Abstract
Picocyanobacteria of the genus Synechococcus are important contributors to marine primary production and are ubiquitous in the world's oceans. This genus is genetically diverse, and at least 10 discrete lineages or clades have been identified phylogenetically. However, little if anything is known about the genetic attributes which characterize particular lineages or are unique to specific strains. Here, we used a suppression subtractive hybridization (SSH) approach to identify strain- and clade-specific genes in two well-characterized laboratory strains, Synechococcus sp. strain WH8103 (clade III) and Synechococcus sp. strain WH7803 (clade V). Among the genes that were identified as potentially unique to each strain were genes encoding proteins that may be involved in specific predator avoidance, including a glycosyltransferase in strain WH8103 and a permease component of an ABC-type polysaccharide/polyol phosphate export system in WH7803. During this work the genome of one of these strains, WH7803, became available. This allowed assessment of the number of false-positive sequences (i.e., sequences present in the tester genome) present among the SSH-enriched sequences. We found that approximately 9% of the WH8103 sequences were potential false-positive sequences, which demonstrated that caution should be used when this technology is used to assess genomic differences in genetically similar bacterial strains.
Marine cyanobacteria of the genus Synechococcus are ubiquitous in the world's oceans and are major contributors to marine primary production (18, 30). This genus is genetically diverse, with at least 10 specific lineages or clades in the marine cluster A Synechococcus group already identified from phylogenetic analyses of 16S rRNA, internal transcribed spacer, and rpoC1 gene sequences (13, 32, 39). It has been suggested that the successful colonization of the marine ecosystem by this genus likely reflects the adaptation of these specific lineages to particular niches (36), which gives rise to a competitive advantage for members of a lineage under a given suite of environmental conditions. Consistent with this idea is the fact that molecular ecological data obtained by using specific oligonucleotides or antibodies have shown that there are obvious differences in the relative abundance of these lineages in particular oceanic areas (7, 8, 13, 14, 40; N. J. Fuller, S. Mazard, and D. J. Scanlan, unpublished data). Furthermore, in these genetic lineages phenotypic traits, such as motility (41) and possibly chromatic adaptation in nonmotile strains (27), show phylogenetic coherence, suggesting that some traits are clade specific. Underlying these phenotypic differences are genomic differences in either single genes or gene clusters, which, combined with differential expression of common genes in strains belonging to different clades, allow specific marine Synechococcus lineages to adapt to different niches.
Genome sequencing has highlighted some of the underlying genomic differences that underpin the physiology of specific niche-adapted groups in the closely related genus Prochlorococcus, in which there are lineages that are adapted to niches in the surface or depths of a water column (12, 33), but much less is known about the more widespread genus Synechococcus. In order to begin to define more specifically differences at the genomic level that have allowed marine Synechococcus lineages to adapt to specific niches, we utilized a suppression subtractive hybridization (SSH) approach to identify genes that might be considered clade or strain specific.
SSH was developed and has been mostly used to identify genetic differences between virulent and avirulent strains of various pathogens (2, 11, 17, 29, 45), but more recently it has also been used to identify genes unique to the saxitoxin-producing cyanobacterium Anabaena circinalis (31), as well as to identify genetic diversity in an environmental metagenome (15). In this technique pools of genomic DNA from a bacterial strain of interest (tester) are depleted, by hybridization and PCR, by sequences that are also in a reference strain (driver). The remaining highly enriched fraction of tester-specific sequences is then cloned and sequenced. We chose this technique over, for example, DNA-DNA hybridization using a microarray, because the latter technique identifies only genes that are present in the reference genome and not genes that are absent.
In order to gauge the utility of the SSH approach for assessing genomic differences in the marine genus Synechococcus, Synechococcus strains WH8103 (clade III) and WH7803 (clade V) were used as the tester and driver (and vice versa), respectively, to identify genes “unique” to each strain. Strain WH8103 is closely related to another clade III strain, strain WH8102. The complete genome of Synechococcus sp. strain WH8102 was recently published (28). Hence, our SSH approach should also identify the genes that are potentially clade specific (i.e., present in both WH8103 and WH8102 but not in WH7803). Synechococcus sp. strains WH7803 and WH8103 are axenic, well-characterized laboratory strains that represent different Synechococcus clades (13, 44). These strains have very similar DNA base ratios, which helps rule out genomic differences due to G+C content. Furthermore, several physiological differences between these strains with regard to the composition of the light-harvesting pigments and the ability to swim or use urea have already been identified (9, 44). Together, these features should allow rigorous testing of the SSH technique for identifying genomic differences which ultimately could define niche-specific genes (i.e., genes important in open-ocean regions or in coastal or up-welling regions) in these important photoautotrophs.
MATERIALS AND METHODS
Genomic DNA isolation.
Genomic DNA was isolated from Synechococcus sp. strains WH8103 and WH7803 by using a previously described protocol (13).
SSH.
Genes unique to either Synechococcus sp. strain WH8103 or Synechococcus sp. strain WH7803 were identified by SSH using first WH8103 as the tester and WH7803 as the driver (to identify WH8103-specific sequences) and then WH7803 as the tester and WH8103 as the driver (to identify WH7803-specific sequences). SSH was performed using a CLONTECH PCR-Select bacterial genome subtraction kit (CLONTECH Laboratories, Inc., Palo Alto, CA) according to the manufacturer's instructions but with the following modifications.
A combination of AluI and HaeIII was used to digest Synechococcus sp. strain WH8103 and Synechococcus sp. strain WH7803 chromosomal DNAs, which yielded fragments that were less than 1 kb long. Adaptor ligation was performed according to the kit protocol, and this was followed by addition of 1 μl T4 DNA ligase and reincubation at 15°C overnight to ensure sufficient ligation of adaptors.
To determine the efficiency of adapter ligation when Synechococcus sp. strain WH8103 was used as the tester, PCR primers were designed to amplify a 1,146-bp fragment of nirA (accession number AF065403), a gene encoding nitrate reductase (5), containing no AluI or HaeIII recognition sites. The gene-specific primers were SSHLEW8103F (5′-CGA CAT CAC CAC AAG GCA AA-3′) and SSHLEWH8103R (5′-TGA CCA ATA GTT GGG TTG CG-3′), while SSHPRIMER1 (5′-CTA ATA CGA CTC ACT ATA GGG C-3′) was the adapter-specific primer. PCRs were carried out in 25-ml mixtures containing each deoxynucleoside triphosphate at a concentration of 200 μM, 2 mM MgCl2, 10 pmol of each primer, 1 μl of template (prepared as instructed in the kit manual), and 0.75 U of Taq polymerase in 1× enzyme buffer (Invitrogen, Carlsbad, CA). The amplification conditions comprised a denaturation step of 94°C for 3 min; 80°C for 1 min, at which time Taq polymerase was added; and 72°C for 2 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min and a final extension for 5 min at 72°C. The reaction mixtures were stored at 4°C prior to analysis. Products (10 μl of 25 μl) were resolved by gel electrophoresis on a 1.5% (wt/vol) agarose gel at 100 V. DNA was stained with ethidium bromide (0.5 μg ml−1), visualized under short-wavelength UV, and photographed using a gel documentation system (UVP Inc., Upland, CA).
When Synechococcus sp. strain WH7803 was used as the tester, a 307-bp fragment of napA (accession number AAG45172), a gene encoding nitrate permease (A. F. Post, D. Lindell, A. Moyal, S. Solomon, and Q. Wang, unpublished data), containing no AluI or HaeIII recognition sites was used to perform a ligation efficiency analysis. The primers used were SSHLEW7803F (5′-GCT TGG CGA ACT TTG GTC ATT T-3′) and SSHLEWH7803R (5′-CTG ATT GAA GTC CTG AGC AGA T-3′). PCRs were performed as described above, but the amplification conditions were 94°C for 3 min; 75°C for 1 min, at which time Taq polymerase was added; and 72°C for 2 min, followed by 27 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min and a final extension for 2 min at 68°C.
For the primary PCR using the adapter-specific primer SSHPRIMER1 (5′-CTA ATA CGA CTC ACT ATA GGG C-3′), PCRs were performed as described above, but the amplification conditions were 94°C for 3 min; 75°C for 1 min, at which time Taq polymerase was added; and 72°C for 5 min, followed by 30 cycles of 94°C for 1 min, 66°C for 1 min, and 72°C for 1.5 min.
For the nested PCR, primers SSHnestpri1 (5′-TCG AGC GGC CGC CCG GGC AGG T-3′) and SSHnestpri2 (5′-AGC GTG GTC GCG GCC GAG GT-3′) were used, and PCRs were again performed as described above, except that the template DNA was 1 μl of a 1:40 dilution of the PCR products from the primary PCR. The amplification conditions consisted of 94°C for 3 min, 75°C for 1 min, at which time Taq polymerase was added, and 72°C for 5 min, followed by 15 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1.5 min.
DNA sequencing.
The nested PCR products were TA cloned into the pCR2.1TOPO vector, and plasmid DNA was extracted using a QIAprep Spin Miniprep kit (QIAGEN GmbH, Hilden, Germany). PCR products were sequenced bidirectionally at the Warwick University sequencing facility using Big Dye Terminator version 3.1 chemistry (Applied Biosystems, Foster City, Calif.) and were examined with a 3100 genetic analyzer. The M13R primer (5′-CAG GAA ACA GCT ATG AC-3′), which annealed to the plasmid 74 bp upstream of the insert, was used as the sequencing primer.
Computer analysis.
SSH-enriched sequences from Synechococcus sp. strains WH8103 and WH7803 were initially analyzed by BLASTX searches (3) against the closed WH7803 genome (F. Partensky, personal communication). Sequences derived from WH8103 were aligned with the complete genome sequence of WH7803 using Washington University BLASTN (W. Gish, 1996 to 2004; http://BLAST.wustl.edu) with gapped alignments. Sequences with more than 85% nucleotide identity to the WH7803 sequence were considered false positives and were removed from the WH8103-specific set. The rest of the specific sequences were analyzed by BLASTX against the GenBank nonredundant database (http://www.ncbi.nlm.nih.gov). Analyses of WH7803 sequences were completed with the aid of the draft annotation of the WH7803 genome based on Glimmer, Critica, and GeneMarks software predictions with manual annotation. All of the WH7803-specific sequences exhibited 100% identity with the WH7803 genome sequence.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under the following accession numbers: for WH8103 SSH-enriched fragments, DU635204 to DU635313 and DU635372; and for WH7803 SSH-enriched fragments, DU635093 to DU635203.
RESULTS AND DISCUSSION
Genomic subtraction between Synechococcus sp. strains WH8103 and WH7803. (i) Genes specific to WH8103 or to WH8103 and WH8102.
Plasmid DNA was extracted from 174 white or light blue colonies obtained from the nested PCR product cloning; 60 of the clones provided insufficient sequence data or had no insert. Of the 114 sequences obtained, 10 were defined as false positives and another 25 were found to be duplicates (i.e., identical to the sequence[s] of another fragment[s]), which left 79 unique sequences for analysis (Table 1). The lengths of the fragments sequenced ranged from 45 bp to 708 bp. The WH8103 fragment sequences obtained were identified by BLASTN comparison to the complete genome sequence of the very closely related organism Synechococcus sp. strain WH8102 (accession number NC_005070) (28). Five fragments (79, 140, 281, 313, and 363 nucleotides) exhibited no homology with the Synechococcus sp. strain WH8102 genome. Either these fragments contained numerous stop codons in all reading frames and hence were likely intergenic regions in WH8103, or the open reading frames exhibited no significant similarity to sequences encoding database proteins.
TABLE 1.
Summary of Synechococcus sp. strain WH8103 SSH-enriched DNA fragments
| No. of sequences | Fragmenta | Length (bp) | Homology to predicted encoded proteinb | e valuec | Identityd | Homolog accession no. | Gene or organism |
|---|---|---|---|---|---|---|---|
| 1 | A001 | 376 | Hypothetical protein | 6e-36 | 72/72 | NP_897845 | SYNW1754 |
| Hypothetical protein | 6e-09 | 29/29 | NP_897846 | SYNW1755 | |||
| 2 | A002 | 708 | Conserved hypothetical protein | 1e-80 | 140/236 | NP_897078 | SYNW0985 |
| A003 | 597 | 2e-44 | 93/98 | ||||
| 1 | A005 | 301 | Putative ABC transporter, ATP binding component | 3e-54e | 100/100 | NP_897205 | SYNW1112 |
| 3 | A006 | 277 | Hypothetical protein | 6e-06 | 28/63 (38/63) | NP_896979 | SYNW0886 |
| 1 | A010 | 462 | Hypothetical protein | 4e-61 | 109/110 | NP_896450 | SYNW0355 |
| 2 | A012 | 274 | Putative asparagine synthetase protein | 2e-47 | 89/90 | NP_898542 | SYNW2453 |
| 5 | A014 | 340 | ABC transporter component, possibly zinc transport | 7e-50 | 97/102 | NP_898568 | SYNW2479 |
| A143 | 437 | 2e-32 | 80/101 | ||||
| A155 | 317 | 1e-09 | 42/66 | ||||
| A051 | 339 | 6e-48 | 98/102 | ||||
| 1 | A018f | 174 | Conserved hypothetical protein | 2e-27 | 57/57 | NP_897305 | SYNW1212 |
| 7 | A020f | 338 | ABC phosphate transporter, ATP binding component, PstB | 2e-59 | 111/112 | NP_897365 | SYNW1270 |
| A077f | 336 | 3e-57 | 110/112 | ||||
| 1 | A022f | 278 | Conserved hypothetical protein | 2e-20 | 47/48 | NP_896965 | SYNW0872 |
| 3 | A024 | 310 | Putative chromate transport protein CHR family | 2e-7 | 21/21 | NP_897416 | SYNW1323 |
| 1 | A025f | 207 | Glutathione S-transferase domain protein | 5e-36 | 68/68 | NP_897024 | SYNW0931 |
| 1 | A026f | 285 | Inositol monophosphate family protein | 8e-16 | 43/44 | NP_898277 | SYNW2186 |
| 4 | A029 | 292 | Putative urea ABC transporter, urea binding protein | 9e-55 | 97/97 | NP_898531 | SYNW2442 |
| A055 | 116 | 8e-17 | 36/37 | ||||
| A097 | 295 | 2e-29 | 61/81 | ||||
| 1 | A030 | 228 | ABC transporter, nitrate/sulfonate/bicarbonate-like substrate binding protein | 1e-38 | 75/76 | NP_897508 | SYNW1415 |
| 1 | A033f | 129 | Conserved hypothetical protein | 2e-15 | 35/36 | NP_896807 | SYNW0714 |
| 1 | A039f | 132 | FtsH ATP-dependent protease homolog | 5e-18 | 44/44 | NP_897393 | SYNW1300 |
| 1 | A040f | 187 | Photolyase family protein | 4e-29 | 62/62 | NP_897337 | SYNW1244 |
| 1 | A045f | 136 | Probable exodeoxyribonuclease V, beta subunit RecB | 7c-18 | 43/44 | NP_897010 | SYNW0917 |
| 2 | A046f | 237 | Putative photosystem II oxygen-evolving complex 23,000-Da protein PsbP | 2e-37 | 77/78 | NP_897020 | SYNW0927 |
| 6 | A048 | 135 | SwmB repeated sequence | 5e-18 | 42/44 | NP_897046 | SYNW0953 |
| A062 | 158 | 1e-24 | 52/52 | ||||
| A113 | 522 | 9e-51 | 65/73 | ||||
| A130 | 463 | 1e-22 | 59/94 | ||||
| A146 | 426 | 2e-40 | 89/100 | ||||
| A169 | 425 | 7e-73 | 140/141 | ||||
| 1 | A049f | 120 | ABC transporter substrate binding protein, phosphate | 5e-14 | 39/39 | NP_897906 | SYNW1815 |
| 1 | A052 | 120 | Carbamoyl phosphate synthase, large subunit | 5e-15 | 39/39 | NP_896923 | SYNW0830 |
| 1 | A059 | 213 | Hypothetical protein | 2e-9 | 26/26 | NP_896473 | SYNW0378 |
| 2 | A061 | 58 | Hypothetical protein | 4e-6 | 18/18 | NP_897910 | SYNW1819 |
| 1 | A065 | 190 | Putative 4′-phosphopantetheinyl transferase | 3e-17 | 37/40 | NP_898252 | SYNW2161 |
| 1 | A070 | 371 | Hypothetical protein | 2e-66 | 121/122 | NP_897657 | SYNW1564 |
| 1 | A075 | 146 | Possible DnaJ domain | 2e-8 | 34/48 | NP_897323 | SYNW1230 |
| 1 | A078f | 339 | Putative deacetylase sulfotransferase | 2e-58 | 107/110 | NP_896178 | SYNW0083 |
| 1 | A079f | 163 | Cell division protein FtsH4 | 2e-17 | 46/54 | NP_897304 | SYNW1211 |
| 3 | A080 | 100 | Putative ATPase, AAA family | 3e-14 | 31/33 | NP_898249 | SYNW2158 |
| 1 | A088f | 172 | Glutathione peroxidase | 2e-9 | 32/33 | NP_896194 | SYNW0099 |
| 2 | A094 | 352 | Hypothetical protein | 9e-26 | 48/48 | NP_896484 | SYNW0389 |
| A095 | 300 | 1e-22 | 48/48 | ||||
| 1 | A099f | 237 | Apocytochrome b6 | 8e-11 | 35/48 | NP_892444 | PMT1649 |
| 1 | A106 | 409 | Hypothetical protein | 2e-10 | 18/22 | NP_897994 | SYNW1903 |
| 1 | A118 | 83 | Porin homolog | 5e-9 | 27/27 | NP_898317 | SYNW2228 |
| 3 | A121 | 103 | Adenylosuccinate synthase | 2e-9 | 31/33 | NP_897864 | SYNW1773 |
| 1 | A135f | 108 | Possible ferredoxin | 2e-13 | 35/35 | NP_897859 | SYNW1768 |
| 1 | A136 | 381 | COG2089; sialic acid synthase | 3e-8 | 27/47 | ZP_00128881 | Desulfovibrio desulfuricans G20 |
| 1 | A141 | 176 | Hypothetical protein | 4e-19 | 30/36 | NP_898389 | SYNW2300 |
| 1 | A144 | 211 | Possible glycosyltransferase | 1e-31 | 55/69 | NP_896511 | SYNW0416 |
| 2 | A150 | 445 | Hypothetical protein | 3e-40 | 82/83 | NP_897748 | SYNW1655 |
| A166 | 283 | 9e-48 | 90/93 | ||||
| 1 | A171 | 112 | Putative succinate dehydrogenase flavoprotein subunit | 5e-17 | 35/36 | NP_896684 | SYNW0591 |
For A004, A009, A021, A023, A027, A028, A035, A036, A042, A050, A056, A063, A091, A098, A102, A103, A105, A114, A115, A129, A134, and A148 the e value was >0.005 or the level of identity was <50% and there were no significant hits.
Where it is known, the product of the gene is indicated. The products of genes without a known product were designated conserved hypothetical if they had a homolog(s) in a different organism(s) or hypothetical if they had no homolog.
The e value indicates the probability of the match. A match with an e value of <0.05 or >50% identity over the entire length of the sequence was considered significant.
Where the level of identity was <50% but there was significant similarity, the percentages of positives are indicated in parentheses.
The e value for WH7803 was 9.9e-36, but since the fragment encodes an ABC transporter, we considered this difference significant in this gene category.
False positives were defined as sequences with >85% nucleotide identity with WH7803 as determined by WU-BLASTN with gapped alignment (Gish, 1996 to 2004; http://blast.wustl.eduref).
The remaining fragments were initially considered to be absent from the WH7803 genome, or at least the level of homology was below the level which allowed DNA-DNA hybridization during the SSH procedure. However, the availability during this work of the complete Synechococcus strain WH7803 genome allowed verification by more rigorous bioinformatic analysis of the absence of these fragments in this strain. Hence, we were able to identify fragments which might be false positives arising from linear amplification of driver-derived DNA, as well as fragments which we defined as false positives because they exhibited more than 85% nucleotide identity with the WH7803 sequence (see below).
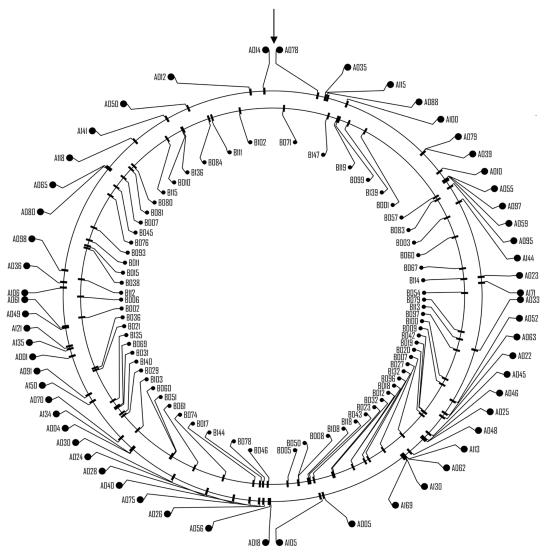
Fifty-seven of the remaining WH8103 and WH8102 enriched fragments had significant hits with gene sequences in the database, and 55 of the hits were best hits in WH8102. Another 22 sequences had no significant hits when tBLASTX was used, indicating that they were derived from intergenic spacer regions or were entirely novel sequences. Thus, the majority of the differences found correspond to genes rather than noncoding DNA. The WH8103 and WH8102 enriched fragments were found to be evenly distributed around the Synechococcus sp. strain WH8102 genome, as shown in Fig. 1. The genes to which the fragments correspond are shown in Table 1. Several of the DNA fragments exhibited homology with the sequence encoding the ABC urea transporter, SYNW2442 (fragments A029, A055, and A097), or the sequence encoding SwmB (fragments A048, A062, A113, A130, A146, and A169); the former is required for high-affinity urea transport (42), and the latter is required for swimming motility (20). Both of these physiological traits (i.e., urea utilization and swimming motility) are known to be absent in WH7803 (9, 41, 44). SYNW2442 appears to be a homolog of urtA, a component of the urtABCDE gene cluster, and hence this component of the high-affinity urea transport system can be added to the urease structural genes as missing genes required for urea utilization by this strain. The genes for several other potential ABC transporter components appear in these WH8103- and WH8102-specific sequences, including the gene encoding a potential nitrate-like substrate binding protein for an ABC transporter, SYNW1415. The presence of such a gene is interesting given the fact that WH8102 contains a reported deletion after nucleotide position 617 (resulting in a frameshift that produces a stop codon 26 codons downstream [see EMBL entry CAE08977]) in the napA (or nrtP) gene encoding a nitrate permease belonging to the major facilitator superfamily (5, 35, 43). Certainly, in WH7803 the napA product seems to be the sole nitrate transporter, since interposon mutagenesis of the gene revealed a strain that could not grow on nitrate (A. F. Post, D. Lindell, A. Moyal, S. Solomon, and Q. Wang, submitted for publication). This leads to speculation about whether nitrate uptake by the napA product is a functional system in Synechococcus sp. strain WH8102 and whether the SYNW1415 product is part of a functional nitrate transport system together with other ABC transporter components encoded by SYNW1416 and SYNW1417 adjacent to SYNW1415 on the genome. Certainly, thus far, nitrate transport via an ABC-type system, encoded by the nrtABCD genes, appears to be restricted to freshwater cyanobacteria (24, 25).
FIG. 1.
Genomic distribution of SSH-enriched DNA fragments. SSH-enriched DNA sequences from Synechococcus sp. strain WH8103 were mapped on the complete genome of Synechococcus sp. strain WH8102 (outer circle), while SSH-enriched sequences from Synechococcus sp. strain WH7803 were mapped on the complete genome sequence of this strain (inner circle). The origin of both sequences is indicated by an arrow.
Glycosyltransferases are involved in carbohydrate modification of the cell envelope, and it has been suggested that they may be required for construction of the swimming motility apparatus in marine Synechococcus, since at least one of the components of this apparatus is glycosylated (6). The presence of a WH8103- and WH8102-specific fragment encoding a putative glycosyltransferase (SYNW0416) (Table 1) may support the hypothesis that this glycosyltransferase is involved in construction of the motility apparatus (28); alternatively, the enzyme may allow these organisms to modify their cell surface characteristics to help evade grazers and other predators, such as phage (28). Certainly, freshwater Synechococcus strains containing a highly glycosylated, paracrystalline surface layer have been shown to be ingested at lower rates than strains lacking this structure are ingested (23), and the same may be true in marine environments (22).
Succinate dehydrogenase (EC 1.3.99.1), encoded by sdhA (SYNW0591), a citric acid cycle enzyme, catalyzes the conversion between succinate and fumarate. It has been known for a long time that cyanobacteria have an incomplete citric acid cycle and lack α-ketoglutarate dehydrogenase (37). Interestingly, several other central carbon metabolism genes appear to be missing entirely in marine Synechococcus and Prochlorococcus genomes; these genes include genes encoding malate dehydrogenase, glucose-1-phosphate dehydrogenase, 6-phosphofructokinase, succinyl coenzyme A synthetase, and NAD- and NADP-dependent alcohol dehydrogenases (16). Succinate dehydrogenase appears to be an exception to this, however; the data presented here show that this enzyme is present in Synechococcus sp. strains WH8103 and WH8102 and Prochlorococcus spp. strains MIT9313 and SS120 but is not present in Synechococcus sp. strain WH7803 and Prochlorococcus sp. strain MED4.
Asparagine synthetase (EC 6.3.5.4), encoded by asnB (SYNW2453), is responsible for the following reaction, which results in the synthesis of asparagine: ATP + l-aspartate + l-glutamine → AMP + diphosphate + l-asparagine + l-glutamate. Several bacteria have been shown to possess a tRNA-dependent transamidation pathway in which aspartyl-tRNAAsn is converted to asparaginyl-tRNAAsn (4, 10). It has been demonstrated that in Deinococcus radiodurans (which lacks asparagine synthetase) this tRNA-dependent transamidation pathway is the sole route for asparagine synthesis (21). Roy et al. (34) have proposed that a truncated archaeal asparaginyl-tRNA synthetase that is responsible for transamidation of aspartyl-tRNAAsn was introduced into bacteria via lateral gene transfer, where it became the evolutionary ancestor of bacterial asparagine synthetase. The presence of asnB (encoding asparagine synthetase) in Synechococcus sp. strain WH8103 but not in strain WH7803 suggests that Synechococcus sp. strain WH7803 is forced to rely on a tRNA-dependent transamidation pathway for the synthesis of asparagine, whereas Synechococcus sp. strain WH8103 possesses an alternative tRNA-independent pathway for asparagine synthesis.
(ii) Genes specific to WH7803.
A total of 150 colonies were cloned into the pCR2.1TOPO vector, and 37 of the clones gave insufficient sequence data or had no insert. Of the remaining 113 sequences, 27 were found to be duplicates (i.e., they had sequences identical to the sequence[s] of another fragment[s]), which left 86 sequences for analysis (Table 2). The lengths of the fragments sequenced ranged from 114 bp to 897 bp. These sequences were identified by BLASTX comparison to the nonredundant database, as well as to the recently derived Synechococcus sp. strain WH7803 genome sequence (A. Dufresne, M. Ostrowski, P. Wincker, D. J. Scanlan, and F. Partensky, unpublished data). Forty of these sequences had significant hits to gene sequences in the database, and 20 had best hits to WH8102. These Synechococcus sp. strain WH7803-specific fragments were found to be evenly distributed around the genome (Fig. 1), suggesting that there are not distinct “islands” of genomic differences for both organisms. Similarly, only two of the fragments were found to consist of only noncoding sequences, while 14 fragments were a mixture of noncoding sequences and coding sequences and 35 fragments consisted of only coding sequences. Thus, the vast majority of the differences found corresponded to genes rather than to noncoding DNA. Many of the unique genes encode hypothetical or conserved hypothetical proteins, while 13 have an assigned function (Table 2). Among the “identifiable” strain-specific genes are genes that may be involved in deterring grazing or phage infection. For example, the fact that fragment B139 encodes a permease component of an ABC-type polysaccharide/polyol phosphate export system suggests that WH7803 may produce an extracellular polysaccharide, although this extracellular polysaccharide as well as being unpalatable to grazers may also be useful as a sink for excess carbon under nitrogen limitation conditions (26); alternatively, one of the fragments, fragment B114, potentially encodes a component of an ABC-type bacteriocin/antibiotic exporter, which may be important in alleviating bacterial competition via production of an antibacterial compound.
TABLE 2.
Summary of Synechococcus sp. strain WH7803 SSH-enriched DNA fragments
| No. of sequences | Fragmenta | Length (bp) | Homology to predicted encoded proteinb | e valuec | Identityd | Homolog accession no. | Gene or organism |
|---|---|---|---|---|---|---|---|
| 3 | B001 | 299 | Rod shape determining protein | 9e-12 | 36/38 | NP_896217 | SYNW0122 |
| 1 | B003e | 155 | tRNA pseudouridine synthetase A | 1e-10 | 30/44 | NP_895584 | PMT1757 |
| 4 | B005e | 143 | Cobalamin biosynthetic protein CobN | 3e-10 | 29/45 | NP_894559 | PMT0727 |
| 1 | B006e | 202 | KaiC | 6e-25 | 56/60 | NP_896645 | SYNW0550 |
| 3 | B007 | 185 | Possible porin | 2e-16 | 35/58 | NP_898219 | SYNW2128 |
| 3 | B008 | 196 | Hypothetical protein | 3e-9 | 30/54 | NP_897352 | SYNW1259 |
| 3 | B010 | 160 | Conserved hypothetical protein | 2e-19 | 44/53 | NP_894988 | PMT1158 |
| 3 | B012e | 293 | Putative methionyl-tRNA synthetase | 2e-28 | 63/89 | NP_894575 | PMT0743 |
| 2 | B017 | 250 | Penicillin binding protein | 2e-15 | 40/75 | ZP_00165017 | S. elongatus PCC7942 |
| 1 | B022 | 225 | Periplasmic phosphate binding protein SphX | 2e-23 | 40/60 | NP_897379 | SYNW1286 |
| 2 | B031 | 395 | Possible neuromiedin U | 3e-19 | 42/61 | NP_895341 | PMT1514 |
| 2 | B036 | 450 | Hypothetical protein | 8e-18 | 49/119 (72/119) | NP_894091 | PMT0258 |
| 2 | B043e | 169 | Deoxyxylulose-5-phosphate synthase | 8e-24 | 54/56 | NP_875320 | Pro0928 |
| B055 | 191 | 1e-23 | 54/56 | ||||
| 2 | B045 | 166 | Hypothetical protein | 2e-17 | 44/47 | NP_898287 | SYNW2196 |
| 1 | B050 | 169 | Hypothetical protein | 2e-11 | 34/49 | NP_897352 | SYNW1259 |
| 1 | B051e | 227 | Diaminopimelate decarboxylase | 2e-28 | 60/75 | NP_897029 | SYNW0936 |
| 1 | B054e | 210 | Possible cystathione gamma-synthase | 8e-08 | 26/32 | NP_894058 | PMT0225 |
| 1 | B056 | 516 | Glycyl-tRNA synthetase beta chain | 1e-5 | 25/31 | NP_924529 | Gloeobacter violaceus |
| 1 | B057e | 314 | Conserved hypothetical protein | 6e-49 | 91/99 | NP_898241 | SYNW2150 |
| 1 | B058e | 412 | Conserved hypothetical protein | 1e-12 | 40/80 | NP_896449 | SYNW0354 |
| 1 | B060e | 315 | Coproporphyrinogen III oxidase | 1e-41 | 73/85 | NP_898131 | SYNW2040 |
| 1 | B067 | 312 | Formamidopyrimidine-DNA glycolase | 2e-32 | 61/94 | NP_894019 | PMT0186 |
| 1 | B075 | 406 | Hypothetical protein | 5e-15 | 45/77 | AAZ57900 | PMN2A_0408 |
| 1 | B076e | 578 | Conserved hypothetical protein | 7e-27 | 60/100 | NP_898274 | SYNW2183 |
| 2 | B079e | 392 | Putative glucosyl-glycerol-phosphate synthase | 9e-27 | 54/67 | NP_897374 | SYNW1281 |
| 1 | B080 | 333 | Septum formation inhibitor-activating ATPase | 2e-22 | 48/79 | ZP_00108431 | Nostoc punctiforme |
| 2 | B081 | 495 | Catabolite gene activator and regulatory subunit of cAMP-dependent protein kinase | 2e-26 | 42/85 | ZP_00163669 | S. elongatus PCC7942 |
| 1 | B084 | 367 | Small mechanosensitive ion channel, MscS family | 1e-34 | 70/110 | NP_895901 | PMT2077 |
| 1 | B096e | 145 | Putative modulator of DNA gyrase; TldD | 8e-17 | 43/46 | NP_897290 | SYNW1197 |
| 1 | B097 | 441 | Conserved hypothetical protein | 4e-19 | 45/58 | NP_897620 | SYNW1527 |
| 1 | B102e | 171 | DAHP synthetase class I | 1e-23 | 53/57 | NP_896072 | PMT2288 |
| 3 | B103e | 261 | Putative ferredoxin like protein | 1e-10 | 25/39 | NP_896790 | SYNW0697 |
| 1 | B108 | 204 | Putative glucosyl-glycerol-phosphate synthase | 2e-27 | 54/67 | NP_897374 | SYNW1281 |
| 1 | B110 | 159 | Hypothetical protein | 1e-18 | 44/52 | NP_897620 | SYNW1527 |
| 1 | B112 | 140 | Putative CaCA family calcium/proton exchanger | 5e-10 | 30/35 | NP_896602 | SYNW0507 |
| 1 | B113 | 415 | Putative CO2 hydration protein ChpX | 2e-67 | 121/138 | NP_897800 | SYNW1709 |
| 1 | B114 | 395 | ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase domain | 2e-8 | 27/80 (46/80) | ZP_00110567 | Nostoc punctiforme PCC 731 |
| 1 | B115 | 225 | Hypothetical protein | 2e-8 | 28/49 | AAQ87420 | Rhizobium sp. strain NGR234 |
| 1 | B118e | 292 | ABC transporter, likely for sugar transport | 2e-18 | 43/56 | NP_894524 | PMT0692 |
| 1 | B139 | 310 | ABC-type polysaccharide/polyol phosphate export systems, permease component | 4e-07 | 53/102 | ZP_00264217 | Pseudomonas fluorescens PfO-1 |
For B002, B009, B011, B014, B015, B021, B023, B025, B026, B027, B028, B029, B033, B035, B038, B039, B041, B044, B046, B048, B053, B059, B061, B065, B066, B069, B071, B072, B074, B082, B083, B090, B093, B099, B101, B107, B110, B111, B132, B135, B136, B140, B144, B147, and B149 the e value was >0.005 or the level of identity was <50% and there were no significant hits (intergenic spacer or unique sequence).
Where it is known, the product of the gene is indicated. The products of genes without a known product were designated conserved hypothetical if they had a homolog(s) in a different organism(s) or hypothetical if they had no homolog.
The e value indicates the probability of the match. A match with an e value of <0.05 and >50% identity over the entire length of the sequence was considered significant.
Where the level of identity was <50% but there was significant similarity, the percentages of positives are indicated in parentheses.
Potential false positives were defined as an SSH sequence conserved in at least five sequenced marine cyanobacterial genomes (MED4, SS120, MIT9313, WH8102, and WH7803), using an e value of <0.05 and a level of identity of >50% at the nucleotide level.
(iii) False positives.
SSH-enriched fragments of Synechococcus sp. strain WH8103 contained no sequences that were false positives resulting from the linear amplification of driver-derived DNA. However, potential false positives that met the criterion of exhibiting more than 85% nucleotide identity with WH7803 (WH8103 enriched) sequences occurred at a rate of 9% of the total number of sequences. Without the complete genome sequence of WH8103 it is not possible to determine the number of false positives in the SSH-enriched WH7803 sequences. However, if we consider that SSH sequences conserved in at least five sequenced marine cyanobacterial genomes (MED4, SS120, MIT9313, WH8102, and WH7803) are potentially false positives, then there would be 15 candidates, a rate of 13% of the total (Table 2). Two examples are the clock gene kaiC (B006) and the ABC transporter component gene pstB (A020, A077), which appear to be present in all marine picocyanobacterial genomes analyzed so far. This apparently high number of false positives may simply reflect the criteria described above (i.e., the stringency used for determining matches and the assumption that a sequence conserved in at least five marine cyanobacterial genomes should also be conserved in WH8103). Alternatively, they may be a result of the fact that even small stretches of unique sequence within a fragment prevent suppression of common genes. Examples of the latter are likely the WH7803 SSH sequences derived from a possible porin gene (B007) and a periplasmic phosphate binding protein gene, sphX (B022). Multiple copies of orthologues of each of these genes are maintained and randomly distributed throughout the genomes of sequenced cyanobacteria, including WH7803. Given that identification of false-positive sequences is critical for correct identification of clade- or strain-specific genes, in future SSH experiments workers should attempt to minimize these types of sequences. A potential approach to do this is to construct SSH libraries generated from a wider variety of restriction endonucleases than the restriction endonucleases used here (1) and to sequence a larger number of clones. With these modifications unique SSH sequences representing truly unique genes would be returned with a higher frequency than false positives and sequences generated from short regions with low levels of identity.
Conclusions.
We describe here identification of several interesting candidate genes that may represent clade-specific characteristics that allow occupation of a specific environmental niche by a particular marine Synechococcus lineage. Although we are cautious about the existence of a plethora of clade-specific traits, it would not be unexpected that some specific physiological traits are shared by clades, given that it is the total suite of traits that correlates with the environmental niche occupied.
Certainly, it is interesting that genes potentially related to predator avoidance have been identified using the SSH approach, and each marine Synechococcus strain apparently has distinct genetic predator avoidance capabilities. It is known that resistance to protist grazing can be influenced by a variety of factors, such as high motility, reduced or increased size, and cell surface masking (for a review see reference 19), while modification of the cell surface is also clearly important with respect to cyanophage infection. Hence, it is not unreasonable to suggest that the high numbers of cyanophage in marine waters (38) and also potentially the grazers themselves are involved in a “biological arms race” that results in strong selective pressures for the host cells to evolve specific mechanisms that make them resistant to predation.
Clearly, the SSH approach used here is a cheap and relatively fast approach to identify strain-specific genes in the marine genus Synechococcus. These organisms differ from most pathogenic organisms that have been targeted previously with the SSH approach because they lack defined “islands” of genomic differences. Certainly, the strain-specific sequences could augment information contained in microarrays, expanding the scope of gene content or gene expression analysis and allowing multistrain chips to be developed. Clearly, strain variation also provides valuable insights into evolutionary processes, while finding sequences that are variable but common in strains should facilitate more precise and reliable taxonomy beyond the 16S rRNA gene nomenclature, as has been initiated using rpoC1 (39). Finally, mutagenesis studies (for an example, see reference 20) of the many conserved hypothetical genes and hypothetical genes identified here as strain specific should lead to a more fundamental understanding of the processes involved in niche adaptation.
Acknowledgments
We are grateful to John Jones for writing the computer program used to plot Fig. 1.
H.J. was the recipient of a BBSRC-funded Ph.D. studentship. This work was also supported in part by the EU FP5 program MARGENES (grant QLRT-2001-0226 to D.J.S.).
REFERENCES
- 1.Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridisation for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, H. D., and D. Kern. 1998. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. USA 95:12832-12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, C., and M. Wyman. 2003. Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl. Environ. Microbiol. 69:7009-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha, B. 1996. An abundant cell surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. USA 93:6504-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, L., and E. J. Carpenter. 1987. Characterisation of phycoerythrin-containing Synechococcus spp. populations by immunofluorescence. J. Plankton Res. 9:1167-1181. [Google Scholar]
- 8.Campbell, L., and R. Iturriaga. 1988. Identification of Synechococcus spp. in the Sargasso Sea by immunofluorescence and fluorescence excitation spectroscopy performed on individual cells. Limnol. Oceanogr. 33:1196-1201. [Google Scholar]
- 9.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH 7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447-459. [DOI] [PubMed] [Google Scholar]
- 10.Curnow, A. W., D. L. Tumbula, J. T. Pelaschier, B. Min, and D. Soll. 1998. Glutamyl-tRNAgln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA 95:12838-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diatchenko, L., Y. F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 100:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller, N. J., N. J. West, D. Marie, M. Yallop, T. Rivlin, A. F. Post, and D. J. Scanlan. 2005. Dynamics of community structure and phosphate status of picocyanobacterial populations in the Gulf of Aqaba, Red Sea. Limnol. Oceanogr. 50:363-375. [Google Scholar]
- 15.Galbraith, E. A., D. A. Antonopoulos, and B. A. White. 2004. Suppressive subtractive hybridisation as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ. Microbiol. 6:928-937. [DOI] [PubMed] [Google Scholar]
- 16.García-Fernández, J. M., and J. Diez. 2004. Adaptive mechanisms of nitrogen and carbon assimilatory pathways in the marine cyanobacteria Prochlorococcus. Res. Microbiol. 155:795-802. [DOI] [PubMed] [Google Scholar]
- 17.Janke, B., U. Dobrindt, J. Hacker, and G. Blum-Oehler. 2001. A subtractive hybridisation analysis of genomic differences between the uropathogenic E. coli strain 536 and the E. coli K-12 strain MG1655. FEMS Microbiol. Lett. 199:61-66. [DOI] [PubMed] [Google Scholar]
- 18.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 19.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 20.McCarren, J., and B. Brahamsha. 2005. Transposon mutagenesis in a marine Synechococcus strain: isolation of swimming motility mutants. J. Bacteriol. 187:4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monger, B. C., M. R. Landry, and S. L. Brown. 1999. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol. Oceanogr. 44:1917-1927. [Google Scholar]
- 23.Müller, H. 1996. Selective feeding of a freshwater chrysomonad, Paraphysomonas sp., on chroococcoid cyanobacteria and nanoflagellates. Arch. Hydrobiol. Spec. Issues Adv. Limnol. 48:63-71. [Google Scholar]
- 24.Omata, T. 1995. Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC7942. Plant Cell Physiol. 36:207-213. [DOI] [PubMed] [Google Scholar]
- 25.Omata, T., X. Andriesse, and A. Hirano. 1993. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC7942. Mol. Gen. Genet. 236:193-202. [DOI] [PubMed] [Google Scholar]
- 26.Otero, A., and M. Vincenzini. 2004. Nostoc (Cyanophyceae) goes nude: extracellular polysaccharides serve as a sink for reducing power under unbalanced C/N metabolism. J. Phycol. 40:74-81. [Google Scholar]
- 27.Palenik, B. 2001. Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 29.Parsons, Y. N., R. Banasko, M. G. Detsika, K. Duangsonk, L. Rainbow, C. A. Hart, and C. Winstanley. 2003. Suppression subtractive hybridisation reveals variations in gene distribution amongst the Burkholderia cepacia complex, including the presence in some strains of a genomic island containing putative polysaccharide production genes. Arch. Microbiol. 179:214-223. [DOI] [PubMed] [Google Scholar]
- 30.Partensky, F., J. Blanchot, and D. Vaulot. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. (Monaco) 19:457-475. [Google Scholar]
- 31.Pomati, F., B. P. Burns, and B. A. Neilan. 2004. Identification of an Na+-dependent transporter associated with saxitoxin-producing strains of the cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 70:4711-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 34.Roy, H., H. D. Becker, J. Reinbolt, and D. Kern. 2003. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl. Acad. Sci. USA 100:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakamoto, T., K. Inoue-Sakamoto, and D. A. Bryant. 1999. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC7002. J. Bacteriol. 181:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scanlan, D. J. 2003. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47:1-64. [DOI] [PubMed] [Google Scholar]
- 37.Smith, A. J., J. London, and R. Y. Stanier. 1967. Biochemical basis of obligate autotrophy in blue-green algae and thiobacilli. J. Bacteriol. 94:972-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo, G., and B. Palenik. 1997. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63:4298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toledo, G., and B. Palenik. 2003. A Synechococcus serotype is found preferentially in surface marine waters. Limnol. Oceanogr. 48:1744-1755. [Google Scholar]
- 41.Toledo, G., B. Palenik, and B. Brahamsha. 1999. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl. Environ. Microbiol. 65:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valladares, A., M. L. Montesinos, A. Herrero, and E. Flores. 2002. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43:703-715. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Q. F., and A. F. Post. 2000. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp strain WH9601. J. Bacteriol. 182:1764-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterisation of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71-120. [Google Scholar]
- 45.Zhang, Y. L., C. T. Ong, and K. Y. Leung. 2000. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology 146:999-1009. [DOI] [PubMed] [Google Scholar]