Abstract
The structure and dynamics of small eukaryotes (cells with a diameter less than 5 μm) were studied over two consecutive years in an oligomesotrophic lake (Lake Pavin in France). Water samples were collected at 5 and 30 m below the surface; when the lake was stratified, these depths corresponded to the epilimnion and hypolimnion. Changes in small-eukaryote structure were analyzed using terminal restriction fragment length polymorphism (T-RFLP) and cloning and sequencing of the 18S rRNA genes. Terminal restriction fragments from clones were used to reveal the dominant taxa in T-RFLP profiles of the environmental samples. Spumella-like cells (Chrysophyceae) did not dominate the small eukaryote community identified by molecular techniques in lacustrine ecosystems. Small eukaryotes appeared to be dominated by heterotrophic cells, particularly Cercozoa, which represented nearly half of the identified phylotypes, followed by the Fungi-LKM11 group (25%), choanoflagellates (10.3%) and Chrysophyceae (8.9%). Bicosoecida, Cryptophyta, and ciliates represented less than 9% of the community studied. No seasonal reproducibility in temporal evolution of the small-eukaryote community was observed from 1 year to the next. The T-RFLP patterns were related to bottom-up (resources) and top-down (grazing) variables using canonical correspondence analysis. The results showed a strong top-down regulation of small eukaryotes by zooplankton, more exactly, by cladocerans at 5 m and copepods at 30 m. Among bottom-up factors, temperature had a significant effect at both depths. The concentrations of nitrogenous nutrients and total phosphorus also had an effect on small-eukaryote dynamics at 5 m, whereas bacterial abundance and dissolved oxygen played a more important structuring role in the deeper zone.
Small phototrophic and heterotrophic eukaryotes (<5 μm) are found throughout the world's oceans and lakes at concentrations between 102 and 104 cells ml−1 in the photic zone (11). Small eukaryotes are known to be essential components in marine trophic food webs (20). The small-eukaryote assemblage is formed by picoalgae, which participate in primary production (55), by colorless heterotrophic cells, mostly flagellates, which are considered to be important grazers of prokaryotic and eukaryotic cells (11) and also play a significant role in the mineralization of organic matter, and finally by some small eukaryotes which can be mixotrophs. Despite the ecological importance of small eukaryotes and the general lack of distinct morphological features of these small cells, they have only recently been studied from a molecular perspective (20, 37). Thanks to these techniques, recent studies, conducted in various environments, have revealed a surprisingly high diversity of small eukaryotes and the existence of novel lineages (39). For example, the genetic diversity of small eukaryotes from coastal waters showed the dominance of novel alveolates (from 36% to 62% of total sequences obtained in their libraries) and the importance of novel stramenopiles, which account for up to 10% of sequences (38, 63). Furthermore, Prasinophyceae generally constituted the most conspicuous photosynthetic group and have been detected in all clone libraries (21, 38, 49). Deep-sea research has shown that novel stramenopiles may represent up to 23% of sequences and that pigmented organisms are dominant (20, 49). Thus, studies have generally focused on marine food webs, and freshwater picoplankton structure and dynamics have received little attention until now (32, 48).
Although these studies clearly provide better information on the diversity of the picoplanktonic community composition, factors involved in the regulation of these communities remain very poorly known. Indeed, only a few attempts have been made to relate the structure of picoplanktonic communities with biological, chemical, and physical variables in a lake. Some studies have reported seasonal changes in heterotrophic nanoflagellate community structure (15) in relation to environmental variables, such as grazing (top down), resources (bottom up), and viruses (25). Organisms such as cladocerans, especially the Daphnia genus, are well known for their high grazing pressure on a wide spectrum of particles (29). Other organisms, including large heterotrophic flagellates, may also belong to top-down regulation factors, consuming bacteria preferably (51) but also small eukaryotic algae (43). With regard to bottom-up regulation, picoplanktonic organisms are characterized by a high surface/volume ratio with a large surface for exchange, which favors nutrient uptake. Studies performed in lakes have shown that the contribution of the picophytoplankton to total phytoplankton biomass decreases with higher trophic status (2).
The aim of this work was to investigate the dynamics and diversity of small eukaryotes (<5 μm) over a 2-year study period in a lacustrine ecosystem (Lake Pavin). Two depths were sampled, corresponding to the epilimnion and hypolimnion, during the thermal stratification period. Changes in small-eukaryote community composition (SECC) were assessed using terminal restriction fragment length polymorphism (T-RFLP). Finally, temporal changes in SECC were related to bottom-up and top-down variables using canonical correspondence analysis (CCA), a direct multivariate analysis.
MATERIALS AND METHODS
Study site and sampling.
The study was conducted in an oligomesotrophic lake located in the Massif Central (France). Lake Pavin is a meromictic lake characterized by a maximum depth of 92 m. Samples were taken monthly from March to November 2001 and from September to November 2002 and every 2 weeks from April to August 2002. Sampling was carried out at a permanent station situated at the deepest zone of the water column. Water samples from 5 and 30 m below the surface, corresponding to epilimnion and hypolimnion in thermal stratification period, were collected with a Van Dorn bottle.
Water samples (from 100 to 120 ml) were prefiltered through 5-μm-pore-size polycarbonate filters (Millipore) at a pressure of <20 mbar in order to eliminate larger cells. It is well known that whatever the aquatic ecosystem, the prefiltration process allows the passage of cells larger than their nominal pore sizes and can lead to the retention of smaller cells if the filters are clogged (20). Using epifluorescence microscopy after primulin staining (10), we compared the abundance of small eukaryotes (diameter, <5 μm) in the nonfiltered and filtered fraction in several samples. We found that the filtration step led to a slight decrease of total abundance (of about 10 to 15%) but no modification of relative abundance of different morphotypes. The microbial biomass was collected on 0.2-μm-pore-size (pressure, <100 mbar) polycarbonate filters (Millipore) and stored at −80°C until nucleic acid extraction. Samples were collected and fixed immediately with a final concentration of 4% formaldehyde for total bacteria and 1% glutaraldehyde for protists. The metazooplankton was fixed in a sucrose/formaldehyde solution (final concentration, 6% and 4%, respectively) (46).
Biotic and abiotic variable measurements.
The water temperature and level of dissolved oxygen were determined with a multiparameter probe (YSI GRANT 3800). Chemical analyses, namely, ammonium (NH4-N), nitrates (NO3-N), nitrites (NO2-N), orthophosphate (PO4-P), and total phosphorus (Pt), were performed using standard methods (1). Chlorophyll a concentrations were obtained by spectrophotometry (57).
Counts of planktonic organisms.
For determining total prokaryotic abundance, 1- to 6-ml samples were filtered onto 0.2-μm black polycarbonate filters (25 mm; Millipore), stained by 1 μg liter−1 (final concentration) of 4,6-diamidino-2-phenylindole. Four hundred to eight hundred bacterial cells were counted under an epifluorescence microscope (45). After being stained with primulin (final concentration, 200 μg ml−1) (10), protists were filtered (5 to 10 ml of samples) onto black polycarbonate membrane of 0.8-μm pore size (Nuclepore) and counted by means of epifluorescence microscopy. A total of 200 to 300 cells were counted per filter and were separated in two size classes: under 5 μm (small eukaryotes) and from 5 to 30 μm (large flagellates). Autotrophs were distinguished from heterotrophs by their difference in color under epifluorescence. The metazoan zooplankton was counted under a binocular microscope (Wild M3 Z) in a Dolfuss chamber. To prevent the plankton from moving about or drying out, a few drops of 10% alcohol glycerin solution were added. Metazooplankton was made more visible by staining with a few drops of rose Bengal. If the density of organisms in a sample was too high, a subsample was taken with a Motoda box (60). Phytoplankton was counted monthly using Utermöhl's method (1958) with a Leitz-type inverted microscope (Wild M40).
T-RFLP analysis.
18S rRNA genes from environmental samples and clones obtained from Lake Pavin (32) were amplified with the eukaryote-specific primers Ek-1F-FAM (CTGGTTGATCCTTGCCAG) and Ek-516r (ACCAGACTTGCCCTCC) (14). The PCR mixture (50 μl) contained about 10 ng of environmental DNA, 200 μM of each deoxynucleoside triphosphate, 2 mM MgCl2, 10 pmol of each primer, 1.5 U of Taq DNA polymerase (Eurobio), and the PCR buffer supplied with the enzyme. Reactions were carried out in an automated thermocycler (PTC 200-cycler; MJ Research) with the following cycle: initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 1 min, annealing at 59°C for 1 min, and extension at 72°C for 1 min 30 s, and a final extension at 72°C for 10 min. PCR products were purified using the QiaQuick PCR purification kit (QIAGEN), visualized on 1% agarose gels, and quantified (DNA quantitation kit; Sigma). Several PCR products (at least four 50-μl samples) were pooled, precipitated with ethanol-sodium acetate, and resuspended in 50 μl of sterile water. Enzymatic digestions were performed by incubating 100 ng of PCR products with 20 U of MspI or RsaI (Sigma) at 37°C overnight. The samples were desalted with Microcon columns (Amicon; Millipore). The terminal restriction fragments (T-RFs) were separated on an automated sequencer (ABI 3700). Terminal restriction fragment sizes between 48 bp and 560 bp with a peak area of >50 fluorescence units were determined using Genescan analytical software. Samples were analyzed in triplicate, and a peak was included in the analysis if it occurred in at least two profiles. To account for small differences in the running time among samples, we considered fragments from different profiles with less than 1 base pair difference to be the same length. The resulting values were rounded up or down to the nearest integer. A program in Visual Basic for Excel was developed to automate these procedures. The results were then expressed either in terms of presence or absence or as a relative percentage area compared to the total area.
T-RF identification.
Environmental DNA extracted from 2 July 2002 was used to construct the 18S rRNA gene clone library (32). To determine the spatio-temporal changes in the sequences, we compared T-RFs obtained experimentally from clones and T-RFs obtained from the environmental DNA. A clone was present at a given date only if the two T-RFs generated by the two restriction enzymes (MspI and RsaI) were also present in the two T-RFLP environmental profiles.
Statistical analysis.
To explain the variation of SECC measured by T-RFLP and expressed as a percentage of area (>2%), CCA was used. Forward selection was performed to select the environmental variables that explained a significant part of changes in SECC (P < 0.05) (59). We tested the following variables: NH4-N, NO2-N, NO3-N, PO4-P, Pt, temperature, dissolved oxygen, water clarity, prokaryotes, chlorophyll a, large pigmented flagellates, large heterotrophic flagellates, and zooplankton (cladocerans, copepods, and rotifers) abundances.
Furthermore, a variation partitioning analysis was performed (4, 27). The variation partitioning analysis distinguished between pure top-down and bottom-up effects on SECC and the proportion explained by interactions between both these effects. These statistics were computed with R software using the Vegan package for the CCA and related methods (http://cran.r-project.org/).
RESULTS
Physicochemical characteristics of the study site.
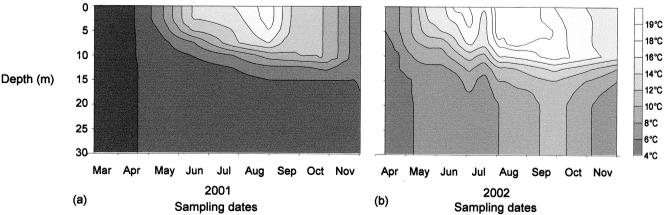
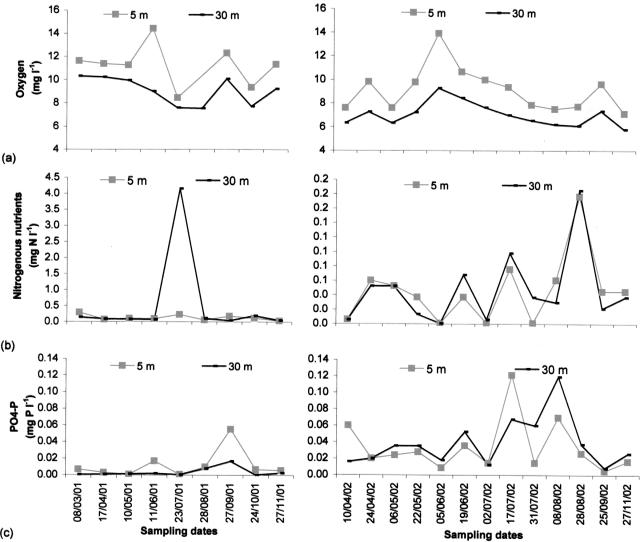
The stratification period in Lake Pavin was well established from mid-June to late October 2001 and early May to late September 2002 (Fig. 1). At a 30-m depth, we observed a warming up of the deep-water layers in September 2002, whereas the temperature had remained stable during the previous year. Dissolved oxygen varied between 5.76 mg liter−1 and 14.37 mg liter−1 during the study, with higher values in both the epilimnion and hypolimnion in 2001 (Fig. 2a). On average, nitrogenous nutrient concentrations were higher in 2001 than in 2002, whereas the PO4-P concentrations were higher in 2002 (Fig. 2b and c). The main physicochemical characteristics of the lake are listed in Table 1.
FIG. 1.
Isotherms of Lake Pavin in 2001 (a) and 2002 (b).
FIG. 2.
Temporal changes in oxygen (a), nitrogenous nutrient (NH4-N and NO3-N) (b), and PO4-P (c) concentrations at 5-m and 30-m depths. mg N l−1 and mg P l−1, mg of N and P per liter.
TABLE 1.
Environmental parameters measured in lake Pavin in 2001 and 2002a
| Parameter | Value for depth(s)
|
||
|---|---|---|---|
| 5 m | 30 m | Variousb | |
| Bacteria (106 cells ml−1) | 3.82 (2.35-5.94) | 1.15 (0.53-2.95) | |
| Chl a (μg liter−1) | 1.27 (ND-2.46) | 1.77 (ND-3.79) | |
| Zooplankton (ind liter−1) | 25 (2-86) | ||
| Cladocerans (ind liter−1) | 1 (ND-3) | ||
| Copepods (ind liter−1) | 3 (ND-16) | ||
| Rotifers (ind liter−1) | 21 (1-69) | ||
| Phytoplankton (106 cells liter−1) | 1.28 (ND-5.57) | ||
| Small eukaryotes (105 cells liter−1) | 3.02 (0.43-8.02) | 2.70 (0.46-6.48) | |
| PF of <5 μm (105 cells liter−1) | 1.10 (ND-2.78) | 0.10 (ND-1.30) | |
| HF of <5 μm (105 cells liter−1) | 2.00 (0.40-7.84) | 2.60 (0.6-7.80) | |
| Large eukaryotes (105 cells liter−1) | 7.32 (ND-41,31) | 1.61 (0.22-7.28) | |
| PF of >5 μm (105 cells liter−1) | 2.71 (ND-10.11) | 0.62 (ND-2.93) | |
| HF of >5 μm (105 cells liter−1) | 4.60 (ND-41.20) | 0.95 (0.21-5.70) | |
| NO3-N (mg N liter−1) | 0.05 (ND-0.06) | 0.26 (ND-3.97) | |
| NH4-N (mg N liter−1) | 0.03 (ND-0.17) | 0.04 (ND-0.18) | |
| PO4-P (mg P liter−1) | 0.02 (ND-0.12) | 0.02 (ND-0.12) | |
| Oxygen (mg liter−1) | 9.88 (7.08-14.37) | 7.83 (5.76-10.29) | |
| Temp (°C) | 11.76 (2.60-21.00) | 4.53 (3.30-6.00) | |
| Water clarity (m) | 6.93 (4.25-10) | ||
Values are means (minimum-maximum). HF, heterotrophic flagellates; PF, pigmented flagellates; ND, not detected; ind, individuals; Chl a, chlorophyll a.
Water column: 0 to 30 m.
Biological characteristics of the study site.
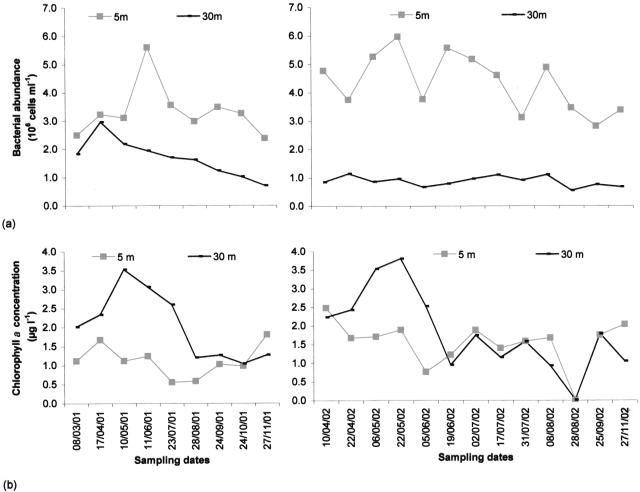
In contrast to the dynamics observed in the hypolimnion, total bacterial abundance fluctuated greatly in the euphotic zone, varying from 2.35 × 106 cells ml−1 to 5.94 × 106 cells ml−1 (maxima in June for 2001 and in May for 2002) (Fig. 3a; Table 1).
FIG. 3.
Temporal changes in bacterial abundance (a) and chlorophyll a concentration (b) at 5-m and 30-m depths. μg l−1, micrograms per liter.
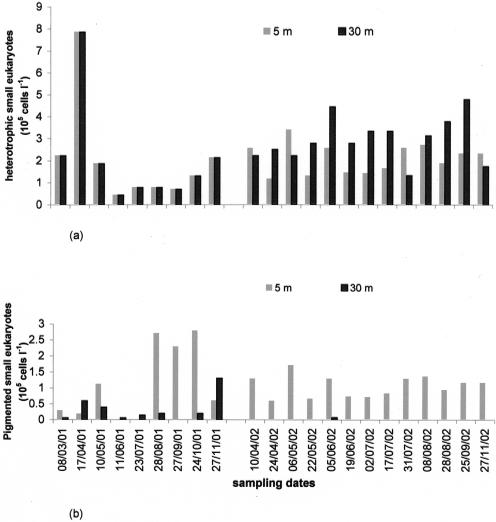
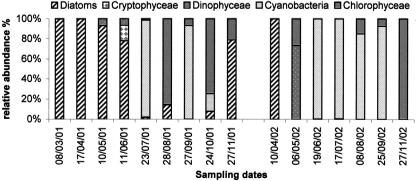
Total large flagellates (>5 μm) were generally less abundant at 30 m than at a 5-m depth (Table 1). Microscopic observations showed that heterotrophic organisms were the most abundant small eukaryotes (Table 1; Fig. 4a and b), especially in the hypolimnion, where choanoflagellates and incertae sedis flagellates accounted for 10.5% (±9.2) and 85.1% (±14.7) of total small-eukaryote community abundance, respectively. Mean chlorophyll a values were slightly higher in the hypolimnion (1.77 μg liter−1) than in the epilimnion (1.27 μg liter−1) (Table 1). The highest values were reached at a 5-m depth during the spring period in 2001 and 2002 (Fig. 3b), which was dominated by diatoms. Autotrophic organisms of the summer period consisted of Cyanobacteria and Chlorophyceae in 2001 and only Cyanobacteria in 2002 (Fig. 5). Large pigmented flagellates (>5 μm) represented on average 64.5% of the total abundance of flagellates (mean abundance, 2.7 × 105 cells liter−1) (Table. 1). The greatest abundance was observed at the end of the summer in 2001 and in spring in 2002. Small pigmented eukaryotes (<5 μm) were mainly represented by Chrysophyceae (37.6% [mean] ± 35.6%) and Cryptophyta (32.0% [mean] ± 28.5%).
FIG. 4.
Temporal changes in abundances of heterotrophic small eukaryotes (a) and pigmented small eukaryotes (b) at 5-m and 30-m depths. Cells l−1, cells per liter.
FIG. 5.
Relative abundances of phytoplankton at 5-m depth.
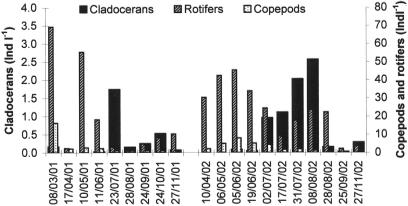
Metazoan zooplankton abundance reached its peak in spring (mean, 52 individuals liter−1) (Fig. 6). In terms of abundance, this community was dominated by rotifers, mainly represented by Polyarthra spp. (40.0%) and Kellicotia spp. (40.0%) throughout the study period. Fifty-three percent of copepods were at the nauplii stage. Cladocerans were represented by two genera: Daphnia spp. and Ceriodaphnia spp.
FIG. 6.
Temporal changes in abundance of zooplankton (cladocerans, copepods, and rotifers) (Ind l−1, individuals per liter).
Structure and dynamics of the small-eukaryote community.
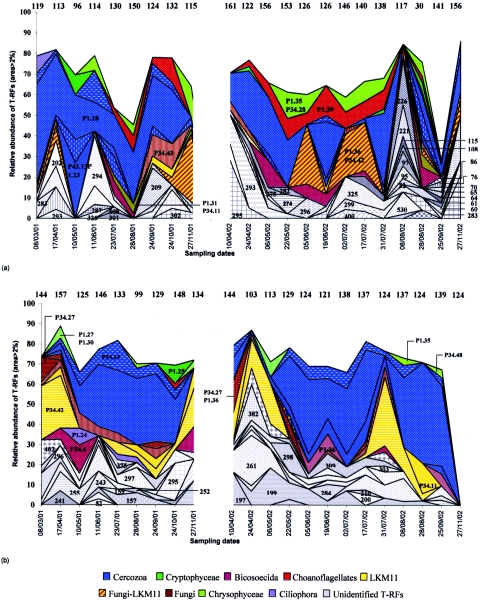
Terminal restriction fragments obtained by enzymatic digestion by MspI and RsaI allowed us to track changes in the SECC. The mean number of T-RFs obtained was lower for RsaI (101 T-RFs) than for MspI (128 T-RFs), regardless of depth. Thus, MspI seems to be more discriminative in terms of diversity. For this reason, we have presented only data obtained with this enzyme. A total of 357 T-RFs were detected for both years. The mean number of T-RFs varied slightly with depth, but in contrast, the numbers of T-RFs fluctuated strongly during the study, from 30 to 161 at 5 m and from 99 to 157 at a 30-m depth (Fig. 7). Most of the T-RFs occurred in both years of the study. Only 5.4% of the T-RFs were specific to 2001 and 4.4% to 2002. Of a total of 357 T-RFs detected, only 94 had a relative area higher than 2% and were thus considered dominant. For example, all the operational taxonomic units (OTUs) detected on 27 November 2002 at 30 m had an area which represented less than 2% of the total area. On average, eight T-RFs represented 67% of the total area. Among the 94 dominant T-RFs, 28 were phylogenetically identified using both restriction enzymes (Table 2). On average, these T-RFs represented 66% of the area determined by the dominant T-RFs at both depths studied. T-RFs 49, 236, 239, 291, and 398 were therefore associated with clones P1.24 (Ciliophora), P1.25 (Cryptophyta), P34.43 (Fungi), P1.39 (choanoflagellates) and P1.18 (Cercozoa), respectively. However, some dominant T-RFs, such as 295, which were found on most sampling dates, could not be identified.
FIG. 7.
Seasonal variations, at 5-m (a) and 30-m (b) depths, in number and relative abundance of the various OTUs detected by T-RFLP analysis of 18S rRNA gene digestion by MspI, representing more than 2% of the total area (sampling date are expressed as day/month/year). The numbers at the top of the environmental T-RFLP profile represent the total number of T-RFs detected for each sampling date by the restriction enzyme MspI. For example, 156 OTUs were detected on 6/05/02 at 5 m, and 8 OTUs (area, >2%) represented nearly 68% of the total area. Numbers on the environmental T-RFLP profiles correspond to the lengths of T-RFs in bp (the lengths of the T-RFs corresponding to the clones are presented in Table 2). On 27/11/02, any T-RFs had an area of >2%.
TABLE 2.
Clones of the Pavin library
| Clone | Division | Phylogenetic affiliation species | Accession no. | T-RF size (bp)a
|
|
|---|---|---|---|---|---|
| MspI | RsaI | ||||
| P34.19 | Cryptophyta | Chrysochromulina throndsenii | AY642708 | 279 | 140 |
| P1.25 | Cryptophyta | Chroomonas sp. | AY642699 | 236 | 250 |
| P1.31 | Cryptophyta | Chroomonas sp. | AY642716 | 389 | 244 |
| P1.30 | Cryptophyta | Geminigera cryophila | AY642715 | 369 | 377 |
| P1.27 | Cryptophyta | Geminigera cryophila | AY642713 | 384 | 559 |
| P34.28 | Chrysophyceae | Oikomonas mutabilis | AY642697 | 245 | 51 |
| P1.35 | Paraphysomonas bandaiensis | AY642717 | 245 | 554 | |
| P34.45 | Spumella elongata | AY642705 | 249 | 552 | |
| P34.48 | Hibberdia magna | AY642709 | 247 | 55 | |
| P34.6 | Bicosoecida | Cafeteria roenbergensis | AY642710 | 392 | 521 |
| P34.38 | Ciliophora | Glaucoma chattoni | AY642718 | 205 | 54 |
| P34.44 | Ciliophora | Prorodon teres | AY642703 | 387 | 554 |
| P1.24 | Ciliophora | Prorodon teres | AY642698 | 49 | 54 |
| P1.23 | Cercozoa | Cercomonas sp. | AY642696 | 250 | 49 |
| P1.18 | Cercozoa | Cercomonas sp. | AY642694 | 398 | 103 |
| P34.13 | Cercozoa | Cercomonas sp. | AY642704 | 243 | 103 |
| P34.14 | Cercozoa | Heteromita globosa | AY642693 | 403 | 104 |
| P1.39 | Choanoflagellates | Diaphanoeca grandis | AY642707 | 291 | 141 |
| P34.27 | Fungi | Spizellomyces acuminatus | AY642695 | 386 | 89 |
| P1.36 | Fungi | Spizellomyces acuminatus | AY642706 | 385 | 71 |
| P34.43 | Fungi | Spizellomyces acuminatus | AY642701 | 239 | 530 |
| P34.42 | Environmental sequences | Unidentified eukaryote LKM11 | AY642700 | 395 | 528 |
| P34.11 | Environmental sequences | Unidentified eukaryote LKM11 | AY642711 | 390 | 89 |
TRF sizes were obtained using a fluorescent forward primer (1F-FAM) and MspI and RsaI restriction enzymes.
Strong variations in dominant T-RFs were recorded in environmental T-RFLP profiles. A few T-RFs had a relative area higher than 2% only at some dates, such as T-RFs 283 (8 March 2001 at 5 m), 202 (17 April 2001 at 5 m), and P1.25 (24 October 2001 at 30 m). Other T-RFs, such as P1.18, were regularly detected during both years and at both depths. Otherwise, some T-RFs were detected at one particular depth, such as T-RFs 202 in the epilimnion and 199 in the deeper zone.
Like the counts conducted by epifluorescence microscopy, the T-RFs identified showed that heterotrophic organisms dominated the small-eukaryote community, especially in the hypolimnion. Cercozoa, represented by genera Cercomonas spp. (48% of total identified area) and Heteromita globosa (6%), accounted for almost half of the organisms identified in both study years and at both depths (Fig. 7). At 30 m, Cercozoa seemed to be dominant, particularly during the period of thermal stratification. The second-largest group, at both depths, was Fungi-LKM11 (average, 25% of areas identified). Moreover, when the Fungi-LKM11 association was present on a given date, then Cercozoa were rare or absent, and the inverse. For example, at 30 m, from 3 August 2001 to 6 November 2001, Fungi (P34.43) and LKM11 (P34.42) dropped from 41.2% to 4.5% of total identified areas, whereas Cercozoa increased from 0% to 38.4% (Fig. 7b). At 5 m, Cercozoa seemed to be associated with the presence of diatoms (Fig. 5), whereas the fungi-LKM11 group was observed in situations where the phytoplankton community was dominated by Chlorophyceae (Fig. 5 and 7a). Cryptophyta (2.8% of identified areas) were only occasionally detected in the epilimnion, on 10 May 2001, 24 April 2002, 31 July 2002, and 28 August 2002 (Fig. 7a). Important clades at 5 m, such as Chrysophyceae (average, 8.9% of areas identified) and choanoflagellates (10.3%), were observed only in very small numbers at 30 m. Cafeteria roenbergensis, belonging to the lineage of Bicosoecida, was present in both zones sampled in 2001 but at different periods. In 2002, this taxon was detected mainly in the epilimnion.
Small-eukaryote community composition in relation to environmental variables.
The T-RFLP patterns of the different samples were analyzed in relation to physical, chemical, and biological data from the lake. A partial CCA was performed to explain the relationship between SECC and explanatory variables. This direct multivariate analysis revealed several significant relationships (P < 0.05) between explanatory variables and SECC (Table 3). The variation partitioning analysis was performed with variables that independently explained a significant amount of the variations in CCA. Pure top-down and pure bottom-up values represented 8.2% and 19.8% in epilimnion and 22.1% and 21.8% of the total inertia in hypolimnion. These results showed that top-down and bottom-up factors were of the same order of magnitude of importance in the hypolimnion, whereas in the euphotic zone, resources had a higher impact on SECC.
TABLE 3.
Results of canonical correspondence analysisa
| Parameter | Value for depth
|
|
|---|---|---|
| 5 m | 30 m | |
| Bottom-up factors (%) | ||
| Bacteria | 11.2 | |
| NO2-N | 7.2 | |
| NO3-N | ||
| NH4-N | 7.8 | |
| PO4-P | ||
| Pt | 7.6 | |
| Temperature | 6.3 | 9.5 |
| Oxygen | 9.4 | |
| Water clarity | 9.5 | |
| Top-down factors (%) | ||
| Cladocerans | 14.4 | 6.2 |
| Copepods | 13.8 | |
| Rotifers | 8.7 | |
| HF of >5 μm | 12.4 | |
| Total inertia | 5.7 | 3.6 |
| Sum of constrained eigenvalues | 3.9 | 2.1 |
Percentage of variation in small eukaryote community composition (expressed by percentage of area) explained by the different environmental variables (HF, heterotrophic flagellates; Pt, total phosphorus).
Among bottom-up factors, temperature appeared to significantly control the composition of small eukaryotes at both depths. Nutrients also played a significant role in the epilimnion, whereas the dissolved-oxygen concentration and water temperature appeared to have a greater effect on SECC in the hypolimnion. Moreover, bacteria (11.2%) were significantly involved in the regulation of SECC in the hypolimnion (Table 3). For example, the dynamics of Cafeteria roenbergensis (P34.6) (Fig. 7) was associated with the dynamics of bacterial abundance in the epilimnion (Fig. 3a), since this organism appeared shortly after an increase in bacterial density, which decreased thereafter.
With regard to top-down regulation factors, the results of this study show that zooplankton seemed to be the main factor associated with variations in SECC at both depths studied. Among the zooplankton community, cladocerans were the most important regulatory factor in the epilimnion. Thus, the peak abundance of these organisms coincided simultaneously with the shift in small-eukaryote structure on 8 August 2002 (Fig. 7a). In contrast, according to the statistical analysis, SECC in the hypolimnion appeared to depend mainly on the abundance of copepods. However, other predators, including large flagellates and rotifers, also played a role in this zone.
DISCUSSION
Methodological aspects.
In this study, the water was prefiltered through 5-μm-pore-size filters according to the methods of Díez et al. (20) and Lòpez-Garcia et al. (34) in order to take into account most of the small eukaryotic cells observable by standard epifluorescence microscopy but usually considered unidentified Protista and which have been reported to represent a large proportion of microorganisms in lakes (12, 60). To study these organisms, we chose T-RFLP (33) as a fingerprinting method. It is considered to have both a high resolving power and reproducibility (40) and the advantage of being semiquantitative (7). T-RFLP provides a tentative identification of the present species by direct comparison with a database of sequences (36). However, such identification can depend on the presence of the fluorescent sequencing dyes and on the purine fragment content (30). The number of T-RFs can nevertheless be biased by the formation of pseudo-T-RFs (23). We therefore chose to identify the T-RFs from clones already characterized in the ecosystems studied (16, 54). The clone library obtained on 2 July 2002 in Lake Pavin (32) allowed us to identify most of the T-RFs considered to be dominant (>2%) for many dates in environmental profiles (Fig. 7). These results showed a good relation between the two techniques. Moreover, according to our results from both molecular analysis and epifluorescence microscopy counts, heterotrophic organisms dominated the small-eukaryote community. This high proportion of heterotrophs is in agreement with results from previous studies showing that the pigmented organisms generally represent only a small proportion of small eukaryotes in lakes of this area (12, 60). In the epilimnion, small pigmented eukaryotes represented 5.2% of the areas identified and were members of the Chrysophyceae and Cryptophyta groups. The presence of Cryptophyta in the smallest planktonic fraction is consistent with previous results obtained from lacustrine ecosystems (28, 56). In the hypolimnion, according to microscopic results, pigmented organisms were present when light penetration was greatest. However, the presence of pigmented organisms in the epilimnion was apparently underestimated by molecular techniques. This may be explained by the fact that the library was built on only one date and did not cover the full diversity of the small-eukaryote community (32). Furthermore, PCR-based methods are susceptible to potential biases, and more specifically, these results could be explained by preferential amplification of templates (3).
Structure and dynamics of the small-eukaryote community.
The average diversity of small eukaryotes as determined by T-RFLP over both years and at both depths was clearly greater than that obtained in marine environments with the same technique. Indeed, the only studies available reported an average of 14 T-RFs in a hypersaline environment (14) and a maximum of 25 T-RFs in a marine environment (Mediterranean Sea) (19). However, the differences observed may have been due to a difference with the latter study in the computer processing applied at data integration (19, 20). Despite considerable diversity, the small-eukaryote community remains dominated by a small number of taxa. Thus, eight T-RFs accounted on average for 67% of the total area. Nanoflagellates (<20 μm) of the genera Spumella/Monas, which are typical colorless Chrysophyceae, have been reported to be generally common in freshwaters (5). Microscopic investigations are often hampered by the sparseness of diagnostic characteristics for taxonomic identification, and this is valid in particular for the small heterotrophic flagellates. In our microscopic investigation, 80% of the small heterotrophs were unidentified cells or cells of uncertain taxonomy, among which Chrysophyceae were probably largely represented; Spumella-like cells, in particular, were observable. Moreover, although the molecular techniques showed that small Chrysophyceae were present (Spumella elongata, Oikomonas, Paraphysomonas, and Hibberdia), particularly at 5 m, they did not dominate the small-eukaryote population. Similar observations were reported by Lefranc et al. (32) and Richards et al. (48), who also identified this group within small-eukaryote communities as recurrent but not dominant.
Among the heterotrophic organisms, the dynamics of dominant T-RFs and the clone library showed that Cercozoa are the most abundant group in Lake Pavin. The Cercozoa group demonstrates huge morphological, ecological, and genetic diversity (31). However, little is known of their very heterogeneous morphology, which makes them particularly difficult to identify by microscopy. Among the organisms identified, Fungi and the environmental clade LKM11 (61) were abundant at both depths and were the second most dominant group after the Cercozoa. LKM11 were strongly associated with Fungi (61) and were not always separated by T-RFLP (Table 2; Fig. 7). These results therefore confirm cloning-sequencing results from various ecosystems demonstrating numerous clones belonging to both lineages (32). The presence of Fungi would therefore appear to be a specific feature of lacustrine ecosystems. Indeed, these sequences have been reported as being either absent or occurring in very low proportions in pelagic marine environments (21, 38, 63). Choanoflagellates were present in the epilimnion and more or less absent in the hypolimnion. This distribution may be explained by several ecological factors. Choanoflagellates are epiphytic organisms that may depend on both the presence of microalgae (13) and the quality of available organic materials, since they are geared to use high-molecular-weight dissolved organic matter and colloidal organic particles (53). Cafeteria roenbergensis, belonging to the Bicosoecida lineage, was detected at both depths. However, when it was present at 5 m it was absent at 30 m and vice versa. This organism has not been identified in abundance in the clone libraries produced in marine environments (38, 49), whereas the protist flagellate counts previously conducted in Lake Pavin showed Bicosoecida as representing 7% of total abundance (13).
Forces regulating the dynamics of SECC.
Seasonal cycles in temperate lakes are driven by the basic physical parameters of light, temperature, and wind, which control the dynamics of all biota via nutrient upwelling and primary production. In Lake Pavin, temperature was a significant explanatory factor in SECC variations at both depths. Water clarity and oxygenation appeared to influence the small-eukaryote composition only in the hypolimnion. For example, Delaney (17) showed that growth rates of Paraphysomonas spp. (heterotrophic flagellates) were temperature dependent, decreasing sixfold between 15°C and 0°C. Although we observed the same taxonomic groups in both 2001 and 2002, no clear seasonal reproducibility between years was identified, suggesting that the physical characteristics of the lake (e.g., stratification of the water column) were not the main factors controlling SECC variation. Some seasonal variations in heterotrophic nanoflagellate composition had previously been reported (15), but studies were mostly conducted over a single year; thus, it is difficult to conclude whether there is a real seasonal reproducibility. Our results showed that SECC variations in the euphotic zone were controlled mainly by bottom-up effects (availability of inorganic resources and prey), and nutrients seemed to be the main factor associated with these variations. More especially, nitrogenous nutrients and total phosphorus play a significant role. Using a cross-factorial experimental design to test resource and predation effects on microbial community composition, we also observed that nutrient levels (NO3-N, NH4-N, and PO4-P) had a significant impact on the epilimnetic small-eukaryote structure (unpublished data). In the hypolimnion, the SECC is significantly related to the prokaryotic abundance, and there is a relationship between bacterial density and the dynamics of bacterivorous Cafeteria roenbergensis (24). The prey-predator-type interactions existing between bacteria and flagellates are now well known. Thus, the main bacterivores in aquatic ecosystems are typically small heterotrophic flagellates, generally <5-μm-size cells (58) or, in some cases, bacterial grazers belonging to potentially mixotrophic species (22). Moreover, by taking into account the fact that bacterivory can be selective (e.g., see reference 26), heterotrophic eukaryote diversity could therefore be linked to prokaryote diversity, which varies in the deepest zone of this ecosystem (6).
Bacterial abundance had been considered to be a bottom-up factor, especially for small heterotrophic flagellates, but for Fungi-LKM11, phytoplankton may represent resources, since they were identified when chlorophyll a concentrations were highest. Organisms belonging to the clade LKM11 seem to be associated with the decomposition of detritus composed of algae and cyanobacteria (61). Moreover, the fungi found in this ecosystem were affiliated with the Chytrids lineage (32), whose members are known to be parasites of green algae (35) and diatoms (9), which may also be regulated by Cercozoa (49). In the epilimnion, Cercozoa were present when diatoms developed, whereas fungi were associated with Chlorophyceae (Fig. 7a; Fig. 5). Furthermore, we observed at both depths that when Fungi-LKM11 were present, Cercozoa were either absent or present at low densities and vice versa. Different hypotheses can explain why these small eukaryotes were not associated within picoplankton assemblages: (i) they may compete for the same host, diatoms, with the Cercozoa proving more efficient parasites; (ii) Fungi may preferentially parasitize Chlorophyceae, whereas Cercozoa could be associated with diatoms.
Finally, changes in small-eukaryote structure were also linked to metazooplankton abundance and structure in both the epilimnion and the hypolimnion. Cladocerans intervened in an important way (14.4%; Table 3) in the epilimnion. Cladocerans are able to consume particles of about 0.5 μm (8) and therefore exert an important impact on the structure of the microbial trophic food web (29). Most of them are considered to be poorly selective, although some studies have reported that some species belonging to the genus Bosmina show taste selectivity (18). In the hypolimnion, the CCA combined with variation partitioning showed that top-down (predation) factors due to copepods and to a lesser extent rotifers were just as important as bottom-up factors (resources). Rotifers are known to preferentially graze particles belonging to the size class 1 to 5 μm (50), with a preference for autotrophic, mixotrophic, or heterotrophic flagellates without protection (44), whereas nauplii stages, which represented a large proportion of the copepods in the present study, are able to graze particles of 4 to 5 μm in diameter (41, 42, 62). Moreover, copepods appear to demonstrate much more prey selectivity than cladocerans in terms of both size and the nature of the particles digested (44). Thus, copepods that consisted essentially of cyclopoids in Lake Pavin had a predatory impact on small eukaryotes preferentially in zones where heterotrophs are predominant, while the less-selective cladocerans may have a larger impact in the euphotic zone, where there is a higher density of indigestible particles. In the hypolimnion, the results of the CCA also showed a regulatory effect of large heterotrophic flagellates, which can consume small eukaryotes in pelagic microbial food webs (47, 52) and may play a role in controlling the diversity in the small-eukaryote community composition.
Acknowledgments
We thank Sébastien Specel for automated sequencer and GENESCAN analysis, Jean-Claude Romagoux for his invaluable collaboration, and Christophe Portelli and Aurélie Thénot for their technical assistance.
REFERENCES
- 1.American Public Health Association. 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 2.Bell, T., and J. Kalff. 2001. The contribution of picophytoplankton in marine and freshwater systems of different trophic status and depth. Limnol. Oceanogr. 46:1243-1248. [Google Scholar]
- 3.Berney, C., J. Fahrni, and J. Pawlowski. 2004. How many novel eukaryotic “kingdoms”? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bocard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 5.Boenigk, J., K. Pfandl, P. Stadler, and A. Chatzinotas. 2005. High diversity of the “Spumella-like” flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, D., L. Jardillier, and D. Debroas. 2005. Succession of bacterial community composition over two consecutive years in two aquatic systems: a natural lake and a lake-reservoir. FEMS Microbiol. Ecol. 55:79-97. [DOI] [PubMed] [Google Scholar]
- 7.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brendelberger, H., and W. Geller. 1985. Variability in eight Daphnia species: mesh sizes and filtering area. J. Plankton Res. 7:473-487. [Google Scholar]
- 9.Canter, H. M., and G. H. Jaworski. 1981. The effect of light and darkness upon infection of Asterionella formosa Hassall by the chytrid Rhizophydium planktonicum Canter emend. Ann. Bot. 47:13-30. [Google Scholar]
- 10.Caron, D. A. 1983. Technique for enumeration of heterotrophic and phagotrophic nanoplankton, using epifluorescence microscopy, and comparison with other procedures. Appl. Environ. Microbiol. 46:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron, D. A., E. R. Peele, E. L. Lim, and M. R. Dennett. 1999. Picoplankton and nanoplankton and their trophic coupling in the surface waters of the Sargasso Sea south of Bermuda. Limnol. Oceanogr. 44:259-272. [Google Scholar]
- 12.Carrias, J. F., C. Amblard, and G. Bourdier. 1996. Protistan bacterivory in an oligomesotrophic lake: importance of attached ciliates and flagellates. Microb. Ecol. 31:249-268. [DOI] [PubMed] [Google Scholar]
- 13.Carrias, J. F., C. Amblard, C. Quiblier-Lloberas, and G. Bourdier. 1998. Seasonal dynamics of free and attached heterotrophic nanoflagellates in an oligomesotrophic lake. Freshw. Biol. 39:91-101. [Google Scholar]
- 14.Casamayor, E. O., R. Massana, S. Benlloch, L. Øvreås, B. Díez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alios. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprint methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 15.Cleven, E. J., and T. Weisse. 2001. Seasonal succession and taxon-specific bacterial grazing rates of heterotrophic nanoflagellates in Lake Constance. Aquat. Microb. Ecol. 23:147-161. [Google Scholar]
- 16.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 17.Delaney, M. P. 2003. Effects of temperature and turbulence on the predator-prey interactions between a heterotrophic flagellate and a marine bacterium. Microb. Ecol. 45:218-225. [DOI] [PubMed] [Google Scholar]
- 18.De Mott, W. R. 1986. The role of taste in food selection by freshwater zooplancton. Oecologia 69:334-340. [DOI] [PubMed] [Google Scholar]
- 19.Diez, B. 2001. Diversity of the eukaryotic marine picoplankton by means of molecular methods. Thesis. University of Barcelone, Barcelona, Spain.
- 20.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblage and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díez, B., C. Pedrós-Alió, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domaizon, I., S. Viboud, and D. Fontvielle. 2003. Taxon-specific and seasonal variations in flagellates grazing on heterotrophic bacteria in the oligotrophic Lake Annecy—importance of mixotrophy. Microb. Ecol. 46:317-329. [DOI] [PubMed] [Google Scholar]
- 23.Egert, F., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenchel, T., and D. J. Patterson. 1988. Cafeteria roenbergensis nov. gen., nov. sp., a heterotrophic microflagellate from marine plankton. Mar. Microb. Food Webs 3:9-19. [Google Scholar]
- 25.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 27.Jardillier, L., D. Boucher, S. Personnic, S. Jacquet, A. Thénot, D. Sargos, C. Amblard, and D. Debroas. 2005. Relative importance of nutrients and mortality factors on prokaryotic community composition in two lakes of different trophic status: microcosms experiments. FEMS Microbiol. Ecol. 55:429-443. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, P. W., and J. M. Sieburth. 1982. In situ morphology and occurrence of eukaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J. Phycol. 8:312-327. [Google Scholar]
- 29.Jürgens, K. 1994. Impact of Daphnia on planktonic microbial food web: a review. Mar. Microb. Food Webs 8:295-324. [Google Scholar]
- 30.Kaplan, C. W., and C. L. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 1776:1-5. [DOI] [PubMed] [Google Scholar]
- 31.Keeling, P. J. 2001. Foraminifera and Cercozoa are related in actin phylogeny: two orphans find a home? Mol. Biol. Evol. 18:1551-1557. [DOI] [PubMed] [Google Scholar]
- 32.Lefranc, M., A. Thénot, C. Lepère, and D. Debroas. 2005. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, W. L., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lòpez-Garcia, P., F. Rodriguez-Valera, C. Pedròs-Aliò, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-llorca, L. V., and P. Hernandez. 1996. Infection of the green alga Oocystis lacustris Chod with the Chytrid fungus Diplochytridium deltanum (Masters) Karling. An SEM study. Micron 27:355-358. [Google Scholar]
- 36.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 37.Massana, R., L. Guillou, B. Diez, and C. Pedros-alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massana, R., V. Balagué, L. Guillou, and C. Pedros-Alio. 2004. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol. Ecol. 50:231-243. [DOI] [PubMed] [Google Scholar]
- 39.Moreira, D., and P. Lopez-Garcia. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 40.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 41.Paffenhöfer, G. A. 1984. Food ingestion by marine planktonic copepod Paracalanus in relation to abundance and size distribution of food. Mar. Biol. 80:328-333. [Google Scholar]
- 42.Paffenhöfer, G. A. 1998. Heterotrophic protozoa and small metazoa: feeding rates and prey-consumer interaction. J. Plankton Res. 20:121-133. [Google Scholar]
- 43.Parslow, J. S., G. J. Doucette, F. J. R Taylor, and P. J. Harisson. 1986. feeding by the zooflagellates Pseudobodo sp. on Micromonas pusilla. Mar. Ecol. Prog. Ser. 29:237-246. [Google Scholar]
- 44.Pont, D. 1995. Le zooplancton herbivore dans les chaînes alimentaires pélagique, p. 515-540. In R. Pourriot and M. Meybeck (ed.), Limnologie générale. Masson, Paris, France.
- 45.Porter, K. J., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 46.Prepas, E. 1978. Sugar frosted Daphnia: an improved fixation technique for cladocera. Limnol. Oceanogr. 23:557-559. [Google Scholar]
- 47.Reckermann, M., and M. J. W. Veldhuis. 1997. Trophic interactions between picophytoplankton and micro- and nano-zooplankton in the western Arabian Sea during the NE monsoon 1993. Aquat. Microb. Ecol. 12:263-273. [Google Scholar]
- 48.Richards, T. A., A. A. Vepritskiv, D. E. Gouliamova, and S. A. Nierzwicki-Bauer. 2005. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ. Microbiol. 7:1413-1425. [DOI] [PubMed] [Google Scholar]
- 49.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784-798. [Google Scholar]
- 50.Ross, P. E., and M. Nunavar. 1981. Preference for nanoplankton size fractions in Lake Ontario zooplankton grazing. J. Great Lakes Res. 7:65-67. [Google Scholar]
- 51.Sakka, A., L. Legendre, M. Gosselin, and B. Delesalle. 2000. Structure of the oligotrophic planktonic food web under low grazing of heterotrophic bacteria: Takapoto Atoll, French Polynesia. Mar. Ecol. Prog. Ser. 197:1-17. [Google Scholar]
- 52.Samuelsson, K., and A. Andersson. 2003. Predation limitation in the pelagic microbial food web in an oligotrophic aquatic system. Aquat. Microb. Ecol. 30:239-250. [Google Scholar]
- 53.Sherr, E., B. Sherr, and L. Fessenden. 1997. Heterotrophic protists in the central Arctic Ocean. Deep Sea Res. II 44:1665-1682. [Google Scholar]
- 54.Stepanauskas, R., M. A. Moran, B. A. Bergamaschi, and J. T. Hollibaugh. 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat. Microb. Ecol. 31:85-98. [Google Scholar]
- 55.Stockner, J. G., and N. J. Antia. 1986. Algal picoplankton from marine and freshwater ecosystems: a multidisciplinary perspective. Can. J. Fish. Aquat. Sci. 43:2472-2503. [Google Scholar]
- 56.Stockner, J. G., and K. S. Shortreed. 1989. Algal picoplankton production and contribution to food-webs in oligotrophic British Columbia Lakes. Hydrobiologia 173:151-166. [Google Scholar]
- 57.Strickland, J. D. H., and R. Parson. 1972. Spectrophotometric determination of chlorophylls and total carotenoïds, section IV.3. In J. C. Stevenson (ed.), A practical handbook for sea water analysis. Fisheries Research Board of Canada, Ottawa, Canada.
- 58.Strom, S. L. 2000. Bacterivory: interactions between bacteria and their grazers, p. 351-386. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, Hoboken, N.J.
- 59.Ter Braak, C. J. F. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167-1179. [Google Scholar]
- 60.Thouvenot, A., D. Debroas, M. Richardot, L. B. Jugnia, and J. Dévaux. 2000. A study of changes between years in the structure of plankton community in a newly-flooded reservoir. Arch. Hydrobiol. 149:131-152. [Google Scholar]
- 61.Van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Von Elert, E., and P. Stampfl. 2000. Food quality for Eudiaptomus gracilis: the importance of particular highly unsaturated fatty acids. Freshw. Biol. 45:189-200. [Google Scholar]
- 63.Yuan, J., M. Chen, S. Peng, H. Zhou, Y. Chen, and L. Qu. 2004. Genetic diversity of small eukaryotes from the coastal waters of Nansha Islands in China. FEMS Microbiol. Lett. 240:163-170. [DOI] [PubMed] [Google Scholar]