Abstract
Tuber spp. are ectomycorrhizal ascomycetes that produce ascocarps known as truffles. Basic aspects of Tuber biology have yet to be fully elucidated. In particular, there are conflicting hypotheses concerning the mating system and the ploidy level of the mycorrhizal and truffle hyphae. We used polymorphic microsatellites to compare the allelic configurations of asci with those from the network of the surrounding hyphae in single Tuber magnatum truffles. We then used these truffles to inoculate host plants and evaluated the microsatellite configurations of the resulting mycorrhizal root tips. These analyses provide direct evidence that T. magnatum outcrosses and that its life cycle is predominantly haploid. In addition to its scientific significance, this basic understanding of the T. magnatum life cycle may have practical importance in developing strategies to obtain and select nursery-produced mycorrhizal plants as well as in the management of artificial plantations of this and other Tuber spp.
Tuber spp. are Ascomycetes fungi that establish an ectomycorrhizal symbiosis with trees and shrubs. As a result of this mutualistic symbiosis, ascocarps known as truffles are produced. Some Tuber spp. produce edible truffles that, given their distinctive taste and aroma, are highly valued by gourmets. Research on these fungi has focused on promoting the cultivation of these fungi to meet increasing worldwide demand and to provide replacements for the catastrophic decline in their natural production (10). Truffle cultivation is no longer an agronomic practice confined to Europe, where the most profitable species are endemic. Truffle plantations have been established in various countries worldwide, including New Zealand and Israel. Nevertheless, the understanding of many basic aspects of truffle biology is still in its infancy, and the ecological requirements for some of these species are still not known. One of the most elusive goals has been discerning the reproductive system of Tuber spp. The reproductive structures of these species in pure cultures have not been reported, and axenic spore germination remains an unresolved problem (26). Furthermore, the mating-type genes have never been characterized in truffles.
Molecular markers are being developed to type each truffle species to overcome the difficulty of identifying these species solely on their morphological traits (1, 8, 11, 17, 18, 19, 22, 23). By combining molecular markers with an appropriate sampling strategy, we may be able to critically evaluate the truffle reproductive system and life cycle even without reproducing the entire life cycle in the lab. To date, Tuber melanosporum Vittad. and Tuber magnatum Pico, the finest black and white truffle species, respectively, have been regarded as selfing species. When codominant markers were evaluated, heterozygous ascocarps were not detected (2, 3, 7, 15, 16). These studies proceed from the assumption that the ascocarps are diploid (dikaryotic) structures.
We recently used simple sequence repeat (SSR) markers and a large survey of natural populations to show that extensive genetic exchange occurs within T. magnatum populations, which suggests that this truffle outcrosses (24). We interpreted the lack of heterozygotes to mean that haploid, maternal tissue is the dominant component of truffle ascocarps, while paternal DNA is not easily recoverable. Such partitioning of genetic material typifies many ascomycetes (4, 12).
Our objectives in this study were to use SSR markers (i) to provide direct genetic evidence of outcrossing in T. magnatum and (ii) to determine the ploidy of the T. magnatum mycorrhizae, gleba, and spores. We tested the hypothesis that truffles are primarily haploid and reproduce by outcrossing. Our results led us to significantly reinterpret the T. magnatum life cycle, which also has implications for other Tuber spp., and to urge further reconsideration of existing data and strategies concerning truffle population genetic studies and growth management.
MATERIALS AND METHODS
Sample source.
Ten fresh mature ascocarps of T. magnatum, each characterized by a gray-brown gleba, were used for DNA isolation, ascus purification, and plant inoculation (Table 1). Asci were isolated by grinding small pieces (10 to 20 mg) of a fresh ascocarp in a ceramic mortar in sterile distilled water. Two hundred microliters of the ground material was layered over a 10 to 50% (wt/vol) sucrose density gradient (total volume, 1.5 ml) in a 2-ml centrifuge tube and centrifuged (800 × g, 10 min, room temperature). The band containing asci, which was free of hyphal fragments, was recovered from the gradient with a Pasteur pipette, and the asci were washed twice in sterile distilled water and used immediately for DNA isolation.
TABLE 1.
SSR patterns of the gleba and asci from single ascocarps
| Sample name | SSR allele size (bp) at indicated locusd
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MA12
|
MA19
|
MA5
|
MA4
|
MA2
|
||||||
| g | a | g | a | g | a | g | a | g | a | |
| ma453a | 130 | 130/134 | 109 | 109/121 | 201 | 201 | 177 | 177 | 168 | 168 |
| ma455a | 134 | 134 | 121 | 121 | 209 | 209 | 157 | 157 | 168 | 168 |
| ma462a | 130 | 130/132 | 121 | 121 | 207 | 205/207 | 159 | 159 | 168 | 168 |
| ma490b | 130 | 130/132 | 109 | 109/121 | 207 | 203/207 | 177 | 177 | 168 | 168 |
| ma493c | 132 | 132 | 109 | 109 | 203 | 203/207 | 177 | 177 | 168 | 168 |
| ma497c | 130 | 130 | 121 | 121 | 203 | 203 | 159 | 159 | 168 | 168 |
| ma500c | 130 | 130 | 121 | 121 | 203 | 203 | 159 | 159/177 | 162 | 162/168 |
| ma501c | 130 | 130/132 | 121 | 109/121 | 203 | 203 | 159 | 159 | 168 | 168 |
| ma508c | 130 | 130 | 121 | 121 | 203 | 203 | 159 | 159 | 162 | 162/168 |
| ma510c | 130 | 130 | 121 | 121 | 203 | 203 | 159 | 159 | 168 | 168 |
Ascocarps sampled in C. di Castello (Umbria, Italy).
Ascocarps sampled in Florence (Tuscany, Italy).
Ascocarps sampled in Valtopina (Umbria, Italy).
g, gleba; a, asci.
The ma453 and ma455 ascocarps were used to inoculate two sets of 8 to 10 Quercus pubescens Willd. plantlets grown under semisterile conditions, as previously described (22). The inoculated plants were grown spaced in the greenhouse under ambient environmental conditions. After ∼6 months, individual T. magnatum ectomycorrhizal root tips were collected and processed for DNA isolation or frozen in liquid N2 and stored in microcentrifuge tubes at −80°C. The position of each mycorrhiza on the root branch was recorded.
DNA isolation.
DNAs were isolated from the following different sources: small portions of internal gleba, pools of purified asci collected from single ascocarps, and a number of single mycorrhizae collected from plants inoculated with genetically typed ascocarps. DNA was isolated from small pieces of gleba basically as described by Paolocci et al. (18), i.e., by grinding the truffle either in a ceramic mortar with liquid N2 or in a microcentrifuge tube with a sterile glass pestle.
DNAs were extracted from pools of purified asci (about 200) with a FastPrep apparatus (Q-BIOgene, Carlsbad, CA) according to the manufacturer's instructions. Spores were disrupted with ceramic spheres (diameter, 1.4 mm) in the presence of 300 μl of NTE buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA). Nucleic acids were precipitated with cold isopropanol and resuspended in sterile distilled water. DNAs from individual mycorrhizae were isolated according to the method of Paolocci et al. (18).
PCR amplification.
The molecular characterization of ascocarps, asci, and mycorrhizae was performed with T. magnatum species-specific internal transcribed spacer (ITS) and universal ITS1/ITS4 primer pairs as previously described (22). The resulting DNA fragments were sequenced directly and analyzed as reported previously (22).
Primer pairs specific to the T. magnatum SSR loci MA2, MA4, MA5, MA7, MA12, and MA19 (25) were used to analyze the gleba and the purified asci from the T. magnatum ascocarp (Table 1). The primers specific to the MA12, MA19, and MA4 loci were also used to amplify DNAs from individual ectomycorrhizal root tips. All of the resulting SSR DNA fragments were separated in an ABI 310 genetic analyzer and analyzed with Genescan and Genotyper software (Applied Biosystems, Foster City, CA).
RESULTS
DNA isolation from asci and the gleba.
Asci are surrounded by a network of hyphae, arranged as a pseudoparenchyma, in truffle ascocarps. In the white truffles we evaluated, the asci contained primarily two or three pale yellow ascospores or, more rarely, one or four ascospores. Asci and ascospores were largely undamaged during the gleba DNA isolation process, regardless of the tissue grinding procedures followed (data not shown). Even if a few ascospores were broken, their contribution in terms of relative amount of DNA was expected to be very low with respect to the DNA derived from the hyphae that composed the ascocarp. Thus, screening aimed at genetically differentiating the spores from the surrounding hyphae could be difficult even if a PCR approach is used.
To analyze DNA from only the spores/asci, the glebae from mature T. magnatum ascocarps were ground and then centrifuged through a sucrose density gradient to selectively recover asci. Purified asci, based on microscopic observation, were layered on top of the sucrose gradient. A pool of purified asci was recovered from each of the 10 ascocarps (Table 1). DNAs from ∼200 asci were then isolated in the presence of ceramic beads with a FastPrep apparatus, which ensured that most of the spores were broken. The resulting DNAs sufficed for several PCRs.
Amplification and direct sequencing analysis of the DNAs isolated from the gleba and purified asci, with both T. magnatum ITS species-specific and universal ITS1/ITS4 primer pairs, confirmed the species identity and the absence of any other fungi.
SSR amplification from ascocarps and purified asci.
For the 10 ascocarps analyzed, the DNAs from the gleba produced a single peak (allele) for each of the six SSR loci, while 7/10 DNA samples from the purified asci and ascospores had two peaks (alleles) for one or more of the SSR loci (Table 1). Note that one of the two alleles found in the ascospores was also always found in the gleba, regardless of the locus analyzed (Table 1). Locus MA7 was monomorphic (allele 211), regardless of the tissue or ascocarp analyzed. Repeated DNA isolation and amplification with material from different portions of the gleba never resulted in multiple alleles/loci, confirming the results previously reported for a larger sample of T. magnatum ascocarps (25).
Plant inoculation and molecular characterization of ectomycorrhizae.
Ascospores from two of the ascocarps, ma453 and ma455, were used to inoculate separate lots of Quercus pubescens plantlets. ma453 was selected because its asci had multiple alleles, while the gleba and ascospore DNA from ma455 was monomorphic.
Successful plant inoculation was confirmed by microscopic observations of all the inoculated plantlets. Sixty-two mycorrhizal root tips from two plants inoculated with ma453 ascospores and 35 root tips from a plant inoculated with ma455 ascospores were collected and molecularly typed. The MA12 and MA19 primer pairs were selected because the loci were both polymorphic in the ma453 ascospores, while the MA4 primer pair amplification product was monomorphic.
Based on amplification with the T. magnatum ITS species-specific primers and the sequences of the DNA fragments resulting from amplification with the universal ITS1/ITS4 primers, the mycorrhizae produced were free of mycelia from other fungal species. The mycorrhizae from plants inoculated with asci from ma455 all had the same SSR pattern and only one allele per locus. This haplotype was the same as that of the gleba of ma455, as expected.
Mycorrhizae from plants inoculated with ascospores from ma453 were individually monomorphic, but collectively all of the alleles present in the ascospore pool were present among the progeny. All four possible genotypes were also represented among the progeny, with the genotype of the gleba (130/109, MA12/MA19) being the most common (46/62 ascospores) and that of the other imputed parent (134/121) being the least common (2/62 ascospores). The two recombinant types, 130/121 and 134/109, were also relatively rare (8/62 and 6/62 ascospores, respectively). Mycorrhizae with different haplotypes could be recovered from the same root branch.
DISCUSSION
The results from this study clearly support the hypothesis that T. magnatum outcrosses and that its mycorrhizae originate from primary monokaryotic mycelia. These genetically different mycelial strains can coexist on the same root branches, but probably not on the same mycorrhizal root tip.
The current model for the truffle life cycle has a very closed mating system, with homothallism or exclusive selfing as the precursor to sexual reproduction. This model was based on evidence that neither SSR, single-nucleotide polymorphism, nor allozyme markers were heterozygous in either T. magnatum or T. melanosporum truffles (2, 3, 7, 15, 16). However, linkage disequilibrium analyses of microsatellite loci showed that extensive gene flow occurs in T. magnatum both within and among geographically discrete populations (24). Extensive genetic exchange without the formation of detectable heterozygotes could occur but would be difficult to model.
The asci and, more importantly, the ascospores within them are largely undamaged by the protocols commonly adopted for isolating nucleic acids from Tuber spp. Thus, the DNAs extracted from the ascocarps are almost exclusively that of the maternal parent. If this maternal tissue is haploid, which is typical of many ascomycetes, then the lack of heterozygosity observed when whole truffles are ground reflects this haploid DNA content and not necessarily a homothallic or self-fertilizing lifestyle. In this model, gleba tissue should always be monomorphic when evaluated with codominant markers. Heterozygosity, however, is expected among the ascospores unless the fertilizing parent is the same as the maternal parent (homothallism or selfing) or the maternal and the fertilizing parents are closely related (inbred). The fact that only 7/10 of the truffles analyzed had progeny that were polymorphic for the SSR markers suggests that either inbreeding or selfing occurs to at least some degree. Our sampling of truffles for this study does not suffice to distinguish between these hypotheses since fungi that reproduce homothallically, e.g., Aspergillus nidulans and Gibberella zeae, can also outcross under both laboratory and field conditions (5).
We followed meiosis from one heterozygotic truffle and characterized 62 mycorrhizal root tips. If each of these root tips represents a single ascospore, then we recovered the four expected progeny classes (two parental and two recombinant). The relative frequencies of these classes should be equal if the two loci are unlinked, Mendelian segregation occurs, and there is no selection for the genotype that colonizes the root tips. The heterokaryotic truffle we analyzed yielded an excess of the maternal genotype and similar numbers of the fertilizing and two recombinant genotypes among the progeny. Thus, we think that the two loci are unlinked and that Mendelian segregation occurs and, for the moment, attribute the excess of the maternal genotype to selection for the ability to form mycorrhizae on the root tips or to foster growth in the soil or along the root (if not all of the root tips were colonized by mycelia from distinct ascospores). As a further hypothesis, we can envisage that the fragments of gleba present in the inocula also contribute to the synthesis of mycorrhizae.
Tuber ascospores are reported to be multinucleate (9, 15, 20). We think that these nuclei are all copies of a single meiotic product, as found in other ascomycetes, and are not representatives of different meiotic products. If the ascospores are initially heterokaryotic, then these nuclei must segregate from one another mitotically during vegetative growth since the mycorrhizal mycelia we recovered from the root tips were all monomorphic. Thus, it is unlikely that truffle mycorrhizae are formed only following the establishment of heterokaryotic mycelia (13, 21). The nonexistence of such heterokaryons suggests that this species may also contain a vegetative compatibility system, as found in many other ascomycetes (14), that serves to keep haploid individuals discrete.
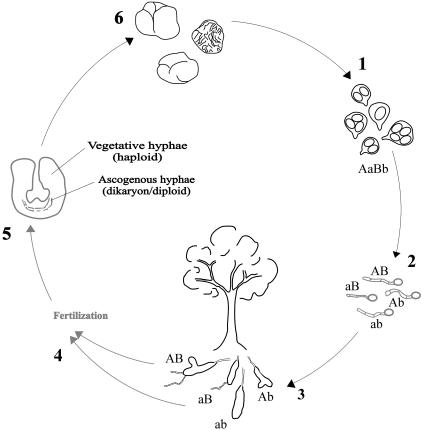
We can now use molecular markers to trace the truffle life cycle (Fig. 1). Existing data show that outcrossing occurs, but the proportion of ascocarps that do so (or are even fertilized) is unknown. The morphology of the fertilization process in the truffle life cycle still remains elusive. Reports of ascogonia within truffles are not common (6), but antheridia have never been described. Interestingly, an anamorphic phase was recently described for Tuber borchii Vittad. and Tuber oligospermum (Tul. and C. Tul.) Trappe (26), and asexual conidia could act as fertilizing agents in Tuber, as also occurs in many other Ascomycetes, e.g., most of the Pyrenomycetes.
FIG. 1.
Revised life cycle of Tuber magnatum. (1) Asci are released following ripening of the ascocarp. (2) In close proximity to host plant root tips, ascospores germinate to produce primary homokaryotic (haploid) mycelia. (3) The primary mycelia colonize the apical root tips and form ectomycorrhizae. The mycorrhizae colonizing roots can arise from different primary mycelia, so different fungal genotypes may colonize a common host root. From the mycorrhizae, extramatrical primary mycelia develop and spread into the soil. (4) Details of the fertilization process are unknown, but by analogy to other Ascomycetes organisms, a spermatization process seems likely. The function of a male gamete may be filled by any detached cell, e.g., a hyphal fragment, conidium, or ascospore. (5) Ascocarp primordia develop. The ascogenous heterokaryotic hyphae resulting from the fertilization process are surrounded by homokaryotic maternal vegetative hyphae. (6) Adult ascocarps are composed of asci and sterile hyphae. Stages not directly observed in this study are indicated in light gray.
In summary, we think that our study substantially deepens our fundamental understanding of Tuber biology and could lead to a reinterpretation of existing data and to altered strategies for the development and management of both artificial Tuber plantations and native populations.
Acknowledgments
This research was supported in part by a grant from the Regione Umbria/CNR-IGV-Perugia.
Footnotes
Contribution no. 67 from the Institute of Plant Genetics.
REFERENCES
- 1.Amicucci, A., A. Zambonelli, G. Giomaro, L. Potenza, and V. Stocchi. 1998. Identification of ectomycorrhizal fungi of the genus Tuber by species-specific ITS primers. Mol. Ecol. 7:273-277. [Google Scholar]
- 2.Bertault, G., M. Raymond, A. Berthomieu, G. Callot, and D. Fernandez. 1998. Trifling variation in truffles. Nature 394:734. [Google Scholar]
- 3.Bertault, G., F. Rousset, D. Fernandez, A. Berthomieu, M. E. Hochberg, G. Callot, and M. Raymond. 2001. Population genetics and dynamics of the black truffle in a man-made truffle field. Heredity 86:451-458. [DOI] [PubMed] [Google Scholar]
- 4.Bistis, G. N. 1981. Chemotropic interactions between trichogynes and conidia of opposite mating-type in Neurospora crassa. Mycologia 73:959-975. [Google Scholar]
- 5.Bowden, R. L., and J. F. Leslie. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182-188. [DOI] [PubMed] [Google Scholar]
- 6.Callot, G. 1999. La truffe, la terre, la vie. INRA Editions, Paris, France.
- 7.Frizzi, G., G. Lalli, M. Miranda, and G. Pacioni. 2001. Intraspecific isozyme variability in Italian populations of the white truffle Tuber magnatum. Mycol. Res. 105:365-369. [Google Scholar]
- 8.Gandeboeuf, D., C. Dupré, P. Roeckel-Drévet, P. Nicolas, and G. Chevalier. 1997. Typing Tuber ectomycorrhizae by polymerase chain amplification of the internal transcribed spacer of rDNA and the sequence characterized amplified region markers. Can. J. Microbiol. 43:723-728. [DOI] [PubMed] [Google Scholar]
- 9.Gross, G. 1987. Zu den europäischen Sippen der Gattung Tuber, p. 79-99. In H. Derbsch and J. A. Schmitt (ed.), Atlas der Pilze des Saarlandes. Teil 2. Nachweise, Ökologie, Vorkommen und Beschreibungen. Derlattinia, Saarbruken, Germany.
- 10.Hall, I. R., W. Yun, and A. Amicucci. 2003. Cultivation of edible ectomycorrhizal mushrooms. Trends Biotechnol. 21:433-438. [DOI] [PubMed] [Google Scholar]
- 11.Henrion, B., G. Chevalier, and F. Martin. 1994. Typing truffle species by PCR amplification of the ribosomal DNA spacers. Mycol. Res. 98:37-43. [Google Scholar]
- 12.Johnson, T. E. 1978. Isolation and characterization of perithecial development mutants in Neurospora. Genetics 88:27-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanfranco, L., M. Arlorio, A. Matteucci, and P. Bonfante. 1995. Truffles: their life cycle and molecular characterization, p. 139-149. In V. Stocchi, P. Bonfante, and M. Nuti (ed.), Biotechnology of ectomycorrhizae. Molecular approach. Plenum Press, New York, N.Y.
- 14.Leslie, J. F. 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31:127-151. [DOI] [PubMed] [Google Scholar]
- 15.Mello, A., C. Murat, A. Vizzini, V. Gavazza, and P. Bonfante. 2005. Tuber magnatum, a species of limited geographical distribution: its genetic diversity inside and outside a truffle ground. Environ. Microbiol. 7:55-65. [DOI] [PubMed] [Google Scholar]
- 16.Murat, C., J. Díez, P. Luis, C. Delaruelle, C. Dupré, G. Chevalier, P. Bonfante, and F. Martin. 2004. Polymorphism at the ribosomal DNA ITS and its relation to postglacial re-colonization routes of the Perigord truffle Tuber melanosporum. New Phytol. 164:401-411. [DOI] [PubMed] [Google Scholar]
- 17.Paolocci, F., P. Angelini, E. Cristofari, B. Granetti, and S. Arcioni. 1995. Identification of Tuber spp. and corresponding ectomycorrhizae through molecular markers. J. Food Sci. Agric. 69:511-517. [Google Scholar]
- 18.Paolocci, F., A. Rubini, B. Granetti, and S. Arcioni. 1999. Rapid molecular approach for a reliable identification of Tuber spp. ectomycorrhizae. FEMS Microbiol. Ecol. 28:23-30. [Google Scholar]
- 19.Paolocci, F., A. Rubini, B. Granetti, and S. Arcioni. 1997. Typing Tuber melanosporum and Chinese black truffle species by molecular markers. FEMS Microbiol. Lett. 153:255-260. [DOI] [PubMed] [Google Scholar]
- 20.Parguey-Leduc, A., M. C. Janex-Favre, and C. Montant. 1987. Formation et évolution des ascospores de Tuber melanosporum Vitt. (truffe noire du Périgord, Discomycètes). Can. J. Bot. 65:1491-1503. [Google Scholar]
- 21.Rouquerol, T., and H. Payre. 1974. Observations sur le comportement de Tuber melanosporum dans un site naturel. Rev. Mycol. 39:107-117. [Google Scholar]
- 22.Rubini, A., F. Paolocci, B. Granetti, and S. Arcioni. 2001. Morphological characterization of molecular-typed Tuber magnatum ectomycorrhizae. Mycorrhiza 11:179-185. [Google Scholar]
- 23.Rubini, A., F. Paolocci, B. Granetti, and S. Arcioni. 1998. Single step molecular characterization of morphologically similar black truffle species. FEMS Microbiol. Lett. 164:7-12. [Google Scholar]
- 24.Rubini, A., F. Paolocci, C. Riccioni, G. G. Vendramin, and S. Arcioni. 2005. Genetic and phylogeographic structure in the symbiotic fungus Tuber magnatum. Appl. Environ. Microbiol. 71:6584-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubini, A., F. Topini, C. Riccioni, F. Paolocci, and S. Arcioni. 2004. Isolation and characterization of polymorphic microsatellite loci in white truffle (Tuber magnatum). Mol. Ecol. Notes 4:116-118. [Google Scholar]
- 26.Urban, A., I. Neuner-Plattner, I. Krisai-Greilhuber, and K. Haselwandter. 2004. Molecular studies on terricolous microfungi reveal novel anamorphs of two Tuber species. Mycol. Res. 108:749-758. [DOI] [PubMed] [Google Scholar]