Abstract
Streptococcus salivarius is a prominent member of the oral microbiota and has excellent potential for use as a probiotic targeting the oral cavity. In this report we document safety data relating to S. salivarius K12, including assessment of its antibiogram, metabolic profiles, and virulence determinants, and we examine the microbial composition of saliva following the dosing of subjects with K12.
Certain lactic acid bacteria (LAB) have had a long history of consumption by humans, either as probiotics or in traditional foods. Proposals for the use of nontraditional species in humans generally evoke greater concern about potential adverse effects than proposals for LAB probiotics (11, 12). Nevertheless, even species generally regarded as safe and with long histories of application can still potentially cause infection in humans. Recent indications are that some of the more exciting new probiotic developments will include a shift in focus toward strains having both their origins and primary mucosal targets in tissues other than the intestinal tract (5).
Although there have been some attempts to use intestinally derived bacteria such as lactobacilli for oral cavity probiotics, it appears more likely that bacteria isolated directly from the oral microbiota will be efficacious for such purposes (5). Streptococcus salivarius K12 (isolated from the saliva of a healthy child) is a probiotic intended for use in the oral cavity. Strain K12 has had a 5-year history of commercial application as a probiotic in New Zealand, with approximately 150,000 doses administered to date. Its in vitro antimicrobial activity against Streptococcus pyogenes and various bacterial species incriminated in the etiology of halitosis appears to be due to the production of lantibiotic bacteriocins (14, 18, 21). Streptococcus salivarius is a prominent member of the oral microbiota of “healthy” humans and is closely related to Streptococcus thermophilus (3), a benign organism used in the manufacture of yogurt. Streptococcus salivarius is known to be a pioneer colonizer of infants, who typically acquire it from their mothers shortly after birth (7, 10, 19). As with lactobacilli, there have been occasional reports of infections involving S. salivarius, though their occurrence (even in adverse medical conditions) is extremely low (1, 2, 6, 8, 16, 20, 23).
What safety considerations should apply to a probiotic intended for application in the oral cavity? Many of the requirements for intestinal probiotics are relevant here, for example, whether the bacterium exhibits (i) antibiotic resistance, (ii) metabolic activities potentially adversely affecting the host, or (iii) inhibitory activity against other commensal microorganisms. Consideration should also be given to the evolutionary origins of probiotic candidates as an indicator of the potential for them to carry particular virulence determinants. For example, the genus Streptococcus includes many species that are largely commensals of the mucosal membranes of the upper respiratory tract, and some species commonly cause disease.
The antibiograms of three samples of strain K12 were tested by the antibiotic disk sensitivity method (conducted according to CLSI [formerly NCCLS] standards) to determine whether they exhibited any differences in profile. Strains tested were (i) the original isolate (K12-J89), stored at −70°C for 15 years, (ii) a laboratory stock culture (K12-Lab) that had been subcultured every 2 weeks for 3 years, and (iii) a commercially prepared batch (K12-BN21) of freeze-dried cells. The antibiograms of the K12 isolates did not differ following long-term storage, recurrent in vitro propagation, or commercial lyophilization (Table 1). Streptococcus salivarius K12 was assessed to be moderately resistant to both gentamicin and ofloxacin. Eight additional S. salivarius isolates from different individuals were also tested for sensitivity to gentamicin and ofloxacin to help determine the level of resistance to these antibiotics in the general S. salivarius population. Each displayed moderate levels of resistance to gentamicin and ofloxacin, similar to that of strain K12 (Table 1). Thus, S. salivarius K12 is sensitive to a variety of commonly utilized antibiotics, including several that are routinely used for the control of upper respiratory tract infections. The low levels of gentamicin and ofloxacin resistance in strain K12 were similar to those of a series of natural S. salivarius isolates, indicating that they are intrinsic resistances.
TABLE 1.
Antibiotic disk sensitivities of S. salivarius isolates
| Antibiotic (concn [μg]) | Inhibition zone size (mm) for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain K12 lineage
|
HD | ToveR | #6 | K30 | HA | HB | HC | K26R | |||
| K12- J89a | K12- Labb | K12- BN21c | |||||||||
| Penicillin (10) | 34 | 34 | 39 | 34 | 29 | 26 | 26 | 26 | 27 | 27 | 28 |
| Amoxicillin (10) | 35 | 32 | 35 | 35 | 28 | 26 | 29 | 26 | 26 | 28 | 27 |
| Ofloxacin (5) | 18d | 18d | 15d | 18d | 19d | 17d | 16d | 16d | 18d | 16d | 18d |
| Tetracycline (30) | 28 | 27 | 26 | 28 | 27 | 27 | 22 | 27 | 28 | 28 | 27 |
| Erythromycin (15) | 30 | 30 | 31 | 30 | 28 | 29 | 26 | 28 | 29 | 27 | 27 |
| Gentamicin (10) | 15d | 14d | 14d | 15d | 15d | 16d | 14d | 12d | 12d | 12d | 14d |
| Clindamycin (2) | 29 | 26 | 28 | 29 | 28 | 26 | 26 | 29 | 30 | 28 | 25 |
Original isolate.
Routinely subcultured isolate.
Isolate from commercial batch.
Moderately resistant.
In order to determine the metabolic profile of strain K12 and its stability, the API 20 Strep and API 50CH systems (bioMérieux, Marcy-l'Etoile, France) were utilized. None of the fermentation or enzymatic reactions of S. salivarius K12 are indicative of deleterious effects for the human host (Table 2). Additionally, the metabolic profiles given by strain K12 following either recurrent propagation or commercial processing were identical to that of the original isolate, indicating that the phenotypic expression of metabolites and fermentation pathways represents stable characteristics of this strain. The ability of S. salivarius K12 to lyse red blood cells was tested on three media: (i) human blood agar (BaCa, consisting of Columbia agar base with 5% [vol/vol] human blood; Fort Richard Laboratories, New Zealand), (ii) sheep blood agar (Columbia agar base with 5% [vol/vol] defibrinated sheep blood), and (iii) buffered (pH 7.5) CNA-P agar (Difco) with 5% defibrinated sheep blood (9). In each case, no hemolytic activity was detected.
TABLE 2.
API 20 Strep and API 50CH positive reactions for Streptococcus salivarius K12 cultures
| Test | Reaction of:
|
||
|---|---|---|---|
| K12-J89a | K12-Labb | K12-BN21c | |
| API 20 Strepd | |||
| Acetoin production | + | + | + |
| β-Glucosidase | + | + | + |
| Alkaline phosphatase | + | + | + |
| Leucine aminopeptidase | + | + | + |
| d-Lactose | + | + | + |
| d-Trehalose | + | + | + |
| Inulin | + | + | + |
| d-Raffinose | + | + | + |
| API 50CH | |||
| d-Galactose | + | + | + |
| d-Glucose | + | + | + |
| d-Fructose | + | + | + |
| d-Mannose | + | + | + |
| N-Acetylglucosamine | + | + | + |
| Arbutine | + | + | + |
| Salicin | + | + | + |
| d-Cellobiose | + | + | + |
| d-Maltose | + | + | + |
| d-Lactose | + | + | + |
| d-Saccharose | + | + | + |
| d-Trehalose | + | + | + |
| Inulin | + | + | + |
| d-Raffinose | + | + | + |
| d-Tagatose | + | + | + |
Original isolate.
Routinely subcultured isolate.
Isolate from commercial batch.
The API 20 Strep code for all three isolates was 5060470.
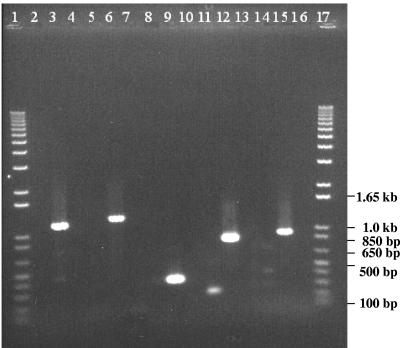
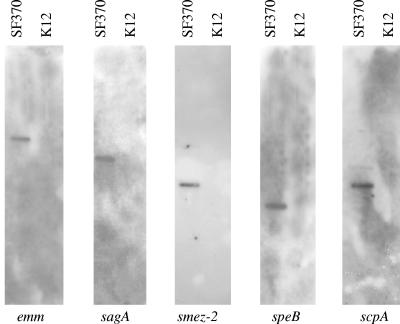
For the detection of known streptococcal virulence determinants, chromosomal DNA was extracted from cultures of S. salivarius strain K12 and S. pyogenes strain SF370 (M-serotype 1, genome strain) using the DNeasy tissue kit (QIAGEN, Valencia, CA). The presence of streptococcal virulence genes in SF370 and K12 was assessed using the specific primers described in Table 3. Amplicons from S. pyogenes strain SF370 DNA were labeled with digoxigenin and then used to probe Southern blots resulting from HindIII digestion of K12 and SF370 chromosomal DNA. None of the selected virulence factor genes were detected in strain K12 by PCR or Southern hybridization (Fig. 1 and 2). Unfortunately, there is as yet no genome sequence available for S. salivarius to facilitate an in-depth bioinformatics analysis for other potential virulence genes. However, the annotated genome sequence of the closely related organism S. thermophilus was recently published, and genes demonstrated in other species to be involved in virulence were either nonfunctional or absent in S. thermophilus (4). Preliminary work in our laboratory has shown that S. salivarius K12 has an sbcD homologue similar to that in S. thermophilus. An interesting difference between streptococci considered pathogenic and dairy streptococci is the presence of sbc genes in the latter. These products reduce the efficiency of recombination, effectively stabilizing the genome (4).
TABLE 3.
Primers used to amplify streptococcal virulence genes
| Virulence determinant | Gene designation | Primer name | Primer sequence (5′-3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| Streptolysin S | sagA | sagA Fwd | ATTGAGCTAGCCTTGTCCTTGT | 1,164 | This study |
| sagB Rev | GTATTCCGCAAAATCTCTAACG | ||||
| C5a peptidase | scpA | scpA Fwd | CGGGTATCATGGGACTGTTGC | 1,259 | This study |
| scpA Rev | TTGCCGATGTTGCGACTTC | ||||
| SMEZ-2 | smez-2 | smez-2 Fwd | GGACGAATATGCAGCCAATGA | 332 | This study |
| smez-2 Rev | GTATGAAAAACCAGTCTACCAC | ||||
| SPE-B | speB | speB Fwd | TGACGCTAACGGTAAAGAAAACA | 819 | This study |
| speB Rev | GCCGCCACCAGTACCAAGAGC | ||||
| M-protein | emm | M-all Fwd | TATTSGCTTAGAAAATTAA | 961 | 13 |
| M-all Rev | GCAAGTTCTTCAGCTTGTTT |
FIG. 1.
Streptococcus salivarius K12 (even-numbered lanes) and S. pyogenes SF370 (odd-numbered lanes) amplification products resulting from PCR using specific primers for the sagA (lanes 2 and 3), scpA (lanes 5 and 6), smez-2 (lanes 8 and 9), speB (lanes 11 and 12), and emm (lanes 14 and 15) genes. Lanes 1 and 17, 1-kb marker (Gibco).
FIG. 2.
Autoradiographs of Southern blots hybridized with amplicons of the different streptococcal virulence factors (given at the bottom). Lanes containing HindIII-digested DNA from S. pyogenes SF370 are labeled SF370. Lanes containing HindIII-digested DNA from S. salivarius strain K12 are labeled K12.
A study approved by the Otago Ethics Committee was conducted to determine whether the use of the K12 strain by humans altered the composition of the oral microbiota. Saliva samples were collected from 14 individuals 24 h prior to the commencement of the colonization protocol and periodically during the study. On the following day, each subject brushed his or her teeth and rinsed with 10 ml of 0.2% chlorhexidine gluconate to reduce the population levels of existing oral microbiota. At 2-h intervals for 8 h, the subjects sucked a lozenge containing ca. 1 × 109 CFU of S. salivarius K12 (BLIS K12 ThroatGuard). This protocol was repeated on days 2 and 3. No adverse symptoms were reported by any of the subjects. Microbial populations in the saliva specimens were evaluated. Saline dilutions were plated in duplicate on the following media: Mitis-Salivarius agar (Difco) (for S. salivarius); CHROMagar Candida, CHROMagar ECC (for Escherichia coli and coliforms), and CHROMagar Staph aureus (all from CHROMagar Microbiology, Paris, France); Pseudomonas isolation medium (Fort Richard Laboratories); TSYCSB selective medium (for Streptococcus mutans) (22); and BaCa. The majority of pathogens and opportunistic microorganisms tested for in the saliva were those suggested for the assessment of adverse effects of chemotherapy on the oral microbiota (15). Total counts of Streptococcus salivarius and facultatively anaerobic bacteria remained stable throughout the study (Table 4). Examination of the saliva of subjects dosed with S. salivarius K12 for 3 days indicated that there was no overt change in its microbial composition. The bacteriocin-like inhibitory substance activity of representative S. salivarius isolates was determined as described previously (17). Two subjects had S. salivarius organisms in their oral cavities that exhibited bacteriocin profiles similar to that of strain K12 prior to the taking of the course of K12 lozenges. After 2 days of lozenge taking, 13 of the 14 subjects had S. salivarius populations in which more than 1% exhibited strain K12-like inhibitory activity, but by day 28, this was reduced to only 4 subjects (Table 5). These bacteriocin-producing cell lines appeared in some cases to persist in the oral cavity for more than 1 month after the completion of the course. Streptococcus salivarius K12 isolates obtained from the saliva of 5 subjects at day 14 were tested by API 20 Strep and 50CH kits, and no metabolic profile changes were detected.
TABLE 4.
Counts of facultatively anaerobic bacteria and S. salivarius in saliva of individuals prior to and in the days following dosing with S. salivarius K12a
| Organism(s) | Mean CFU/ml (SD) at the following time of sampling:
|
Maximum CFU/ml detected in any single sample | ||||
|---|---|---|---|---|---|---|
| Predosing | Day 3 | Day 7 | Day 14 | Day 28 | ||
| Facultatively anaerobic bacteria | 3.11e7 (2.4e7) | 3.09e7 (2.1e7) | 3.98e7 (2.3e7) | 3.93e7 (2.9e7) | 3.32e7 (2.1e7) | 1.1e8 |
| S. salivarius | 1.54e7 (2.4e7) | 7.80e6 (1.3e7) | 1.33e7 (1.1e7) | 6.58e6 (5.9e6) | 7.2e6 (7.1e6) | 7.7e7 |
Subjects included 4 males and 10 females; mean age, 19 years. P values for time point differences for counts of facultatively anaerobic bacteria and S. salivarius were not significant (>0.5 by nonparametric analysis of variance).
TABLE 5.
Detection of specific microorganisms in saliva of individuals prior to and in the days following dosing with Streptococcus salivarius K12a
| Organism(s) | No. of subjects in which the indicated class of microbe was detected/total no. of subjects at the following sampling time:
|
Maximum CFU/ml detected in any single sample | ||||
|---|---|---|---|---|---|---|
| Predosing | Day 3 | Day 7 | Day 14 | Day 28 | ||
| S. salivarius K12-like BLISc profile | 2/14 | 13/14 | 12/14 | 9/14 | 4/14 | NAd |
| S. salivarius K12-like BLIS profile >50% | 2/14 | 7/14 | 7/14 | 3/14 | 1/14 | NA |
| S. mutansb | 9/14 | 11/14 | 9/14 | 11/14 | 9/13 | 6.3e5 |
| S. mutans >1e4 CFU/ml | 4/14 | 4/14 | 4/14 | 5/14 | 4/14 | NA |
| S. mutans >1e5 CFU/ml | 2/14 | 2/14 | 1/14 | 1/14 | 1/14 | NA |
| Lactobacilli | 6/14 | 5/14 | 4/14 | 7/14 | 2/14 | 2.8e5 |
| Candida | 1/14 | NDe | 1/14 | 1/14 | 1/14 | 4.4e4 |
| Coliforms | 1/14 | 2/14 | 0/14 | 1/14 | 0/14 | 140 |
| Pseudomonas | 11/14 | 8/14 | 10/14 | 9/14 | 5/14 | 100 |
| Staphylococcus aureus | 3/14 | 4/14 | 1/14 | 2/14 | 2/14 | 1,000 |
Subjects included 4 males and 10 females; mean age, 19 years.
P values for time point differences for S. mutans counts were not significant (>0.5 by nonparametric analysis of variance).
BLIS, bacteriocin-like inhibitory substance.
NA, not applicable.
ND, not determined.
The data presented in this study, demonstrating the absence of adverse reactions in subjects actively ingesting S. salivarius K12, combined with the results of analysis of the biochemical, antibiogram, and virulence gene profiles of this bacterium, indicate that it has very low pathogenic potential and is unlikely to cause disease in healthy humans.
REFERENCES
- 1.Afek, S., A. D. Sperber, and Y. Almog. 2004. Carcinoma of the colon presenting as Streptococcus salivarius sepsis. J. Clin. Gastroenterol. 38:86-87. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, R., T. Hassall, B. Morland, and J. Gray. 2003. Viridans streptococcus bacteremia in children on chemotherapy for cancer: an underestimated problem. Pediatr. Hematol. Oncol. 20:439-444. [PubMed] [Google Scholar]
- 3.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, J., C. Chilcott, and J. Tagg. 2005. The rationale and potential for the reduction of oral malodour using Streptococcus salivarius probiotics. Oral Dis. 11:29-31. [DOI] [PubMed] [Google Scholar]
- 6.Cannon, J. P., T. A. Lee, J. T. Bolanos, and L. H. Danziger. 2005. Pathogenic relevance of Lactobacillus: a retrospective review of over 200 cases. Eur. J. Clin. Microbiol. Infect. Dis. 24:31-40. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, J., H. Grahnen, G. Jonsson, and S. Wikner. 1970. Early establishment of Streptococcus salivarius in the mouth of infants. J. Dent. Res. 49:415-418. [DOI] [PubMed] [Google Scholar]
- 8.Corredoira, J. C., M. P. Alonso, J. F. Garcia, E. Casariego, A. Coira, A. Rodriguez, J. Pita, C. Louzao, B. Pombo, M. J. Lopez, and J. Varela. 2005. Clinical characteristics and significance of Streptococcus salivarius bacteremia and Streptococcus bovis bacteremia: a prospective 16-year study. Eur. J. Clin. Microbiol. Infect. Dis. 24:250-255. [DOI] [PubMed] [Google Scholar]
- 9.Dierksen, K. P., N. L. Ragland, and J. R. Tagg. 2000. A new alkaline pH-adjusted medium enhances detection of β-hemolytic streptococci by minimizing bacterial interference due to Streptococcus salivarius. J. Clin. Microbiol. 38:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz, C. M., M. E. Stiles, K. H. Schleifer, and W. H. Holzapfel. 2003. Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 88:105-122. [DOI] [PubMed] [Google Scholar]
- 12.Lund, B., and C. Edlund. 2001. Probiotic Enterococcus faecium strain is a possible recipient of the vanA gene cluster. Clin. Infect. Dis. 32:1384-1385. [DOI] [PubMed] [Google Scholar]
- 13.Podbielski, A., B. Melzer, and R. Lutticken. 1991. Application of the polymerase chain reaction to study the M protein(-like) gene family in beta-hemolytic streptococci. Med. Microbiol. Immunol. (Berlin) 180:213-227. [DOI] [PubMed] [Google Scholar]
- 14.Ross, K. F., C. W. Ronson, and J. R. Tagg. 1993. Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl. Environ. Microbiol. 59:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandham, H. J. 1994. Criteria for the assessment of adverse effects of chemotherapy on the oral microflora. J. Dent. Res. 73:692-694. [DOI] [PubMed] [Google Scholar]
- 16.Smith, A., M. S. Jackson, and H. Kennedy. 2004. Antimicrobial susceptibility of viridans group streptococcal blood isolates to eight antimicrobial agents. Scand. J. Infect. Dis. 36:259-263. [DOI] [PubMed] [Google Scholar]
- 17.Tagg, J. R., and L. V. Bannister. 1979. “Fingerprinting” beta-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J. Med. Microbiol. 12:397-411. [DOI] [PubMed] [Google Scholar]
- 18.Tagg, J. R., and K. P. Dierksen. 2003. Bacterial replacement therapy: adapting ‘germ warfare’ to infection prevention. Trends Biotechnol. 21:217-223. [DOI] [PubMed] [Google Scholar]
- 19.Tagg, J. R., V. Pybus, L. V. Phillips, and T. M. Fiddes. 1983. Application of inhibitor typing in a study of the transmission and retention in the human mouth of the bacterium Streptococcus salivarius. Arch. Oral Biol. 28:911-915. [DOI] [PubMed] [Google Scholar]
- 20.Trautmann, M., P. M. Lepper, and F. J. Schmitz. 2002. Three cases of bacterial meningitis after spinal and epidural anesthesia. Eur. J. Clin. Microbiol. Infect. Dis. 21:43-45. [DOI] [PubMed] [Google Scholar]
- 21.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Palenstein Helderman, W. H., M. Ijsseldijk, and J. H. Huis in 't Veld. 1983. A selective medium for the two major subgroups of the bacterium Streptococcus mutans isolated from human dental plaque and saliva. Arch. Oral Biol. 28:599-603. [DOI] [PubMed] [Google Scholar]
- 23.Watanakunakorn, C., and J. Pantelakis. 1993. Alpha-hemolytic streptococcal bacteremia: a review of 203 episodes during 1980-1991. Scand. J. Infect. Dis. 25:403-408. [DOI] [PubMed] [Google Scholar]