Abstract
Ostreococcus is a genus of widely distributed marine phytoplankton which are picoplanktonic in size (<2 μm) and capable of rapid growth. Although Ostreococcus has been detected around the world, little quantitative information exists on its contribution to planktonic communities. We designed and implemented a genus-specific TaqMan-based quantitative PCR (qPCR) assay to investigate the dynamics and ecology of Ostreococcus at the USC Microbial Observatory (eastern North Pacific). Samples were collected from 5 m and the deep chlorophyll maximum (DCM) between September 2000 and August 2002. Ostreococcus abundance at 5 m was generally <5.0 × 103 cells ml−1, with a maximum of 8.2 × 104 cells ml−1. Ostreococcus abundance was typically higher at the DCM, with a maximum of 3.2 × 105 cells ml−1. The vertical distribution of Ostreococcus was examined in March 2005 and compared to the distribution of phototrophic picoeukaryotes (PPE) measured by flow cytometry. The largest contribution to PPE abundance by Ostreococcus was ∼70% and occurred at 30 m, near the DCM. Despite its relatively low abundance, the depth-integrated standing stock of Ostreococcus in March 2005 was ∼30 mg C m−2. Our work provides a new technique for quantifying the abundance of Ostreococcus and demonstrates the seasonal dynamics of this genus and its contribution to picoeukaryote biomass at our coastal sampling station.
Picoeukaryotes (algae and protozoa of <2 μm in diameter) from marine and freshwater ecosystems have increasingly become the focus of ecological, physiological, and genomic studies. The discovery of their widespread distribution, large genetic diversity, and periodically high abundance has caused a reassessment of their importance in microbial food webs. In particular, picoeukaryotic phototrophs contribute significantly to microbial biomass and total primary productivity (39, 40, 70). As a consequence, phototrophic picoeukaryotes (PPE) are an important component of the base of pelagic microbial food webs. Picoeukaryotes serve as prey for nanoplanktonic (2 to 20 μm) phagotrophic protists (14), whose grazing activities provide a link to higher trophic levels (57) and a mechanism for nutrient remineralization (56).
Many picoeukaryotes have only recently been described in the scientific literature. These discoveries include both phototrophic (3, 16, 18, 31) and heterotrophic (30) taxa. The global distribution of many picoeukaryotes remains to be determined. However, Ostreococcus isolates have been documented at numerous sites around the world, including the Mediterranean Sea (22, 67), Long Island Sound, New York (48), the English Channel (32, 54), the Arabian Sea (9), and the North Pacific Ocean (13, 70; S. Suda et al., unpublished data).
The discovery of many picoeukaryotes has been aided by advances in instrumentation (e.g., flow cytometry [FCM]), improved culturing techniques, and the development of culture-independent molecular assays. Culture-independent molecular tools such as environmental cloning/sequencing of rRNA genes (49) and DNA fragment-based techniques such as denaturing gradient gel electrophoresis (DGGE) (45) and terminal restriction fragment length polymorphism analysis (41) have become particularly important for revealing the presence of previously unknown taxa (21, 23, 30, 43) and documenting the geographic ranges of both newly and previously described taxa (42, 53). In addition, the accumulation of DNA sequence information in public databases has facilitated the development of taxon-directed approaches to identify and count species of interest. Fluorescent in situ hybridization (FISH) and quantitative PCR (qPCR) have proven effective for the detection and quantification of specific microbial eukaryotes from complex natural assemblages (47, 71).
The use of qPCR for targeting eukaryotic microbes in environmental samples has received increased attention in recent years. A number of studies employing quantitative genetic methods for the study of bloom-forming algae that pose a threat to the health of humans or marine organisms have recently emerged (5, 8, 15, 28, 29, 50, 55). These approaches have detection accuracies over a large dynamic range and have substantially shorter sample processing times than more traditional techniques. Increasingly, qPCR methods have been developed for carrying out ecological studies of cryptic marine microbes.
We designed and validated a qPCR assay specific to members of the genus Ostreococcus and applied this technique to samples collected at the University of Southern California (USC) Microbial Observatory site, located in coastal waters of the eastern North Pacific. Subsurface samples from 5 m and the deep chlorophyll maximum (DCM) were analyzed for a 2-year period (September 2000 to August 2002) to reveal Ostreococcus dynamics. Our estimates constitute some of the highest abundances ever reported for Ostreococcus (up to 3.2 × 105 cells ml−1) at a coastal oceanic site. Additionally, Ostreococcus comprised a substantial proportion of the total number of phototrophic picoeukaryotes in March 2005. Overall, this minute prasinophyte was almost always present and occasionally abundant at our study site off the coast of Southern California.
MATERIALS AND METHODS
Collection and processing of environmental samples.
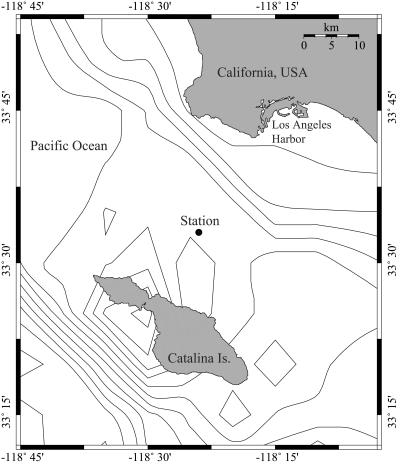
Samples were collected at approximately monthly intervals from the San Pedro Ocean Time-Series Station (latitude, 33°33′N; longitude, 118°24′W) (Fig. 1). Water samples were collected with Niskin bottles during conductivity-temperature-depth (CTD) casts from 5 m and from the DCM between September 2000 and August 2002. The depth of the DCM was determined by examination of real-time in situ fluorometry data.
FIG. 1.
Station location (33°33′N, 118°24′W), site of sample collection for the USC Microbial Observatory and San Pedro Ocean Time Series (SPOTS) projects, San Pedro Channel, Pacific Ocean. The map was drawn with Online Map Creation (http://www.aquarius.geomar.de/omc/).
Individual water samples were prescreened through a 47-mm inline filter apparatus (Pall Corp., East Hills, NY) containing 200-μm Nitex fabric (Sefar Filtration Inc., Monterey Park, CA) via gravity filtration directly from Niskin sampling bottles into carboys. All sample collection and processing were conducted with acid-washed (5% HCl) lab equipment. Two liters of the <200-μm filtrate from each depth was filtered onto a GF/F filter (Whatman Inc., Florham Park, NJ), using a gentle vacuum (<10 mm Hg), to collect planktonic biomass. Filters were loosely rolled and placed into 15-ml Falcon tubes (BD Biosciences, San Jose, CA). Two milliliters of lysis buffer (100 mM Tris [pH 8], 40 mM EDTA [pH 8], 100 mM NaCl, 1% sodium dodecyl sulfate) was added to each tube and frozen in liquid nitrogen. All samples were stored at −80°C until processing.
A vertical profile of Ostreococcus at 10 depths through the water column was obtained during our March 2005 time-series cruise. Samples were collected at 10-m intervals from near the surface to a depth of 80 m, with one additional sample at 100 m. The purpose of this cast was to examine the vertical distribution of Ostreococcus at a finer scale than was possible with our two-depth sample archive. Samples collected from the fine-scale vertical profile were prescreened through both 200- and 80-μm Nitex fabric. Size fractionation was performed to investigate whether Ostreococcus was associated with particles in the 80- to 200-μm size fraction.
Additional samples were collected from unfiltered seawater for analysis of the protistan community (in particular diatoms) by light microscopy. Seawater was sampled directly from Niskin bottles and preserved with Lugol's solution in amber glass collection jars (63). Lugol's solution-preserved samples were allowed to settle overnight onto counting chambers and examined by inverted light microscopy for microplankton enumeration.
qPCR sample processing.
Environmental samples and Ostreococcus culture-based standards were processed as lysates to minimize sample-to-sample variability due to losses associated with traditional DNA extraction procedures. Frozen samples and standards were thawed in a 70°C water bath prior to the addition of 200 μl of 0.5-mm zirconia/silica beads (BioSpec Products, Bartlesville, OK). Cells were lysed by bead beating (vortexing at highest setting for 30 to 60 s) followed by heating in a 70°C water bath for 3 to 5 min. Two additional rounds of bead beating and heating were conducted to ensure complete cell lysis. Lysates were filtered through 0.22-μm, low-binding cellulose acetate syringe-tip filters (Corning Life Sciences, Acton, MA) to remove cellular debris, filter remnants, and beads. Filtered lysates were collected in sterile cryotubes (Nalge Nunc, Rochester, NY) and frozen at −20°C.
qPCR probe and primer design.
A TaqMan (Roche Molecular Systems, Inc.)-based qPCR assay (34, 35) was developed to enumerate members of the genus Ostreococcus. An 18S rRNA gene probe (Ostreo-670F; Table 1) specific to positions 670 to 697 (relative to Ostreococcus tauri [GenBank accession no. YI5814]) (17) was designed by manually searching a ClustalX (66) alignment of prasinophyte 18S rRNA gene sequences retrieved from GenBank (6). Only two Ostreococcus sequences (Y15814 [17] and AB058376 [S. Suda et al., unpublished data]) were available in GenBank at the time of probe and primer design. A full-length Ostreococcus 18S rRNA gene sequence (GenBank accession no. DQ007077) from our study site aided in the development of our qPCR probe and primers. This clone was selected for full-length sequencing (∼6× coverage) based on its prior identification as Ostreococcus by single-pass DNA sequencing (∼500 bp). The full-length Ostreococcus clone was 98.7% similar to Ostreococcus tauri (Y15814) across 1,738 common nucleotide positions revealed by pairwise sequence comparison in BioEdit, version 7.0.1 (33).
TABLE 1.
Properties of Ostreococcus-specific TaqMan probe and primersa
| Name | Sequence (5′→3′) | Length (nucleotides) | Tm (°C) | % GC |
|---|---|---|---|---|
| Ostreo-670F | CAGCTTCCTGGTGAGGAGGTGTGCTTCA | 28 | 69.4 | 57.1 |
| Ostreo-636F | GTCGGTCCGCCGTTAGGTGTG | 21 | 63.8 | 66.7 |
| Ostreo-822R | CCCGTCCCGAGACCAACGAAA | 21 | 62.9 | 61.9 |
The Ostreococcus TaqMan probe (Ostreo-670F) contained a 5′ modification with the reporter molecule 6-carboxyfluorescein and a 3′ modification with Black Hole Quencher 1. The high-performance liquid chromatography-purified probe and primers were diluted to stock concentrations of 150 μM and stored as 20-μl aliquots at −20°C until use. Stock solutions were diluted 1:10 with sterile water prior to reaction setup. The melting temperatures (°C) of the probe and primers were estimated using Beacon Designer software (Premier Biosoft International, Palo Alto, CA).
The three Ostreococcus sequences described above were 100% identical in the selected probe region (see Table S1 in the supplemental material). This region displayed a 6-nucleotide gap (among other mismatches) between the C and the T at a position four nucleotides from the 3′ end of the probe relative to all other prasinophytes in an alignment of Mamiellales sequences. A recent (January 2006) BLAST search (1) indicated that 11 additional Ostreococcus 18S rRNA gene sequences had been submitted to GenBank since the time of our probe and primer design (see Table S1 in the supplemental material). The TaqMan probe showed 100% sequence identity to all additional Ostreococcus sequences, with one exception, AY425313 (32), which displayed five mismatches compared to the probe and all other Ostreococcus sequences (see Table S1 in the supplemental material). Such mismatches would likely render the Mediterranean Ostreococcus strain represented by the AY425313 sequence undetectable. In addition to Ostreococcus sequences, the probe matched a number of “uncultured environmental” 18S rRNA gene clone sequences in GenBank, suggesting the widespread distribution of this genus. Not et al. (46) designed an Ostreococcus FISH probe targeting a similar region of the 18S rRNA gene to that targeted by our TaqMan probe to successfully detect Ostreococcus in environmental samples (46).
Forward and reverse qPCR primers (Table 1) were generated with Beacon Designer 2.1 (Premier Biosoft International, Palo Alto, CA), which predicted a 187-bp PCR product (see Table S1 in the supplemental material). The forward primer (Ostreo-636F) was located at nucleotide positions 636 to 656, and the reverse primer (Ostreo-822R) was located at positions 802 to 822 relative to the Ostreococcus tauri sequence (YI5814) (see Table S1 in the supplemental material). The 3′ end of the forward primer was designed to anneal as close to the 5′ end of the Ostreococcus probe as possible to ensure rapid hydrolysis of the probe and nearly instantaneous fluorescence detection during the combined annealing/extension phase (11). Additionally, the forward primer was designed for specificity to Ostreococcus, thus enhancing the stringency of detection and decreasing the probability of primer hybridization to nonspecific targets. The reverse primer was not unique to Ostreococcus but was specific to phytoplankton within the order Mamiellales. Four of the 11 new Ostreococcus sequences noted above displayed either 1- or 2-bp mismatches with our Ostreococcus primers (see Table S1 in the supplemental material). It is conceivable that such mismatches would render these strains undetectable by our method; however, it should be noted that these sequences were recovered from the Atlantic Ocean and the Mediterranean Sea (32; M. Viprey and D. Vaulot, unpublished data) and, to our knowledge, have not been detected in the Pacific Ocean. The Ostreococcus primers and probe (Ostreo-670F) were synthesized by QIAGEN (Alameda, CA). The TaqMan probe included a 6-carboxyfluorescein fluorescent reporter molecule at the 5′ end of the oligonucleotide and Black Hole Quencher 1 (Biosearch Technologies, Inc., Novato, CA) at the 3′ end.
Calibration of Ostreococcus qPCR method.
The qPCR method was calibrated by two methods, with plasmid DNA from an Ostreococcus 18S rRNA gene clone and with cell lysates from an Ostreococcus culture. The cloned 18S rRNA gene was used to test reaction conditions and assay sensitivity, while cell lysate reactions were the primary reference for calibrating the abundance of Ostreococcus in environmental samples. A 3.2 ng μl−1 stock of the cloned Ostreococcus 18S rRNA gene was serially diluted to create a standard curve spanning 8 orders of magnitude. One-microliter aliquots of each dilution were used for each qPCR (see details below).
An actively growing culture of Ostreococcus sp. (MBIC-10636) was used to establish a standard curve for calibration of the qPCR assay by direct comparison of cell numbers to the qPCR threshold cycle (CT). Flow cytometry indicated an abundance of 3.2 × 107 (±0.1 × 107) cells ml−1 in a stock culture of Ostreococcus. Standards were prepared by adding 100, 10, and 1 ml of undiluted Ostreococcus culture to triplicate 1-liter aliquots of filtered seawater (FSW) for the three highest concentration standards. The three lowest concentration standards were constructed by adding 1 ml of serially diluted stock culture (10−1, 10−2, and 10−3) to triplicate 1-liter aliquots of FSW. Ostreococcus-spiked FSW aliquots were filtered onto 47-mm GF/F filters and prepared as lysates as described above. The preparation of standards by spiking cultured cells into 1 liter of FSW simulated the processing procedure applied to natural samples.
qPCR optimization and reaction conditions.
Optimization reaction mixtures and plasmid standards for qPCR were prepared with 1 μl of the cloned Ostreococcus 18S rRNA gene (3.2 ng μl−1) plus 49 μl of reagent master mix (see below) and run on a Bio-Rad iCycler machine (Hercules, CA). Initial testing of thermal protocols was performed with a Mg2+ concentration of 2.5 mM and an annealing temperature of 55°C. Two- and three-step thermal protocols generated similar amounts of the 187-bp product, as determined by the intensity of SYBR gold (Molecular Probes, Eugene, OR)-stained DNA bands on a 1.2% agarose gel. Further optimization tests were performed using a two-step amplification protocol (described below) typical of most TaqMan assays.
The annealing/extension temperature was optimized with the gradient feature of the iCycler across a temperature range of 60 to 70°C. The 60°C reaction reached the CT ∼2 cycles earlier than the reaction at 70°C (13.6 versus 15.4) and attained higher relative fluorescence unit (RFU) values at the plateau phase of amplification (>1,100 RFU at 60°C versus ∼500 RFU at 70°C). This result was expected since the TaqMan probe is required to anneal to the template before the primers and to remain hybridized until hydrolysis by Taq polymerase, a condition which is favored at lower annealing/extension temperatures (10).
The optimal Mg2+ concentration was determined by testing reactions prepared with 2.5 to 5.0 mM Mg2+. Average CT values decreased slightly from 12.7 to 11.4 with increasing Mg2+ concentrations; however, there was no significant difference between the average CT values for reactions at 4.5 and 5.0 mM Mg2+ (P > 0.10). DNA amplification reached higher RFU values with increasing Mg2+ concentrations (1,100 RFU at 2.5 mM versus ∼2,000 RFU for 4.5 and 5.0 mM). The lowest concentration of Mg2+ that produced maximum RFU values and the earliest CT was 4.5 mM and was therefore selected as the optimal concentration for subsequent reactions. Higher RFU values enhance the ability to distinguish amplification over baseline fluorescence.
The Ostreococcus primer set was tested with Bio-Rad's iQ SYBR green supermix to determine the specificity of amplification, as indicated by postamplification melting-curve analysis. Melting-curve analysis indicated a single PCR product. Secondary PCR products and primer dimers were not detected in the melting-curve profile (see Fig. S1 in the supplemental material).
Sample and cell standard lysates were diluted 1:100 with sterile Milli-Q water to minimize PCR inhibition due to the presence of lysis buffer and residual cellular contents. The optimal volume of diluted lysate for use in qPCR assays was determined because increasing the lysate volume increased the reaction sensitivity only to a certain point. A lysate for this test was created by spiking 50 ml of a dense Ostreococcus culture (∼2.0 × 107 cells ml−1) into 1 liter of FSW and processing the sample as described above. Various volumes of 1:100 diluted lysate were tested in 50-μl reaction mixtures to determine the appropriate lysate volume (see Fig. S2 in the supplemental material). Lysate volumes of >10 μl had no further effect on the reduction of qPCR CT values, as evaluated by unpaired t tests assuming equal variance. Specifically, the average CT values decreased significantly (P < 0.05) with increasing volumes until 15 μl, which did not have a significantly different CT value from the average value obtained for 10 μl (P = 0.07).
The final reaction conditions in 50-μl volumes included 500 nM of each primer and 250 nM of TaqMan probe. Reaction mixtures also contained the following reagents: 250 μM of each deoxynucleoside triphosphate, 4.5 mM Mg2+, 1× buffer B, and 2.5 units of Taq polymerase in buffer B (Promega, Madison, WI). Bovine serum albumin (A7030; Sigma) was added to each reaction to a final concentration of 300 ng μl−1 to enhance the PCR (37). All reagents, samples, and standards were prepared on ice prior to thermal cycling. Ten microliters of each lysate was loaded into a well of a 96-well PCR plate, followed by the addition of 40 μl of qPCR master mix. Plates were sealed with optically clear tape (Bio-Rad), centrifuged briefly to remove bubbles, and transferred to the iCycler machine.
Cell-based standards were comprised of triplicate independent lysates for each of six Ostreococcus concentrations and were run with every set of unknown samples to facilitate the conversion of CT values of unknowns to equivalent Ostreococcus abundances. Three individual reactions were set up for each sample to control for reaction variability. Samples and standards were cycled with the following thermal protocol: 95°C for 90 s (1×), 70 cycles of 95°C for 15 s and 60°C for 30 s, and a hold at 4°C. Real-time data were collected during the annealing/extension step.
Cross-reactivity testing with protistan cultures.
Fifty-two clonal cultures of marine and freshwater protists were grown to high abundance in 250-ml sterile culture flasks for Ostreococcus qPCR cross-reactivity testing. Marine phytoplankton cultures were grown in K medium (36) modified with 36 μM PO42−, while freshwater phytoplankton cultures were grown in DY-IV medium (2). Heterotrophic protists were grown in sterile seawater or freshwater amended with sterile yeast extract (final concentration, 0.005%) plus several rice grains. Abundances of cultured protists were checked by light microscopy after 1 to 2 weeks of growth to confirm a minimum abundance of ∼1.0 × 103 cells ml−1. Cell cultures were filtered onto Whatman GF/F filters, lysed, and stored at −20°C. Selected culture lysates were amplified with universal eukaryote-specific PCR primers to ensure the presence of the 18S rRNA gene prior to cross-reactivity testing with Ostreococcus primers and probe. Lysates for qPCR testing were diluted 1:100 with sterile water, and 10 μl of diluted lysate was used in duplicate 50-μl qPCR assays. An Ostreococcus lysate served as the positive control.
Forty-eight of the 52 lysates returned negative results for duplicate qPCR amplifications (see Table S2 in the supplemental material). Two of the 52 lysates (Asterionellopsis glacialis and Paraphysomonas vestita) were positive in one of two reactions, while both duplicates of Aureoumbra lagunensis were positive. These nonspecific amplifications displayed much lower RFU values than the Ostreococcus positive control and occurred late in the thermal protocol (CT ≥ 50). The three “positive” lysates were tested a second time and returned negative results for Ostreococcus cross-reactivity. The initial amplifications of nontarget taxa were attributed to aerosol contamination from the cloned Ostreococcus 18S rRNA gene, which initially was run as a secondary positive control but was omitted from later runs.
Comparison of picoeukaryotes by flow cytometry.
Environmental samples were collected in triplicate from the March 2005 vertical profile for analysis by flow cytometry (described above). Samples were prescreened through 80-μm Nitex fabric, preserved with formalin (final concentration, 1%), and frozen in liquid nitrogen. Analysis by FCM provided the putative abundance of Ostreococcus as a subset of total PPE.
An actively growing culture of Ostreococcus (MBIC-10636) was used to calibrate the flow cytometer for environmental detection of this genus. Ostreococcus cells were grown for approximately 1 week at 20°C on a 12-h light/dark cycle with an average irradiance of 165 microeinsteins m−2 s−1 (in water). FCM gate parameters for counting Ostreococcus were established with a four-color, dual-laser FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Cultured cells formed a discrete FCM region based on forward-angle light scatter and red fluorescence (FL3 channel), detection parameters that have been used in previous studies (16, 18, 48). Micromonas pusilla (CCMP-487) and Micromonas sp. (a local isolate) were also analyzed by FCM to investigate potential overlap with the Ostreococcus FCM gate parameters. Micromonas and Ostreococcus are similarly sized prasinophytes that co-occur at the USC Microbial Observatory.
Nucleotide sequence accession number.
An environmental Ostreococcus sp. clone from the USC Microbial Observatory (North Pacific Ocean) was sequenced in both directions to yield a full-length 18S rRNA gene construct. The sequence was deposited in GenBank and assigned the accession number DQ007077.
RESULTS
Calibration of qPCR.
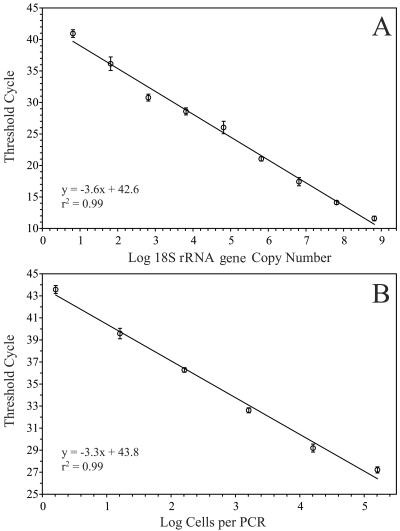
Initial testing of the TaqMan reaction sensitivity with the 18S rRNA gene cloned from Ostreococcus indicated linearity across 8 orders of magnitude (r2 = 0.99) (Fig. 2A). This range corresponded to 6.5 × 100 to 6.5 × 108 copies of the Ostreococcus 18S rRNA gene per reaction. Standardization of the qPCR assay with the cloned 18S rRNA gene indicated a reaction sensitivity of ∼10 copies for highly purified templates like plasmid DNA containing cloned inserts.
FIG. 2.
Calibration of Ostreococcus qPCR method with a dilution series of plasmid DNA (A) and a dilution series of an Ostreococcus culture, processed as cell lysates (B).
PCR analysis of lysates from known numbers of cultured Ostreococcus cells was used to convert CT values for unknown environmental samples directly into cell abundances. Cell lysate standards prepared from an Ostreococcus culture were analyzed on each plate with environmental samples. The cell-based standard curve displayed linearity over 5 orders of magnitude, ranging from the genomic equivalents of 16 to 160,000 cells per reaction (r2 = 0.99) (Fig. 2B). The cell-based standard curve depicted in Fig. 2B represents an average of three standard curves analyzed on three different days using the same set of lysates (stored frozen between runs). Error bars represent standard deviations of pooled determinations. This dynamic range bracketed the upper range of Ostreococcus abundance in all (diluted 1:100) environmental samples. In practice, the lowest reliably detected standard (equivalent to 16 cells per reaction) translated into the detection of 320,000 cells in 2 ml of undiluted lysate (e.g., 16 cells in 10 μl of 1:100 diluted lysate, equivalent to 160 cells μl−1 of undiluted lysate, × 2,000 μl lysis buffer = 320,000 cells). Thus, the limit of detection was approximately 100 Ostreococcus cells ml−1 in natural samples for typical sample collection volumes (2 to 4 liters).
Vertical profile of Ostreococcus.
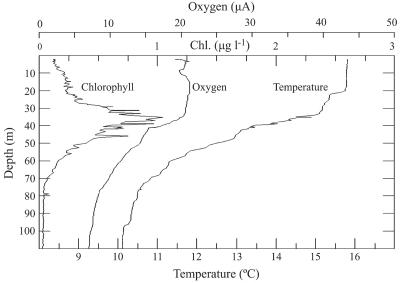
Profiles of chlorophyll a, oxygen, and temperature in March 2005 revealed a mixed surface layer down to a depth of 20 m at our study site (Fig. 3). The DCM at ∼35 m corresponded to the top of the oxycline and the middle of the main water column thermocline. This water column profile was fairly typical of profiles observed at our sampling site throughout much of the year (data not shown). The vertical profile of total PPE abundance by FCM reflected the profiles of the physical and chemical measurements described above. PPE abundances were fairly uniform in the upper water column down to a depth of 40 m, where they reached a maximum value of nearly 1.0 × 104 cells ml−1 just below the depth of the DCM (Fig. 4). PPE abundances decreased to ∼6.0 × 103 cells ml−1 at 50 to 60 m and dropped precipitously to <200 cells ml−1 at 80 m and below.
FIG. 3.
CTD profile at the USC Microbial Observatory during March 2005 showing in situ chlorophyll fluorescence, oxygen content, and temperature through the upper 100 m of the water column. Relatively invariant properties in the upper 20 m indicated a well-mixed surface layer.
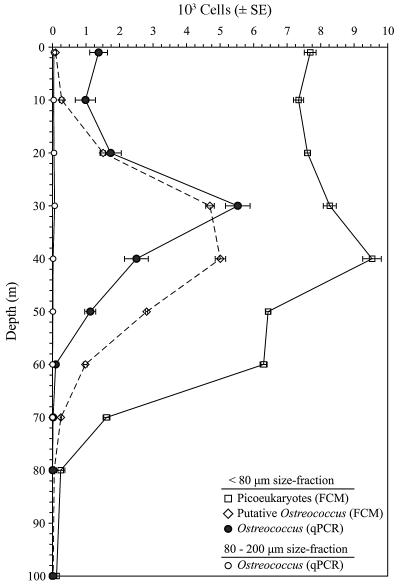
FIG. 4.
Vertical distribution of total picoeukaryotes (open squares) and Ostreococcus sp. in the upper water column at the USC Microbial Observatory site during March 2005. Ostreococcus abundances were determined for two size classes (<80 μm and 80 to 200 μm) by qPCR (open and closed circles, respectively) and by FCM (open diamonds). Error bars represent the standard errors of the estimates.
Ostreococcus abundance in the <80-μm size fraction was determined by qPCR and FCM and compared to the total PPE abundance (Fig. 4). Ostreococcus abundances determined by qPCR were fairly uniform from the surface to 20 m, ranging from 1.0 × 103 to 2.0 × 103 cells ml−1, while a distinct subsurface maximum of 5.5 × 103 cells ml−1 was observed at 30 m, just above the DCM, at the top of the primary water column thermocline (Fig. 3 and 4). Ostreococcus abundances declined to <100 cells ml−1 at 60 m and below. Overall, the qPCR-based Ostreococcus abundance estimates followed the trends in the FCM-based PPE profile, with qPCR estimates generally constituting 13 to 26% of the total PPE abundance at particular depths. A notable exception occurred at 30 m, where the qPCR estimate of Ostreococcus comprised nearly 70% of the total PPE abundance, indicating that Ostreococcus can dominate PPE assemblages. Ostreococcus abundances in the 80- to 200-μm size fraction were generally insignificant (<100 cells ml−1) at all depths tested (1 to 70 m).
In addition to measurement by qPCR, the putative abundance of Ostreococcus was measured by FCM, using gate parameters established with an Ostreococcus culture. Cytometry-based estimates were <200 cells ml−1 for depths down to 10 m and then increased to a broad peak of ∼5.0 × 103 cells ml−1 between 30 and 40 m before decreasing to <200 cells ml−1 at 70 m and below. Ostreococcus abundances determined by FCM differed from qPCR-based estimates in comparisons for 7 of the 10 depths (P < 0.05). Cytometry-based abundance estimates of Ostreococcus were lower than qPCR estimates for surface water samples and higher than qPCR estimates for depths below 30 m. However, cytometry and qPCR estimates were not significantly different at 10, 20, and 30 m (P = 0.08, 0.53, and 0.10, respectively; α = 0.05).
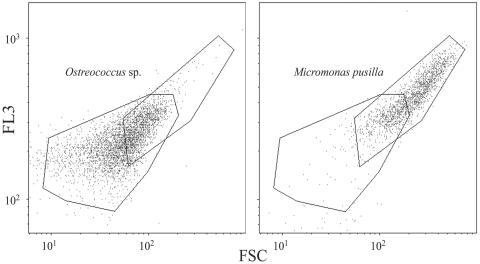
In an effort to reconcile differences between qPCR estimates of Ostreococcus and putative abundances determined by FCM, the FCM characteristics of two Micromonas cultures were compared to those of a single Ostreococcus culture to examine the potential contribution of other small prasinophytes to the cytometric signature of Ostreococcus. Approximately 30% of the cytogram events detected with the Micromonas culture overlapped with the cytogram for Ostreococcus (Fig. 5). These results indicated a substantial overlap in Ostreococcus and Micromonas cytograms for the tested cultures. This test indicated the inability to accurately distinguish these genera by FCM and further illustrated the need for taxon-specific molecular approaches to establish species-specific abundances and distributions.
FIG. 5.
Flow cytometric signatures of Ostreococcus sp. (left panel) and Micromonas pusilla (right panel) indicating overlap in detection regions (polygons) that could lead to an overestimate of Ostreococcus abundance of ∼30% when both genera are present.
Abundance and distribution of Ostreococcus through time.
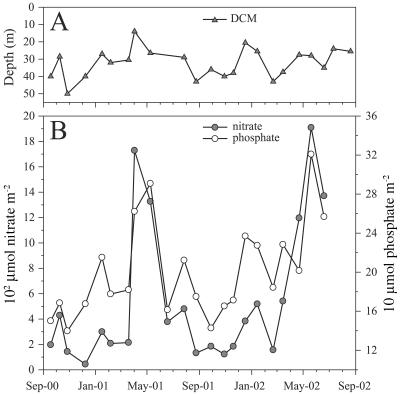
The depth of the DCM at the USC Microbial Observatory varied between 14 and 50 m during the 2-year study period (Fig. 6A). The shallowest depth of the DCM occurred during winter to early spring of 2001, while the deepest DCM was observed in November 2000. The overall shoaling or deepening of the DCM appeared to be more dependent on episodic events (vertical and horizontal advection) than seasonal trends. The 23-month average depth of the DCM was 32 m and varied within a relatively narrow range (±8 m [standard deviation]), which provided a stable water column feature to sample throughout the study. At times, the study site displayed relatively “open-ocean” characteristics (e.g., low nutrient concentrations, low planktonic biomass in net tows, and high light transmission) (data not shown), and at other times, the waters were much more coastal in nature, being visibly greener and more characteristic of nearshore (<1 km from the coast) water masses and phytoplankton communities.
FIG. 6.
Depth variability of the DCM over the period of sample collection (A) and depth-integrated (1 to 40 m) nitrate (shaded circles) and depth-integrated phosphate (open circles) contents for the same period (B). Nutrient data for the last two sampling dates were not available.
Depth-integrated profiles of nitrate and phosphate (courtesy of the USC Wrigley Institute for Environmental Studies) were examined for seasonal trends to help explain the observed patterns of Ostreococcus abundance (Fig. 6B). Nutrients were expressed as depth-integrated values over the upper 40 m of the water column based on discrete measurements at 1, 10, 20, 30, and 40 m. Integrated nitrate and phosphate values were positively correlated (P < 0.01) at the USC Microbial Observatory site due to the dominance of coastal upwelling as the primary influence on nutrient delivery at this location. Two major nutrient upwelling events between March and June 2001 and February and June 2002 were revealed by the profiles (Fig. 6B).
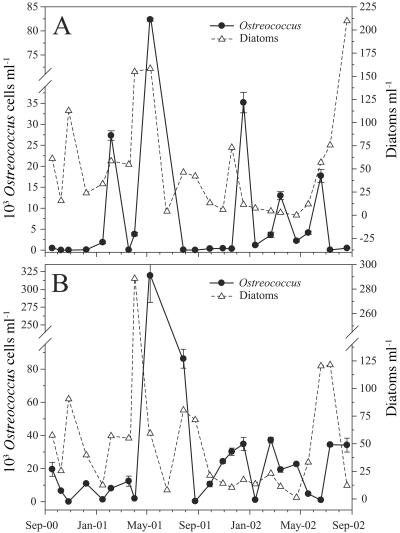
Ostreococcus abundances determined by qPCR indicated that this genus was nearly ubiquitous at our study site, as it was detected at one or both depths on all dates except late October 2001 (Fig. 7). Month-to-month abundances of Ostreococcus during this study period were highly variable, ranging over 4 orders of magnitude at 5 m (from <100 cells ml−1 to 8.2 × 104 cells ml−1; Fig. 7A) and over 5 orders of magnitude at the DCM (from <100 cells ml−1 to 3.2 × 105 cells ml−1; Fig. 7B). Ostreococcus abundances were generally higher at the DCM than at 5 m, reaching values of >1.0 × 104 cells ml−1 on 14 of 23 sampling dates, in contrast to only 5 of 23 dates for 5 m. The 23-month average abundance of Ostreococcus for samples collected at the DCM (3.1 × 104 cells ml−1) was approximately four times greater than the average for samples collected at 5 m (8.5 × 103 cells ml−1). Abundance often oscillated from <100 to several tens of thousands of cells ml−1 between consecutive monthly samples. Ostreococcus was below the detection limit (<100 cells ml−1) in 6 of 23 samples from 5 m and in 1 sample from the DCM.
FIG. 7.
Ostreococcus and total diatom abundances at 5 m (A) and the deep chlorophyll a maxima (B) during a 2-year period at the USC Microbial Observatory sampling site.
Episodic peaks in the abundance of Ostreococcus occurred in all seasons and at both depths, in particular during February and May 2001 and December, March, and June 2002 (Fig. 7). Two prolonged periods of low Ostreococcus abundance (<1.0 × 103 cells ml−1) at 5 m were apparent, between September 2000 and January 2001 and between July and November 2001. In contrast, abundances of Ostreococcus of <1.0 × 103 cells ml−1 at the DCM were detected on only three dates (Fig. 7B). There was no significant correlation between Ostreococcus abundance and the depth of the DCM (P > 0.05; data not shown). The average DCM depth on dates with the 11 highest Ostreococcus abundances and the 12 lowest abundances was 32 m.
A major bloom of Ostreococcus was detected at both 5 m and the DCM (27 m) in May 2001 (Fig. 7). Ostreococcus abundance increased from 3.8 × 103 to 8.2 × 104 cells ml−1 at 5 m and from 1.9 × 103 to 3.2 × 105 cells ml−1 at the DCM between April and May 2001. The Ostreococcus bloom was preceded by high abundances of diatoms in April 2001 (∼200 cells ml−1 at 5 m and ∼300 cells ml−1 at the DCM). The peak in diatom abundance in April 2001 corresponded to high nitrate and phosphate concentrations in the upper 40 m of the water column (Fig. 6B). Abundances of Ostreococcus were among the lowest values detected during peak diatom blooms (Fig. 7). Nutrient upwelling events of similar magnitude occurred in April-May 2001 and May-June 2002 (Fig. 6B). Diatom abundances at 5 m and the DCM increased near the end of the study period, but Ostreococcus abundances increased only modestly at the DCM during this time frame (Fig. 7A and B).
DISCUSSION
Quantitative PCR for ecological research.
Real-time qPCR analysis is increasingly the method of choice for quantifying abundances of protistan taxa in environmental samples (5, 8, 15, 28, 44, 50, 69, 71). This approach combines extreme sensitivity with the ability to process large numbers of samples rapidly. It is conceivable that global surveys of particular taxa could be completed in a matter of days to help address the question, “Is everything everywhere?” (24-26). Although most qPCR assays have approximately the same sensitivity (detection of several to tens of target copies), we developed a TaqMan assay because the TaqMan probe provides another level of reaction specificity compared to primer-only SYBR green assays.
Several methods for relating CT values from environmental samples to gene copy numbers or cell abundance have been applied in qPCR-based studies of protistan taxa. Plasmid DNAs containing cloned target sequences have been used in a number of studies to create standard curves (28, 69, 71), and this is presently the only available method for calibrating qPCR detection of uncultured taxa (65). Calibration with plasmid DNA is advantageous in that exact numbers of target genes can be calculated by measuring the concentration of a DNA standard. However, a notable disadvantage when using plasmid standards is that environmental samples should be fully extracted and purified to ensure that amplification is directly comparable to the amplification of purified plasmid DNA. Complete extraction and purification may lead to high variability among replicate extracts and to spurious results due to inconsistent DNA recovery. The alternative to plasmid-based calibration involves calibration with a cultured target organism. Variations on cell-based standardizations include dilution of a single genomic DNA extract from a known number of cells (5), dilution of a cell culture of known concentration followed by multiple DNA extractions (8, 15), spiking specific volumes of target organisms directly onto filters for collection and extraction (50), and spiking cultured cells into filtered seawater aliquots for subsequent collection by filtration and preparation as lysates (44). Cell-based calibration approaches (when possible) are the most direct means of relating CT values from qPCR assays to cell abundance. Also, the use of lysates prepared simultaneously from both standards and unknowns minimizes variability due to slight variances in DNA extraction efficiency and/or reagent quality.
Several caveats should be considered before the application of cell-based qPCR standardizations. Specifically, cell constituents released by lysis and the lysis buffer itself can act as PCR inhibitors leading to reductions in reaction sensitivity or accuracy. We minimized inhibition problems in our TaqMan assay by diluting all lysates 1:100 with sterile Milli-Q water and optimizing the volume of diluted lysate used in 50-μl reaction mixtures (see Fig. S2 in the supplemental material). Cell-based qPCR calibrations can also be affected by the accuracy of microscopy- or FCM-based cell counts, thereby requiring replicate determinations of cell abundances in cultures. We quantified triplicate samples of live Ostreococcus cells by FCM immediately before dilutions of stock culture and subsequent collection by filtration to ensure the accuracy of standard curves. Finally, variability in the rRNA gene copy number per cell may affect the interpretation of cell-based calibrations. Recent evidence suggests that the copy number of 18S rRNA genes in Ostreococcus is relatively small, ranging from two to four copies per cell (71). However, there have been no comprehensive studies examining the variability in 18S rRNA gene copy number in Ostreococcus cells during different stages of growth. We did not attempt to control for variability in the 18S rRNA gene copy number of our cultured cells but assumed that a culture in nonsynchronous, exponential growth would provide a reasonable average copy number per cell. Furthermore, we assumed that individuals in natural populations of Ostreococcus expressed a similar range of 18S rRNA gene copy numbers as cultured cells.
Our Ostreococcus TaqMan assay was performed for 70 cycles of amplification to maximize the sensitivity of the assay. The small genome size of Ostreococcus (20, 52), its low 18S rRNA gene copy number (71), and the low concentration of DNA in lysates were factors contributing to the requirement of a large number of amplification cycles. Most PCR assays are carried out using 35 or fewer amplification cycles to prevent the introduction of PCR bias to the final pool of amplicons (with overrepresentation of particular taxa) when amplifying environmental samples for cloning and sequencing (64) and to avoid the amplification of nonspecific targets, primer dimers, or contaminant DNA. Amplification of nonspecific 18S rRNA gene fragments and the formation of primer dimers were not apparent in our qPCR assays because of the high specificity of our probe and primers for the target region (see Fig. S1 in the supplemental material). Additionally, interference of Ostreococcus-specific rRNA gene amplification by the DNAs of other protistan taxa in environmental lysates was not likely a significant factor given the results of our taxonomically diverse cross-reactivity tests (see Table S2 in the supplemental material). PCR contamination was adequately controlled, as indicated by the results of our negative controls (Milli-Q water), which remained below the amplification threshold for all sample runs (data not shown). Contamination was primarily controlled by isolating preparation steps during the qPCR setup and avoiding the use of plasmid DNAs containing 18S rRNA gene inserts in the vicinity of qPCR lysate preparation or reaction setup.
Molecular detection has become an essential tool for documenting the presence and abundance of Ostreococcus because of the diminutive size of species within this genus and the lack of definitive morphological features. A variety of molecular approaches have begun to reveal its ecological dynamics and global distribution. Ostreococcus has been quantified by 18S rRNA probes (47) and detected in environmental clone libraries from a variety of marine ecosystems (7, 13, 32, 46, 54). Most recently, qPCR methods including SYBR green (71) and TaqMan (this study) have provided quantitative estimates of Ostreococcus abundances in environmental samples collected from time-series stations. Ostreococcus appears to be a significant component of PPE assemblages in some of the ecosystems where it has been detected. For example, Ostreococcus constituted ∼10% of the clones in an 18S rRNA gene library from the USC Microbial Observatory (13).
Vertical and temporal distribution of Ostreococcus.
Ostreococcus has been isolated and cultured previously from depths across the euphotic zone (68); however, our qPCR-based vertical profile from March 2005 constitutes the first discrete profile of Ostreococcus abundance through the euphotic zone at an oceanic station. This profile revealed the highest abundance of Ostreococcus near the depth of the DCM (Fig. 3 and 4). Ostreococcus comprised a substantial fraction of the PPE abundance across the euphotic zone and dominated the picoeukaryotic assemblage near the DCM. The maximum qPCR-based abundance of Ostreococcus in March 2005 reached 5.5 × 103 cells ml−1 (Fig. 4), which was low compared to abundance estimates from time-series samples (Fig. 7) but higher than Ostreococcus abundances from other locations (46, 71). The vertical distribution of Ostreococcus in the euphotic zone (Fig. 4, open circles) is typical for phototrophic picoeukaryotes (12, 39) and showed a strong correspondence to the physical structure of the water column (Fig. 3).
Ostreococcus abundances estimated by qPCR and FCM were significantly different for some depths but not significantly different for other depths (see Results) (Fig. 4). Samples for which FCM-based estimates were greater than qPCR-based estimates (e.g., samples from 40 to 70 m) were likely the result of non-Ostreococcus detections in the Ostreococcus FCM gate. The prasinophyte Micromonas was identified as a potential cause of discrepancies between qPCR- and FCM-based measurements. Micromonas and Ostreococcus regularly co-occur at other coastal sites (46, 71). The overlap of Micromonas and Ostreococcus detection by FCM (∼30%) may have lead to an overestimation of the Ostreococcus abundance by FCM. Additionally, the lower qPCR estimates could have been due to the probe-primer mismatches noted above; however, this is extremely unlikely since none of the other strains have been detected in extensive clone libraries from the same site (P. D. Countway and D. A. Caron, unpublished data). FCM-based estimates of Ostreococcus abundance at the two shallowest depths in the vertical profile were lower than the corresponding qPCR estimates, which was possibly related to differences in the chlorophyll contents of near-surface Ostreococcus assemblages. Ostreococcus cells collected from near-surface assemblages may have been high-light-adapted ecotypes (53) with less chlorophyll per cell than the Ostreococcus culture used for FCM calibrations.
Monthly sample collections at our study site indicated that Ostreococcus abundances varied by several orders of magnitude at both 5 m and the DCM, with higher abundances typically occurring at the DCM (Fig. 7). To our knowledge, this is the first study to reveal the abundance and distribution of Ostreococcus across multiple depths for a multiyear time series. Prior investigations have reported abundances of Ostreococcus in the upper 1 to 2 m of the water column, where Ostreococcus was a lower percentage of total picoeukaryotes (46, 71) than that observed in the present study. Not et al. (46) conducted a survey of prasinophytes at a time-series station in the English Channel, using FISH-tyramide signal amplification (47). Picoeukaryotes belonging to the order Mamiellales (<1.0 × 104 cells ml−1) were detected in low abundance, with only 3% of the count attributed to Ostreococcus (46). Zhu et al. (71) reported the relative abundances of Ostreococcus from a time-series study in the Mediterranean Sea. Their qPCR-based Ostreococcus detections never accounted for more than 1% of qPCR-based PPE detections (71). These low contributions contrast with our findings that Ostreococcus may constitute a significant fraction of total phototrophic picoeukaryotes, especially near the DCM (Fig. 4). However, Ostreococcus comprised a relatively low percentage of the total PPE assemblage at the shallowest depths of our study site (Fig. 4), which is consistent with previous reports. These results lead us to speculate that previous studies from other sites may have underestimated the importance of Ostreococcus, which appears to exhibit subsurface abundance maxima.
High concentrations of phototrophic picoeukaryotes (>1 × 105 cells ml−1), as determined by flow cytometry, have previously been reported from nearshore locations along the Mediterranean coast of France (18, 67) and Long Island, N.Y. (48). The authors of those studies reported that Ostreococcus was a significant component of PPE assemblages. Despite occasionally high abundances (>1 × 105 cells ml−1), PPE from the Mediterranean site averaged 3.5 × 104 cells ml−1 over a 27-month time series at a depth of 0.5 m (67), a value similar to the interannual average DCM abundance of Ostreococcus at our study site (3.1 × 104 cells ml−1) but greater than the average at 5 m (8.5 × 103 cells ml−1) (Fig. 7). Ostreococcus at our study site was frequently present at abundances of >1.0 × 104 cells ml−1 (14 of 23 samples from the DCM, but only 5 of 23 samples from 5 m) (Fig. 7).
Discrete measurements of qPCR-based Ostreococcus abundance from the vertical profile during March 2005 yielded a depth-integrated carbon biomass of 26.6 mg C m−2 based on a cell-specific carbon conversion factor of 212 fg C cell−1 (70). The use of this carbon conversion factor is justified because the culture from which this value was derived (CCE9901) was isolated from a location (Scripps Pier) relatively close (∼130 km) to our sampling site. Furthermore, CCE9901 is a clade A Ostreococcus ecotype (32), which is the only Ostreococcus ecotype that has been detected at our site, sharing ≥99% 18S rRNA gene sequence identity with all Ostreococcus clones detected at the USC Microbial Observatory (Countway and Caron, unpublished data).
The biomass of Ostreococcus reached 17.5 μg C liter−1 at 5 m and 67.7 μg C liter−1 at the DCM. The upper range of carbon biomass estimated for total phototrophic picoeukaryotes at Scripps Pier, a location where PPEs dominated the biomass of the picoplankton assemblage, was 33.3 μg C liter−1 (70). Thus, Ostreococcus appears to be an important component of the picoeukaryote community at our coastal Pacific location, particularly at the depth of the DCM. The DCM is a common feature of the water column off the coast of Southern California and has been shown to occur near the depth of the nitracline and the 10% light level (19). It is probable that Ostreococcus plays an important role in microbial food webs within the Southern California Bight region, given its prevalence on the northern (this study) and southern (70) sides of the Bight.
Ostreococcus populations displayed tremendous resiliency at our station, showing numerous but variable peaks in abundance over 2 years (Fig. 7). Reports of growth rates for control and nutrient-amended field samples dominated by Ostreococcus range from 2 to 8 day−1, with grazing losses in the range of 1.5 to 6.5 day−1 (27). High growth rates and grazing losses could explain the large month-to-month variability that we observed for Ostreococcus abundance. Rapid fluctuations between high and low abundances are a normal feature of picoeukaryote populations (46, 67, 70) and suggest that when conditions become favorable for growth (e.g., sudden nutrient availability or relief from grazing pressure), phototrophic picoeukaryote populations increase rapidly.
Two spring upwelling events were observed during our study, with one occurring between March and June 2001 and the other occurring between April and July 2002 (Fig. 6B). Nutrient concentrations in the upper 40 m of the water column increased rapidly during the early spring of 2001, while increases were more gradual and temporally shifted in 2002. Nutrient input corresponded to a threefold increase in diatom abundance at 5 m and a fivefold increase at the DCM in April 2001, while the Ostreococcus abundance did not increase until the following month (Fig. 7). Diatom abundance at the DCM returned to prebloom levels during the interval between sample collections, while Ostreococcus abundance increased >2 orders of magnitude, to 3.2 × 105 cells ml−1, in May 2001. A substantial increase in Ostreococcus abundance was also observed at 5 m between April and May 2001, but the diatom abundance remained unchanged. The nutrient upwelling in 2002 was of similar magnitude to the upwelling in 2001, but the highest nutrient concentrations occurred later in the year, shifting the spring diatom bloom into the summer. Ostreococcus cells did not bloom to high abundance at either depth following the upwelling in 2002. Factors controlling the abundance of Ostreococcus remain to be investigated at our study site.
The pattern of bloom succession from diatoms to other phytoplankton has been observed at many locations (4, 38, 58, 61). Smayda and Villareal (62) suggested that an “open phytoplankton niche” develops after a typical spring diatom bloom, which would allow nondiatom phytoplankton to become relatively more abundant. This concept was extended to the “picoalgal niche” hypothesis, where particular picoplankton become dominant depending on the nutrient regime of the environment and the relative differences in grazing pressure experienced by various taxa that could occupy this niche (59, 60). Picoeukaryotic phytoplankton are thought to be abundant in Thau Lagoon because of the relatively higher grazing pressures exerted upon larger-sized classes of phytoplankton and the removal of picoeukaryote grazers by cultured oysters (22, 67). In general, picoeukaryotic phytoplankton in pelagic systems are thought to be more susceptible to grazing losses than larger phytoplankton because of the higher growth rates of the grazers (ciliates and flagellates) that feed upon the smallest phytoplankton (51). Many of these hypotheses remain to be adequately tested for populations of Ostreococcus; however, our newly developed method and preliminary observations may help to advance this endeavor. The sensitivity of our qPCR method will help to answer questions about the Ostreococcus dynamics at low cell abundance (prior to bloom formation), and its speed will permit the analysis of large numbers of samples.
In summary, we have developed and applied a real-time TaqMan-based qPCR assay for the detection and ecological study of Ostreococcus sp. This dynamic prasinophyte was detected at the USC Microbial Observatory on 22 of 23 sampling dates spanning a 24-month time series. Ostreococcus formed a large bloom at our study site in May 2001, reaching abundances of 8.2 × 104 cells ml−1 at 5 m and 3.2 × 105 cells ml−1 at the DCM. Our qPCR approach readily detected low abundances of Ostreococcus, revealing a vertical distribution profile that was relatively typical for other phytoplankton species, with an abundance maximum near the DCM. The detection of Ostreococcus at such low concentrations shows promise for investigating “prebloom” dynamics of this picoeukaryote in the future.
Supplementary Material
Acknowledgments
This work was funded by National Science Foundation grants MCB-0084231 and OCE-9818953, Environmental Protection Agency grant RD-83170501, graduate fellowship support from the University of Southern California to P.D.C., and the USC Wrigley Institute for Environmental Studies.
We thank Tony Michaels for his support of the USC Microbial Observatory project. We thank the members of the Caron lab who participated in various aspects of this project, including Stephanie Moorthi, Julie Rose, Rebecca Schaffner, Astrid Schnetzer, Beth Stauffer, and Patrick Vigil. We give additional thanks to Ian Hewson, Josh Steele, Mike Schwalbach, and other members of Jed Fuhrman's lab for their collaboration on the USC Microbial Observatory project. We also thank Reni and Gerry Smith for help with sample collection and CTD deployments. We thank Kenny Kivett, David Reynoso, and crew members of the R/V Sea Watch for many successful cruises.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, R. A., S. L. Morton, and J. P. Sexton. 1997. Provasoli-Guillard National Center for Culture of Marine Phytoplankton 1997 list of strains. J. Phycol. 33(Suppl.):1-75. [Google Scholar]
- 3.Andersen, R. A., G. W. Sauders, M. P. Paskind, and J. P. Sexton. 1993. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J. Phycol. 29:701-715. [Google Scholar]
- 4.Andersson, A., P. Haecky, and A. Hagstroem. 1994. Effect of temperature and light on the growth of micro-, nano- and pico-plankton: impact on algal succession. Mar. Biol. 120:511-520. [Google Scholar]
- 5.Audemard, C., K. S. Reece, and E. M. Burreson. 2004. Real-time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl. Environ. Microbiol. 70:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Nucleic Acids Res. 32:D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biegala, I. C., F. Not, D. Vaulot, and N. Simon. 2003. Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry. Appl. Environ. Microbiol. 69:5519-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, S. L., M. R. Landry, S. Christensen, D. Garrison, M. M. Gowing, R. R. Bidigare, and L. Campbell. 2002. Microbial community dynamics and taxon-specific phytoplankton production in the Arabian Sea during the 1995 monsoon seasons. Deep-Sea Res. Part II 49:2345-2376. [Google Scholar]
- 10.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 11.Bustin, S. A., and T. Nolan. 2004. Primers and probes, p. 279-328. In S. A. Bustin (ed.), A-Z of quantitative PCR (IUL Biotechnology Series no. 5). International University Line, La Jolla, Calif.
- 12.Campbell, L., and D. Vaulot. 1993. Photosynthetic picoplankton community structure in the subtropical North Pacific-Ocean near Hawaii (Station Aloha). Deep-Sea Res. Part I 40:2043-2060. [Google Scholar]
- 13.Caron, D. A., P. D. Countway, and M. V. Brown. 2004. The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J. Eukaryot. Microbiol. 51:38-48. [DOI] [PubMed] [Google Scholar]
- 14.Caron, D. A., E. R. Peele, E. L. Lim, and M. R. Dennett. 1999. Picoplankton and nanoplankton and their trophic coupling in surface waters of the Sargasso Sea south of Bermuda. Limnol. Oceanogr. 44:259-272. [Google Scholar]
- 15.Casper, E. T., J. H. Paul, M. C. Smith, and M. Gray. 2004. Detection and quantification of the red tide dinoflagellate Karenia brevis by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 70:4727-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrétiennot-Dinet, M. J., C. Courties, A. Vaquer, J. Neveux, H. Claustre, J. Lautier, and M. C. Machado. 1995. A new marine picoeucaryote—Ostreococcus tauri gen. et sp.-nov. (Chlorophyta, Prasinophyceae). Phycologia 34:285-292. [Google Scholar]
- 17.Courties, C., R. Perasso, M. J. Chretiennot-Dinet, M. Gouy, L. Guillou, and M. Troussellier. 1998. Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Prasinophyceae). J. Phycol. 34:844-849. [Google Scholar]
- 18.Courties, C., A. Vaquer, M. Troussellier, J. Lautier, M. Chretiennot-Dinet, J. Neveux, C. Machado, and H. Claustre. 1994. Smallest eukaryotic organism. Nature 370:255. [Google Scholar]
- 19.Cullen, J. J. 1982. The deep chlorophyll maximum—comparing vertical profiles of chlorophyll-a. Can. J. Fish. Aquat. Sci. 39:791-803. [Google Scholar]
- 20.Derelle, E., C. Ferraz, P. Lagoda, S. Eychenie, R. Cooke, F. Regad, X. Sabau, C. Courties, M. Delseny, J. Demaille, A. Picard, and H. Moreau. 2002. DNA libraries for sequencing the genome of Ostreococcus tauri (Chlorophyta, Prasinophyceae): the smallest free-living eukaryotic cell. J. Phycol. 38:1150-1156. [Google Scholar]
- 21.Diez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupuy, C., A. Vaquer, T. Lam-Hoai, C. Rougier, N. Mazouni, J. Lautier, Y. Collos, and S. Le Gall. 2000. Feeding rate of the oyster Crassostrea gigas in a natural planktonic community of the Mediterranean Thau Lagoon. Mar. Ecol. Prog. Ser. 205:171-184. [Google Scholar]
- 23.Fawley, M. J., K. P. Fawley, and M. A. Buchheim. 2004. Molecular diversity among communities of freshwater microchlorophytes. Microb. Ecol. 48:489-499. [DOI] [PubMed] [Google Scholar]
- 24.Fenchel, T., and B. J. Finlay. 2004. The ubiquity of small species: patterns of local and global diversity. Bioscience 54:777-784. [Google Scholar]
- 25.Finlay, B. J., and T. Fenchel. 2004. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 155:237-244. [DOI] [PubMed] [Google Scholar]
- 26.Foissner, W. 1999. Protist diversity: estimates of the near-imponderable. Protist 150:363-368. [DOI] [PubMed] [Google Scholar]
- 27.Fouilland, E., C. Descolas-Gros, C. Courties, Y. Collos, A. Vaquer, and A. Gasc. 2004. Productivity and growth of a natural population of the smallest free-living eukaryote under nitrogen deficiency and sufficiency. Microb. Ecol. 48:103-110. [DOI] [PubMed] [Google Scholar]
- 28.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garces, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray, M., B. Wawrik, J. Paul, and E. Casper. 2003. Molecular detection and quantitation of the red tide dinoflagellate Karenia brevis in the marine environment. Appl. Environ. Microbiol. 69:5726-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillou, L., M. J. Chretiennot-Dinet, S. Boulben, S. Y. Moon-van der Staay, and D. Vaulot. 1999. Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist 150:383-398. [DOI] [PubMed] [Google Scholar]
- 31.Guillou, L., M. J. Chretiennot-Dinet, L. K. Medlin, H. Claustre, S. Loiseaux-de Goer, and D. Vaulot. 1999. Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta). J. Phycol. 35:368-381. [Google Scholar]
- 32.Guillou, L., W. Eikrem, M. J. Chretiennot-Dinet, F. Le Gall, R. Massana, K. Romari, C. Pedrós-Alió, and D. Vaulot. 2004. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155:193-214. [DOI] [PubMed] [Google Scholar]
- 33.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 34.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 35.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller, M. D., R. C. Selvin, W. Claus, and R. R. L. Guillard. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23:633-638. [Google Scholar]
- 37.Kirchman, D. L., L. Y. Yu, B. M. Fuchs, and R. Amann. 2001. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26:13-22. [Google Scholar]
- 38.Larsen, A., G. A. F. Flaten, R. A. Sandaa, T. Castberg, R. Thyrhaug, S. R. Erga, S. Jacquet, and G. Bratbak. 2004. Spring phytoplankton bloom dynamics in Norwegian coastal waters: microbial community succession and diversity. Limnol. Oceanogr. 49:180-190. [Google Scholar]
- 39.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton—measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 40.Li, W. K. W., D. V. S. Rao, W. G. Harrison, J. C. Smith, J. J. Cullen, B. Irwin, and T. Platt. 1983. Autotrophic picoplankton in the tropical ocean. Science 219:292-295. [DOI] [PubMed] [Google Scholar]
- 41.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massana, R., J. Castresana, V. Balague, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedrós-Alió. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massana, R., L. Guillou, B. Diez, and C. Pedrós-Alió. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moorthi, S. D., P. D. Countway, B. Stauffer, and D. A. Caron. Use of quantitative real-time PCR to investigate the dynamics of the red tide dinoflagellate Lingulodinium polyedrum. Microb. Ecol., in press. [DOI] [PubMed]
- 45.Muyzer, G., E. de Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Not, F., M. Latasa, D. Marie, T. Cariou, D. Vaulot, and N. Simon. 2004. A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the western English Channel. Appl. Environ. Microbiol. 70:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Not, F., N. Simon, I. C. Biegala, and D. Vaulot. 2002. Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microb. Ecol. 28:157-166. [Google Scholar]
- 48.O'Kelly, C. J., M. E. Sieracki, E. C. Thier, and I. C. Hobson. 2003. A transient bloom of Ostreococcus (Chlorophyta, Prasinophyceae) in West Neck Bay, Long Island, New York. J. Phycol. 39:850-854. [Google Scholar]
- 49.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 50.Popels, L. C., S. C. Cary, D. A. Hutchins, R. Forbes, F. Pustizzi, C. J. Gobler, and K. J. Coyne. 2003. The use of quantitative polymerase chain reaction for the detection and enumeration of the harmful alga Aureococcus anophagefferens in environmental samples along the United States East Coast. Limnol. Oceanogr. 1:92-102. [Google Scholar]
- 51.Riegman, R., B. R. Kuipers, A. A. M. Noordeloos, and H. J. Witte. 1993. Size-differential control of phytoplankton and the structure of plankton communities. Netherlands J. Sea Res. 31:255. [Google Scholar]
- 52.Robbens, S., B. Khadaroo, A. Camasses, E. Derelle, C. Ferraz, D. Inze, Y. Van de Peer, and H. Moreau. 2005. Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol. Biol. Evol. 22:589-597. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez, F., E. Derelle, L. Guillou, F. Le Gall, D. Vaulot, and H. Moreau. 2005. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7:853-859. [DOI] [PubMed] [Google Scholar]
- 54.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784-798. [Google Scholar]
- 55.Saito, K., T. Drgon, J. A. F. Robledo, D. N. Krupatkina, and G. R. Vasta. 2002. Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl. Environ. Microbiol. 68:5394-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selph, K. E., M. R. Landry, and E. A. Laws. 2003. Heterotrophic nanoflagellate enhancement of bacterial growth through nutrient remineralization in chemostat culture. Aquat. Microb. Ecol. 32:23-37. [Google Scholar]
- 57.Sherr, E. B., and B. F. Sherr. 1991. Planktonic microbes—tiny cells at the base of the ocean's food webs. Trends Ecol. Evol. 6:50-54. [DOI] [PubMed] [Google Scholar]
- 58.Sherr, E. B., B. F. Sherr, P. A. Wheeler, and K. Thompson. 2003. Temporal and spatial variation in stocks of autotrophic and heterotrophic microbes in the upper water column of the central Arctic Ocean. Deep-Sea Res. Part I 50:557-571. [Google Scholar]
- 59.Sieracki, M. E., C. J. Gobler, T. L. Cucci, E. C. Thier, I. C. Gilg, and M. D. Keller. 2004. Pico- and nanoplankton dynamics during bloom initiation of Aureococcus in a Long Island, N.Y. bay. Harmful Algae 3:459-470. [Google Scholar]
- 60.Sieracki, M. E., M. D. Keller, T. L. Cucci, and E. Their. 1999. Plankton community ecology during the bloom initiation period of the Brown Tide organism Aureococcus anophagefferens in coastal embayments of Long Island, N.Y. EOS 80:285. [Google Scholar]
- 61.Sieracki, M. E., P. G. Verity, and D. K. Stoecker. 1993. Plankton community response to sequential silicate and nitrate depletion during the 1989 North Atlantic spring bloom. Deep-Sea Res. Part II Top. Stud. Oceanogr. 40:213-225.
- 62.Smayda, T. J., and T. A. Villareal. 1989. The 1985 “brown tide” and the open phytoplankton niche in Narragansett Bay during summer, p. 159-188. In E. M. Cosper, E. J. Carpenter, and V. M. Bricelj (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms. Coastal and Estuarine Studies no. 35. Springer-Verlag, Berlin, Germany.
- 63.Stoecker, D. K., D. J. Gifford, and M. Putt. 1994. Preservation of marine planktonic ciliates—losses and cell shrinkage during fixation. Mar. Ecol. Prog. Ser. 110:293-299. [Google Scholar]
- 64.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaquer, A., M. Troussellier, C. Courties, and B. Bibent. 1996. Standing stock and dynamics of picophytoplankton in the Thau Lagoon (northwest Mediterranean coast). Limnol. Oceanogr. 41:1821-1828. [Google Scholar]
- 68.Vaulot, D., F. Le Gall, D. Marie, L. Guillou, and F. Partensky. 2004. The Roscoff Culture Collection (RCC): a collection dedicated to marine picoplankton. Nova Hedwigia 79:49-70. [Google Scholar]
- 69.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of RbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worden, A. Z., J. K. Nolan, and B. Palenik. 2004. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol. Oceanogr. 49:168-179. [Google Scholar]
- 71.Zhu, F., R. Massana, F. Not, D. Marie, and D. Vaulot. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.