Abstract
Members of the genus Arsenophonus comprise a large group of bacterial endosymbionts that are widely distributed in arthropods of medical, veterinary, and agricultural importance. At present, little is known about the role of these bacteria in arthropods, because few representatives have been isolated and cultured in the laboratory. In the current study, we describe the isolation and pure culture of an Arsenophonus endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. We propose provisional nomenclature for this bacterium in the genus Arsenophonus as “Candidatus Arsenophonus arthropodicus.” Phylogenetic analyses indicate that “Candidatus Arsenophonus arthropodicus” is closely related to the Arsenophonus endosymbionts found in psyllids, whiteflies, aphids, and mealybugs. The pure culture of this endosymbiont offers new opportunities to examine the role of Arsenophonus in insects. To this end, we describe methods for the culture of “Candidatus Arsenophonus arthropodicus” in an insect cell line and the transformation of this bacterium with a broad-host-range plasmid.
Many members of the class Insecta maintain mutualistic associations with one or more specialized symbiotic bacteria (2). Bacteria that participate in these associations are classified either as primary (P) or secondary (S) endosymbionts, because they often coexist in a single insect host. The P-endosymbionts are predicted to be ancient in origin because their phylogenies are concordant with those of their host insects over a substantial period of evolutionary time, indicating long-term coevolution. On the other hand, the S-endosymbionts are predicted to be recent in origin because their phylogenies show little or no concordance with their insect hosts, indicating recent acquisition.
While the ancient P-endosymbionts are known to have defined mutualistic functions in their insect hosts, the role of the S-endosymbionts is not yet well understood. From an evolutionary standpoint it seems likely that S-endosymbionts have beneficial (mutualistic) roles in their insect hosts because they are maintained predominantly through a maternal (vertical) transmission strategy. Several recent studies have provided experimental evidence for a number of beneficial effects conferred by the S-endosymbionts of aphids, which recently received new nomenclature (20). These benefits include host plant specialization (17, 27), increased resistance to hymenopteran parasitoids (21, 22), and increased tolerance to heat stress (5, 19). In addition, there is evidence indicating that S-endosymbionts can provide some level of functional compensation for the loss of P-endosymbionts in a laboratory population of aphids (16). While these studies are both exciting and encouraging, the ability to perform experimentation in these systems would be greatly enhanced with the opportunity to genetically manipulate S-endosymbionts. The application of recombinant DNA technology would permit functional analysis of individual genes in the endosymbiont genomes, providing a platform to explore the molecular mechanisms involved in symbiotic interactions. Ultimately, it is the lack of availability of culture systems for insect endosymbionts that limits the application of these techniques in the laboratory.
To date, several S-endosymbionts have been cultured successfully outside of their natural hosts in insect cell lines (9, 15, 29). These laboratory cell lines have proved extremely useful for the establishment of monoseptic S-endosymbiont cultures. Since S-endosymbionts are not readily isolated in large numbers from their host insects, the establishment of these insect cell cultures provides a source of material that is useful both for genome studies and for the subsequent development of pure culture systems. The isolation of bacteria in pure culture is important because it facilitates the application of recombinant DNA technology through the isolation of clones. To date only two insect endosymbionts, Arsenophonus nasoniae and Sodalis glossinidius, have been isolated in pure culture (7, 11). S. glossinidius currently serves as a model system for the investigation of insect-symbiont interactions because tools are available to manipulate the bacterial genome (6, 8).
In the current study we describe the isolation, culture, and characterization of an S-endosymbiont that resides in the tissues of a pigeon louse fly, Pseudolynchia canariensis. We propose the provisional name “Candidatus Arsenophonus arthropodicus” for this newly discovered S-endosymbiont based on the fact that it shows a close phylogenetic relationship to other members of the genus Arsenophonus. Since close relatives of “Candidatus Arsenophonus arthropodicus” are found in a wide range of arthropod taxa, the isolation, pure culture, and genetic transformation of this bacterium should prove useful in future efforts directed towards elucidating the role of this group of insect S-endosymbionts.
MATERIALS AND METHODS
PCR amplification, cloning, and sequencing of 16S rRNA gene sequences from P. canariensis.
Universal bacterial 16S rRNA gene primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) were used to establish an inventory of 16S rRNA gene sequences in DNA isolated from newly deposited P. canariensis pupae, obtained from a colony of hippoboscid flies maintained at the University of Utah. Five pupae were washed in 0.5% Triton X-100 to remove any surface contamination prior to DNA extraction. For this and all other total DNA extractions we used the DNeasy tissue extraction kit (QIAGEN), following the supplier's instructions. PCR was performed on isolated DNA using primers 27F and 1492R under standard reaction conditions (14) with 10 ng of template DNA, 300 nM of each primer, 200 μM of each deoxynucleoside triphosphate, 2.5 mM MgCl2, and 0.02 U of Taq DNA polymerase (Promega, Madison, Wis.) per microliter of reaction mix. Cycling conditions consisted of an initial denaturation step (95°C, 4 min) followed by 30 rounds of amplification involving denaturation (95°C, 1 min), annealing (50°C, 1 min), and extension (72°C, 2 min), followed by a final extension step at 72°C for 4 min. The PCR product was analyzed by gel electrophoresis. Gels were stained with SYBR Gold (Molecular Probes, Invitrogen, Carlsbad, Calif.) and visualized on a Dark Reader (Clare Chemical Research, Delores, Colo.) to prevent DNA damage. The 1.5-kbp 16S rRNA gene PCR product was excised and purified with a QIAGEN MinElute gel extraction kit (QIAGEN, Valencia, Calif.). The PCR product was then cloned into the pTOPO 2.1 vector (Invitrogen) according to the supplier's instructions. Forty-eight recombinant clones were sequenced to completion at the University of Utah sequencing facility using vector-specific primers and internal sequencing primers that were designed as sequence information became available.
PCR detection of “Candidatus Arsenophonus arthropodicus.”
Specific PCR primers CAIF (5-GCC TGA TGC AGC CAT GCC GCG TGT ATG-3′) and CAIR (5-GTC ATC CCC ACC TTC C-3′) were designed to amplify a 500-bp fragment of the “Candidatus Arsenophonus arthropodicus” 16S rRNA gene sequence. These primers were then used to detect “Candidatus Arsenophonus arthropodicus” both in tissues isolated from P. canariensis and in samples of medium collected throughout the culturing procedure. For tissue-specific PCR, insect tissues (hemocytes, gut, fat body, and reproductive tissues) were isolated by dissection and rinsed three times in sterile 0.85% (wt/vol) saline. DNA was isolated from individual tissues using the QIAGEN DNeasy tissue kit (QIAGEN). For both liquid and solid-phase cultures, DNA was extracted directly from culture material using the same procedure. The detection of “Candidatus Arsenophonus arthropodicus” was performed in PCRs with 10 ng of template DNA, 300 nM of each primer, 200 μM of each deoxynucleoside triphosphate, 2.5 mM MgCl2, and 0.02 U of Taq DNA polymerase (Promega) per microliter of reaction mix. Cycling conditions consisted of an initial denaturation step (95°C, 4 min) followed by 30 rounds of amplification involving denaturation (95°C, 30 s), annealing (65°C, 1 min), and extension (72°C, 1 min), followed by a final extension step at 72°C for 4 min.
Phylogenetic methods.
We analyzed the 16S rRNA gene sequence of “Candidatus Arsenophonus arthropodicus” alongside 16S rRNA gene sequences from a number of other arthropod endosymbionts and free-living members of the family Enterobacteriaceae obtained from the GenBank database. Sequences were aligned using ClustalX, and the alignments were checked manually and adjusted for accuracy. The phylogenetic analysis was conducted using the PAUP3 4.0 package (25). Initially, a neighbor-joining tree was constructed based on the F84 model of sequence evolution. Maximum likelihood (ML) parameters were then estimated from the neighbor-joining tree and used to search for the best ML tree with the heuristic tree-bisection-reconnection (TBR) branch-swapping algorithm. Once the ML tree was obtained, ML parameters were reestimated from the tree and used to search again using the TBR algorithm. This process was repeated until there was no significant improvement in the log likelihood score of the best ML tree. ML bootstrap analysis was then conducted using the same parameters and TBR algorithm.
Isolation and culture of bacteria from P. canariensis.
Initially, bacteria were isolated from newly deposited pupae derived from the laboratory colony of P. canariensis. All steps in the isolation and culture procedure were carried out in a sterile cabinet under laminar flow conditions. In preparation for culture, five pupae were rinsed for 5 min in at least 20 volumes of sterile water containing 0.5% Triton X-100, with vigorous shaking to remove surface contaminants. Pupae were then washed for 5 min in 20 volumes of 5% sodium hypochlorite in order to kill any remaining surface contaminants. Following surface sterilization, pupae were rinsed five times in sterile water and twice in sterile Mitushashi and Maramorosch basal medium without fetal calf serum (MM medium) (7). Pupae were then homogenized in 10 ml MM medium supplemented with antibiotics (100 μg/ml polymyxin B and 500 μg/ml vancomycin) to prevent growth of contaminating bacteria. The 10-ml culture was maintained in a 25-cm2 ventilated tissue culture flask at 25°C and checked at daily intervals for growth.
Pure culture isolation of “Candidatus Arsenophonus arthropodicus.”
For pure culture isolation, we used methods described previously for the solid-phase culture of S. glossinidius (7). Briefly, 100 to 200 μl of liquid from the 5-day-old pupal culture (described above) was streaked onto sterile agar plates containing MM medium supplemented with 0.7% agar. Inoculated plates were quickly transferred to a microbiological gas jar and flushed with at least 20 volumes of a commercially prepared gas mixture comprising 5% oxygen, 10% carbon dioxide, and 85% nitrogen. The gas jar was sealed to maintain microaerophilic conditions and transferred to a 25°C incubator. Single colonies were picked and grown in MM medium to establish pure liquid cultures. Pure liquid cultures were cryopreserved following the addition of a cryoprotectant (15% [vol/vol] glycerol final concentration) by snap-freezing in a dry ice-ethanol bath, prior to storage at −80°C.
Culture of “Candidatus Arsenophonus arthropodicus” in an Aedes albopictus cell line.
A. albopictus cell line C6/36 was obtained from the American Type Culture Collection. The cell line was maintained according to the supplier's instructions and established procedures (29). Aliquots of “Candidatus Arsenophonus arthropodicus” were inoculated into synchronized confluent monolayers of insect cells at a low multiplicity of infection (<50). Insect cells were gently removed at intervals following infection using a cell scraper to provide material for microscopic examination.
Microscopy.
The deconvolution microscope was used to visualize “Candidatus Arsenophonus arthropodicus” in samples of hemolymph isolated from teneral adult hippoboscids, in cultured insect cell lines, and in pure culture. For hemolymph and pure culture samples, freshly isolated material was stained in a three-step procedure using a lipophilic styryl dye, FM4-64 (Molecular Probes/Invitrogen), in combination with 4′,6′-diamidino-2-phenylindole (DAPI). In this procedure material was first stained with FM4-64 (10 μg/ml in MM medium) for 30 min, then stained with DAPI (6 μg/ml in MM medium) for 20 min, and finally destained in MM medium for 20 min. Material was pelleted at 2,000 × g for 5 min between each staining step to permit removal and replacement of stains. Material from the insect cell lines was stained using the same procedure, except in this case DAPI was omitted to permit visualization of bacteria in sections throughout the insect cells. Following staining, specimens were examined on an Applied Precision Deltavision inverted deconvolution microscope under the DAPI and rhodamine channels. Three-dimensional Z-projections were collected using automated procedures and deconvolved using the Softworx package (Applied Precision, Issaquah, Wash.).
Determination of genome size and structure.
We used a CHEF DR-II pulsed-field gel electrophoresis (PFGE) system (Bio-Rad, Hercules, Calif.) to determine the size and organization of the genome of “Candidatus Arsenophonus arthropodicus.” Chromosomal DNA plugs were prepared from 3-day-old cultures of “Candidatus Arsenophonus arthropodicus” (optical density at 600 nm [OD600], ≈0.1) using the Bio-Rad bacterial genomic DNA plug kit (Bio-Rad), following the supplier's instructions. At 4 h prior to the harvesting of cells, 12.5 μg/ml chloramphenicol was added to the symbiont cultures to ensure synchronization of chromosome replication. To obtain a suitable concentration of chromosomal DNA for restriction digestion and PFGE, each 100 μl of agarose plug contained bacteria from 2 ml of symbiont culture (OD600, 0.2). When we checked the integrity of the chromosomal DNA by performing PFGE on undigested plug DNA, we observed multiple small DNA species representing extrachromosomal elements. Subsequent to further analyses these extrachromosomal elements were removed by preparative PFGE according to previously described methods (1). For chromosome size determination, 1- to 2-mm slices of chromosomal DNA plugs were digested in separate reactions with restriction enzymes that cut infrequently in the genomes of enteric bacteria. Prior to digestion, chromosomal plugs were incubated overnight in 250-μl aliquots of 1× restriction buffer. Following equilibration, restriction buffers were replaced and supplemented with 20 U of restriction enzyme. The digestion reaction mixtures were stored at 4°C for 1 h to permit diffusion of restriction enzymes into the agarose plugs. The reactions were then maintained at the appropriate temperature for optimal digestion of each enzyme overnight. Digested plugs and appropriate size standards (New England Biolabs) were subjected to PFGE using a range of separation conditions. The resulting gels were stained and analyzed using the Quantity One software package (Bio-Rad).
Plasmid transformation of “Candidatus Arsenophonus arthropodicus.”
The broad-host-range plasmid pCM66 (18) was used for the transformation of “Candidatus Arsenophonus arthropodicus” using standard procedures. Briefly, 100-ml aliquots of bacteria from a 5-day-old culture (OD600, 0.2) were made competent for heat shock transformation using the CaCl2 method (13). Approximately 50 ng of plasmid DNA was then added to a 1-ml aliquot of competent cells (approximately 108 to 109 cells/ml). The cells were maintained on ice for 30 min and then subjected to a brief heat shock (42°C, 90 s). Following heat shock the bacteria were chilled on ice for 90 seconds and then transferred to 20 ml of MM medium for overnight recovery at 25°C. Antibiotic selection was applied the following morning by supplementing the culture with 20 μg/ml kanamycin to select for bacteria harboring pCM66. Kanamycin-resistant transformants were maintained by serial passage in the laboratory using standard procedures.
Restriction enzyme analysis of extrachromosomal DNA isolated from wild-type and transformed symbionts.
Circular extrachromosomal DNA was isolated from 25-ml cultures (OD600, 0.2) of wild-type and pCM66-transformed “Candidatus Arsenophonus arthropodicus,” using the Promega Wizard miniprep system (Promega), according to the supplier's instructions. One-microgram aliquots of the resulting extrachromosomal DNA samples were digested with a range of restriction enzymes (Promega and New England BioLabs) under appropriate reaction conditions. The digested extrachromosomal DNA samples were then analyzed by conventional agarose gel electrophoresis.
Nucleotide sequence accession numbers.
The sequences of PC1 to PC4 have been submitted to GenBank and their accession numbers are DQ115535 to -8.
RESULTS
Analysis of 16S rRNA gene sequences amplified from P. canariensis pupal DNA.
The sequencing of 48 clones from the P. canariensis 16S rRNA gene library led to the identification of four distinct 16S rRNA gene sequences, represented approximately equally within the library. The four 1.5-kbp consensus 16S rRNA gene sequences were submitted for BLAST search with the NCBI database to establish their identity. The results are presented in Table 1. The first sequence (PC1) was unique among the four identified because it did not share a high level of sequence identity with any 16S rRNA gene sequences in the GenBank database. Instead, it shared a relatively low level of sequence identity (<95%) with the 16S rRNA genes of a number of insect endosymbionts, including Sodalis glossinidius. This result suggests that PC1 is derived from the primary endosymbiont of P. canariensis. P-endosymbionts typically have 16S rRNA gene sequences that are substantially diverged from those of other insect endosymbionts and related free-living bacteria because they have evolved in isolation for long periods of time in their hosts. The second 16S rRNA gene sequence, PC2, shared the highest level of identity (99%) with a number of 16S rRNA gene sequences from the Arsenophonus group of S-endosymbionts that are found in a number of unrelated insect hosts. The remaining two P. canariensis 16S rRNA gene sequences, PC3 and PC4, each shared a high level of sequence identity (99%) with 16S rRNA gene sequences reported for Wolbachia spp. Notably, PC3 and PC4 shared only 97% sequence identity with one another, indicating that P. canariensis harbors two distinct strains of Wolbachia. For the remainder of the current study we focus on the Arsenophonus endosymbiont that harbors 16S rRNA gene sequence PC2. The details relating to the characterization of the putative P-endosymbiont and Wolbachia spp. will be published in a separate manuscript.
TABLE 1.
Sequences in the GenBank database that share the highest identity with 16S rRNA gene sequences from P. canariensis (PC1 to -4)
| Query | Accession no. | % Identity | Description |
|---|---|---|---|
| PC1 | AY861701 | 94 | Sodalis glossinidius (tsetse fly symbiont) |
| AF476100 | 94 | Symbiont of Amonostherium lichtensiodes | |
| AF476109 | 93 | Symbiont of Paracoccus nothofagicola | |
| PC2 | AY264665 | 99 | Symbiont of Aleuroplatus gelatinosus |
| AB038366 | 99 | Symbiont of Diaphorina citri | |
| AY264673 | 99 | Symbiont of Australiococcus greville | |
| PC3 | M85267 | 99 | Wolbachia sp. from Rhinocyllus conicus |
| AY833061 | 98 | Wolbachia sp. from Culex pipiens | |
| AY007548 | 98 | Wolbachia sp. from Acalymma blandulum | |
| PC4 | M84646 | 99 | Wolbachia sp. from Nasonia vitripennis |
| U83096 | 99 | Wolbachia sp. from Gryllus integer | |
| AY754820 | 99 | Wolbachia sp. from Metaseiulus occidentalis |
Tissue-specific distribution of “Candidatus Arsenophonus arthropodicus.”
Primers CAIF and CAIR were used in PCRs to specifically detect the Arsenophonus 16S rRNA gene sequence in host insect tissues. We detected the Arsenophonus 16S rRNA gene sequence in hemocytes, gut, fat body, and reproductive tissues isolated from newly emerged P. canariensis flies. The widespread distribution of this bacterium in tissues of the host insect is not unusual given that other S-endosymbionts are known to share similar widespread tissue distributions (7, 15, 29).
Phylogenetic analysis.
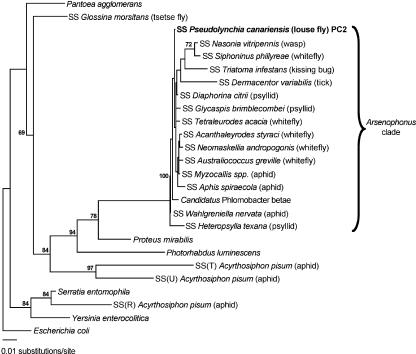
According to the maximum likelihood analysis presented in the current study (Fig. 1), the Arsenophonus group of arthropod endosymbionts forms a robust clade supported by 100% of bootstrap resamples. Only one member of this clade, “Candidatus Phlomobacter betae,” a pathogen of sugar beets closely related to “Candidatus Phlomobacter fragariae” (31), is not known to be associated with an arthropod host. The PC2 sequence, derived from the 16S rRNA gene library of P. canariensis, is tightly clustered within the Arsenophonus clade. Although some phylogenetic structure does exist within the Arsenophonus clade, it is not well resolved by analysis of the 16S rRNA gene, which evolves relatively slowly. The important point to note is that very closely related Arsenophonus endosymbionts are found in distantly related arthropod hosts. This can only be explained by the recent lateral transfer of Arsenophonus between distantly related arthropod hosts.
FIG. 1.
16S rRNA gene phylogeny based on S-endosymbionts of arthropods and some closely related free-living bacteria. The S-endosymbionts have the prefix SS followed by the proper name of their host, whereas free-living bacteria have proper binomial nomenclature. Only bootstrap values greater than 60% are shown adjacent to each node. The GenBank accession numbers for the 16S rRNA gene sequences are as follows: P. agglomerans, AF373196; E. coli, U00096; S. entomophila, AJ233427; Y. enterocolitica, AF366378; “Candidatus Phlomobacter betae,” AY057392; P. mirabilis, AF008582; P. luminescens, BX571859; SS Glossina morsitans, AY861704; SS(R) Acyrthosiphon pisum, AY620432; SS Pseudolynchia canariensis, DQ115536; SS Glycaspis brimblecombei, AF263561; SS Nasonia vitripennis, M90801; SS Triatoma infestans, U91786; SS Dermacentor variabilis, AY265347; SS Diaphorina citrii, AB038366; SS Tetraleurodes acacia, AY264670; SS Acanthaleyrodes styraci, AY264663; SS Neomaskellia andropogonis, AY264668; SS Australiococcus greville, AY264673; SS Heteropsylla texana, AF263562; SS Wahlgreniella nervata, AY136168; SS Myzocallis spp., AY136153; SS Siphoninus phillyreae, AY264669; SS Aphis spiraecola, AY136142; SS(T) Acyrthosiphon pisum, AY462101; SS(U) Acyrthosiphon pisum, AY462102.
Establishment of a primary Arsenophonus culture from P. canariensis pupae.
The primary culture of the P. canariensis Arsenophonus endosymbiont was obtained from surface-sterilized P. canariensis pupae. Initially, cultures were obtained from homogenates derived from five pupae, although we subsequently found that cultures could be established from single pupae. To restrict the growth of contaminating bacteria prior to pure culture isolation, the primary culture medium was supplemented with vancomycin and polymyxin B. Whereas vancomycin prevents the growth of a wide range of gram-positive bacteria, polymyxin B restricts the growth of gram-negative bacteria that are sensitive to the bactericidal effects of antibacterial cationic peptides (10). Given that cationic peptides are an important component of the insect innate immune system (3), it is logical to assume that many insect S-endosymbionts, especially those residing in hemolymph, would display resistance to these peptides. Bacterial growth was first observed in the P. canariensis pupal cultures at 5 days following establishment. Microscopic examination revealed the presence of uniform gram-negative rods in all cultures. The identity of the cultured bacterium was then established by 16S rRNA gene sequencing. All 16 clones examined from a library of 16S rRNA gene PCR products derived from the bacteria in culture had sequences that were 100% identical to the PC2 (Arsenophonus) sequence from P. canariensis. All subsequent culturing was performed without antibiotics.
Establishment of a pure culture on solid medium.
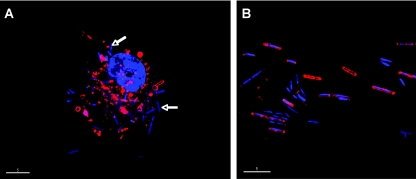
Pure cultures were established from the primary liquid cultures on MM agar plates under an artificial microaerobic atmosphere in a sealed gas jar. Colonies were first visible on the agar plates after 5 days of incubation at 25°C. After 10 days, colonies had reached 2 to 3 mm in diameter and plates were removed from the gas jar. All colonies were irregular and raised with an undulate margin, light brown pigmentation, and a dull, moist surface. Ten colonies were picked at random from plates and inoculated into MM medium to establish pure liquid cultures. Following growth, we examined the morphology of cells from these pure cultures under the deconvolution microscope (Fig. 2). We also examined hemocytes collected from newly emerged adult flies for comparison. All of the pure cultures contained uniform rod-shaped cells, measuring 2 to 5 μm in length and 0.3 μm in diameter. Morphologically similar forms were found in the insect hemocytes, although many of these intracellular forms did not stain well with FM4-64. To confirm the identity of bacteria in the pure cultures, DNA was isolated from bacterial cells in each culture and subjected to PCR analysis. PCR was performed using both universal 16S rRNA gene primers and primers CAIF and CAIR, designed to specifically amplify the Arsenophonus 16S rRNA gene sequence obtained initially from the P. canariensis clone library. The 16S rRNA gene sequences obtained from each pure culture were found to be identical to the PC2 (Arsenophonus) sequence from P. canariensis. DNA from each culture also tested positive with the CAIF and CAIR primers in PCRs. We therefore conclude that we obtained pure cultures of the Arsenophonus S-endosymbiont of P. canariensis. Pure cultures were cryopreserved, stored at −80°C for several weeks, and successfully resuscitated into culture in MM medium.
FIG. 2.
“Candidatus Arsenophonus arthropodicus” in a P. canariensis hemocyte (A) and in pure culture in the laboratory (B). The deconvolved images were generated from material stained with FM4-64 and DAPI. The majority of bacteria (indicated by arrows) in the hemocyte were not stained with FM4-64, indicating that they lack cell walls in this intracellular form. Bars, 5 μm.
Provisional nomenclature.
Following pure culture isolation, we propose the provisional name “Candidatus Arsenophonus arthropodicus” for the S-endosymbiont of P. canariensis. Provisional placement in the genus Arsenophonus is proposed based on the high level of 16S rRNA gene sequence identity between the bacterium obtained in pure culture in this study and other members of the genus Arsenophonus. Since it is likely that the majority of the Arsenophonus S-endosymbionts found in arthropods will belong to the same species (based on the fact that many share 99% sequence identity in the 16S rRNA gene), we propose the use of the species epithet arthropodicus (Latin for belonging to arthropods) Formal nomenclature will be proposed for “Candidatus Arsenophonus arthropodicus” following the application of phenotypic tests to establish the biochemical properties of this bacterium. Attempts will also be made to obtain pure cultures of related bacteria harbored by other arthropods.
Culture of “Candidatus Arsenophonus arthropodicus” in an insect cell line.
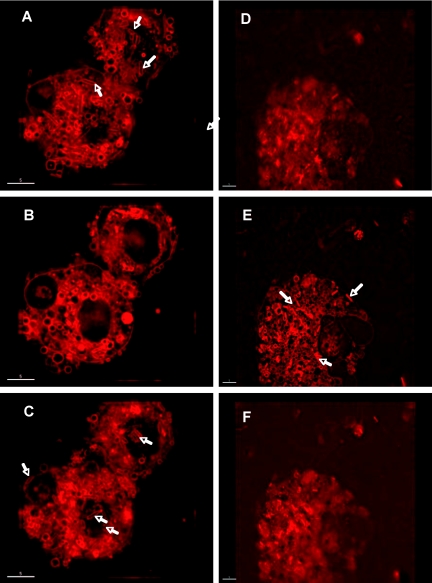
The pure culture isolate of “Candidatus Arsenophonus arthropodicus” was inoculated at a low multiplicity of infection into a confluent A. albopictus C6/36 cell culture. We then used a deconvolution microscope to monitor the course of the infection process. The deconvolution microscope was used to visualize sections (Z-projections) of insect cells at 4 h and 48 h postinfection. At 4 hours postinfection, bacteria were observed adhering to the surface of insect cells (Fig. 3A and C, showing the Z-projections corresponding to the cell surface) but not in the intracellular section (Fig. 3B). At 48 h postinfection, bacteria were observed in the cytoplasm of insect cells (Fig. 3E, showing the intracellular Z-projection) but not on the surfaces of insect cells (Fig. 3D and F). These results indicate that “Candidatus Arsenophonus arthropodicus” is capable of establishing an intracellular infection in insect cells in vitro.
FIG. 3.
“Candidatus Arsenophonus arthropodicus” in the Aedes albopictus cell line C6/36, stained with FM4-64. Micrographs were obtained at 4 h (A, B, and C) and 48 h (D, E, and F) following infection of the cell line. Plates A and D are sections obtained from the top surface of insect cells, plates B and E are intracellular sections, and plates C and F are sections obtained from the bottom surface of insect cells. Note that bacteria (indicated by arrows) adhered to the top and bottom surfaces of insect cells at 4 h postinfection (A and C) but are absent in the intracellular section (B). At 48 h postinfection, bacteria are visible in the intracellular section as they divide in the cytoplasm of insect cells (E).
“Candidatus Arsenophonus arthropodicus” genome size and composition.
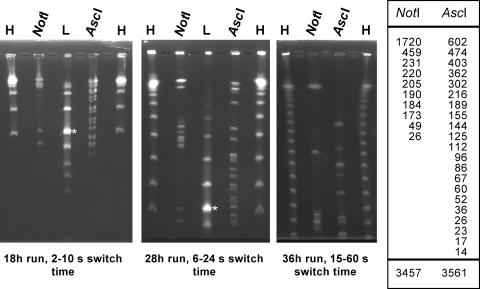
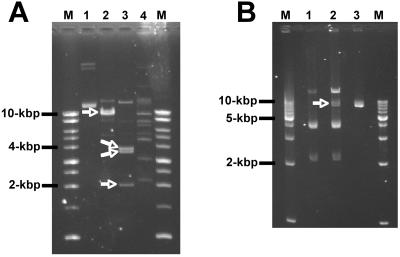
The genome of “Candidatus Arsenophonus arthropodicus” is comprised of a single chromosome and multiple extrachromosomal elements. To obtain an accurate estimate of chromosome size, it was necessary to remove extrachromosomal DNA from agarose plug preparations of chromosomal DNA prior to restriction analysis and PFGE. In the absence of extrachromosomal DNA, we analyzed intact chromosomal DNA using two rare-cutting restriction enzymes, NotI and AscI. Each of these enzymes yielded a reasonable number of fragments for chromosome size estimation (Fig. 4). The chromosome size estimates based on the NotI and AscI digests were 3,457 kbp and 3,561 kbp, respectively. The stoichiometry of fragments resolved on the pulsed-field gels was consistent with the presence of a single chromosome. Following the estimation of chromosome size, we attempted to estimate the size and composition of extrachromosomal elements in the genome of “Candidatus Arsenophonus arthropodicus.” Extrachromosomal DNA was isolated from cultures by using an alkaline lysis procedure and digested with a range of restriction enzymes. Restriction fragments were then separated by conventional agarose gel electrophoresis (Fig. 5A). The stoichiometry of restriction fragments indicated that at least two species of extrachromosomal DNA are present in “Candidatus Arsenophonus arthropodicus.” The smallest of these species (designated pARS1) is a circular plasmid with a size of 9.9 kbp with a single EcoRI site and at least three XbaI sites. One or more additional, larger plasmids are also present. At this stage we cannot provide an accurate estimate of the number and sizes of the larger extrachromosomal elements in “Candidatus Arsenophonus arthropodicus.”
FIG. 4.
PFGE analysis of the “Candidatus Arsenophonus arthropodicus” genome following removal of extrachromosomal elements. Restriction fragments and markers were resolved under three distinct sets of electrophoresis conditions, as indicated below each gel photograph. The lanes are labeled as follows: H, high-range PFGE marker (New England BioLabs), a concatemer of a 48.5-kbp λ DNA fragment; L, low-range PFGE marker (New England BioLabs), a HindIII digest of λ DNA mixed with 48.5-kbp λ DNA concatemers (asterisk marks the 48.5-kbp λ DNA fragment); N and A, NotI and AscI digests, respectively, of chromosomal DNA isolated from “Candidatus Arsenophonus arthropodicus.” The smallest λ concatemer visible in lane H on the gel from the 36-h run is the 145.5-kbp fragment. The sizes of restriction fragments from the NotI and AscI digests are presented in the adjacent table. The total sizes of the NotI and AscI fragments were estimated to be 3,457 kbp and 3,561 kbp, respectively.
FIG. 5.
Restriction enzyme digestion of extrachromosomal elements in untransformed and pCM66-transformed pure culture isolates of “Candidatus Arsenophonus arthropodicus.” (A) Extrachromosomal DNA from untransformed bacteria before digestion (lane 1) and after digestion with EcoRI (lane 2), XbaI (lane 3), and CspCI (lane 4). Arrows indicate EcoRI and XbaI fragments of the 10-kbp plasmid pARS1. (B) Extrachromosomal DNA from untransformed bacteria digested with XbaI (lane 1), extrachromosomal DNA from pCM66-transformed bacteria digested with XbaI (lane 2), and plasmid pCM66 DNA digested with XbaI (lane 3). The XbaI-linearized fragment of pCM66 (7.6 kbp) is highlighted with an arrow. Lanes labeled M contain the Promega 1-kbp benchtop ladder (A) and Promega 1-kbp step ladder (B), with fragment sizes indicated adjacent to the gel photographs.
Plasmid transformation of “Candidatus Arsenophonus arthropodicus.”
We used the broad-host-range plasmid pCM66 (18) to transform the pure culture isolate of “Candidatus Arsenophonus arthropodicus.” Transformation was achieved with high efficiency after rendering bacteria competent for plasmid transformation by treatment with CaCl2. Following selection for pCM66 transformants, extrachromosomal DNA was isolated from a cloned transformant and analyzed by digestion with the restriction enzyme XbaI. The restriction profile (Fig. 5B) confirmed the presence of both pCM66 and endogenous extrachromosomal DNAs in the transformed bacteria. Plasmid pCM66-transformed bacteria have now been maintained for over 3 months in continuous culture in the laboratory.
DISCUSSION
Members of the provisional genus Arsenophonus have been identified in a wide range of arthropods, including parasitoid wasps (11), triatomine bugs (15), ticks (12), whiteflies (26), aphids (23, 28), and psyllids (24). In the current study, we demonstrated that P. canariensis, a blood-feeding hippoboscid louse fly, also maintains an endosymbiotic bacterium closely allied to the Arsenophonus group. Based on the 16S rRNA gene phylogeny, the Arsenophonus group forms a distinct and robust monophyletic clade of arthropod endosymbionts in the gamma subdivision of Proteobacteria. Only one member of the Arsenophonus clade is not formally known to be associated with an arthropod host (31).
Although many arthropod taxa are known to maintain intimate symbiotic relationships with bacteria, in most cases there is a high degree of specialization between hosts and their endosymbionts. In other words, it is unusual to find extremely closely related endosymbionts in phylogenetically unrelated hosts. This holds true for both the primary (long-established) and secondary (facultative) endosymbionts of arthropods. One obvious exception to this rule occurs with Wolbachia, the most widely distributed symbiont found in association with arthropods and nematodes (4). Aside from Wolbachia, the only other group of closely related arthropod endosymbionts that shares a wide host distribution is the Arsenophonus group.
Although members of the Arsenophonus group have been identified in a wide range of arthropod species, little is known about their role in the context of symbiosis. In the parasitoid wasp Nasonia vitripennis, Arsenophonus nasoniae is known to induce a female-biased sex ratio distortion, similar to that observed with some strains of Wolbachia spp. (11, 30). However, in Triatoma infestans, a blood-sucking bug that harbors a closely related member of the Arsenophonus group (“Candidatus Arsenophonus triatominarum”), no sex ratio distortion effects have been detected in laboratory animal colonies (15). Similarly, there was no sex ratio distortion in our laboratory colony of P. canariensis; we recently determined the sex of a sample of 200 newly emerged flies and found 100 males and 100 females. Given that members of the Arsenophonus group are found in arthropod hosts that persist on a variety of diets (including vertebrate blood and plant sap), it seems unlikely that symbioses involving Arsenophonus have a nutritional basis. However, it is likely that they serve some important role in insects because they are so widely distributed.
In the current study, we obtained a pure culture isolate of “Candidatus Arsenophonus arthropodicus,” a newly described species from the louse fly P. canariensis. According to the phylogenetic analysis, the pure culture isolate of “Candidatus Arsenophonus arthropodicus” is most closely related to the Arsenophonus spp. described from aphids, whiteflies, and psyllids (23, 24, 26). Together, these bacteria share >99% sequence identity in their 16S rRNA genes, most likely indicating recent acquisition of Arsenophonus among these distantly related insects.
The pure culture isolation of “Candidatus Arsenophonus arthropodicus” provides new opportunities to explore the role of Arsenophonus spp. in insects. In this study we demonstrated pure culture isolation, cryopreservation, and artificial infection of an insect cell line with “Candidatus Arsenophonus arthropodicus.” We also determined the composition and size of the “Candidatus Arsenophonus arthropodicus” genome. The size of the “Candidatus Arsenophonus arthropodicus” chromosome was estimated to be 3.51 Mbp, only slightly reduced relative to the 4.06-Mbp chromosome of the closest free-living relative, Proteus mirabilis (whole genome sequence currently undergoing annotation at the Sanger Center). We also discovered a broad-host-range plasmid vector suitable for the high-frequency transformation of “Candidatus Arsenophonus arthropodicus.” Together, the availability of these culture and transformation systems provides a new platform to explore the associations between Arsenophonus and insects.
Acknowledgments
We thank Dale Clayton for the provision of insects from his laboratory colony of P. canariensis. We also thank Mary Lidstrom, who provided the plasmid vector pCM66.
Financial support for the maintenance of this colony is provided by NSF grant DEB-0107947 to D.C.
REFERENCES
- 1.Akman, L., R. V. Rio, C. B. Beard, and S. Aksoy. 2001. Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J. Bacteriol. 183:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, N.Y.
- 3.Bulet, P., C. Hetru, J. L. Dimarcq, and D. Hoffmann. 1999. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23:329-344. [DOI] [PubMed] [Google Scholar]
- 4.Charlat, S., G. D. Hurst, and H. Mercot. 2003. Evolutionary consequences of Wolbachia infections. Trends Genet. 19:217-223. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D. Q., C. B. Montllor, and A. H. Purcell. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95:315-323. [Google Scholar]
- 6.Dale, C., T. Jones, and M. Pontes. 2005. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22:758-766. [DOI] [PubMed] [Google Scholar]
- 7.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49:267-275. [DOI] [PubMed] [Google Scholar]
- 8.Dale, C., S. A. Young, D. T. Haydon, and S. C. Welburn. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. USA 98:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby, A. C., S. M. Chandler, S. C. Welburn, and A. E. Douglas. 2005. Aphid-symbiotic bacteria cultured in insect cell lines. Appl. Environ. Microbiol. 71:4833-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galizzi, A., G. Cacco, A. G. Siccardi, and G. Mazza. 1975. Mode of action of polymyxin B: physiological studies with Bacillus subtilis-resistant mutant. Antimicrob. Agents Chemother. 8:366-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gherna, R. L., J. H. Werren, W. Weisburg, R. Cote, C. R. Woese, L. Mandelco, and D. J. Brenner. 1991. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int. J. Syst. Bacteriol. 41:563-565. [Google Scholar]
- 12.Grindle, N., J. J. Tyner, K. Clay, and C. Fuqua. 2003. Identification of Arsenophonus-type bacteria from the dog tick Dermacentor variabilis. J. Invertebr. Pathol. 83:264-266. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 14.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hypsa, V., and C. Dale. 1997. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47:1140-1144. [DOI] [PubMed] [Google Scholar]
- 16.Koga, R., T. Tsuchida, and T. Fukatsu. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B 270:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonardo, T. E., and G. T. Muiru. 2003. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc. Biol. Sci. 7:S209-S212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 19.Montllor, C. B., A. Maxmen, and A. H. Purcell. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189-195. [Google Scholar]
- 20.Moran, N. A., J. A. Russell, R. Koga, and T. Fukatsu. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver, K. M., N. A. Moran, and M. S. Hunter. 2003. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. USA 102:12795-12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver, K. M., J. A. Russell, N. A. Moran, and M. S. Hunter. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 100:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell, J. A., A. Latorre, B. Sabater-Munoz, A. Moya, and N. A. Moran. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12:1061-1075. [DOI] [PubMed] [Google Scholar]
- 24.Subandiyah, S., N. Nikoh, S. Tsuyumu, S. Somowiyarjo, and T. Fukatsu. 2000. Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zoolog. Sci. 17:983-989. [Google Scholar]
- 25.Swofford, D. L. 1998. PAUP3: phylogenetic analysis using parsimony (3 and other methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 26.Thao, M. L., and P. Baumann. 2004. Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Curr. Microbiol. 48:140-144. [DOI] [PubMed] [Google Scholar]
- 27.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchida, T., R. Koga, H. Shibao, T. Matsumoto, and T. Fukatsu. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11:2123-2135. [DOI] [PubMed] [Google Scholar]
- 29.Welburn, S. C., I. Maudlin, and D. S. Ellis. 1987. In vitro cultivation of rickettsia-like organisms from Glossina spp. Ann. Trop. Med. Parasitol. 81:331-335. [DOI] [PubMed] [Google Scholar]
- 30.Werren, J. H., S. W. Skinner, and A. M. Huger. 1986. Male-killing bacteria in a parasitic wasp. Science 231:990-992. [DOI] [PubMed] [Google Scholar]
- 31.Zreik, L., J. M. Bove, and M. Garnier. 1998. Phylogenetic characterization of the bacterium-like organism associated with marginal chlorosis of strawberry and proposition of a Candidatus taxon for the organism, “Candidatus phlomobacter fragariae.” Int. J. Syst. Bacteriol. 48:257-261. [DOI] [PubMed] [Google Scholar]