Abstract
The distribution of log counts at a given time during the exponential growth phase of Listeria innocua measured in food samples inoculated with one cell each was applied to estimate the distribution of the single-cell lag times. Three replicate experiments in broth showed that the distribution of the log counts is a linear mapping of the distribution of the detection times measured by optical density. The detection time distribution reflects the lag time distribution but is shifted in time. The log count distribution was applied to estimate the distributions of the lag times in a liquid dairy product and in liver paté after different heat treatments. Two batches of ca. 100 samples of the dairy product were inoculated and heated at 55°C for 45 min or at 62°C for 2 min, and an unheated batch was incubated at 4°C. The final concentration of surviving bacteria was ca. 1 cell per sample. The unheated cells showed the shortest lag times with the smallest variance. The mean and the variance of the lag times of the surviving cells at 62°C were greater than those of the cells treated at 55°C. Three batches of paté samples were heated at 55°C for 25 min, 62°C for 81 s, or 65°C for 20 s. A control batch was inoculated but not heated. All paté samples were incubated at 15°C. The distribution of the lag times of the cells heated at 55°C was not significantly different from that of the unheated cells. However, at the higher temperatures, 62°C and 65°C, the lag duration was longer and its variance greater.
Nowadays, there is a remarkable increase in consumer demand for minimally processed foods. These products are characterized by better sensorial attributes and higher nutritive values than those produced conventionally, but their manufacture and consumption usually imply higher microbiological risks as a consequence of the minimization of processing (20). Food quality optimization may sometimes force new approaches to maintain the required level of safety of the products.
Growth studies usually concentrate on the effect of the actual environment on microbial growth. However, both the previous environment and the condition (or physiological state) of the bacterial cells have an effect on their growth in the current environment. These effects gradually diminish after inoculation as a result of an adjustment (or adaptation) process. The adjustment period is the lag time (3, 4). The lag time, or period prior to bacterial division, has been studied using rather high inoculum levels, but in reality, foods are often contaminated with low cell numbers (7). The mean value of the single-cell lag times is not necessarily the same as the population lag time, and information on the variability of the lag phase among the single cells within a population cannot be deduced from the lag time of that population (11, 12). It has been shown by different authors that the inoculum size affects the duration and variability of the lag time, and the effect is noticeable mainly with low concentrations of cells (1, 13, 15, 16). Hence, the extrapolation of population growth parameters to low-level contaminated food products could give inaccurate results.
To study single-cell lag times, a large number of replicate observations are needed. Automated turbidity measurements or microscopy-based systems generate a great amount of data (2, 5-8, 11, 12, 17, 19), but their application is restricted to a small range of products because most foods are either solid or, if liquid, inherently turbid. Hence, there is a need to determine the lag phase of single cells in food. The lag times of populations initiated with small inocula cannot be measured accurately with traditional microbiology techniques such as bacterial counts. In this work, we present an alternative way of estimating the lag times of single cells in food, and we validate and show its application with real food products.
The time to reach a given microbial concentration is affected by the initial inoculum, the lag time, and the maximum specific growth rate. Assuming that the growth rate is constant during the exponential growth phase and that the initial number of cells is homogeneous, the lag time distribution can be estimated from the distribution of the detection times (7, 11, 19). These two distributions, of lag and detection times, are denominated horizontal distributions, in contrast to the vertical distribution of the log counts at a given time during exponential growth, which, under the same assumption, is a linear mapping of the horizontal distributions, as explained previously (10).
The aim of this paper is to show the use of the distribution of log counts determined during the exponential growth phase in food samples inoculated with a single cell each to estimate the distribution of the lag times of single cells in food. To date, this is the first attempt to measure single-cell growth parameters in food. A first set of experiments was set up in broth to validate the use of the log counts, or vertical distribution, to estimate the detection times, or horizontal distribution. Finally, the log count distribution was applied to estimate the distribution of the single-cell lag times in a liquid dairy product and in liver paté after different heat treatments.
MATERIALS AND METHODS
Strain and inoculum preparation.
Listeria innocua NCTC 11288 was maintained at −20°C. Immediately before the experiments, it was revitalized by being subcultured twice consecutively in tryptic soy broth (TSB; Oxoid/Unipath CM129) at 30°C for 24 h.
Broth experiments.
The bacterial culture was diluted in TSB to a final concentration of approximately a single viable cell per 300 μl, the volume of bacterial suspension dispensed into wells of four sterile multiwell plates of 100 wells each. Two plates were incubated at 19°C in a conventional incubator, and two plates were incubated in Bioscreen C equipment (Labsystems, Helsinki, Finland) at the same temperature.
For the two plates incubated in the Bioscreen equipment, the optical density at 600 nm was measured in each well automatically every 15 min. The detection time was calculated as the time required for the optical density to reach 0.2, which was equivalent to a concentration of ca. 108 cells/ml.
The contents of each well of the two plates maintained in a conventional incubator were plated out on tryptic soy agar (TSA; Oxoid/Unipath CM131) during the exponential growth phase of the cultures, using a spiral plater (model Eddy Jet; IUL Instruments, Barcelona, Spain). The incubation times after which each well was sampled were recorded. Plates were incubated at 35°C for 24 h.
The average number of cells per well (m) was estimated by assuming that the number of cells per well (N) followed a Poisson distribution. Hence, the following equation was used:
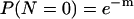 |
(1) |
where P(N = 0) is the probability of having no cells in a well and is estimated as the proportion of wells without growth of the 400 wells inoculated. The number of wells inoculated with exactly 1 cell was approximated from the equation P(N = 1) = me−m.
To compare the horizontal or detection time distribution with the vertical or log count distribution, the log counts were transformed into detection times as follows:
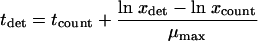 |
(2) |
where tdet is the detection time, tcount is the sampling time at which the well was plated out, ln xdet is the natural logarithm of the detection level or bacterial concentration corresponding to an optical density at 600 nm of 0.2 (in this case, 108 cells/ml), ln xcount is the natural logarithm of the cell concentration detected at time tcount, and μmax is the maximum specific growth rate of L. innocua at 19°C, which is equal to 0.4378 (±0.02641), as calculated from a growth curve.
The whole experiment was repeated three times.
Food experiments.
Samples of a Spanish dairy product, “Natillas,” a dessert whose main ingredients are eggs, sugar, and milk, and samples of liver paté were bought from local shops.
Aliquots (500 ml) of the dairy product were inoculated with ca. 104 to 105 CFU/ml and dispensed into plastic sachets, with 5 ml in each. The sachets were heat sealed, submerged in a water bath preheated to the target temperature, and held for the time required to reach a final concentration of ca. 1 cell per sachet (45 min at 55°C and 2 min at 62°C). Immediately after the heat treatment, the sachets were cooled in an ice bath. An unheated control batch of sachets was prepared from 500 ml of product inoculated with ca. 1 cell per 5 ml. All sachets, both heated and unheated, were stored at 4°C. Every 2 to 3 days, one to two samples were plated out to check the growth phase and cell concentration. When the concentration was ca. 100 to 1,000 CFU ml−1, all sachets were plated out on TSA, and the sampling times were recorded.
Ten bags, each containing 140 g paté, were inoculated with 102 to 103 CFU g−1 and heat sealed. The bags were submerged in a water bath set at the target temperature for the time required to reach a final concentration of ca. 1 cell per 13 g of paté (25 min at 55°C, 81 s at 62°C, and 20 s at 65°C). After heat treatment, the paté was cooled in an ice bath, and 13-g samples were dispensed into individual plastic sachets. An unheated control batch of samples was prepared from paté inoculated with ca. 1 cell per 13 g. All the sachets were stored at 15°C, and the bacterial concentration was measured daily. When the concentration was ca. 100 to 1,000 CFU ml−1, all sachets were plated out on TSA.
All food samples were plated out by using a spiral plater, and the plates were incubated at 35°C for 24 h.
The average number of cells per food portion (m) able to grow at 4°C in the dairy product or at 15°C in the paté was calculated by assuming that m is the parameter of a Poisson distribution as described above. The probability of having no cells in a food sachet was estimated as the proportion of sachets without growth from the total number inoculated. The Poisson probability density function was used to estimate the number of food portions inoculated with exactly 1 cell, as described above.
The lag time was calculated from the log counts as follows:
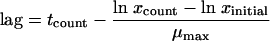 |
(3) |
where “lag” is the lag time, tcount is the time at which the food sample was plated out, ln xcount is the natural logarithm of the number of cells detected at time tcount, ln xinitial is the natural logarithm of the initial number of bacteria (when the average number of cells per sachet was <1, xinitial was fixed to 1), and μmax is the maximum specific growth rate of L. innocua determined by growth curves under the experimental conditions used, equal to 0.0259 (±0.00352) at 4°C for the dairy product and 0.180 (±0.0106) at 15°C for the paté.
Simulation of bacterial growth in a batch of samples with Poisson-distributed initial numbers of cells.
All simulations were carried out according to a stochastic process that was described previously (15).
The effect of the average number of cells per food portion on the detection time distribution was studied by simulating the growth of bacteria on 200 portions of food, assuming that the number of bacteria per portion was Poisson distributed. Thirty-nine simulations were carried out with different values for the Poisson parameter, calculated as −ln(Ni/200) (N1 = 195, N2 = 190…N39 = 5), where Ni is the number of food portions that contain no bacteria.
These distributions were compared with that obtained when the simulation was initiated with exactly 1 cell per food portion.
The interdivisional times were randomly sampled from previously described single-cell measurements observed at 32°C (15). In that work, the average time to first division of the single cells was 1 h, with a standard deviation of 0.5 h; the average second-generation time was 0.7 h, with a 0.2-h standard deviation, and 0.5 and 0.2 h were the average and standard deviation of the third-generation times, respectively. To assess the results, the simulation was repeated with theoretical distributions for the division intervals. The single-cell lag times were generated from a gamma distribution with an expected value of 10 h and a standard deviation of 4.5 h. The generation times followed an exponential distribution of parameters equal to 0.5 h−1.
Statistical analysis.
Distributions were compared by a χ2 test.
For the broth experiments, the detection time distributions obtained with the Bioscreen instrument were compared with those obtained from the transformation of the bacterial counts at a given time.
For food experiments, the lag time distributions of the unheated and heated cells were compared for each product.
A regression analysis was carried out to study the relationship between the logarithms of the average and the standard deviation of the lag times.
RESULTS
The distribution of detection times simulated from a single cell was not significantly different from the distributions simulated with Poisson-distributed initial numbers of cells if the Poisson parameter was smaller than 1.6 ± 0.08. This result was consistent for both cases when the simulations were based on measured division intervals at the single-cell level (15) and cases when the simulations were based on theoretical distributions. Thus, to measure the distribution of the single-cell detection times, the percentage of samples positive for growth must be smaller than 80%. This means that >40% of the positive samples will originate from exactly 1 cell.
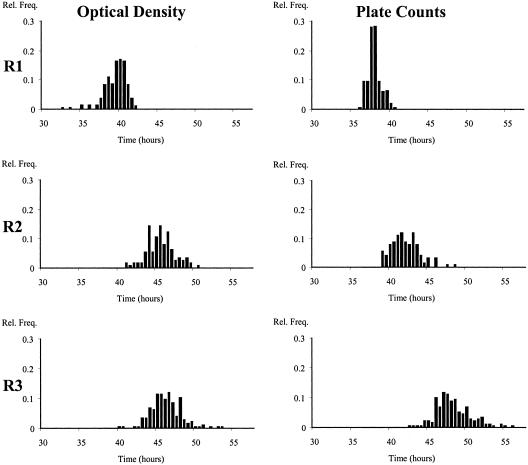
Figure 1 shows the distributions of the detection times obtained in laboratory medium directly from the Bioscreen instrument and indirectly from the distribution of the bacterial counts of the populations growing exponentially. The maximum specific growth rate is a highly reproducible parameter, with a relative deviation of <3% (11). The standard error of the plate counts was experimentally estimated here to be 0.211 log10 CFU/ml. In spite of this error, the distributions of the detection times measured by optical density and by bacterial counts were not significantly different in any of the three replicate experiments.
FIG. 1.
Distributions of detection times estimated directly by optical density measurements in broth inoculated with ca. 1 cell/well and by transforming the log plate counts of detection times in three replicate experiments (R1 to R3).
In experiment 1, the average initial number of cells per well was ca. 16, while in experiments 2 and 3, it was very close to 1. The proportions of wells with only one cell of the total number of positive wells were 0%, 69%, and 62% for experiments 1, 2, and 3, respectively. In fact, in experiment 1, there were no wells with fewer than 5 cells, and only 7% of the wells had fewer than 10 cells.
A χ2 test indicated a lack of homogeneity between the three distributions for the replicate experiments in broth. The distributions of the detection times measured either by optical density measurements or by plate counts in experiment 1 were significantly different from those obtained in the other two experiments, in which the number of cells per well was 1 for most of the wells (Table 1). The average detection time observed in experiment 1 was shorter, and its variance was smaller as a consequence of the higher initial number of cells per well.
TABLE 1.
Means, standard deviations, and P values associated with a χ2 test to compare the distributions of detection times measured by optical densities and estimated from bacterial plate counts in three replicate experiments
| Replicate | No. of cells/well | Techniquea | Mean (h) | SD (h) | P value |
|---|---|---|---|---|---|
| 1 | 16 | OD | 39.14 | 1.435 | 0.8887 |
| PC | 37.63 | 0.8191 | |||
| 2 | 0.70 | OD | 45.27 | 1.828 | 0.4756 |
| PC | 41.69 | 1.811 | |||
| 3 | 0.88 | OD | 44.80 | 1.155 | 0.8466 |
| PC | 47.72 | 2.531 |
OD, optical density measurement; PC, bacterial plate counts.
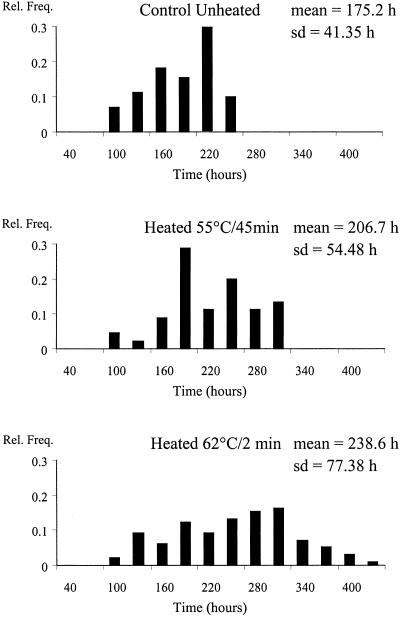
Figure 2 shows the distributions of the lag times of the heated and unheated cells growing in the dairy product stored at 4°C. The number of bacteria per food portion able to grow under those conditions was very close to 1 in the control batch and in the samples heated at 55°C (Table 2). The average number of cells in the batch heated at 62°C was ca. 4 cells per portion, which means that ca. 62% of the food samples positive for growth had fewer than 5 cells and that portions with 10 or more cells were very unlikely. The unheated cells showed the shortest lag times with the smallest variance. The decimal reduction in the bacterial population was approximately the same (ca. 4 to 5) for the two heat treatments; however, the mean lag time of the cells heated at the highest temperature, 62°C, was longer and its standard deviation was greater (see Fig. 4).
FIG. 2.
Distributions of lag times estimated from an unheated and heated dairy product stored at 4°C with a final load of ca. 1 cell per portion.
TABLE 2.
Numbers of cells per portion able to grow in paté at 15°C and in a dairy dessert at 4°C in unheated control batches and after different heat treatments
| Sample | Temp/time (°C/min) | No. of cells/portion |
|---|---|---|
| Liver paté (13-g portions) | Unheated | 2.4 |
| 55/25 | 0.84 | |
| 62/1.35 | 0.64 | |
| 65/0.33 | 1.0 | |
| Dairy dessert (5-ml portions) | Unheated | 0.73 |
| 55/45 | 0.60 | |
| 62/2 | 4 |
FIG. 4.
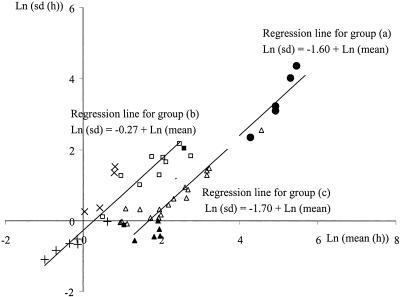
Relationship between natural logarithms of the standard deviation and the mean value of the single-cell lag times estimated for the following groups: preheated cells grown in liver paté stored at 15°C and in a dairy product stored at 4°C, as presented in this paper (•) (a); previously stressed cells grown either in a Bioscreen instrument, as described previously (9 [□] and 14 [▪]), or in a flow chamber, as reported previously (6 [×] and 18 [+]) (b); and healthy cells grown in laboratory media as described previously (11 [▵] and 21 [▴]).
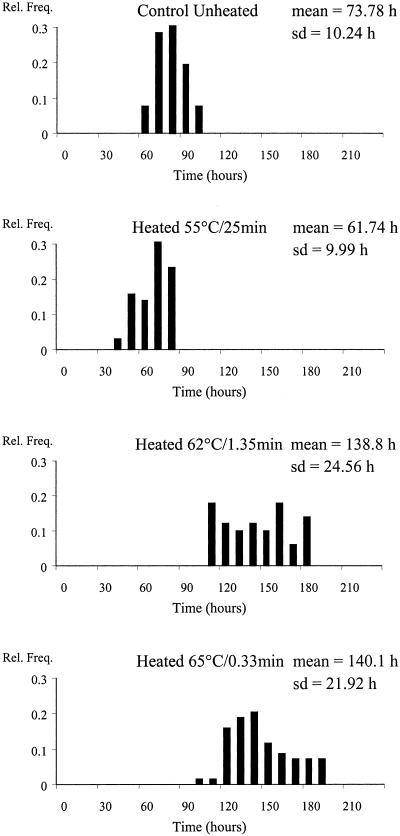
The observed lag times in paté were shorter and had less variability than those observed with the dairy product. This was mainly due to the higher incubation temperature of the heated and unheated samples (15°C). The distributions measured in paté are shown in Fig. 3. The heat treatments caused a 2- to 3-decimal reduction in the population of L. innocua. The final number of cells per paté portion able to initiate growth at 15°C was ca. 1 cell (Table 2) for the heated batches and ca. 2.4 cells for the control batch. Hence, of the unheated paté samples, ca. 30% initiated growth from a single cell. The distribution of the lag times of the cells heated at 55°C for 25 min was not significantly different from that of the unheated cells; however, at the higher temperatures, 62°C and 65°C, the lag duration was longer and its variance was greater. There was no significant difference between the lag times of the survivors in paté after applying the two higher temperatures (Fig. 4).
FIG. 3.
Distributions of lag times estimated from unheated and heated liver paté stored at 15°C with a final load of ca. 1 cell per portion.
DISCUSSION
The three replicate experiments in broth showed that the detection time distribution (horizontal) can be reliably obtained from the distribution of the log counts (vertical) during the exponential growth phase. This result validates the use of the vertical distribution to estimate the detection time distribution of single cells growing in food. It is often assumed that the detection time distribution is identical to the lag time distribution (7, 11) but shifted in time; thus, if the maximum specific growth rate is known, the distributions can be easily transformed into each other. This is true only if the bacterial growth rate (or doubling time) is constant during the growth process. In the case of a crowded bacterial population, this is ensured by keeping the environment constant. However, if growth is initiated from one or a small number of cells, the disturbing effect of the heterogeneity of the single cells may be observed at the start of growth, when the population is small. Recent work has shown that a bacterial cell does not divide immediately at its maximum exponential growth rate but that the intervals between divisions decrease during the first generations after the lag phase (6, 12, 14, 15), so the growth rate increases. Even if after the lag time the cells divide at their maximum rate, the distribution of the single-cell generation times may distort the connection between lag and detection times. For example, if the variance of the generation times was greater than the variance of the lag times, then the lag time distribution would not be reflected in the first division time. In spite of all these concerns, it has been proven that the detection time distribution reflects the distribution of the first division times of single cells of Escherichia coli growing at 25°C (15). However, the concerns pointed out above must be taken into account when transforming detection times into lag times.
The number of cells per well or food portion was not exactly 1 in all cases but was assumed to follow a Poisson distribution. In fact, the lag time is not measured with single cells but with cultures initiated with different numbers of cells depending on the Poisson parameter. The effect of the inoculum size on the lag time duration has been reported when growth is initiated from a small number of cells, i.e., small inoculum sizes or pretreatments that give very small numbers of cells able to grow (1, 13, 16). This effect is due to the fact that the population lag time is the result of the growth of the cells with the shortest lag times. In consequence, the population lag time is shorter with greater inoculum sizes, and this effect is already remarkable between the lag times of cultures initiated with one and with two cells (3, 15). Thus, the observed distribution for wells or food portions inoculated with small numbers of cells may not reflect the single-cell lag times. From the simulation results, it was concluded that the distribution of the single-cell lag times was significantly different from those obtained for cultures initiated with Poisson-distributed numbers of cells when the Poisson parameter was >1.6, i.e., when <40% of the positive wells were initiated from a single cell. This is why the first experiment with broth which was inoculated with ca. 16 cells per well showed much shorter lag times than the other two replicate experiments. Also, with the dairy product heated at 62°C, only 8% of the positive samples started from one cell, and thus the real average and variance of the single-cell lag times are likely to be greater.
The effect of sublethal heat shocks on the distribution of the times to the first division of single cells of L. innocua at 52°C for 1, 2, or 5 min in laboratory medium has recently been reported (6). In that article, greater average lag times and variances were reported for the longer heating times. The different temperature-time treatments applied in our work had lethal effects of 4 to 5 and 2 to 3 decimal reductions of the population in the dairy product and the paté samples, respectively. For each food product, different heat treatments with identical bactericidal effects yielded surviving cells characterized by different lag time distributions. The mean and standard deviation of the lag time were greater at the higher temperatures for both food products. With paté, the heat treatment at 55°C did not change the distribution of the lag times of the surviving cells with respect to the unheated samples, while the lag times after the treatments at 62°C and 65°C were notably increased. The heating time at 55°C for the dairy product was longer than that for paté and resulted in a significantly greater bactericidal effect and longer lag times of the survivors. With the dairy product, the lag times were longer when samples were heated at 62°C than at 55°C. Thus, for the same bactericidal effect, the higher the temperature of the heat treatment, the longer the bacterial lag time and, consequently, the product shelf life and the lower the level of pathogens at the time of consumption.
The standard deviations of the lag times increased as the mean lag times increased. An increase in the absolute deviation, but not necessarily in the relative deviation (relative to the mean), is expected for time measurements (14). Thus, analysis of the relative deviations informs on the effect of the environmental conditions on the variability of the lag times of single cells. Figure 4 shows a regression analysis between the natural logarithms of the standard deviations and the mean values of the single-cell lag times presented in this work and observed by other authors (6, 9, 11, 14, 18, 21). The first division times measured at the single-cell level by others (6, 14) were transformed into lag times by subtracting a generation time estimated from the maximum specific growth rate under those conditions. Measurements were classified into three categories, and a regression analysis was carried out separately for each of them, as follows: (i) heated cells grown in food (presented here); (ii) cells previously subjected to different stresses and grown under optimum laboratory conditions in either a Bioscreen instrument (9, 18) or a flow chamber (6, 14); and (iii) unstressed cells grown under laboratory conditions with different pH values and NaCl contents (11, 21). Figure 4 shows consistently that the regression slope between the logarithms of the standard deviation and the mean was not significantly different from 1 for any data set. This means that the standard deviation is proportional to the average lag time. The regression intercept is a transformation of the ratio between these two quantities for that group of observations. This ratio is equivalent to the relative deviation (to the mean) or coefficient of variation. The relative deviation can be considered fairly constant within the observations of each group (Fig. 4). For the lag times of group i, the ratio of the standard deviation to the mean was equal to an e1.61 of 0.2. A much greater relative deviation, 0.76, was observed for cells growing under optimum laboratory conditions but previously subjected to stressing conditions (group ii). The smallest relative error, 0.18, was that of the lag times of previously unstressed cells, i.e., group iii, although for some observations in this group, the current growing environment was not optimum (11). This comparative analysis must be interpreted very cautiously since the compared data sets were obtained by very different technical approaches and since none of them provided direct measurements of the lag period. In any case, it is important that the traditionally fitted lag of a growth curve is a convenient parameter only, from which one cannot draw conclusions on either the lag time distribution of the single cells or the effects of any controlled variables on the lag time, since the population has an averaging effect that can mask the effects of these variables. For these reasons, analysis of the lag time distribution of single cells will certainly improve the accuracy of predictive models and contribute to the development of tools for risk analysis.
This work shows for the first time that the effects of different pretreatments or the cell's prehistory on the distribution of single-cell lag times can be studied in foods by means of the vertical distribution, which is measured by enumerating bacteria during the exponential growth phase (10). In this way, food processes prolonging the expected lag time or reducing its variance can be identified to improve the control of the process. Also, this methodology provides the necessary measurements to approach the bacterial lag time as a function of the prehistory of the cells and to improve and validate the risk analysis of food products with low levels of contamination.
Acknowledgments
We thank Yolanda García Mesa for her valuable help in carrying out the experiments.
This paper was prepared under the funding of the EU program Quality of Life and Management of Living Resources project no. QLK1-CT-2001-01145 (BACANOVA). G.D.G.D.F. is grateful for the support of the CICYT (Comisión Interministerial de Ciencia y Tecnología, Spain) under project numbers AGL2000-0692 and AGL2005-01239.
REFERENCES
- 1.Augustin, J. C., A. Brouillaud-Delattre, L. Rosso, and V. Carlier. 2000. Significance of inoculum size in the lag time of Listeria monocytogenes. Appl. Environ. Microbiol. 66:1706-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranyi, J., and C. Pin. 1999. Estimating bacterial growth parameters by means of detection times. Appl. Environ. Microbiol. 65:732-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranyi, J., and C. Pin. 2003. Modelling the history effect on microbial growth and survival, p. 285-302. In R. C. McKellar and X. Lu (ed.), Modelling microbial responses in food. CRC Press, Boca Raton, Fla.
- 4.Baranyi, J., and T. A. Roberts. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277-294. [DOI] [PubMed] [Google Scholar]
- 5.Dalgaard, P., and K. Koutsoumanis. 2001. Comparison of maximum specific growth rates and lag times estimated from absorbance and viable count data by different mathematical models. J. Microbiol. Methods 43:183-196. [DOI] [PubMed] [Google Scholar]
- 6.Elfwing, A., Y. LeMarc, J. Baranyi, and A. Ballagi. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl. Environ. Microbiol. 70:675-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francois, K., F. Devlieghere, K. Smet, A. R. Standaert, A. H. Geeraerd, J. F. Van Impe, and J. Debevere. 2005. Modelling the individual cell lag phase: effect of temperature and pH on the individual cell lag distribution of Listeria monocytogenes. Int. J. Food Microbiol. 100:41-53. [DOI] [PubMed] [Google Scholar]
- 8.Francois, K., F. Devlieghere, A. R. Standaert, A. H. Geeraerd, J. F. Van Impe, and J. Debevere. 2003. Modelling the individual cell lag phase. Isolating single cells: protocol development. Lett. Appl. Microbiol. 37:26-30. [DOI] [PubMed] [Google Scholar]
- 9.Guillier, L., P. Pardon, and J. C. Augustin. 2005. Influence of stress on individual lag time distributions of Listeria monocytogenes. Appl. Environ Microbiol. 71:2940-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutalik, Z., S. George, M. Razaz, and J. Baranyi. Submitted for publication.
- 11.Metris, A., S. M. George, M. W. Peck, and J. Baranyi. 2003. Distribution of turbidity detection times produced by single cell-generated bacterial populations. J. Microbiol. Methods 55:821-827. [DOI] [PubMed] [Google Scholar]
- 12.Metris, A., Y. Le Marc, A. Elfwing, A. Ballagi, and J. Baranyi. 2005. Modelling the variability of lag times and the first generation times of single cells of E. coli. Int. J. Food Microbiol. 100:13-19. [DOI] [PubMed] [Google Scholar]
- 13.Pascual, C., T. P. Robinson, M. J. Ocio, O. O. Aboaba, and B. M. Mackey. 2001. The effect of inoculum size and sublethal injury on the ability of Listeria monocytogenes to initiate growth under suboptimal conditions. Lett. Appl. Microbiol. 33:357-361. [DOI] [PubMed] [Google Scholar]
- 14.Pin, C., and J. Baranyi. 2004. Distribution of the lag times of individual cells as a function of the age of the cells in the inoculum, p. 93. In P. Raspor, S. Smole Mozina, and A. Cencic (ed.), Food Micro 2004: new tools for improving microbial food safety and quality. Conference proceedings of the Slovenian Microbiology Society. Slovenian Microbiology Society, Ljubljana, Slovenia.
- 15.Pin, C., and J. Baranyi. 2006. Kinetics of single cells: observation and modeling of a stochastic process. Appl. Environ. Microbiol. 72:2163-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson, T. P., O. O. Aboaba, A. Kaloti, M. J. Ocio, J. Baranyi, and B. M. Mackey. 2001. The effect of inoculum size on the lag phase of Listeria monocytogenes. Int. J. Food Microbiol. 70:163-173. [DOI] [PubMed] [Google Scholar]
- 17.Standaert, A. R., A. H. Geeraerd, K. Bernaerts, K. Francois, F. Devlieghere, J. Debevere, and J. F. Van Impe. 2005. Obtaining single cells: analysis and evaluation of an experimental protocol by means of a simulation model. Int. J. Food Microbiol. 100:55-66. [DOI] [PubMed] [Google Scholar]
- 18.Stephens, P. J., J. A. Joynson, K. W. Davies, R. Holbrook, H. M. Lappin-Scott, and T. J. Humphrey. 1997. The use of an automated growth analyser to measure recovery times of single heat-injured Salmonella cells. J. Appl. Microbiol. 83:445-455. [DOI] [PubMed] [Google Scholar]
- 19.Stringer, S. C., M. D. Webb, S. M. George, C. Pin, and M. W. Peck. 2005. Heterogeneity of times required for germination and outgrowth from single spores of nonproteolytic Clostridium botulinum. Appl. Environ. Microbiol. 71:4998-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woteki, C. E., and B. D. Kineman. 2003. Challenges and approaches to reducing foodborne illness. Annu. Rev. Nutr. 23:315-344. [DOI] [PubMed] [Google Scholar]
- 21.Wu, Y., M. W. Griffiths, and R. C. McKellar. 2000. A comparison of the Bioscreen method and microscopy for the determination of lag times of individual cells of Listeria monocytogenes. Lett. Appl. Microbiol. 30:468-472. [DOI] [PubMed] [Google Scholar]