (A), “Bosea sp. strain laus-1” (B), “Craurococcus-related sp. strain laus-1” (C), and “Rhodoplanes sp. strain laus-1” (D) in A. castellanii after 5 days of coculture. Numerous “Bosea sp. strain laus-1” cells are present within vacuoles (B). Only a few cells of the other strains are present within amoebae (arrows). Bar = 2 μm.
Coculture from water and swab samples: alphaproteobacterial strain related to Craurococcus.
Another alphaproteobacterial strain was recovered from one CYE agar plate. This strain formed pinkish colonies that grew within 5 to 7 days at both 32°C and 37°C. The API 20NE results are shown in Table 3. All API 50CH results were negative. In intra-amoebal growth experiments, the number of CFU increased only at day 7, after a slight decrease at day 2 (Fig. 2). Intra-amoebal bacteria were rarely observed by electron microscopy, and there was a maximum of two bacteria per amoeba (see Fig. 4). These observations suggest that this strain might resist phagocytosis by amoebae, possibly inducing amoebal lysis and growing saprophytically thanks to nutrients released by lysed amoebae. As determined by BLAST analysis, the sequence of this strain exhibited only 93.4% 16S rRNA gene similarity (1,454 bp) with the sequence of Craurococcus roseus (accession no. D85828). As determined by genetic analysis, “Craurococcus-related sp. strain laus-1” exhibited 96.4% sequence homology with Craurococcus roseus (accession no. D85828) and 96.5% sequence homology with Paracraurococcus ruber (accession no. D85827). All phylogenetic analyses confirmed that this strain belongs to the Acetobacteraceae family (order Rhodospirillales) and that it belongs to a new genus related to Craurococcus. Thus, bootstrap values of 92%, 99%, and 94% in the neighbor-joining, minimum-evolution, and maximum-parsimony trees, respectively, supported the fork separating this strain from C. roseus (Fig. 1). This strain has been deposited in the Collection de l'Institut Pasteur as Neocraurococcus lausannensis strain CIP 108887T.”
Coculture from water and swab samples: alphaproteobacterial strains related to Afipia.
Five Afipia sp. isolates were recovered from CYE agar plates, and these isolates were initially identified as Afipia birgiae based on partial 16S rRNA sequences. Since it has been demonstrated that 16S rRNA is not discriminating enough to allow precise identification at the species level for the genera Afipia and Bosea (25), we completed the identification by sequencing the discriminative rpoB region (Fig. 3). This allowed us to clearly identify two A. birgiae strains (two isolates of each strain), one exhibiting 99.0% sequence homology with the A. birgiae accession no. AY242821 sequence and the other exhibiting 100% identity. The last isolate exhibited only 93.7% rpoB gene sequence similarity with the most closely related previously described species (Afipia massiliensis), whereas the 16S rRNA gene sequence (1,455 nucleotides; GenBank accession no. DQ123622) was 99.6% similar to the A. birgiae accession no. AF288304 sequence. For this strain, bootstrap values of 87%, 94%, and 40% in the neighbor-joining, minimum-evolution, and maximum-parsimony trees, respectively, supported the fork separating this strain from A. massiliensis (Fig. 3). This isolate, which grew at 32°C on sheep blood agar and CYE agar in an aerobic atmosphere, was oxidase positive, able to reduce nitrates to nitrites, able to hydrolyze gelatin, and able to assimilate potassium gluconate, adipate, and malate (Table 3). With API 50CH strips, acidification was detected only in the presence of d-glucose. Thus, like all the other members of the genus Afipia, this isolate is a gram-negative, oxidase-positive, esculin-positive rod. It is noteworthy that this phenotypic pattern is different from those of all other members of the genus and is more similar to that of Afipia clevelandensis. Intra-amoebal growth of “Afipia sp. strain laus-1” was limited, and there was a 1.7-log increase at day 7 (Fig. 2). As determined by electron microscopy, a maximum of three bacteria per amoeba were detected (Fig. 4). This strain has been deposited in the Collection de l'Institut Pasteur as “Afipia lausannensis strain CIP 108885T.”
FIG. 3.
Phylogenetic neighbor-joining tree showing the relationships of Afipia and Bosea. The tree was derived from an alignment of partial rpoB sequences. Taxonomic names and GenBank accession numbers are indicated at the ends of the branches. Best BLAST with known-species isolates recovered in this study, as well as new species recovered in this study, are indicated by boldface type and underlining. The support for each branch, as determined with 250 bootstrap samples, is indicated by the value at the node (expressed as a percentage). Sinorhizobium meliloti was used as the outgroup.
Coculture from water and swab samples: alphaproteobacterial strains related to Bosea.
Both sequencing of 16S rRNA and sequencing of rpoB were used to accurately identify four Bosea spp. isolated from three tap water samples and one shower swab. Three isolates corresponded to a single strain of Bosea eneae and had a partial rpoB sequence identical to that of a B. eneae strain (accession no. AY242841). The third Bosea isolate had a partial 16S rRNA gene sequence (1,441 nucleotides) identical to that of Bosea vestrisii (accession no. AF288302) and exhibited 96.5% and 94.2% partial rpoB sequence similarity with sequences of B. eneae (accession no. AY242841) and B. vestrisii (accession no. AY242839), respectively. For this strain, bootstrap values of 98%, 98%, and 92% in the neighbor-joining, minimum-evolution, and maximum-parsimony trees, respectively, supported the fork separating “Bosea sp. strain laus-1” from B. eneae (Fig. 3). Like most other Bosea species, the Lausanne isolate did not oxidize any sugar in API 50CH tests. The other phenotypic traits of this new Bosea species are shown in Table 3. “Bosea sp. strain laus-1” grew very well within A. castellanii; there was a 2.8-log increase within the first 4 days of incubation in the presence of amoebae, whereas there was only a 0.7-log increase in the absence of cells. Furthermore, transmission electron microscopy after day 5 of coculture showed that there were highly vacuolated amoebae that contained many bacteria (Fig. 4). This strain has been deposited in the Collection de l'Institut Pasteur as “Bosea lausannensis strain CIP 108888T.”
Coculture from water and swab samples: Legionella.
Species belonging to the Gammaproteobacteria were also found in the coculture experiments. All the Legionella species that were detected in coculture by PCR were also recovered on BCYE agar plates, and we did not detect DNA from obligate intracellular Legionella species. Eleven isolates of Legionella spp. were recovered. Legionella anisa was the predominant organism, accounting for 9 of the 11 isolates. A strain of L. pneumophila was also recovered from two water samples. It exhibited 100% sequence homology with the L. pneumophila strain present within an H. vermiformis cell isolated by amoebal enrichment. Isolation of Legionella spp. was more frequent from samples from which an amoeba was also recovered (5/15 samples, 33.3%) than from samples from which no amoebae were isolated (6/185 samples, 3.2%) (P < 0.001) (Table 2).
Coculture from water and swab samples: other species.
We recovered one strain of Pseudomonas aeruginosa from two tap water swabs. No Chlamydiales were recovered by amoebal coculture or detected by PCR.
DISCUSSION
In this study, we investigated the biodiversity of amoebae and ARB in the water network of a hospital with no recent history of epidemics involving such microorganisms. We used a broad range of staining methods, culture approaches, and PCR tools to identify a variety of ARB isolated by coculture with axenic amoebae.
We recovered amoebae from 11.5% of the water samples and 5.7% of the swabs taken from taps and showerheads. The prevalence of amoebae was lower than the prevalence found in a previous study (45), in which approximately 50% of the samples were found to be colonized by amoebae. The lower rate of amoeba colonization that we observed could have been due to the high temperature of the hospital hot water network (in order to prevent colonization by Legionella spp., the temperature had been maintained at 65°C since 2000) (7). In our study, the mean temperature of water samples was 56°C (range, 42 to 68°C). This may also explain why we recovered almost exclusively H. vermiformis (86.7%), a species reported to be tolerant to high temperatures (45).
Two different H. vermiformis strains were isolated, one of which exhibited an intron in its 18S rRNA gene sequence and was resistant to high temperatures; it was even able to grow slightly at 47°C. The other strain showed only limited growth at 37°C and was not able to survive exposure to 44°C for 3 days. Thus, these two strains could have different ecological niches; one could be able to colonize distal (cooler) points of a water network, and the other could be preferentially localized at proximal points. L. pneumophila, which is also resistant to high temperatures (40), was present within 1 of the 13 H. vermiformis isolates. Furthermore, 5 of the 11 samples found to be Legionella sp. positive were also colonized with H. vermiformis (four samples) or the unidentified protist (one sample); thus, there was a statistically significant association (P < 0.001) between the presence of Legionella and the presence of amoebae in the water networks, which confirmed previous results (48).
We were unable to detect any Chlamydia-related organism in any of the 200 samples investigated. This was unexpected since Chlamydia-like organisms, such as Simkania negevensis, have been found to be widely distributed in water networks (23). It is possible that some Chlamydia-like organisms that might have been present (i) in water samples or (ii) within the isolated indigenous amoebae (H. vermiformis and A. polyphaga) could not grow within A. castellanii strain ATCC 30010. A restricted host range with no growth within Acanthamoeba spp. has been described for Neochlamydia hartmannellae (21). Restricted host ranges with no growth within specific free-living amoebae have also been reported for S. negevensis and Waddlia chondrophila (36, 37) and for legionellae other than L. pneumophila. To circumvent the problem of restricted host specificity, we performed Chlamydiales- and Legionellaceae-specific PCR with all cocultures from water and biofilm samples, as well as with indigenous amoebae. Thus, if any Chlamydiales were present inside the indigenous amoebae, we would probably have detected them by PCR. Moreover, if a given Chlamydiales strain not able to grow inside A. castellanii was present in the water samples, there should have been enough DNA after 6 days of coculture (F0) to allow it to be detected by PCR. Thus, altogether, we think that we probably failed to isolate Chlamydia-like organisms because the level of Chlamydiales was too low or Chlamydiales were not present in the heavily cleaned hospital water network system that we examined.
Amoebal coculture proved to be very efficient for recovering mycobacteria. Mycobacteria were easily detected using Ziehl-Neelsen staining, suggesting that the multiplication rate of mycobacteria in the cocultures was high. As observed for Legionella sp., there was a significant association between the presence of amoebae and the presence of mycobacteria (P = 0.009). To our knowledge, this is the first clear evidence that mycobacteria are associated with free-living amoebae in water networks. It has been shown in vitro that mycobacteria can grow in amoebae (10), and amoebal coculture has recently been successfully used to isolate a new species, “Mycobacterium massiliense,” from the sputum of a patient (2). However, our study was the first systematic use of amoebal coculture to investigate the mycobacterial diversity in a water network. It has recently been reported that hospital swimming pools used for physical therapy can be colonized by various mycobacterial species, including some species that cannot be recovered on traditional culture media (4). Thus, the amoebal coculture method might be a valuable tool for recovering such new fastidious mycobacterial species. Whether the species isolated (M. gordonae, M. kansasii, and M. xenopi) can grow inside amoebae and/or saprozoically on products secreted by the amoebae, as described previously for Mycobacterium avium (10, 51), remains to be determined. The presence of these three species in hospital water networks has been reported previously (15, 56), and these organisms may occasionally be involved in human infections (3, 5, 54). Moreover, M. kansasii type 1 is generally considered more pathogenic than types 2 and 3 (52). Although no cases of nosocomial infections due to these mycobacteria occurred during the study period, the presence of potentially pathogenic atypical mycobacteria in the water network and their association with the presence of free-living amoebae underline the importance of considering amoebae when water control measures are designed.
We also recovered various alphaproteobacteria. M. extorquens, which was frequently isolated in this study, was previously isolated using amoebal coculture from the nasal mucosa of a hospitalized patient (17). Methylobacteria are slow-growing, pink-pigmented organisms that have been reported to be opportunistic pathogens in immunocompromised patients (24). Methylobacteria have also been isolated from immunocompetent patients with bacteremia (22). Methylobacteria are commonly found in water distribution networks (28) and are resistant to chlorine disinfection (20). It has been suggested that their presence in the hospital environment should be monitored (42).
B. japonicum has also been isolated previously by amoebal coculture (32). Like M. extorquens, this organism is a plant endophyte, but to date it has not been implicated in human infections. The Rasbo bacterium has previously been recovered by cell culture (Vero cells) from plasma samples of a septic patient (8) and by coculture from 5.2% of hospital water samples (32). Roseomonas gilardii has been involved in cases of bacteremia, catheter-related infections, and ventriculitis (12, 41, 43), and a closely related species, Roseomonas massiliae, has been recovered by amoebal coculture from a nasal swab (17). Interestingly, when constructing phylogenetic trees based on the partial 16S rRNA gene sequences (1,200 nucleotides) of more than 500 alphaproteobacteria (data not shown) (see Fig. 1 for representative tree with 93 sequences), we observed that the Roseomonas group seems to be affiliated with the Acetobacteraceae family (order Rhodospirillales) and not with the Methylobacteriaceae family (order Rhizobiales), as initially proposed (43). The genus Roseomonas was still classified in this family in the 2004 edition of Bergey's Taxonomy Outline of the Prokaryotes (http://141.150.157.80./bergeysoutline/main.html) and in the NCBI Taxonomy tool (http://www.ncbi.nlm.nih.gov/Taxonomy/). However, our observations and the fact that this genus was classified in the order Rhodospirillales in the Ribosomal Database Project (http://rdp.cme.msu.edu) that provided aligned and annotated rRNA gene sequences (35) suggest that this classification may need to be reexamined. We also isolated M. roseus, R. elegans, and C. crescentus by amoebal coculture. To our knowledge, this is the first time that these species were isolated using this technique, and we propose that they should be added to the growing list of ARB. None of these species has been associated with human infections yet.
The amoebal coculture technique allowed us to isolate four new alphaproteobacterial isolates. For the two isolates belonging to the genera Afipia and Bosea, sequencing of the partial hypervariable rpoB region described by Khamis et al. (25) was useful, since it allowed us to differentiate these two isolates from previously identified species, whereas the 16S rRNA gene sequences were identical to that of a previously described species. Both Afipia spp. and Bosea spp. were shown to be common in hospital water networks, and seroconversion of as many as 20% of the patients admitted to an intensive care unit and requiring intubation or mechanical ventilation has been reported for Bosea massiliensis (32). Bosea species have also been described as organisms that are able to replicate and produce high numbers within A. polyphaga (32). For Afipia species there is less evidence that high levels of the bacteria are present within infected cells. An exception is the pathogenic species Afipia felis; an infected HeLa cell can contain 50 bacteria (6). A. felis is also able to grow within A. polyphaga, and there was a 10-fold increase within 3 days (34). Thus, the intra-amoebal growth observed here for “Afipia sp. strain laus-1” is the same order of magnitude as the growth reported for A. felis.
Due to their resistance to amoebae and potential resistance to human macrophages, the new strains are good candidates for agents of pneumonia whose etiology is unknown. Given the high rate of recovery of “Rhodoplanes sp. strain laus-1,” which was present in as many as 11.5% (23/200) of the samples, we especially intend to investigate the exposure of patients to this new species by serology.
This work does not provide a complete report of the biodiversity of ARB and amoebae in the hospital water network, since some ARB that are resistant to specific amoebae other than A. castellanii may not have been detected. Moreover, amoebae that do not feed on E. coli, such as the protist that we detected because of its growth on S. yanoikuyae present in the same sample, also would not have been detected. Compared to other techniques used for detection of noncultivable organisms, such as fluorescent in situ hybridization or PCR and cloning, amoebal coculture has the additional advantage that it potentially detects amoeba-resisting lytic viruses, such as the recently described mimivirus that leads to amoebal lysis in less than 72 h (30), and provides living microorganisms that may be used as antigens for serological studies.
Acknowledgments
This work was supported by Swiss National Science Foundation (SNSF) grant 3200BO-105885.
We thank the PFMU at the Medical Faculty of Geneva for assisting with electron microscopy analysis. We also thank Jacques Bille and Christian Durussel for helpful discussions and Philip Tarr for reviewing the manuscript.
REFERENCES
- 1.Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 2.Adekambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afessa, B. 2001. Mycobacterial and nonbacterial pulmonary complications in hospitalized patients with human immunodeficiency virus infection: a prospective, cohort study. BMC Pulm. Med. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angenent, L. T., S. T. Kelley, A. S. Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arend, S. M., E. Cerda De Palou, P. De Haas, R. Janssen, M. A. Hoeve, E. M. Verhard, T. H. Ottenhoff, D. Van Soolingen, and J. T. Van Dissel. 2004. Pneumonia caused by Mycobacterium kansasii in a series of patients without recognised immune defect. Clin. Microbiol. Infect. 10:738-748. [DOI] [PubMed] [Google Scholar]
- 6.Birkness, K. A., V. G. George, E. H. White, D. S. Stephens, and F. D. Quinn. 1992. Intracellular growth of Afipia felis, a putative etiologic agent of cat scratch disease. Infect. Immun. 60:2281-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanc, D. S., P. Carrara, G. Zanetti, and P. Francioli. 2005. Water disinfection with ozone, copper and silver ions, and temperature increase to control Legionella: seven years of experience in a university teaching hospital. J. Hosp. Infect. 60:69-72. [DOI] [PubMed] [Google Scholar]
- 8.Blomqvist, G., L. Wesslen, C. Pahlson, E. Hjelm, B. Pettersson, T. Nikkila, U. Allard, O. Svensson, M. Uhlen, B. Morein, and G. Friman. 1997. Phylogenetic placement and characterization of a new alpha-2 proteobacterium isolated from a patient with sepsis. J. Clin. Microbiol. 35:1988-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bollin, G. E., J. F. Plouffe, M. F. Para, and B. Hackman. 1985. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl. Environ. Microbiol. 50:1128-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collingro, A., S. Poppert, E. Heinz, S. Schmitz-Esser, A. Essig, M. Schweikert, M. Wagner, and M. Horn. 2005. Recovery of an environmental chlamydia strain from activated sludge by co-cultivation with Acanthamoeba sp. Microbiology 151:301-309. [DOI] [PubMed] [Google Scholar]
- 12.De, I., K. V. Rolston, and X. Y. Han. 2004. Clinical significance of Roseomonas species isolated from catheter and blood samples: analysis of 36 cases in patients with cancer. Clin. Infect. Dis. 38:1579-1584. [DOI] [PubMed] [Google Scholar]
- 13.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J. P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, J., N. Nanki, K. Negayama, S. Tsutsui, T. Taminato, and T. Ishida. 2002. Nosocomial contamination by Mycobacterium gordonae in hospital water supply and super-oxidized water. J. Hosp. Infect. 51:65-68. [DOI] [PubMed] [Google Scholar]
- 16.Gimenez, D. F. 1964. Staining Rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 17.Greub, G., B. La Scola, and D. Raoult. 2004. Amoebae-resisting bacteria isolated from human nasal swabs by amoebal coculture. Emerg. Infect. Dis. 10:470-477. [DOI] [PubMed] [Google Scholar]
- 18.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greub, G., and D. Raoult. 2002. Parachlamydiaceae: potential emerging pathogens. Emerg. Infect. Dis. 8:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraishi, A., K. Furuhata, A. Matsumoto, K. A. Koike, M. Fukuyama, and K. Tabuchi. 1995. Phenotypic and genetic diversity of chlorine-resistant Methylobacterium strains isolated from various environments. Appl. Environ. Microbiol. 61:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn, M., M. Wagner, K. D. Muller, E. N. Schmid, T. R. Fritsche, K. H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 22.Imbert, G., Y. Seccia, and B. La Scola. 2005. Methylobacterium sp. bacteraemia due to a contaminated endoscope. J. Hosp. Infect. 61:268-270. [DOI] [PubMed] [Google Scholar]
- 23.Kahane, S., N. Platzner, B. Dvoskin, A. Itzhaki, and M. G. Friedman. 2004. Evidence for the presence of Simkania negevensis in drinking water and in reclaimed wastewater in Israel. Appl. Environ. Microbiol. 70:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye, K. M., A. Macone, and P. H. Kazanjian. 1992. Catheter infection caused by Methylobacterium in immunocompromised hosts: report of three cases and review of the literature. Clin. Infect. Dis. 14:1010-1014. [DOI] [PubMed] [Google Scholar]
- 25.Khamis, A., P. Colson, D. Raoult, and B. L. Scola. 2003. Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl. Environ. Microbiol. 69:6740-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilvington, S., and J. Price. 1990. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 27.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kressel, A. B., and F. Kidd. 2001. Pseudo-outbreak of Mycobacterium chelonae and Methylobacterium mesophilicum caused by contamination of an automated endoscopy washer. Infect. Control Hosp. Epidemiol. 22:414-418. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 30.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J. M. Claverie, and D. Raoult. 2003. A giant virus in amoebae. Science 299:2033. [DOI] [PubMed] [Google Scholar]
- 31.La Scola, B., L. Barrassi, and D. Raoult. 2000. Isolation of new fastidious alpha-Proteobacteria and Afipia felis from hospital water supplies by direct plating and amoebal co-culture procedures. FEMS Microbiol. Ecol. 34:129-137. [DOI] [PubMed] [Google Scholar]
- 32.La Scola, B., I. Boyadjiev, G. Greub, A. Khamis, C. Martin, and D. Raoult. 2003. Amoeba-resisting bacteria and ventilator-associated pneumonia. Emerg. Infect. Dis. 9:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Scola, B., L. Mezi, P. J. Weiller, and D. Raoult. 2001. Isolation of Legionella anisa using an amoebic coculture procedure. J. Clin. Microbiol. 39:365-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Scola, B., and D. Raoult. 1999. Afipia felis in hospital water supply in association with free-living amoebae. Lancet 353:1330. [DOI] [PubMed] [Google Scholar]
- 35.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel, R., M. Muller, L. Zoller, J. Walochnik, P. Hartmann, and E. N. Schmid. 2005. Free-living amoebae serve as a host for the Chlamydia-like bacterium Simkania negevensis. Acta Protozool. 44:113-121. [Google Scholar]
- 37.Michel, R., M. Steinert, L. Zoller, B. Hauroder, and K. Henning. 2004. Free-living amoebae may serve as hosts for the Chlamydia-like bacterium Waddlia chondrophila isolated from an aborted bovine foetus. Acta Protozool. 43:37-42. [Google Scholar]
- 38.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niederman, M. S., L. A. Mandell, A. Anzueto, J. B. Bass, W. A. Broughton, G. D. Campbell, N. Dean, T. File, M. J. Fine, P. A. Gross, F. Martinez, T. J. Marrie, J. F. Plouffe, J. Ramirez, G. A. Sarosi, A. Torres, R. Wilson, and V. L. Yu. 2001. Guidelines for the management of adults with community-acquired pneumonia. Diagnosis, assessment of severity, antimicrobial therapy, and prevention. Am. J. Respir. Crit. Care Med. 163:1730-1754. [DOI] [PubMed] [Google Scholar]
- 40.Niedeveld, C. J., F. M. Pet, and P. L. Meenhorst. 1986. Effect of rubbers and their constituents on proliferation of Legionella pneumophila in naturally contaminated hot water. Lancet ii:180-184. [DOI] [PubMed] [Google Scholar]
- 41.Nolan, J. S., and K. B. Waites. 2005. Nosocomial ventriculitis due to Roseomonas gilardii complicating subarachnoid haemorrhage. J. Infect. 50:244-251. [DOI] [PubMed] [Google Scholar]
- 42.Rice, E. W., D. J. Reasoner, C. H. Johnson, and L. A. DeMaria. 2000. Monitoring for methylobacteria in water systems. J. Clin. Microbiol. 38:4296-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rihs, J. D., D. J. Brenner, R. E. Weaver, A. G. Steigerwalt, D. G. Hollis, and V. L. Yu. 1993. Roseomonas, a new genus associated with bacteremia and other human infections. J. Clin. Microbiol. 31:3275-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 45.Rohr, U., S. Weber, R. Michel, F. Selenka, and M. Wilhelm. 1998. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 47.Rowbotham, T. J. 1998. Isolation of Legionella pneumophila serogroup 1 from human feces with use of amebic cocultures. Clin. Infect. Dis. 26:502-503. [DOI] [PubMed] [Google Scholar]
- 48.Sanden, G. N., W. E. Morrill, B. S. Fields, R. F. Breiman, and J. M. Barbaree. 1992. Incubation of water samples containing amoebae improves detection of legionellae by the culture method. Appl. Environ. Microbiol. 58:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serra, M. 2003. Pharm. D. thesis. 18S rRNA broad range PCR for the identification of free-living amoebae present in a water network system. Université de la Méditerranée, Marseille, France.
- 50.Sopena, N., and M. Sabria. 2005. Multicenter study of hospital-acquired pneumonia in non-ICU patients. Chest 127:213-219. [DOI] [PubMed] [Google Scholar]
- 51.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taillard, C., G. Greub, R. Weber, G. E. Pfyffer, T. Bodmer, S. Zimmerli, R. Frei, S. Bassetti, P. Rohner, J. C. Piffaretti, E. Bernasconi, J. Bille, A. Telenti, and G. Prod'hom. 2003. Clinical implications of Mycobacterium kansasii species heterogeneity: Swiss National Survey. J. Clin. Microbiol. 41:1240-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, V., T. Bouchez, V. Nicolas, S. Robert, J. F. Loret, and Y. Lévi. 2004. Amoebae in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence. J. Appl. Microbiol. 97:950-963. [DOI] [PubMed] [Google Scholar]
- 54.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright, E. P., C. H. Collins, and M. D. Yates. 1985. Mycobacterium xenopi and Mycobacterium kansasii in a hospital water supply. J. Hosp. Infect. 6:175-178. [PubMed] [Google Scholar]