Abstract
The occurrence, prevalence, and distribution patterns of acute bee paralysis virus (ABPV), black queen cell virus (BQCV), chronic bee paralysis virus (CBPV), deformed wing virus (DWV), Kashmir bee virus (KBV), and sacbrood virus (SBV) were investigated in 90 Austrian honeybee colonies suffering from symptoms of depopulation, sudden collapse, paralysis, or dark coloring by employing reverse transcription-PCR. Infestation with parasites was also recorded. The samples originated from all parts of Austria. The most prevalent virus was DWV, present in 91% of samples, followed by ABPV, SBV, and BQCV (68%, 49%, and 30%, respectively). CBPV was detected in 10% of colonies, while KBV was not present in any sample. In most samples, more than one virus was identified. The distribution pattern of ABPV, BQCV, CBPV, and SBV varied considerably in the different geographic regions investigated, while DWV was widespread in all Austrian federal states. In bees that showed dark coloring and disorientation, CBPV was always detected. Simultaneous infections of DWV and ABPV were most frequently observed in colonies suffering from weakness, depopulation, and sudden collapse. Bees obtained from apparently healthy colonies within the same apiaries showed a similar distribution pattern of viruses; however, the relative virus load was 10 to 126 times lower than in bees from diseased colonies. A limited number of bee samples from surrounding central European countries (Germany, Poland, Hungary, and Slovenia) were also tested for the presence of the above viruses. Variances were found in the distribution of BQCV and SBV.
The scientific interest in viral diseases of the honeybee (Apis mellifera L.) has been increasing considerably during the past few years. At least 18 different viruses have been detected in honeybees so far. Although usually not associated with clinical symptoms, viruses in certain cases may cause serious or even lethal disease in individual bees or the collapse of entire colonies (1). Infestation with the ectoparasitic mite Varroa destructor is the major predisposing factor (6, 14); however, a variety of other weakening circumstances may play a role in clinical manifestation of bee virus infections (e.g., Nosema apis infestation, intoxications, environmental pollution, and cold weather) (1).
In practical terms, six viruses are considered to be able to cause severe disease in honeybees, and hence they are most important in beekeeping. These are sacbrood virus (SBV), chronic bee paralysis virus (CBPV), black queen cell virus (BQCV), deformed wing virus (DWV), acute bee paralysis virus (ABPV), and Kashmir bee virus (KBV).
SBV primarily affects the brood of the honeybee and results in larval death (17). Infected larvae fail to pupate and ecdysial fluid aggregates around the integument, forming the “sac” for which the disease is named. Larvae change in color from a pearly white to pale yellow; after death, they dry out and change to a dark brown ship-shaped scab. Infection of adult bees is possible; the viruses are able to propagate in them, but the bees remain apparently healthy. Sacbrood virus appears mainly in spring, when the brood season begins and large numbers of infected young adults are present (17).
CBPV is transmitted through food or wounds. Two manifestations of the infection have been noted, as follows. (i) The bees are unable to fly, they tremble and crawl, and the wings are asymmetrically outspread. They often suffer from dysentery and die within a few days. (ii) The bees look black because of hair loss. The guard bees do not recognize them and dismiss them because of their altered look. Although in some cases up to 30% of worker bees are affected, CBPV infection sometimes remains undetected, and the colonies usually recover spontaneously from the disease (2).
BQCV is common in adult bees; however, it clinically affects mainly prepupae or pupae of the queen, especially in spring and early summer (13). The symptoms are similar to those of sacbrood. Infected queen pupae die and darken, and the cell walls get black.
DWV is mostly detected in Varroa-infected bees (8). The virus propagates slowly, and pupae infected at the white-eye stage of development may have malformed wings. The majority of infected bees do not show any symptoms.
ABPV commonly appears in apparently healthy bees; however, it has been presumed that this virus plays a role in cases of sudden collapse of honeybee colonies infested with V. destructor. Due to the spread of the varroa mite in Europe during the last decades, ABPV has gained more and more importance (5). On one hand, the mite is a possible vector for the virus; on the other hand, it weakens the bees and activates viral infections (14).
KBV affects brood and adult bees. Adult bees usually die within a few days after infection, but larvae may survive following ingestion of the virus, and some of them become inapparently infected adults. KBV is closely related to ABPV; however, it has been reported in Europe only rarely (1, 18).
Due to difficulties with the classical diagnostic methods in bee virology (i.e., absence of bee tissue cultures, dependence on the season to obtain pupae for experimental infection, or requirement of type-specific serum for agar gel immunodiffusion tests), scientific interest has turned towards molecular techniques. Since complete or at least partial genome sequence information on the aforementioned viruses is available in gene bank databases, diagnostic methods based on reverse-transcription PCR (RT-PCR) have been developed for the detection of viral RNA in honeybee samples (4, 7, 11, 15, 19, 20).
In Austria, the presence of two honeybee viruses, SBV and ABPV, has been reported so far (3, 11). Recently, beekeepers have more frequently observed clinical symptoms in honeybee colonies such as depopulation, morphological changes (dark color and deformed wings), central nervous system symptoms, or the sudden collapse of entire colonies. Besides various levels of varroa infestation, other pathogenic microbes or parasites were rarely detected in these samples, and noninfectious effects such as poisoning or bad weather were usually not recorded in those cases. Thus, virus infections were suspected to cause or contribute to the symptoms observed.
The aim of this study was to survey the occurrence and frequency of SBV, CBPV, BQCV, DWV, ABPV, and KBV in diseased honeybee colonies in different federal states of Austria and to compare these results to a limited extent with results obtained with healthy colonies of the same apiaries, as well as with data collected in bee samples originating from diseased colonies in selected surrounding European countries.
MATERIALS AND METHODS
Samples.
Beekeepers from all nine federal states of Austria submitted bee samples from colonies suffering from symptoms of depopulation, sudden collapse, paralysis or dark color, and varroa infestation. Approximately 100 adult dead worker bees were sent in each case, but occasionally we received broods from certain colonies as well. Altogether, samples from 90 honeybee colonies were collected in different Austrian apiaries and submitted for analysis between January 2003 and January 2004; the majority of them originated from the largest Austrian federal state of Lower Austria (31 samples), followed by Vienna (12 samples), Styria (9), Vorarlberg (7), Tyrol (8), Carinthia (7), Burgenland (6), Salzburg (5), and Upper Austria (5). To compare the virus loads in diseased and apparently healthy colonies, another 15 bee samples were obtained from healthy colonies of selected apiaries.
For a limited comparison with some surrounding countries, we also analyzed dead bees originating from diseased colonies from Poland (12 samples, kindly provided by Gražyna Topolska, Department of Infectious Diseases, Microbiology, and Parasitology, Faculty of Veterinary Medicine, Warsaw Agricultural University), Germany (5 samples, courtesy of Wolfgang Ritter, Department of Bee Pathology, Institute of Animal Health, Freiburg), Hungary (5 samples), and Slovenia (4 samples, kindly provided by Aleš Gregorc, Veterinary Faculty, University of Ljubljana).
The 131 honeybee specimens were stored at −20°C until processed.
Parasitological investigations.
From every sample, 60 bees were investigated for Nosema apis spores and Malpighamoeba mellifica cysts. The abdomens were separated from the thoraces; they were crushed and homogenized in 3 ml of water. Three drops (∼100 μl) of the suspension was placed onto a slide, covered by a slip, and examined under a light microscope, initially at a magnification of ×200, followed by a magnification of ×400 (the procedure was carried out as described in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, Nosemosis of Bees [http://www.oie.int/eng/normes/mmanual/A_00123.htm]). To detect infestation with the tracheal mite Acarapis woodi, the thoraces of 50 bees from each sample were dissected. The heads and forelegs were removed, and the thoraces were cut in front of the middle pair of legs and at the base of the forewings. These thin disks were placed into glass vials containing 8% KOH solution and heated in a boiling water bath for approximately 20 min until the muscle tissues were macerated. After heat treatment, the exposed first pair of thoracic trachea was examined under a dissecting microscope (magnification, ×20 to 40) (Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, Acariosis of Bees [http://www.oie.int/eng/normes/mmanual/A_00120.htm]). In case suspicious color changes were found in the trachea, the affected trachea was removed from the thorax and examined at a magnification of ×200 under a light microscope to detect infestation with Acarapis woodi.
All bee samples were visually examined for Varroa destructor mites, and some beekeepers gave additional information on the level of mite infestation in the sampled colonies and on treatments.
Virological investigations.
The presence of bee viruses was demonstrated by amplification of virus-specific nucleic acid by employing RT-PCR methods. After adults or larvae collected from the same colony were pooled, bees were homogenized in ceramic mortars with sterile sand and suspended in diethylpyrocarbonate-treated water. The homogenates were centrifuged at 20,000 × g for 1 min, and 140 μl of the supernatant was used for RNA extraction employing the QIAmp Viral RNA Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Oligonucleotide primers were designed, based on the genome sequences of SBV, CBPV, BQCV, DWV, ABPV, and KBV by using the Primer Designer 4 program for Windows 95, version 5.20 (Scientific & Educational Software). The sequences, orientations, and locations of the six primer pairs are shown in Table 1. Viral RNA was reverse transcribed and amplified by a continuous RT-PCR method with the One Step RT-PCR kit (QIAGEN, Germany), following the manufacturer's recommendations. Amplifications were performed in a GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer). The reverse transcription at 50°C for 30 min was followed by a denaturation and polymerase activation step at 95°C for 15 min and by 40 cycles of PCR, each consisting of 30 s at 94°C, 50 s at 55°C, and 1 min at 72°C. Reactions were completed by a final elongation step for 7 min at 72°C. The PCR products were electrophoresed in a 1.2% Tris-acetate-EDTA-agarose gel and stained with ethidium bromide. Bands were photographed under UV light with Kodak Digital Science 1D software. Fragment sizes were determined with reference to a 100-bp ladder (Promega).
TABLE 1.
Oligonucleotide primer pairs employed in RT-PCR assaysa
| Primer name | Primer sequence (5′-3′) | Primer position on the genome | Length of amplified product |
|---|---|---|---|
| ABPV 23f | GTG CTA TCT TGG AAT ACT AC | 7928-7947 | 618 |
| ABPV 24r | AAG GYT TAG GTT CTA CTA CT | 8527-8546 | |
| BQCV 3f | AGT AGT TGC GAT GTA CTT CC | 252-277 | 472 |
| BQCV 4r | CTT AGT CTT ACT CGC CAC TT | 710-729 | |
| CBPV 111f | TGT CGA ACT GAG GAT CTT AC | 111-130 | 315 |
| CBPV 426r | GAC CTG ATT AAC GAC GTT AG | 407-426 | |
| DWV 2345f | ATT GTG CCA GAT TGG ACT AC | 2345-2364 | 434 |
| DWV 2779r | AGA TGC AAT GGA GGA TAC AG | 2760-2997 | |
| KBV 5406f | GAT GAA CGT CGA CCT ATT GA | 5406-5425 | 395 |
| KBV 5800r | TGT GGG TTG GCT ATG AGT CA | 5781-5800 | |
| SBV 1f | ACC AAC CGA TTC CTC AGT AG | 221-240 | 487 |
| SBV 2r | CCT TGG AAC TCT GCT GTG TA | 689-708 |
Nucleotide positions refer to the published sequences of ABPV (GenBank accession number AF150629), BQCV (accession number AF125252), CBPV (accession number AF375659), DWV (accession number AJ489744), KBV (accession number AY275710), and SBV (accession number AF092924). f, forward primer; r, reverse primer.
Tenfold serial dilutions of the extracted RNA of selected positive samples were tested with the different primer pairs by RT-PCR to determine the sensitivity of the assays. The specificity and sensitivity of the RT-PCR assay used for the detection of KBV were determined on KBV-positive bee samples kindly provided by Reinhold Siede (Hessisches Dienstleistungszentrum für Landwirtschaft, Gartenbau und Naturschutz, Bieneninstitut Kirchhain, Germany).
In addition, the relative virus load (relative amount of viral RNAs) was comparatively evaluated with 15 selected bee samples from diseased colonies and with 15 selected bee specimens from apparently healthy colonies originating from the same apiaries.
RESULTS
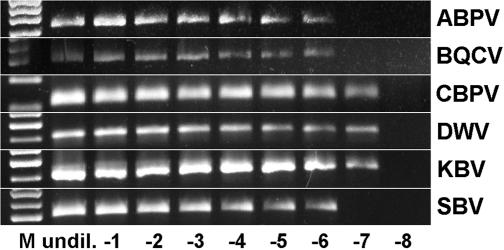
A total of 131 honeybee samples originating from all nine Austrian federal states, as well as from surrounding countries, were investigated by RT-PCR for the presence of the six most important honeybee viruses. For SBV, ABPV, and KBV, the specificity of the reactions had been evaluated in previous studies (3, 11, 19), while amplification products of BQCV, DWV, and CBPV were sequenced in this study to verify the selectivity of the primers (data not shown). All reactions were found to be specific. The RT-PCR assays were able to detect viral RNAs at dilutions of up to 10−6 (ABPV, BQCV, and SBV) and 10−7 (CBPV, DWV, and KBV) (Fig. 1).
FIG. 1.
Determination of the sensitivities of the RT-PCR assays described and employed in this study. M, molecular weight marker; undil. to −8, tenfold dilution steps of positive reference samples, from undiluted to 10−8.
Altogether, five different bee viruses were detected in samples of sick Austrian honeybees. DWV was found to be the most prevalent, present in 91% of samples and detected in samples from every federal state. ABPV exhibited a prevalence of 68%, and it was also present in samples from all Austrian federal states. SBV was detected in 49% of samples; the samples from Upper Austria and Tyrol did not contain this virus. The average prevalence of BQCV was 30%. However, it showed a great diversity in distribution; while 67% of the investigated samples from Vienna were positive for BQCV and a high percentage of samples from Carinthia, Vorarlberg, Styria, and Lower Austria also proved positive for BQCV, this virus was not detected in bees from the three other federal states (Burgenland, Upper Austria, and Salzburg). CBPV was, with the exception of KBV, found to be the least prevalent of the investigated viruses present in Austria; it was identified in only 10% of the samples, and the majority of CBPV-positive bees were found in Lower Austria (six samples) and in Vorarlberg (two samples), and one positive sample was detected in Styria. KBV nucleic acid was not present in any of the Austrian bee samples investigated.
Because relatively small numbers of samples were received from other European countries, only the occurrence of the studied viruses is mentioned, but not their prevalence. In specimens from Poland, nucleic acid from DWV, ABPV, SBV, BQCV, and CBPV was identified. Bees from Germany contained DWV, ABPV, SBV, and BQCV. The Hungarian samples were positive for DWV, ABPV, and BQCV, while the Slovenian specimens exhibited DWV, ABPV, and SBV. KBV was not detected in any of the samples originating from the above-mentioned countries.
Within the Austrian samples, only one did not contain any of the investigated viruses. In this case, intoxication of the colony was suspected by the beekeeper. On the other hand, simultaneous multiple infections have been detected in several colonies (Table 2). Bee samples containing two viruses were found in 50% of the cases, coinfections with three viruses were found in 27% of the samples, and four viruses were present at the same time in 13% of cases. One specimen contained all investigated viruses except KBV.
TABLE 2.
Frequencies of simultaneous virus infections in honeybee samples
| No. of viruses | Type of infection | No. of samples | Percentage | Origina |
|---|---|---|---|---|
| 0 | 1 | 1 | St | |
| 1 | DWV | 3 | 3 | Bu, Vi, Ty |
| SBV | 3 | 3 | LA, Vi, Vo | |
| 2 | DWV, ABPV | 32 | 35 | Bu 3, Ty 6, LA 8, UA 5, St 4, Sa 4, Ca 1, Vo 1 |
| DWV, BQCV | 1 | 1 | Vo | |
| DWV, SBV | 8 | 9 | LA 4, Vi 3, Vo 1 | |
| DWV, CBPV | 1 | 1 | LA | |
| ABPV, BQCV | 1 | 1 | LA | |
| BQCV, SBV | 3 | 3 | Vi, St 2 | |
| 3 | DWV, ABPV, BQCV | 5 | 6 | Ca 3, LA, Ty |
| DWV, ABPV, SBV | 11 | 12 | Bu 2, Ca 2, LA 5, Sa, St | |
| DWV, BQCV, SBV | 8 | 9 | Vi 7, Vo | |
| 4 | DWV, ABPV, BQCV, SBV | 6 | 7 | Vi, Ca, LA 4 |
| DWV, ABPV, BQCV, CBPV | 2 | 2 | Vo, St | |
| DWV, ABPV, SBV, CBPV | 4 | 4 | LA 4 | |
| DWV, BQCV, SBV, CBPV | 1 | 1 | LA | |
| 5 | DWV, ABPV, BQCV, SBV, CBPV | 1 | 1 | Vo |
Abbreviations: Bu, Burgenland; Ca, Carinthia; LA, Lower Austria; Sa, Salzburg; St, Styria; Ty, Tyrol; UA, Upper Austria; Vi, Vienna; Vo, Vorarlberg.
Another aspect of the investigation was the presence of parasites and viruses in the same samples. According to the reports of the beekeepers, all investigated colonies were infested with varroa mites. In 18% of the specimens actually analyzed, both V. destructor and viruses were found. The main reason for the rather low prevalence of varroa in the analyzed samples is that, following the death of bees, the varroa mites usually leave the bee; in some cases, the colonies had been recently treated by an acaricide, which also lowered the number of mites (nonetheless, the bees died). The samples containing V. destructor were mostly infected with ABPV, BQCV, and DWV. N. apis, together with viruses, was found in 12% of the samples; usually ABPV, BQCV, SBV, and DWV were associated with nosema infestation. Malpighamoeba mellifica was identified in only one sample, which was also positive for SBV, BQCV, and DWV.
The honeybee samples investigated in this study were collected from problematic colonies suffering from unspecific symptoms such as weakness, depopulation, or sudden collapse of the colony. Typical symptoms of CBPV (black coloring and disorientation) were observed in some apiaries; CBPV was demonstrated in each of these samples. During the course of the year, different disease symptoms were observed in different seasons: depopulation was mostly noticed in March, sudden collapse of colonies was more frequently seen during autumn and winter, while black coloring and disorientation of the bees occurred mainly during July and August. On the other hand, no significant seasonal variance was identified in the detection frequency of the investigated viruses.
The relative virus loads of samples from diseased colonies were compared to those from apparently healthy colonies of the same apiaries. The samples from diseased colonies contained 20 times more ABPV and/or BQCV RNA than bees from asymptomatic colonies. The mean viral loads of DWV and SBV were 126 times and 10 times higher in diseased colonies than in apparently healthy colonies, respectively. CBPV nucleic acid was not detected in any of the samples obtained from symptomless colonies.
DISCUSSION
Although viral infections of the honeybee have been known for a long time, their significance is still not fully appreciated. Before the emergence and spread of V. destructor in Europe, clinical manifestations of some bee virus infections were sporadically observed; however, at that time they did not lead to significant economic losses (1). The situation has dramatically changed following varroa mite infestation of bee colonies. Besides the direct damage due to the parasitism of the mite, V. destructor also acts as a vector and activator of other pathogens, viruses in particular. ABPV, DWV, and KBV have already been detected in V. destructor; presumably, SBV can also be transmitted by the mite (21). Furthermore, besides the depopulation and collapse of varroa-infested colonies (also called bee parasitic mite syndrome), the direct effect of virus infections is supposed to be the most important factor (12).
In this survey, we investigated the occurrence, prevalence, and distribution pattern of six honeybee viruses in bee samples collected from sick colonies of selected Austrian apiaries. The samples were sent to our Institute for causal diagnosis of health problems in affected colonies, in which the symptoms could not be explained by mere parasite infestation, bacterial or fungal infections, or noninfectious reasons. Therefore, viral infections were presumed to contribute significantly to the observed diseases. In our opinion, analysis of problematic colonies gives a more informative picture of the real pathological impact of viruses, although by this approach the sample collection cannot be planned beforehand, and the number of samples may in some cases be not high enough for statistical analysis. Several studies established that bee viruses are widespread and often cause inapparent, multiple infections (1, 2, 4, 6, 10, 14, 16) in seemingly healthy bee colonies. A comprehensive survey for the aforementioned six viruses has been performed recently in France, involving randomly collected samples from apparently healthy colonies (21). The authors found high prevalences of the viruses without any clear geographic correlation, but seasonal variations were reported. In our study, besides a practical approach for the application of RT-PCR-based diagnostic methods on affected colonies of problematic apiaries, we attempted to provide data for the estimation of a connection between the presence of viruses and manifest diseases of bee colonies. Although diseased honeybee colonies may represent only the “tip of the iceberg” of bee virus infections, the appearance of clinical symptoms is a deliberate basis for sample selection, as we wanted to evaluate the practical (clinical) relevance of bee viruses.
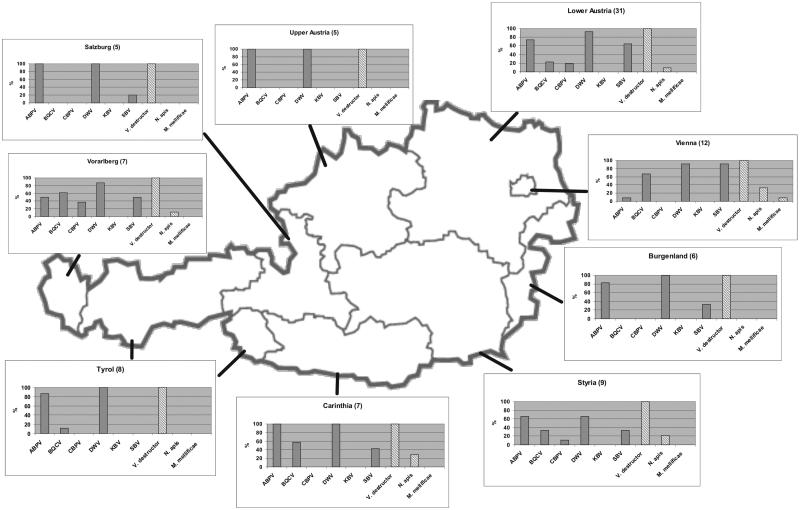
In Austria, the spread of viruses in different regions may also be influenced by Austrian geography: the eastern federal states (Lower Austria, Burgenland, and Vienna) show more plain landscape and continental climate. In these areas, the distribution of the apiaries is relatively even. Thus, the different bee populations have several possibilities for contact or mixing, and hence the spread of infectious agents could be easier. The western federal states are located in the mountains of the Alps, with a cooler alpine climate in higher altitudes; there, bees are mainly kept in valleys. In these areas, the density of bee colonies is lower than that in plain areas, and because the high ridges separate the populations, a direct spread of viruses by flying bees between apiaries is not that effective. For some bee viruses, interesting differences in their occurrence were identified in the different geographic regions (Fig. 2).
FIG. 2.
Distribution pattern of six investigated viruses and three parasites in honeybee colonies from the nine different federal states of Austria. Sample numbers are indicated in brackets.
DWV is considered to be the most widespread bee virus in Europe. It was also found in high percentages in all Austrian federal states. Since its emergence in 1983, V. destructor quickly spread in the Austrian apiaries; thus, to date essentially all apiaries are infested with the mite, although the application of an intensive acaricide treatment usually reduces the direct damage by the parasites to the bee colonies. The presence of DWV is often associated with V. destructor infestation, and the role of the mite in transmission of the virus has already been experimentally demonstrated (8). DWV was also found to be the most prevalent virus in France and was detected in 97% of the apiaries (21). Therefore, it is not surprising that DWV is widespread in Austrian apiaries, too.
The second-most-prevalent virus identified in Austria was ABPV. It was also present in every federal state and usually in high percentages, except in Vienna, where only 8% of the samples were positive (compared to the average frequency of 69%). This virus is also widespread in Europe; 58% of the apiaries investigated in France and 67% of the apiaries investigated in Hungary were found to be infected with ABPV (4, 21). In colonies weakened by the varroa mite, cold weather, nutrition problems, or intoxication, ABPV is considered to be the main causative agent of depopulation and collapse of the colonies (14). Because the sampled colonies showed similar symptoms, our findings support the role of ABPV in the bee parasitic mite syndrome.
Considerable differences have been found in the occurrence and prevalence of BQCV in Austria. Sixty-nine percent of the samples from Vienna were positive for this virus, and it was also present in the federal states of Lower Austria, Styria, and Carinthia. On the other hand, BQCV was not present in the samples from the eastern federal state of Burgenland or in the specimens from two federal states in midwestern Austria (Upper Austria and Salzburg). In the samples from the westernmost federal state of Vorarlberg, however, BQCV was again present in more than half of the samples and in one specimen from Tyrol. It was previously reported that BQCV is usually associated with N. apis infestation (17). In this study, we found that the protozoan was also present in 78% of the BQCV-positive samples; 75% of the nosema-infested colonies were infected with BQCV.
The distribution pattern and frequency of SBV showed similarities to those of BQCV. It was highly prevalent in samples from Vienna (92%) and Lower Austria (65%), and it was present in Vorarlberg (57%), Carinthia (43%), and Styria (33%). In the federal state of Burgenland, SBV was found in two out of the six investigated samples, and in one out of five samples in Salzburg; however, in the bees from Upper Austria and Tyrol (similar to BQCV), this virus was not detected. In the case of BQCV and SBV, the high densities of bee colonies in certain geographic regions and the intensive trade and transport of bee colonies, queens, hives, or equipment should also be taken into consideration as possible important factors for transmission and spread of these viruses between apiaries. In France, where the intensity of beekeeping and trade is high, SBV and BQCV were found to be the second-most-prevalent bee viruses, detected in 86% of the investigated apiaries (21).
CBPV was detected only in three federal states of Austria. In Lower Austria, 19% of the samples were positive for the virus, while only sporadic cases were found in Styria and Vorarlberg. It is an important observation that the CBPV-positive colonies consistently showed disease symptoms. In the French survey, CBPV was the least frequent virus in the investigated samples after KBV (21), but in another survey of diseased bee samples collected in apiaries in the south of France (16), the frequency of CBPV infection was two times higher than in symptomless bees.
KBV was not present in any of the Austrian samples. This virus is widespread in the United States and Australia, but so far it has rarely been detected in Europe (1). The emergence of KBV in Hesse, Germany, was reported recently (18), and the virus was found in 2002 in several colonies in France (10, 21). Since ABPV and KBV share a high level of genetic similarity and are also antigenically related, it is supposed that the two viruses originate from a common ancestor and evolved independently in secluded geographic regions (9). The widespread distribution of ABPV and the lack of KBV in Austria also support this hypothesis. The recent emergence of KBV strains in certain areas of Europe is most probably the result of a recent introduction of the virus to these regions by importation from overseas.
The prevalence and distribution pattern of the five detected viruses in the different geographic regions show interesting tendencies: while in Vienna, Lower Austria, Styria, Carinthia, and Vorarlberg most of the five viruses have been detected, in Upper Austria, Salzburg, Tyrol, and Burgenland practically only DWV and ABPV were present, but in general with higher prevalences than the previously mentioned federal states.
The presence of bee viruses in Austria was compared to that of four surrounding central European countries. Unfortunately, only small sample numbers were received from abroad; therefore, the negative results for some viruses are less informative in these cases. While the presence of DWV and ABPV was detected in the majority of samples and in each country, the distribution of BQCV and SBV again exhibited differences. In the samples from Germany and Poland, both BQCV and SBV were detected; however, in the Slovenian samples only SBV was identified, and in the Hungarian samples only BQCV was found. (Very recent investigations of a larger number of samples detected SBV in Hungary as well, but only at a very low prevalence). Only one CBPV-positive sample was found among the Polish specimens, but when the prevalence of this virus in other countries was as low as it is in Austria, then the number of samples from the surrounding countries was too small to comment on the CBPV prevalence. KBV was not detected in the surrounding countries either, supporting the theory that this is principally an exotic bee virus in central Europe.
The most important aspect of this investigation is the correlation between the presence of bee viruses, parasites, and diseases in bee colonies. In this study, besides viruses, V. destructor, N. apis, and M. mellifica were detected in the tested bee samples. Although V. destructor is probably the most important predisposing factor for virus diseases, its direct effect is hard to demonstrate, because practically all Austrian apiaries are infested with the mite; acaricide treatment has its limitations, and treated colonies quickly become reinfested with mites. Tentcheva et al. (21) reported one geographically isolated apiary (Ouessant Island, France), which was free of V. destructor, and none of the investigated viruses could be detected. In 46% of the samples, sudden collapse, depopulation, or weak colonies were observed. In samples from colonies suffering from these unspecific symptoms, simultaneous infection of DWV and ABPV was identified most frequently (37%), followed by DWV, ABPV, and SBV (13%); a coinfection of DWV, ABPV, and BQCV was found in only 5% of cases. SBV was often associated with DWV (15%) and less frequently with BQCV (5%), and the three viruses were found together in 7% of the colonies exhibiting unspecific symptoms. While single infections of DWV, SBV, and ABPV were detected in 5, 5, and 1% of colonies, respectively, a coinfection of DWV, ABPV, SBV, and BQCV was identified in 7% of the samples.
A limited comparison of the relative virus loads of honeybees originating from diseased and apparently healthy colonies revealed that pools of bees with symptoms contained at least 10 times more viral nucleic acid than bees from asymptomatic colonies. A more appropriate quantitative method (i.e., real-time PCR) could provide even more detailed data on the connection between virus content and disease symptoms in bee infections. These questions will be addressed in further investigations.
To summarize, we have looked into the occurrence of the six most important honeybee viruses in diseased bee colonies and identified remarkable differences in the distribution pattern of the viruses in the different geographic regions of Austria. These differences may be partly explained by differences in climate, landscape, and density of the bee populations; however, trade and exchange of infected animals, contaminated equipment, and bee products between apiaries, regions, or even countries may be of greater importance in the spread of viruses. This suggestion is also supported by the fact that in those countries that recently joined the European Union and previously had less intensive international trade activity in bees and bee products, the prevalence of viruses such as SBV and BQCV was lower than in “old” European Union member states. Therefore, virological and parasitological investigations should be considered by beekeepers before they purchase and import possibly infected bees and bee products. The RT-PCR techniques described in this paper proved to be appropriate methods for this kind of investigation.
Acknowledgments
This study was partially supported by grants OTKA F 043155 and D 048647.
Also, we thank the reviewers for their valuable suggestions.
REFERENCES
- 1.Allen, M., and B. V. Ball. 1996. The incidence and world distribution of honey bee viruses. Bee World 77:141-162. [Google Scholar]
- 2.Bailey, L. 1967. The incidence of virus diseases in the honey bee. Ann. Appl. Biol. 60:43-48. [DOI] [PubMed] [Google Scholar]
- 3.Bakonyi, T., E. Grabensteiner, J. Kolodziejek, M. Rusvai, G. Topolska, W. Ritter, and N. Nowotny. 2002. Phylogenetic analysis of acute bee paralysis virus strains. Appl. Environ. Microbiol. 68:6446-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakonyi, T., R. Farkas, A. Szendroi, M. Dobos-Kovacs, and M. Rusvai. 2002. Detection of acute bee paralysis virus by RT-PCR in honey bee and Varroa destructor field samples: screening of representative Hungarian apiaries. Apidologie 33:63-74. [Google Scholar]
- 5.Ball, B. V. 1997. Varroa and viruses, p. 11-15. In P. Munn and R. Jones (ed.), Varroa! Fight the mite. International Bee Research Association, Cardiff, Wales.
- 6.Ball, B. V., and M. Allen. 1988. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 113:237-244. [Google Scholar]
- 7.Benjeddou, M., N. Leat, M. Allsopp, and S. Davison. 2001. Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase PCR. Appl. Environ. Microbiol. 67:2384-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invert. Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 9.de Miranda, J. R., M. Drebot, S. Tyler, M. Shen, C. E. Cameron, D. B. Stoltz, and S. M. Camazine. 2004. Complete nucleotide sequence of Kashmir bee virus and comparison with acute bee paralysis virus. J. Gen. Virol. 85:2263-2270. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier, L., D. Tentcheva, F. Cousserans, M. E. Colin, and M. Bergoin. 2004. A survey of six bee viruses in France using quantitative PCR, p. 97-98. In Abstracts of the First European Conference of Apidology, 19-24 September 2004, Udine, Italy.
- 11.Grabensteiner, E., W. Ritter, M. J. Carter, S. Davison, H. Pechhacker, J. Kolodziejek, O. Boecking, I. Derakshifar, R. Moosbeckhofer, E. Licek, and N. Nowotny. 2001. Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 8:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung, A. C. F., H. Shimanuki, and D. A. Knox. 1996. The role of viruses in bee parasitic mite syndrome. Am. Bee J. 136:731-732. [Google Scholar]
- 13.Laidlaw, H. H. 1979. Contemporary queen rearing. Dadant and Sons, Hamilton, Ill.
- 14.Nordstom, S., I. Fries, A. Aarhus, H. Hansen, and S. Korpela. 1999. Virus infection in Nordic honey bee colonies with no, low or severe Varroa jacobsoni infestations. Apidologie 30:475-484. [Google Scholar]
- 15.Ribière, M., C. Triboulot, L. Mathieu, C. Aurières, J. P. Faucon, and M. Pépin. 2002. Molecular diagnostic of chronic bee paralysis virus infection. Apidologie 33:339-351. [Google Scholar]
- 16.Ribière, M., J. P. Faucon, and M. Pépin. 2000. Detection of chronic honey bee (Apis mellifera L.) paralysis virus infection: application to a field survey. Apidologie 31:567-577. [Google Scholar]
- 17.Ritter, W. 1996. Diagnostik und Bekämpfung von Bienenkrankheiten, p. 104-114. Gustav Fischer Verlag Jena, Stuttgart, Germany.
- 18.Siede, R., and R. Buchler. 2004. First detection of Kashmir bee virus in Hesse, Germany. Berl. Munch. Tierarztl. Wochenschr. 117:12-15. [PubMed] [Google Scholar]
- 19.Stoltz, D., X. R. Shen, C. Boggis, and G. Sisson. 1995. Molecular diagnosis of Kashmir bee virus infection. J. Apic. Res. 34:153-165. [Google Scholar]
- 20.Tentcheva, D., L. Gauthier, S. Jouve, L. Canabady-Rochelle, B. Dainat, F. Cousserans, M. E. Colin, B. V. Ball, and M. Bergoin. 2004. Polymerase chain reaction detection of deformed wing virus (DWV) in Apis mellifera and Varroa destructor. Apidologie 35:431-439. [Google Scholar]
- 21.Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M. E. Colin, and M. Bergoin. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 70:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]