Abstract
Enterobacter sakazakii is an emerging, infant formula-borne pathogen that causes severe meningitis, meningoencephalitis, sepsis, and necrotizing enterocolitis in neonates and infants, with a high fatality rate. Traditional detection methods take up to 7 days to identify E. sakazakii. The outer membrane protein A gene (ompA), along with its flanking sequences from E. sakazakii (ATCC 51329), was cloned in the pGEM-T Easy vector and sequenced. Comparison of the nucleotide and deduced amino acid sequences of the ompA gene with other sequences available in the GenBank database revealed a high degree of homology with ompA genes of other gram-negative bacteria belonging to the Enterobacteriaceae. Based on regions of the ompA gene unique to E. sakazakii, two primers were synthesized to develop and optimize an E. sakazakii-specific PCR. The PCR amplified a 469-bp DNA product from all E. sakazakii strains tested but not from other bacteria. Experiments to determine the sensitivity of the PCR indicated that it could detect as few as 103 CFU/ml of E. sakazakii bacteria in infant formula directly and 10−1 CFU/ml after an 8-h enrichment step. We conclude that this PCR, combined with enrichment culturing, has the potential to be used as a rapid tool for detecting the presence of E. sakazakii in infant formula.
Enterobacter sakazakii is an emerging food-borne pathogen that causes severe meningitis, meningoencephalitis, sepsis, and necrotizing enterocolitis in neonates and infants (10, 14, 17, 22). Neonatal infections caused by E. sakazakii have been reported from different parts of the world, including the United States (5, 19). E. sakazakii infections in neonates are characterized by a fatality rate of about 14% (11) and often lead to permanent impairment in mental and physical capabilities in surviving patients (2). The epidemiology and reservoir of E. sakazakii are still unknown, and most strains have been isolated from clinical specimens such as cerebrospinal fluid, blood, skin, wounds, urine, and respiratory and digestive tract samples (11). The organism has also been isolated from a variety of foods, including cheese, meat, milk, vegetables, grains, spices, and herbs (7, 12, 20). Recently, Kandhai et al. (8, 9) isolated E. sakazakii from household and food production facility environmental samples and proposed that the organism could be more widespread in the environment than previously thought. Although the environmental source of E. sakazakii has not been conclusively established, epidemiological studies implicate dried infant formula as the route of transmission to infants (1, 17, 22, 25). The bacterium has been isolated from powdered infant formula by numerous investigators (2, 7, 15). Further, there have been many recalls of E. sakazakii-contaminated infant formula in the United States. In November 2002, a nationwide recall of more than 1.5 million cans of dry infant formula contaminated with E. sakazakii was reported (3).
The FDA-recommended methods for isolation and identification of E. sakazakii from dehydrated powdered infant formula are time consuming and labor intensive, taking up to 7 days for results (21). Identification of E. sakazakii by these methods relies on the pigment production and biochemical profile of isolates and is often hampered by false-negative results in the presence of other, related organisms (4). Molecular methods such as PCR provide powerful tools for rapid, specific, and sensitive detection of food-borne pathogens and are considered reliable alternatives to traditional bacteriological methods (13, 18). One of the recommendations of the joint FAO/WHO workshop on E. sakazakii and other microorganisms in powdered infant formula conducted in 2004 was to promote the use of internationally validated detection and molecular typing methods for identifying E. sakazakii (27).
In this study, we report the identification and molecular cloning of the gene encoding outer membrane protein A (OmpA) in E. sakazakii and development of a PCR based on the ompA gene sequence for specific identification of E. sakazakii in pure cultures as well as in reconstituted infant formula.
MATERIALS AND METHODS
Bacterial media and cultures.
The various bacterial strains used in the study and their respective sources are listed in Table 1. A total of 17 isolates of E. sakazakii, including clinical and infant formula isolates, were used to study the specificity of the primers used in PCR. As negative controls, 51 strains of common species of Enterobacteriaceae and meningitis-causing and infant formula-contaminating bacteria were included. All bacteriological media used in the study were purchased from Difco (Sparks, Md.). The cultures were maintained in tryptic soy broth (TSB) containing 15% glycerol at −20°C and were streaked on tryptic soy agar plates before the experiment. The purity of each culture was confirmed by biotyping (API 20E; bioMérieux, Marcy l'Etoile, France).
TABLE 1.
Bacterial strains used in this study
| No. | Species | Strain | No. | Species | Strain | |
|---|---|---|---|---|---|---|
| 1 | E. sakazakii | ATCC 51329 | 35 | S. enterica serovar Typhi | 372d | |
| 2 | E. sakazakii | ATCC 29004 | 36 | Shigella flexneri | 387d | |
| 3 | E. sakazakii | Gd. St. 8a | 37 | Shigella sonnei | 388d | |
| 4 | E. sakazakii | SMA 13a | 38 | Yersinia enterocolitica | ATCC 23715 | |
| 5 | E. sakazakii | LCDC 513a | 39 | Y. pseudotuberculosis | 399d | |
| 6 | E. sakazakii | LCDC 648a | 40 | Serratia marcescens | 361d | |
| 7 | E. sakazakii | LCDC 674a | 41 | Moraxella bovis | 419d | |
| 8 | E. sakazakii | CDC A1a | 42 | Erwinia carotovora | 351d | |
| 9 | E. sakazakii | CDC 415c | 43 | Citrobacter freundii | 239d | |
| 10 | E. sakazakii | Type 54a | 44 | Klebsiella pneumoniae | 344d | |
| 11 | E. sakazakii | 2879a | 45 | Xanthomonas multophila | 159d | |
| 12 | E. sakazakii | 4581b | 46 | Proteus vulgaris | 365d | |
| 13 | E. sakazakii | 4583b | 47 | Proteus mirabilis | 366d | |
| 14 | E. sakazakii | 4586b | 48 | Morganella morganii | 329d | |
| 15 | E. sakazakii | 4593b | 49 | Providencia alcalifaciens | 368d | |
| 16 | E. sakazakii | 4603b | 50 | Chromobacterium violaceum | 294d | |
| 17 | E. sakazakii | 98-308b | 51 | Pantoea stewartii | DE283f | |
| 18 | Enterobacter aerogenes | 341d | 52 | Edwardsiella ictalurii | 92/132f | |
| 19 | Enterobacter aerogenes | NRRL B-115g | 53 | Edwardsiella tarda | 296d | |
| 20 | Enterobacter aerogenes | NRRL B-407g | 54 | Hafnia sp. | 342d | |
| 21 | Enterobacter aerogenes | NRRL B-410g | 55 | Alcaligenes faecalis | 297d | |
| 21 | Enterobacter aerogenes | NRRL B-494g | 56 | Aeromonas hydrophila | 191d | |
| 23 | Enterobacter cloacae | 343d | 57 | Neisseria catarrhalis | 483d | |
| 24 | Enterobacter cloacae | NRRL B-411g | 58 | Vibrio parahaemolyticus | ATCC 17802 | |
| 25 | Enterobacter cloacae | NRRL B-412g | 59 | Vibrio cholerae (DNA) | ATCC 51394D | |
| 26 | Enterobacter cloacae | NRRL B-413g | 60 | Grimontia hollisae | ATCC 33564 | |
| 27 | Enterobacter cloacae | NRRL B-414g | 61 | Burkholderia cepacia | ATCC 25608 | |
| 28 | Enterobacter cloacae | NRRL B-425g | 62 | Pseudomonas fluorescens | 105d | |
| 29 | Enterobacter dissolvens | NRRL B-41145g | 63 | Pseudomonas fragi | 110d | |
| 30 | Escherichia coli K-12 | NP1f | 64 | Pseudomonas putida | 107d | |
| 31 | E. coli O157:H7 | E 6e | 65 | Listeria monocytogenes | ScottAe | |
| 32 | E. coli K88 | 842f | 66 | Streptococcus pneumoniae | 508d | |
| 33 | Salmonella enterica serovar Enteritidis | SE 90f | 67 | Bacillus cereus | F3802A/84f | |
| 34 | S. enterica serovar Typhimurium DT104 | ST 43f | 68 | Staphylococcus aureus | ATCC 35556 |
J. M. Farber, Bureau of Microbial Hazards, Sir Frederick Banting Research Centre, Ottawa, Canada.
W. Ingrid, Department of Microbiology, Academisch Ziekenhuis Vrije Universiteit Brussel, Brussels, Belgium.
M. J. Arduino, Division of Health Care Quality Promotion, CDC, Atlanta, Ga.
Presque Isle Cultures, Presque Isle, Pa.
M. P. Doyle, Center for Food Safety, University of Georgia, Griffin, Ga.
Culture Collection, Food Microbiology Laboratory, University of Connecticut, Storrs, Conn.
A. P. Rooney, Microbial Genomics and Bioprocessing Research Unit, Agricultural Research Service Culture Collection, U.S. Department of Agriculture, Peoria, Ill.
Preparation of genomic DNA.
Genomic DNA from gram-negative bacteria was extracted with the AquaPure genomic DNA extraction kit (Bio-Rad, Hercules, Calif.). DNA was extracted from 1 ml of overnight-grown cultures in TSB and reconstituted in 100 μl of DNA hydration buffer. Genomic DNA from gram-positive bacteria was extracted by the miniprep method described by Wilson (26). The concentration and purity of the DNA samples were estimated spectrophotometrically, and the samples were stored at −20°C until use.
Cloning and sequencing of the ompA gene from E. sakazakii.
The genetic organization of the ompA gene and its flanking genes, ycbG and sulA, is highly conserved among Escherichia coli, Salmonella, and Shigella spp. Based on the E. coli sequence information available in the GenBank database, primers ESLOCF and ESLOCR (Table 2) were designed and synthesized (Integrated DNA Technologies, Coralville, Iowa) to amplify a 2,047-bp DNA fragment from E. sakazakii ATCC 51329. The PCR mix consisted of 1× Easy-A reaction buffer containing 2 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate (dNTP), 1 μM (each) primer, 2.5 U of Easy-A high-fidelity PCR cloning enzyme (Stratagene, La Jolla, Calif.), 50 ng of template DNA, and sterile, deionized water to make the volume up to 50 μl. PCR was carried out in a programmable thermocycler (PTC 100; MJ Research, Waltham, Mass.) under the following conditions. Initially, the reaction mixtures were heated at 92°C for 2 min, and then the PCR progressed through 30 cycles of melting at 94°C for 30 s, annealing at 52°C for 1 min, and extension at 72°C for 2 min. A final extension for 10 min at 72°C was included at the end of the 30th cycle. Reaction mixtures were stored at 4°C until analysis by electrophoresis on a 1% agarose gel in 1× Tris-acetate-EDTA (0.04 M Tris-acetate, 0.001 M EDTA) buffer at a field strength of 8 V/cm. The amplicons were detected by staining with ethidium bromide (0.5 μg/ml) and were photographed under a UV transilluminator.
TABLE 2.
Primers used for PCR and sequencing
| Primer | Sequence | Function | Position |
|---|---|---|---|
| ESLOCF | CCGGGCTAAAAATTCACTCAAGAATGG | Sequencing | 1-27 |
| ESLOCR | CGAAGGCGAGCTGATAACCCGCTATGT | Sequencing | 2021-2047 |
| OMPAF | TAGACTTTACATCGCCAGGG | Sequencing | 246-265 |
| OMPAR | GAGCTTTCACGTTGTCACAG | Sequencing | 1326-1345 |
| SEQF1 | CCGGCGTTTCTCCTGTATTC | Sequencing | 806-825 |
| SEQF2 | TCTGTAGTGGTTCTGGGCTT | Sequencing | 1156-1175 |
| SEQR1 | GGTGTAAACGTCCAGATCGT | Sequencing | 722-741 |
| SEQR2 | CTCGTTATCATCCAAAAAGG | Sequencing | 365-384 |
| ESSF | GGATTTAACCGTGAACTTTTCC | PCR | 325-346 |
| ESSR | CGCCAGCGATGTTAGAAGA | PCR | 775-793 |
| IAC1F | GGATTTAACCGTGAACTTTTCCTCTTCCGCTTCCTCG | PCR | |
| IAC1R | CGCCAGCGATGTTAGAAGACACCGCCTACATACCTC | PCR |
The PCR product was purified with the QIAquick PCR purification kit (QIAGEN, Valencia, Calif.) and cloned into the pGEM-T Easy cloning vector (Promega, Madison, Wis.) by the TA cloning strategy, according to the manufacturer's protocol. The cloned product was sequenced by primer walking using automated fluorescent dye terminator sequencing on ABI Prism 3730xl sequencers (SeqWright, Houston, Tex.). The sequences of the primers used for primer walking are provided in Table 2. The sequences generated were analyzed by Sequencher 4.1.4 (Gene Codes Corporation, Ann Arbor, Mich.), BPROM (http://www.softberry.com), SignalP (http://www.cbs.dtu.dk/services/SignalP), ClustalW (EMBnet), and Blastn (NCBI).
Internal amplification control for E. sakazakii-specific PCR.
An internal amplification control (IAC) was designed to be included as a positive control in every reaction mixture, to ensure that a negative result would be due to the absence of target sequences rather than to inhibition of the PCR (6). The IAC target DNA consisted of a 618-bp fragment of the pGEM-T Easy vector flanked by the target sequences of the ESSF and ESSR primers. The IAC target DNA was PCR amplified from pGEM-T Easy with primers IAC1F and IAC1R (Table 2). The PCR mix consisted of 1× Easy-A reaction buffer containing 2 mM MgCl2, 200 μM (each) dNTP, 1 μM (each) primer, 2.5 U of Easy-A high-fidelity PCR cloning enzyme, 25 ng of template DNA, and sterile, deionized water to make the volume up to 50 μl. The samples were subjected to PCR with a program that consisted of an initial denaturation at 94°C for 2 min, followed by 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s for 30 cycles, and a final extension at 72°C for 5 min. The amplicons were detected by 1% agarose gel electrophoresis and ethidium bromide (0.5 μg/ml) staining. The PCR product was then purified with the QIAquick PCR purification kit, its concentration was estimated spectrophotometrically, and it was cloned into the pGEM-T Easy cloning vector and transformed into JM109 cells. Transformed JM109 cells carrying the recombinant plasmid were selected by blue-white selection. Recombinant plasmid carrying the IAC template sequence was extracted with the QIAprep Spin Miniprep kit (QIAGEN), linearized by digestion with SalI (New England Biolabs, Beverly, Mass.), and purified with the QIAquick PCR purification kit. The concentration of the IAC plasmid was estimated spectrophotometrically, and serial dilutions were made in 10 mM Tris-HCl (pH 8.5). The concentration of IAC plasmid to be added to PCR mixtures was optimized from the different template preparations used in the individual experiments to allow efficient amplification of the E. sakazakii-specific PCR product.
Primers for E. sakazakii-specific PCR.
Based on the sequence data generated and the results of the BLAST analysis, primers ESSF and ESSR (Table 2) were designed to amplify a 469-bp fragment of the ompA gene specific to E. sakazakii. The PCR mix consisted of 1× Gene Amp PCR buffer II (50 mM potassium chloride and 10 mM Tris-HCl, pH 8.3), 2.5 mM MgCl2, 200 μM (each) dNTP, 1 μM (each) primer, 1 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, Calif.), 50 ng of template DNA, 2 fg (corresponding to approximately 500 copies) of IAC template, and sterile, deionized water to make the volume up to 50 μl. The samples were subjected to PCR with a program that consisted of denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. A 5-μl aliquot of the amplified product was characterized by electrophoresis on a 1.5% agarose gel, as described above.
Sensitivity of E. sakazakii detection.
E. sakazakii ATCC 51329 was cultured in 10 ml of TSB for 8 h at 37°C, and serial 10-fold dilutions were made in sterile, deionized water. One-milliliter aliquots of each dilution were transferred to 1.5-ml microcentrifuge tubes, boiled at 100°C in a heating block for 10 min, and centrifuged in a tabletop microcentrifuge (Marathon 16KM; Fisher scientific, Pittsburgh, Pa.) at 1,500 × g for 30 s. A 10-μl aliquot of each dilution containing 108, 107, 106, 105, 104, 103, 102, 101, or 100 CFU/ml was used as a template in a reaction mix containing 1× Gene Amp PCR buffer II, 2.5 mM MgCl2, 200 μM (each) dNTP, 1 μM (each) primer, 2 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, Calif.), 0.2 fg (corresponding to approximately 50 copies) of IAC template, and sterile, deionized water to make the volume up to 50 μl. Amplification was carried out for 50 cycles under the same cycling conditions as described for PCR using genomic DNA as the template. A 10-μl volume of the amplified product was characterized by electrophoresis on a 1.5% agarose gel, as described above.
Detection of E. sakazakii in infant formula.
Three commercially available brands of powdered infant formula were used in this study. The infant formulas were reconstituted by mixing 1-g aliquots in 8 ml of sterile distilled water in 15-ml Falcon tubes (BD, Franklin Lakes, N.J.). Serial 10-fold dilutions of E. sakazakii (ATCC 51329) culture were made in sterile distilled water, and 1-ml volumes were added to 9 ml of reconstituted formula to obtain final concentrations of E. sakazakii ranging from 108 to 10−2 CFU/ml. The samples were subjected to PCR directly or after an 8-h enrichment step at 37°C. One-milliliter aliquots of the formula samples were centrifuged at 16,000 × g for 10 min, and the pellets were resuspended in 1 ml of sterile distilled water. The samples were boiled in a heating block for 10 min and centrifuged at 1,500 × g for 30 s, and 10 μl of the supernatant was used as a template in PCR for 50 cycles. The PCR and electrophoresis conditions were the same as those described for PCR using E. sakazakii cells resuspended in deionized water.
Detection of E. sakazakii by PCR in the presence of S. enterica serovar Typhimurium.
To investigate whether the presence of other bacteria along with E. sakazakii in the infant formula would have any influence on the sensitivity of the PCR, we used formula samples containing different levels of E. sakazakii and Salmonella enterica serovar Typhimurium as a template for PCR. In one set of experiments, 10-ml aliquots of reconstituted infant formula samples containing E. sakazakii 51329 at levels ranging from 108 to 101 CFU/ml were inoculated with serovar Typhimurium DT 104 at 108 CFU/ml. In another set of experiments, infant formula containing 103 CFU/ml of E. sakazakii 51329 were inoculated with serovar Typhimurium DT 104 at levels ranging from 108 to 101 CFU/ml. One-milliliter aliquots were taken from each sample and processed as described above for use as a template for PCR.
Nucleotide sequence accession number.
The nucleotide sequence of ompA of E. sakazakii ATCC 51329 has been deposited in the GenBank database under accession number DQ000206.
RESULTS
ompA gene sequence analysis.
A 2,047-bp fragment of E. sakazakii chromosomal DNA encompassing the ompA gene and its flanking sequences was amplified with primers complementary to the regions conserved in E. coli, Salmonella, and Shigella spp. The putative open reading frame (ORF), as predicted by the ORF Finder (NCBI), was 1,077 bp long, extending from base 361 to base 1437 of the cloned fragment. However, a BLAST search and alignment of the predicted ORF with other sequences in the GenBank database indicated that translation from the start codon ATG at position 394 could be more likely to occur, resulting in an ORF of 1,044 bp. Alignment and sequence comparison of the complete ompA sequence (1,044 bp) from E. sakazakii 51329 with that of E. coli K-12 (1,041 bp) revealed a sequence identity of 86%. We also compared the deduced amino acid sequence of the E. sakazakii OmpA protein with that of E. coli K-12. The proteins were found to have an identity of 89% and a sequence conservation of 94% at the amino acid level. Similarly, the ompA gene of E. sakazakii had sequence identities of 86% and 88% at the nucleic acid and amino acid levels, respectively, with Enterobacter aerogenes, 87% and 90% with serovar Typhimurium, and 85% and 88% with Shigella flexneri, respectively. An NCBI conserved-domain search of the deduced protein revealed the presence of a conserved ompA-like, N-terminal, eight-stranded, beta barrel transmembrane domain and a C-terminal ompA domain. A signal peptide cleavage site prediction analysis revealed a 21-amino-acid N-terminal signal sequence similar to those in E. coli, E. aerogenes, S. flexneri, and S. enterica serovar Typhi. A multiple alignment of the amino acid sequences is shown in Fig. 1.
FIG. 1.
Optimal alignment of the deduced amino acid sequence of E. sakazakii 51329 OmpA with those from other bacteria. E. saka, E. sakazakii 51329; E. aero, E. aerogenes; E. coli, E. coli K-12; S. flex, S. flexneri 2a strain 2457T; S. Typhi, S. enterica serovar Typhimurium LT2.
A partial ORF in the −2 frame was also detected between bases 2047 and 1660, with the stop codon at 1660. BLAST analysis of this ORF revealed high sequence identities with ycbG of E. coli K-12, S. flexneri (79%), and serovar Typhimurium (75%). The deduced amino acid sequence of the E. sakazakii YcbG protein was found to have a high degree of sequence identity with orthologues in E. coli K-12 (85%), serovar Typhimurium (87%), and S. flexneri (85%). YcbG in E. coli is 150 amino acids long and is a putative dehydrogenase. The partial ORF detected included sequences that encode the last 128 amino acids of the C terminus of the E. sakazakii ycbG gene.
Development of E. sakazakii-specific PCR.
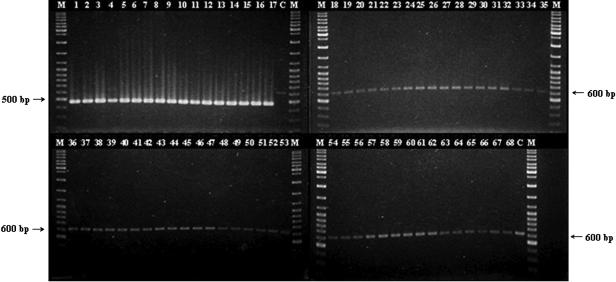
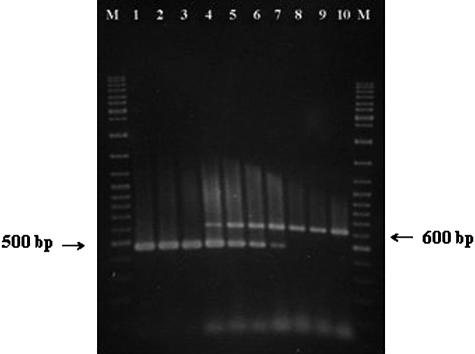
The primer pair ESSF and ESSR was designed based on the nucleotide sequence of ompA and its flanking regions determined in this study. Primer ESSF was complementary to a region between the promoter and transcription start site of ompA, whereas ESSR was complementary to an internal region less conserved among species within the Enterobacteriaceae. Primer pair ESSF-ESSR was used to amplify the target genomic DNA sequence from 17 strains of E. sakazakii and 51 strains of other gram-negative and -positive bacteria. All of the E. sakazakii strains yielded the expected product of 469 bp, and none of the other strains tested amplified this product (Fig. 2). However, all of the negative control strains used amplified the 618-bp IAC product, ruling out false-negatives due to PCR inhibition. The detection limit of the PCR, when applied to E. sakazakii 51329 cells suspended in sterile deionized water, was found to be 102 CFU/ml after 50 cycles of amplification (Fig. 3).
FIG. 2.
Detection of E. sakazakii by PCR amplification of the ompA gene. Primer pair ESSF and ESSR and the amplification conditions and gel electrophoresis method described in Materials and Methods were employed. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 17, PCR products from E. sakazakii strains 1 to 17, as listed in Table 1; 18 to 68, PCR products from negative control DNA from bacterial strains 18 to 68, as listed in Table 1; C, control PCR run without any template DNA.
FIG. 3.
Sensitivity of the PCR for detection of E. sakazakii in pure cultures. Primer pair ESSF and ESSR and the template preparation, amplification conditions, and gel electrophoresis method described in Materials and Methods were employed. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 9, PCR products amplified from samples containing 108 to 100 CFU/ml of E. sakazakii (ATCC 51329); 10, control PCR run without any template DNA.
Detection of E. sakazakii in reconstituted infant formula.
Direct amplification of infant formula samples spiked with E. sakazakii was found to detect as few as 103 CFU/ml of infant formula after 50 cycles of amplification (Fig. 4). Incorporation of an enrichment step before amplification was found to enhance the sensitivity of the PCR. Enrichment for 4 h was found to increase the sensitivity of E. sakazakii detection to 101 CFU/ml (data not shown). PCR carried out after 8 h of enrichment was able to detect 10−1 CFU of E. sakazakii/ml of reconstituted infant formula, which is equivalent to 1 CFU/g of powdered infant formula (Fig. 5). In samples containing E. sakazakii at levels below the detection limit of the PCR, amplification of the IAC product alone was observed (Fig. 4 and 5).
FIG. 4.
Direct detection of E. sakazakii in reconstituted infant formula by PCR. Primer pair ESSF and ESSR and the template preparation, amplification conditions, and gel electrophoresis method described in Materials and Methods were employed. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 9, PCR products amplified from infant formula containing 108 to 100 CFU/ml of E. sakazakii (ATCC 51329); 10, control PCR run with uninoculated, reconstituted infant formula.
FIG. 5.
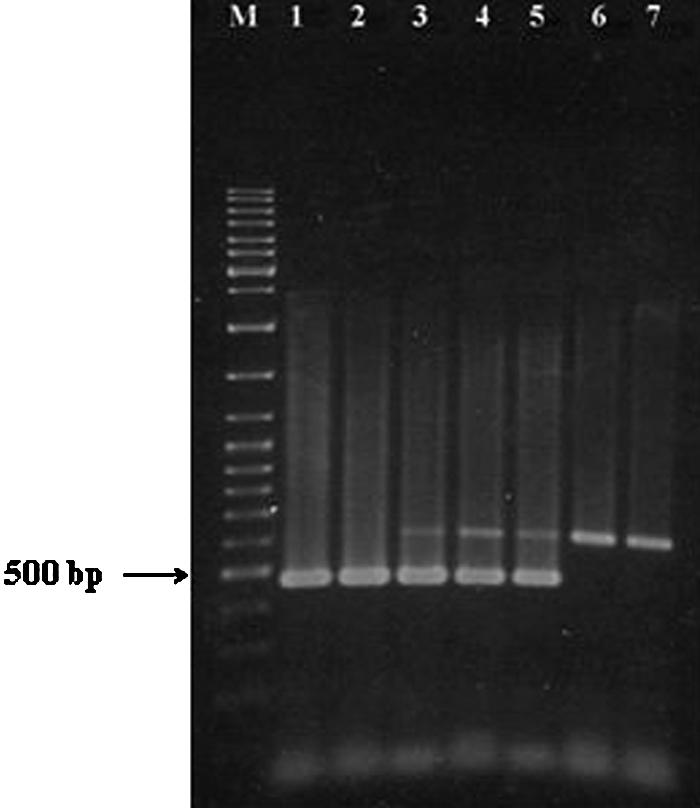
Detection of E. sakazakii in reconstituted infant formula by PCR after an enrichment step of 8 h. Primer pair ESSF and ESSR and the template preparation, amplification conditions, and gel electrophoresis method described in Materials and Methods were employed. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 6, PCR products amplified from infant formula containing 103 to 10−2 CFU/ml of E. sakazakii (ATCC 51329) after 8 h of enrichment; 7, control PCR run with uninoculated, reconstituted infant formula after 8 h of enrichment.
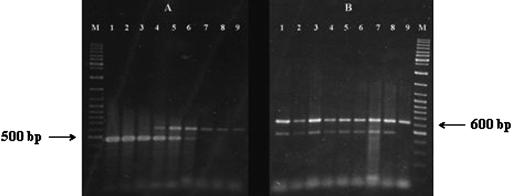
Detection of E. sakazakii in the presence of S. enterica serovar Typhimurium.
The presence of serovar Typhimurium did not affect the sensitivity and specificity of the PCR for detecting E. sakazakii in infant formula. In the presence of 108 CFU/ml of serovar Typhimurium DT 104, the target ompA sequence of E. sakazakii was amplified from infant formula samples containing 108 to 103 CFU/ml of E. sakazakii 51329 (Fig. 6A, lanes 1 to 6). However, samples containing 102 and 101 CFU/ml of E. sakazakii and negative control samples (infant formula with 108 CFU/ml of serovar Typhimurium but no E. sakazakii) yielded only the IAC product; no amplification of the 469-bp ompA product was observed in these samples (Fig. 6A, lanes 7 to 9). Amplification of the target ompA sequence as well as the IAC product was observed in all of the formula samples containing 103 CFU/ml of E. sakazakii 51329 in the presence of 108 to 101 CFU/ml of serovar Typhimurium (Fig. 6B, lanes 1 to 8), whereas the negative control sample amplified the IAC product alone (Fig. 6B, lane 9).
FIG. 6.
Detection of E. sakazakii in reconstituted infant formula by PCR in the presence of S. enterica serovar Typhimurium. Primer pair ESSF and ESSR and the template preparation, amplification conditions, and gel electrophoresis method described in Materials and Methods were employed. (A) PCR amplifications carried out on infant formula samples containing 108 CFU/ml serovar Typhimurium along with 108 to 101 CFU/ml of E. sakazakii. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 8, PCR products amplified from infant formula containing 108 to 101 CFU/ml of E. sakazakii (corresponding samples on each of the lanes also contained 108 CFU/ml serovar Typhimurium); 9, control PCR run with infant formula containing 108 CFU/ml serovar Typhimurium alone. (B) PCR amplifications carried out on infant formula samples containing 103 CFU/ml E. sakazakii along with 108 to 101 CFU/ml of serovar Typhimurium. Lanes: M, molecular weight marker (GeneRuler DNA Ladder Mix; Fermentas, Hanover, Md.); 1 to 8, PCR products amplified from infant formula containing 108 to 101 CFU/ml of serovar Typhimurium (corresponding samples on each of the lanes also contained 103 CFU/ml E. sakazakii); 9, control PCR run with infant formula containing 108 CFU/ml serovar Typhimurium alone.
DISCUSSION
In this study, we cloned and sequenced the ompA homologue of E. sakazakii. An analysis of the genomic sequences of members of the Enterobacteriaceae available in the GenBank database, such as E. coli, Salmonella spp., S. flexneri, and Erwinia carotovora, revealed that the orientation of the ompA gene is highly conserved within the genome, located between sulA, encoding a cell division inhibitor, and ycbG, encoding a hypothetical protein. Based on the results of multiple sequence alignments, we identified regions of sulA and ycbG that are conserved among these bacteria, and primers were designed to amplify an approximately 2,047-bp region encompassing the ompA gene from the E. sakazakii 51329 genome. Over all, ompA of E. sakazakii was found to be highly related (85 to 90% sequence conservation at the nucleic acid and amino acid levels) to ompA orthologues from other Enterobacteriaceae. Being a highly conserved structural membrane protein, this high level of sequence conservation was expected. However, a BLAST search of the E. sakazakii ompA sequence with other GenBank sequences identified regions specific to E. sakazakii, thereby yielding primers for amplification of a specific DNA fragment by PCR.
The primary goal of this study was to develop a PCR assay for rapid and specific detection of E. sakazakii from infant formula samples. The currently approved procedure for detection of E. sakazakii contamination of infant formula is laborious, taking up to 7 days for completion (21). However, even with the additional enrichment step, the PCR developed in this study can detect E. sakazakii in about 12 h. The specificity of the PCR was ensured by selecting primers complementary to the sequences at the regulatory and coding regions of the ompA gene specific to E. sakazakii. PCR amplified a 469-bp DNA sequence from all E. sakazakii strains tested, with no amplification of this PCR product from any of the negative controls, indicating that the assay is specific to E. sakazakii. The negative controls included other major species within the genus Enterobacter; other gram-negative pathogens and spoilage bacteria, especially those that belong to the family Enterobacteriaceae; and other neonatal meningitis-causing pathogens, such as Citrobacter freundii, Listeria monocytogenes, and Streptococcus pneumoniae. We also included the other common infant pathogens potentially encountered in infant formula, as classified by the WHO into three categories based on the strength of evidence of a causal association between their presence in powdered infant formula and illness in infants. These include category A organisms (clear evidence of causing neonatal infections) such as S. enterica; category B organisms (causality plausible but not yet demonstrated) such as Hafnia alvei, Klebsiella pneumoniae, Citrobacter freundii, and Enterobacter cloacae; and category C organisms (causality less plausible or not yet demonstrated) such as Bacillus cereus, Staphylococcus aureus, and Listeria monocytogenes (27).
IAC is a critical component of diagnostic PCR methodologies, since it helps to distinguish negative responses due to the absence of target sequence in the reaction mixture from negative responses due to PCR inhibition resulting from a multitude of reasons, including thermal cycler malfunction, inefficient DNA polymerase activity, incorrect PCR mixture preparation, or the presence of PCR inhibitors in the reaction mixture (16). In this study, we designed a competitive IAC template (6) in which both the target and IAC were amplified by the same primer set. Due to the competition by IAC for the primers, we have designed the IAC product to be larger than the target product (618 bp versus 469 bp) so as to drive the reaction kinetics towards the smaller, target DNA (6). Moreover, in order to prevent any potential reduction in PCR amplification efficiency due to this competitiveness, we have detected the lowest reproducible IAC concentration that did not interfere with amplification of the ompA target sequence from the different template preparations used in individual PCR experiments. Therefore, in PCR involving genomic DNA as the template, 2 fg of IAC template (corresponding to approximately 500 copies) was used in the reaction mixtures. At this concentration, owing to the competition, samples containing E. sakazakii genomic DNA yielded only the species-specific ompA product and not the IAC product. In this case, since the target DNA is present in proportionately higher concentrations than the IAC template, IAC amplification is unnecessary for validation of the results (6). Moreover, the IAC product was amplified in all samples containing non-E. sakazakii DNA, ruling out false-negative results due to amplification failure in these samples. Similarly, in experiments to evaluate the sensitivity of E. sakazakii detection in reconstituted infant formula samples and in deionized water, 0.2 fg of IAC template (corresponding to approximately 50 copies) was used in the reaction mixtures. At this concentration, it was observed that the IAC product coamplified with the species-specific ompA product at template concentrations of 104 and 103 CFU/ml of E. sakazakii in the samples derived from infant formula (105, 104,103, and 102 CFU/ml of E. sakazakii in deionized water). However, in samples containing E. sakazakii at levels higher than 104 CFU/ml (higher than 105 CFU/ml for E. sakazakii in deionized water), only the 469-bp species-specific product was amplified, not the IAC product, due to the competition for the primers. In samples containing E. sakazakii at levels below the detection limits of these PCR assays, only the IAC product was amplified (Fig. 3 and 4). The detection limits observed in these experiments were similar to those observed when PCR was carried out in the absence of IAC template (data not shown).
The sensitivity of PCR detection depends on the number of target nucleic acid copies present in the sample and the method of template preparation (23). After 50 cycles, the PCR detected as few as 102 CFU of E. sakazakii/ml in sterile deionized water and 103 CFU/ml in reconstituted infant formula. When the experimentally inoculated formula preparation was boiled and used directly as the template, the detection limit was 104 CFU/ml (data not shown). Alternatively, when the inoculated formula was pelleted, resuspended in sterile distilled water, and boiled, and the supernatant taken after a brief (30 s), low-speed centrifugation (1,500 × g) was used as the template, the PCR was able to detect as few as 103 CFU/ml. However, when the inoculated formula samples were enriched at 37°C for 8 h before being subjected to PCR, the sensitivity of detection was increased to 10−1 CFU/ml, which is equivalent to 1 CFU of E. sakazakii/g of powdered infant formula.
The sensitivity of PCR may also be affected by high levels of nontarget, background microflora, wherein a simple inhibition of contact between primers, polymerase, and target DNA could result in reduced sensitivity (24). Different formula preparations could have different indigenous microflora owing to the differences in ingredients and manufacturing protocols. Therefore, we used three different brands of commercially available infant formula preparations, from three different manufacturers, in our study. The detection limit of the PCR when applied to the reconstituted formula preparations containing E. sakazakii was found to not differ among the three brands. The inhibitory effect of background microflora on PCR sensitivity is especially important when the PCR assay is combined with an enrichment step, which could have a negative impact on the PCR due to a simultaneous increase in the number of background bacteria. To ascertain this, the reconstituted formula preparations were enriched at 37°C for 24 h, allowing the indigenous microflora to flourish, before inoculation with E. sakazakii. PCR carried out on such samples had the same level of sensitivity as with formula preparations inoculated with E. sakazakii immediately after reconstitution (data not shown). Moreover, our results also show that even in the presence of competing, nontarget bacteria such as serovar Typhimurium at levels as high as 108 CFU/ml, the sensitivity of the assay was not reduced (Fig. 6). Serovar Typhimurium was chosen as the background bacterium because it is classified, along with E. sakazakii, as a category A organism that is encountered in infant formula and shows clear evidence of causing neonatal infections (27). It is currently assumed that very low levels of E. sakazakii in infant formula (<3 CFU/100g) can lead to infection in neonates (27). This PCR, in combination with an enrichment step, can be used to detect the presence of E. sakazakii at levels that violate the current microbiological specification for coliform counts in powdered infant formula, as specified by the Codex Committee on Food Hygiene (fewer than 3 CFU/g).
REFERENCES
- 1.Bar-Oz, B., A. Preminger, O. Peleg, C. Block, and I. Arad. 2002. Enterobacter sakazakii infection in the newborn. Acta Paediatr. 90:356-358. [PubMed] [Google Scholar]
- 2.Biering, G., S. Karlsson, N. C. Clark, K. E. Jonsdottir, P. Ludvigsson, and O. Steingrimsson. 1989. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J. Clin. Microbiol. 27:2054-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FSNET. 8. November 2002, posting date. Recalled baby formula found in Colorado stores. Colorado Department of Public Health and Environment. [Online.] http://archives.foodsafetynetwork.ca/fsnet/2002/11-2002/fsnet_november_8-2.htm#RECALLED%20BABY.
- 4.Guillaume-Gentil, O., V. Sonnard, M. C. Kandhai, J. D. Marugg, and H. Joosten. 2005. A simple and rapid cultural method for detection of Enterobacter sakazakii in environmental samples. J. Food Prot. 68:64-69. [DOI] [PubMed] [Google Scholar]
- 5.Himelright, I., E. Harris, V. Lorch, M. Anderson, T. Jones, A. Craig, M. Kuehnert, T. Forster, M. Arduino, B. Jensen, and D. Jernigan. 2002. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. Morbid. Mortal. Wkly. Rep. 51:297-300. [PubMed] [Google Scholar]
- 6.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iversen, C., and S. Forsythe. 2004. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 21:771-777. [Google Scholar]
- 8.Kandhai, M. C., M. W. Reij, L. G. Gorris, O. Guillaume-Gentil, and M. van Schothorst. 2004. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363:39-40. [DOI] [PubMed] [Google Scholar]
- 9.Kandhai, M. C., M. W. Reij, K. van Puyvelde, O. Guillaume-Gentil, R. R. Beumer, and M. van Schothorst. 2004. A new protocol for the detection of Enterobacter sakazakii applied to environmental samples. J. Food Prot. 67:1267-1270. [DOI] [PubMed] [Google Scholar]
- 10.Kleiman, M. B., S. D. Allen, P. Neal, and J. Reynolds. 1981. Meningoencephalitis and compartmentalization of the cerebral ventricles caused by Enterobacter sakazakii. J. Clin. Microbiol. 14:352-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai, K. K. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113-122. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq, A., C. Wanegue, and P. Baylac. 2002. Comparison of fecal coliform agar and violet red bile lactose agar for fecal coliform enumeration in foods. Appl. Environ. Microbiol. 68:1631-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malorny, B., and M. Wagner. 2005. Detection of Enterobacter sakazakii strains by real-time PCR. J. Food Prot. 68:1623-1627. [DOI] [PubMed] [Google Scholar]
- 14.Nazarowec-White, M., and J. M. Farber. 1997. Incidence, survival, and growth of Enterobacter sakazakii in infant formula. J. Food Prot. 60:226-230. [DOI] [PubMed] [Google Scholar]
- 15.Postupa, R., and E. Aldova. 1984. Enterobacter sakazakii: a Tween 80 esterase-positive representative of the genus Enterobacter isolated from powdered milk specimens. J. Hyg. Epidemiol. Microbiol. Immunol. 28:435-440. [PubMed] [Google Scholar]
- 16.Rådström, P., C. Löfström, M. Lövenklev, R. Knutsson, and P. Wolffs. 2003. Strategies for overcoming PCR inhibition, p. 149-161. In C. W. Diefenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Sanders, W. E., Jr., and C. C. Sanders. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin. Microbiol. Rev. 10:220-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo, K. H., and R. E. Brackett. 2005. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J. Food Prot. 68:59-63. [DOI] [PubMed] [Google Scholar]
- 19.Simmons, B. P., M. S. Gelfand, M. Haas, L. Metts, and J. Ferguson. 1989. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10:398-401. [DOI] [PubMed] [Google Scholar]
- 20.Skladal, P., M. Mascini, C. Salvadori, and G. Zannoni. 1993. Detection of bacterial contamination in sterile UHT milk using an l-lactate biosensor. Enzyme Microb. Technol. 15:508-512. [Google Scholar]
- 21.U.S. Food and Drug Administration. August. 2002. Isolation and enumeration of Enterobacter sakazakii from dehydrated powdered infant formula. [Online.] http://www.cfsan.fda.gov/∼comm/mmesakaz.html.
- 22.van Acker, J., F. de Smet, G. Muyldermans, A. Bougatef, A. Naessens, and S. Lauwers. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Way, J. S., K. L. Josephson, S. D. Pillai, M. Abbaszadegan, C. P. Gerba, and I. L. Pepper. 1993. Specific detection of Salmonella spp. by multiplex polymerase chain reaction. Appl. Environ. Microbiol. 59:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver, J. W., and M. T. Rowe. 1997. Effect of non-target cells on the sensitivity of the PCR for Escherichia coli O157:H7. Lett. Appl. Microbiol. 25:109-112. [DOI] [PubMed] [Google Scholar]
- 25.Weir, E. 2002. Powdered infant formula and fatal infection with Enterobacter sakazakii. CMAJ 166:1570. [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 2.4.1. In F. M. Ausubel, R. Brent, R. E. Kingston, R. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 27.World Health Organization. 2004. Enterobacter sakazakii and other microorganisms in powdered infant formula: meeting report. Microbiological risk assessment series, no. 6. [Online.] http://www.who.int/foodsafety/publications/micro/enterobacter_sakazakii/en/.