Abstract
In this paper we describe construction of a luciferase-based vector, pPL2lux, and use of this vector to study gene expression in Listeria monocytogenes. pPL2lux is a derivative of the listerial integration vector pPL2 and harbors a synthetic luxABCDE operon encoding a fatty acid reductase complex (LuxCDE) involved in synthesis of the fatty aldehyde substrate for the bioluminescence reaction catalyzed by the LuxAB luciferase. We constructed pPL2lux derivatives in which the secA and hlyA promoters were translationally fused to luxABCDE and integrated as a single copy into the chromosome of L. monocytogenes EGD-e. Growth experiments revealed that hlyA was expressed predominantly in the stationary phase in LB medium buffered at pH 7.4, whereas secA expression could be detected in the exponential growth phase. Moreover, the correlation between luciferase activity and transcription levels, as determined by reverse transcriptase PCR, was confirmed using conditions known to lead to repression and activation of hemolysin expression (addition of cellobiose and activated charcoal, respectively). Furthermore, hemolysin expression could be monitored in real time during invasion of an intact monolayer of C2Bbe1 (Caco-2-derived) cells. Finally, hemolysin expression could be detected in the livers, spleens, and kidneys of mice 3 days postinfection. These experiments clearly established the effectiveness of pPL2lux as a quantitative reporter system for real-time, noninvasive evaluation of gene expression in L. monocytogenes.
Listeria monocytogenes is a gram-positive intracellular pathogen that is capable of withstanding a variety of hostile environmental conditions, including the many stresses encountered during the production, preparation, and storage of food. After contaminated food is consumed, L. monocytogenes effectively survives the extreme conditions in the human gastrointestinal tract, including the gastric acidity encountered in the stomach and the presence of bile salts in the duodenum (13). Several virulence factors that play a role during the subsequent intracellular cycle of L. monocytogenes have been determined, and they include hemolysin, encoded by hlyA, which allows escape of listeria from the phagosomal compartment and intracytoplasmic growth (15, 44). Notably, L. monocytogenes is the causative agent of listeriosis, which is estimated to account for up to 35% of all deaths caused by known bacterial food-borne pathogens in the United States (24), which justifies the increased effort that is put into analysis of listerial gene expression signals and the characterization of regulatory elements (4, 20, 36, 41, 47). Moreover, it has been suggested that in order to fully investigate the expression of virulence genes, the development of new tools that allow noninvasive imaging and in situ determination of quantitative temporal changes in gene expression during infection of relevant hosts will become increasingly important in listerial research in the coming years (9).
Various reporter systems have been used to identify regulatory sequences and to monitor gene expression. Historically, bacterial promoters have been studied by insertion of random or defined chromosomal DNA fragments upstream of promoterless reporter genes, such as genes encoding chloramphenicol acetyltransferase (1, 27, 42), alanine racemase (7, 8), and different sugar hydrolases (19, 27, 32). These reporter systems are convenient tools for semiquantitative, plate-based assessment of promoter activities. However, more accurate quantification of promoter strength usually requires relatively laborious enzymatic assays, which typically involve bacterial cell disruption and addition of a substrate to drive the enzymatic reaction, followed by measurement of the optical density (27, 32).
Another group of reporter systems is based on the emission of light. Variants of green fluorescent protein from Aequorea victoria have been used to monitor bacterial promoter activity during infection of eukaryotic cell lines (14, 35). Unfortunately, naturally occurring fluorescence can lead to high background levels during in vitro and in vivo measurements. Alternative strategies for both gram-negative and gram-positive bacteria have involved the luciferase-encoding luxAB genes, typically derived from Vibrio fischeri, Vibrio harveyi, or Photorhabdus luminescens (25). Although the level of the background bioluminescence is extremely low compared to the level of endogenous background fluorescence, luxAB systems require addition of an exogenous aldehyde as a substrate in the light emission reaction (17, 28, 30, 33). This aldehyde requirement can be overcome by using the synthetic operon luxCDABE, as luxCDE encodes a fatty acid reductase complex involved in synthesis of the fatty aldehyde substrate for the bioluminescence reaction catalyzed by the luxAB-encoded luciferase (25). The luxCDABE system has been utilized in a noninvasive, real-time fashion for tagging and assessing promoter activities of several gram-negative bacteria, including Escherichia coli (6, 43, 45), Salmonella enterica serovar Typhimurium (3), Citrobacter rodentium (46), and Campylobacter jejuni (2). Moreover, after introduction of gram-positive ribosome binding sites and shuffling of the gene order to luxABCDE, the luciferase system also appeared to be functional in gram-positive bacteria (11, 34). Transposon mutagenesis strategies have been employed to integrate the luxABCDE system into the chromosome of several gram-positive bacteria, including Staphylococcus aureus (11), L. monocytogenes (18), and Streptococcus pneumoniae (12). These tagged pathogens could be monitored in intact animal models, which allowed temporal and spatial assessment of the infection processes.
Here we describe construction of a luciferase-based reporter system, pPL2lux. This vector is based on the listerial site-specific integration vector pPL2 (22) and the luxABCDE system (34). Derivatives of pPL2lux harboring the listerial secA and hlyA promoters translationally fused to luxABCDE were constructed and integrated into the L. monocytogenes chromosome. The strains that were constructed allowed us to assess the functionality of pPL2lux as a reporter system in three model systems used routinely in listerial research: growth in laboratory media, invasion assays with a Caco-2-derived cell line, and animal experiments using BALB/c mice. The experiments demonstrated the utility and advantages of pPL2lux as a real-time, noninvasive reporter system for quantitative assessment of listerial promoters.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The strains, plasmids, and primers used in this study are listed in Table 1. E. coli strain DH10B (Invitrogen, Paisley, United Kingdom) was used as a cloning host during construction of the pPL2lux derivatives (see below) and was grown aerobically at 37°C in LB medium (39). L. monocytogenes EGD-e (16) was routinely grown aerobically at 37°C in brain heart infusion (BHI) medium (Oxoid, Basingstoke, United Kingdom). However, during the bioluminescence assays LB medium was used. When indicated below, LB medium was buffered with 100 mM 3-(N-morpholino)propanesulfonic acid (pH 7.4). When appropriate, antibiotics were added to the media as follows: for E. coli, ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml); and for L. monocytogenes, chloramphenicol (7.5 μg/ml).
TABLE 1.
Strains, plasmids, and primers used in this study and their relevant characteristics
| Strain, plasmid, or primer | Relevant feature(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | Cloning host | Invitrogen |
| L. monocytogenes strains | ||
| EGD-e | Wild type of serotype 1/2a for which the genome sequence is available | 16 |
| EGD-e::pPL2lux | Cmr, contains pPL2lux integrated at tRNAArg locus on the L. monocytogenes EGD-e chromosome | This study |
| EGD-e::pPL2lux-PsecA | Cmr, contains pPL2lux-PsecA integrated at tRNAArg locus on the L. monocytogenes EGD-e chromosome | This study |
| EGD-e::pPL2lux-PhlyA | Cmr, contains pPL2lux-PhlyA integrated at tRNAArg locus on the L. monocytogenes EGD-e chromosome | This study |
| Plasmids | ||
| pSB2025 | Apr, contains synthetic luxABCDE operon | 34 |
| pPL2secA2-FLAG | Cmr, L. monocytogenes site-specific integration vector harboring secA2 | 23 |
| pPL2a | Cmr, pPL2secA2-FLAG derivative without secA2 | This study |
| pPL2SwaI− | Cmr, pPL2a without SwaI | This study |
| pPL2luxSwaI+ | Cmr, pPL2SwaI harboring luxABCDE derived from pSB2025 | This study |
| pPL2lux | Cmr, pPL2luxSwaI+ with SwaI removed from luxC and SwaI inserted overlapping the start codon of luxA, allowing exact translational promoter fusions to luxABCDE | This study |
| pPL2lux-PsecA | Cmr, pPL2lux derivative containing 0.5-kb promoter region of secA translationally fused to luxABCDE | This study |
| pPL2lux-PhlyA | Cmr, pPL2lux derivative containing 0.5-kb promoter region of hlyA translationally fused to luxABCDE | This study |
| Primers | ||
| IM101 | 5′-GAATTGGCATACTGATTATATGAAGG-3′ | This study |
| IM102 | 5′-GATACACCAAGGAAAGTCTACAC-3′ | This study |
| IM103 | 5′-CCATCCAAGTCGACATTTAAATTTGGAAACTTTTTGCTTACATAC-3′ | This study |
| IM104 | 5′-GAATGATGACTCCCAAGGAAAAATAG-3′ | This study |
| IM105 | 5′-CCATCCAACTCGAGAAGTTCCGCGACAAAGGTGCAGAAAG-3′ | This study |
| IM106 | 5′-CATTTAAATTCTCCTCTACTGTTTTC-3′ | This study |
| IM107 | 5′-CCATCCAACTCGAGTTTGAGCTAGTGGTTTGGTTAATG-3′ | This study |
| IM108 | 5′-CATGGGTTTCACTCTCCTTCTAC-3′ | This study |
| IM109 | 5′-GTAAGTTGGGGATAACTCCG-3′ | This study |
| IM110 | 5′-GCGCTTTACGCCCAATAAATC-3′ | This study |
| IM111 | 5′-AAAAGGACGATTTCGGTTTGG-3′ | This study |
| IM112 | 5′-CCAATGCCCCAGAAATTTCC-3′ | This study |
| IM113 | 5′-CCAATTGCGCAACAAACTGAAG-3′ | This study |
| IM114 | 5′-TTCCACTAATGTATTTACTGCG-3′ | This study |
| PL95 | 5′-ACATAATCAGTCCAAAGTAGATGC-3′ | 22 |
| PL102 | 5′-TATCAGACCTAACCCAAACCTTCC-3′ | 22 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance. Underlining indicates restriction sites used in subsequent cloning procedures.
DNA techniques.
Plasmid DNA was isolated from E. coli using a QIAprep Spin Miniprep kit according to the manufacturer's instructions (QIAGEN, Crawley, United Kingdom). Genomic DNA was isolated from L. monocytogenes EGD-e using a Genelute bacterial genomic DNA kit (Sigma, Steinheim, Germany) as recommended by the manufacturer. Transformation of L. monocytogenes was performed as described previously (29). Standard procedures were used for DNA manipulation in E. coli (39). Restriction endonucleases (Roche Diagnostics, Mannheim, Germany), T4 DNA ligase (Roche), and 2× PCR mixture (Promega, Madison, WI) were used as recommended by the manufacturers. Primers were purchased from Sigma Genosys (Haverhill, United Kingdom). PCR products required for cloning were obtained with KOD hot-start high-fidelity DNA polymerase (Merck, Nottingham, United Kingdom) using either 10 ng L. monocytogenes genomic DNA or 50 ng plasmid DNA as the template.
Construction of pPL2lux and derivatives.
Plasmid pPL2secA2-FLAG (23) was digested with PstI, and the 6.1-kb fragment containing the pPL2 backbone was self-ligated, yielding pPL2a. Except for the modified multiple cloning site, pPL2a is otherwise identical to pPL2 (22). The unique SwaI site present in the PSA integrase-encoding gene of pPL2a was removed by site-directed mutagenesis in a PCR using pPL2a as the template and primers IM101 and IM102. The resulting 0.94-kb amplicon was digested with SphI, and the 0.73-kb fragment was ligated into SphI-SwaI-digested pPL2a, yielding pPL2SwaI−. Compared to parental plasmid pPL2a, the newly constructed plasmid had one point mutation in the SwaI site (introduced by the IM102 primer during the PCR), which did not change the protein sequence of the PSA integrase. Subsequently, pSB2025 (34) was digested with SalI and PstI, and the resulting 5.6-kb fragment harboring the synthetic luxABCDE operon was cloned into complementary digested pPL2SwaI−, yielding pPL2luxSwaI+. Finally, the SwaI site in the luxC gene harbored by pPL2luxSwaI+ was removed using a site-directed mutagenesis approach essentially as described above to introduce a point mutation into the SwaI site, using pPL2luxSwaI+ as the template and primers IM103 and IM104, as well as exchange of the native and mutated fragment by cloning the SalI-digested amplicon into the SalI-SwaI-digested backbone of pPL2luxSwaI+. The resulting vector was designated pPL2lux (Fig. 1) and contains a unique SwaI restriction site that overlaps the start codon of luxA (introduced by the IM103 primer), which allows exact translational fusion of listerial promoters to the luxABCDE operon (see below).
FIG. 1.
Plasmid map of pPL2lux. The luxABCDE operon was derived from pSB2025 (34) with a blunt-end SwaI restriction site introduced overlapping the ATG start codon of luxA. Cloning of PCR-amplified promoter elements into the SalI and SwaI restriction sites allowed construction of exact translational fusions of listerial promoters to luxABCDE.
To test the functionality of the luciferase reporter system, the secA and hlyA promoters were cloned into pPL2lux. Chromosomal DNA from L. monocytogenes EGD-e was used as the template in PCRs using KOD polymerase (Merck) and primer pairs IM105-IM106 and IM107-IM108. The resulting 0.5-kb PCR products harbored the regions immediately upstream of secA and hlyA, respectively. Notably, the start codons of both genes were included at the ultimate 3′ end of the reverse primers. The PsecA and PhlyA amplicons were digested with XhoI and cloned into SwaI-SalI-digested pPL2lux, yielding pPL2lux-PsecA and pPL2lux-PhlyA, respectively. Subsequently, pPL2lux, pPL2lux-PsecA, and pPL2lux-PhlyA were transformed into L. monocytogenes EGD-e, and candidate integrants were checked for site-specific integration by PCR, using template DNA obtained directly from colonies by a microwave treatment (3 min, 600 W) and primers PL95 and PL102 (22). From each transformation one colony having the anticipated genotype was selected for bioluminescence analysis using a Xenogen IVIS 100 system (Xenogen, Alameda, CA).
RNA isolation.
Levels of luciferase and hemolysin transcription were determined in EGD-e::pPL2lux-PhlyA grown with or without 25 mM d-(+)-cellobiose (Calbiochem, San Diego, CA) or 0.2% activated charcoal (0.3 to 0.5 mm; Merck, Nottingham, United Kingdom). Full-grown LB medium cultures were diluted 50-fold in fresh medium and grown statically at 37°C. Growth was monitored by determining the optical density at 600 nm (OD600), and at the beginning of the stationary phase (OD600, 0.3) cells were harvested from 10 ml after addition of 40 ml of quench buffer (60% methanol, 66.7 mM HEPES [pH 6.5]; −40°C). Following quenching (31), the cells were immediately pelleted by centrifugation at 3,220 × g for 10 min, and the cell pellets were resuspended in 0.2 ml of ice-cold distilled H2O. The cell suspensions were added to ice-cold tubes containing MagNA Lyser green beads (Roche Diagnostics), 400 μl of acidified phenol (Sigma), 100 μl of chloroform (Sigma), 30 μl of 10% sodium dodecyl sulfate (Sigma), and 30 μl of 3 M sodium acetate (pH 5.2) (Merck). The cells were disrupted with three 40-s treatments in a MagNA Lyser (Roche Diagnostics) separated by 1 min on ice. After centrifugation, 100 μl of the aqueous phase was used for RNA isolation with QIAGEN RNeasy columns (Crawley, United Kingdom). After elution, 0.5 μl of protector RNase inhibitor was added to all RNA samples (Roche Diagnostics), and 1 μg of RNA was subjected to DNase I treatment according to the manufacturer's instructions (Ambion, Huntingdon, United Kingdom).
cDNA synthesis and reverse transcriptase PCR.
cDNA was synthesized in a 40-μl reaction mixture containing 0.2 nmol of random hexamer primer, 10 mM dithiothreitol, 1× Expand reverse transcriptase (RT) buffer, 50 U Expand reverse transcriptase (Roche Diagnostics, Mannheim, Germany), each deoxynucleoside triphosphate at a concentration of 0.25 mM, and 0.7 μg DNase I-treated RNA. Reverse transcription was initiated by 10 min at 30°C, followed by 45 min at 42°C and inactivation of the reverse transcriptase by incubation at 95°C for 2 min. Subsequently, 1 μl of the cDNA synthesized was used in PCR mixtures containing 1× PCR mixture (Promega) and 1 pmol of the 16S RNA (IM109-IM110), luxA (IM111-IM112), or hlyA (IM113-IM114) primer pair (Table 1).
Cell invasion assay.
C2Bbe1 cells (CRC-2102; American Type Culture Collection), a clone of the Caco-2 human adenocarcinoma cell line, were used for invasion assays. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter Glutamax (Gibco Laboratories, Grand Island, NY), 10% fetal bovine serum (Gibco), 1% (vol/vol) nonessential amino acids (Gibco), 1% (vol/vol) penicillin-streptomycin (Gibco), and 0.01 mg/ml human transferrin (Calbiochem) at 37°C in a 5% CO2 atmosphere. For cell invasion assays, C2Bbe1 cells were trypsinized (Gibco), harvested by centrifugation at 400 × g for 8 min, and resuspended in 1 ml antibiotic-free DMEM containing 10% fetal bovine serum. Cells were seeded onto 24-well flat-bottom tissue culture plates (Sarstedt, Leicester, United Kingdom) at a concentration of 3 × 105 cells per well. The plates were incubated for 72 h at 37°C in a 5% CO2 atmosphere until confluence was reached. An overnight BHI medium culture of EGD-e::pPL2lux-PhlyA or EGD-e::pPL2lux was diluted 1:20 in fresh BHI medium and incubated until the OD600 was 1.0 (1 × 109 CFU/ml). Subsequently, bacterial cells were washed twice in DMEM and added at a multiplicity of infection of 100:1, and the plates were incubated at 37°C in a 5% CO2 atmosphere. At several times gentamicin was added to a final concentration of 100 μg/ml, and the cells were incubated for 30 min (21). Subsequently, the cells were washed three times with phosphate-buffered saline (PBS), and the levels of bioluminescence were determined using a Xenogen IVIS 100 system (Xenogen, Alameda, CA). After this monolayers were lysed with 1 ml of ice-cold distilled H2O, and serial dilutions were plated on BHI agar to assess the number of viable intracellular bacteria.
Animal experiments.
Overnight cultures of either L. monocytogenes EGD-e:: pPL2lux-PhlyA or L. monocytogenes EGD-e::pPL2lux were pelleted by centrifugation (7,000 × g for 5 min), washed with PBS, and used to inoculate 8- to 12-week-old BALB/c mice intraperitoneally with 2 × 106 CFU in 200 μl of PBS. Three days after infection, the mice were sacrificed by cervical dislocation, and individual organs were examined for bioluminescence using a Xenogen IVIS 100 system. Organs were then homogenized in PBS, and serial dilutions were plated onto BHI agar, which was followed by overnight incubation at 37°C. The resulting colonies were used to calculate the number of bacterial cells per organ.
RESULTS
Functionality of the luciferase reporter system (pPL2lux).
The main goal of the work described here was construction and functional analysis of pPL2lux (Fig. 1). This vector is a derivative of pPL2 that contains a PSA listeriophage integrase gene and attachment site which directs site-specific, single-copy integration into the tRNAArg locus on the L. monocytogenes chromosome (22). In addition, pPL2lux harbors luxABCDE, a synthetic operon that contains optimized gram-positive translational initiation sequences upstream of luxA, luxC, and luxE (34). The luxABCDE operon is divergently oriented with respect to flanking genes on pPL2lux to minimize any potential read-through from upstream promoters. Importantly, the use of the SalI and SwaI restriction sites for orientation-specific cloning of PCR-amplified promoters into pPL2lux allowed exact translational fusion to the luxABCDE operon. Moreover, the fact that the start codon of luxA was not present in pPL2lux enabled inclusion of the start codon of the gene under investigation at the 3′ end of the primer used during amplification of the promoter region, thereby allowing direct identification of positive clones by their bioluminescent phenotype in E. coli.
To demonstrate the functionality of the luciferase reporter system in L. monocytogenes, the expression profiles of hlyA and secA were assessed. The latter gene encodes SecA, which is essential in L. monocytogenes as it is required for the initial binding of preproteins to the inner membrane and their subsequent translocation across this structure (23). Hence, this gene should be expressed in all growth phases and was used primarily as a positive control. hlyA, encoding hemolysin, plays an important role during listerial escape from the phagosomal compartment and intracytoplasmic growth (15, 44). The original vector, pPL2lux, and its derivatives, pPL2lux-PsecA and pPL2lux-PhlyA, were constructed and integrated into the chromosome of L. monocytogenes EGD-e as described in Materials and Methods.
The stability of the integrated pPL2lux derivatives in the absence of antibiotic selection pressure was evaluated. Cultures of L. monocytogenes EGD-e::pPL2lux, EGD-e::pPL2lux-PsecA, and EGD-e::pPL2lux-PhlyA were grown for 50 generations in BHI medium without antibiotic selection, dilutions of the cultures were plated, and the presence of the integrated plasmids in the resulting colonies was assessed by scoring for the chloramphenicol resistance encoded by the pPL2 backbone. All 100 colonies tested for each of the three strains were chloramphenicol resistant after 50 generations of growth (data not shown), indicating that the integrated plasmids were stably maintained without antibiotic selection pressure. This high level of stability allowed use of the luciferase reporter system in experiments in which antibiotic selection pressure could not be maintained, such as cell invasion assays and animal experiments (see below).
Overnight cultures of the constructed strains were diluted 1:50 in LB medium buffered at pH 7.4, and growth and bioluminescence were monitored over time (Fig. 2). Notably, relatively low levels of hemolysin-coupled bioluminescence were observed in our initial experiments using the luciferase reporter system and unbuffered BHI and LB media. In contrast, higher levels of bioluminescence were observed in BHI and LB media buffered at pH 7.4; the levels of expression in LB medium were the highest, while the secA transcription profiles appeared to be similar in the unbuffered and buffered LB and BHI media (data not shown). The growth rates of EGD-e::pPL2lux-PsecA and EGD-e::pPL2lux-PhlyA in the different media were not affected compared to the growth rate of EGD-e::pPL2lux (data not shown). Apparently, luciferase expression at the levels reached during the experiments described here does not influence the growth rate of L. monocytogenes. No bioluminescence was detected at any time for the negative control strain EGD-e::pPL2lux, indicating that there was no background signal (data not shown). For strain EGD-e::pPL2lux-PsecA, bioluminescence was detected throughout the growth curve, as expected for this essential system (Fig. 2). However, bioluminescence declined during the late logarithmic phase of growth and decreased considerably after the strain entered the stationary phase, until it was eventually undetectable. In contrast, bioluminescence was not present in EGD-e::pPL2lux-PhlyA in the early logarithmic growth phase and was detected only from the mid-logarithmic growth phase onward (Fig. 2). Higher levels of expression were observed at the end of the logarithmic phase and upon entry into the stationary phase. Although the hemolysin levels fluctuated during the stationary phase, equally high levels of expression were detected after 24 h of growth compared to the levels at the beginning of the stationary phase (Fig. 2). The bioluminescence profiles for EGD-e::pPL2lux-PsecA and EGD-e::pPL2lux-PhlyA differed significantly throughout the different growth phases, suggesting that differential expression can be monitored using the luciferase reporter system.
FIG. 2.
Hemolysin expression profiles during growth at 37°C in LB medium buffered at pH 7.4. The triangles and diamonds indicate the average growth of quadruplicate cultures and the gray and black bars indicate the average luciferase expression profiles for EGD-e::pPL2lux-PsecA and EGD-e::pPL2lux-PhlyA, respectively. BLC, bioluminescence counts; OD600nm, optical density at 600 nm. The data are representative of the data from three independent experiments. At times 1, 2, and 3, samples were taken from the cultures for RNA isolation. Subsequently, luciferase transcript levels were assessed by RT-PCR analysis (inset).
To investigate whether the levels of bioluminescence observed with the EGD-e::pPL2lux-PsecA and EGD-e::pPL2lux-PhlyA stationary-phase cultures resulted from active transcription or accumulation of luciferase, an RT-PCR analysis was performed. RNA was isolated from the cultures during the log phase, at the beginning of the stationary phase, and after a prolonged time in the stationary phase (times 1, 2, and 3 in Fig. 2). After cDNA synthesis, control PCRs were performed with 16S RNA primers (Table 1), which confirmed that the cDNA concentrations were comparable in all samples (Fig. 2, inset). Moreover, the amplicons obtained using the luxA primers (Table 1) demonstrated that there was active luciferase transcription in EGD-e::pPL2lux-PsecA and EGD-e::pPL2lux-PhlyA during the log phase and at the beginning of the stationary phase. In contrast, the intensity of the amplicon obtained from cDNA derived from the EGD-e::pPL2lux-PsecA culture after a prolonged time in the stationary phase was extremely low compared to the intensity of the amplicon obtained from cDNA derived from the EGD-e::pPL2lux-PhlyA culture. These experiments demonstrated that the levels of bioluminescence accurately reflected the levels of luciferase transcription during all growth phases and that luciferase accumulation did not make a major contribution to the bioluminescence signals obtained.
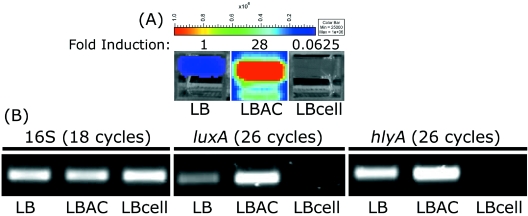
Previous studies have demonstrated that hlyA transcription is induced in the presence of activated charcoal and is repressed by cellobiose (10, 26, 37). To further investigate the potential of pPL2lux as a quantitative reporter system, EGD-e::pPL2lux-PhlyA was grown in media with and without added cellobiose or activated charcoal. At the beginning of stationary phase the levels of bioluminescence were determined, and the results revealed that addition of cellobiose and charcoal to the medium resulted in a 16-fold decrease and a 28-fold increase, respectively, in the levels of bioluminescence compared to growth in the control medium (Fig. 3A). Despite the fact that the levels of bioluminescence in LB medium containing activated charcoal appeared to be 448-fold higher than the levels in LB medium containing cellobiose, both levels could be conveniently measured, indicating the suitability of the luciferase reporter system for assessing levels of hemolysin expression in a wide dynamic range. Simultaneously, RNA was isolated from the cultures, and after cDNA synthesis the correlation between bioluminescence and luxA or hlyA levels was evaluated by PCR analysis. Control PCRs with cDNA derived from the cultures were performed with 16S RNA primers (Table 1), and the results confirmed that the cDNA concentrations were comparable in all samples (Fig. 3B). The relatively high intensity of the amplicon obtained using the luxA primers (Table 1) and cDNA derived from the culture to which activated charcoal had been added indicates that the levels of luxA mRNA were significantly higher than the levels in the control culture. Moreover, the intensity of the amplicon from the culture grown in LB medium containing cellobiose was significantly lower, indicating that the levels of luxA mRNA were decreased in the presence of cellobiose. These experiments revealed that the levels of bioluminescence obtained using the pPL2lux reporter system strongly correlated with the levels of luxA mRNA in the same cultures. Using the same cDNA samples with primers specific for hlyA (Table 1), very similar relative expression profiles were obtained, namely, decreased and increased levels of hlyA mRNA in the cultures containing cellobiose and charcoal, respectively (Fig. 3B). From these data, we concluded that at least in this instance, measuring bioluminescence provides an accurate depiction of transcription of the single-copy, chromosomally located luxABCDE operon, which in turn correctly reflects the transcription profile of the native promoter in its normal chromosomal locus. Taken together, the experiments described above demonstrate that pPL2lux can be used effectively as a quantitative reporter system for real-time, noninvasive detection of promoter activities during in vitro growth of L. monocytogenes.
FIG. 3.
Correlation between bioluminescence and mRNA levels. EGDe::pPL2lux-PhlyA was grown in LB medium, LB medium containing 0.2% activated charcoal (LBAC), or LB medium containing 25 mM cellobiose (LBcell). Upon entry into the stationary phase, 1-ml portions of the cultures were imaged using the IVIS 100 system (A), and cDNA was synthesized from RNA extracted from 10-ml portions of the cultures, which was followed by PCR analysis to assess levels of 16S RNA, luxA, and hlyA transcription (B). The images in panel A were obtained using an IVIS 100 system with 5 min of exposure and a binning value of 8. The color bar indicates the bioluminescence signal intensity (in photons s−1 cm−2). Fold induction indicates the levels of bioluminescence relative to the levels observed in LB medium.
Hemolysin expression during cell invasion.
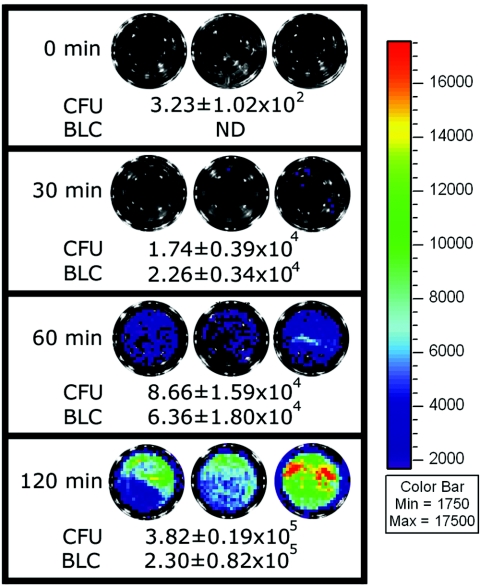
We also investigated the possibility of using pPL2lux to monitor hemolysin expression in real time during a cell invasion assay. EGD-e::pPL2lux and pPL2lux-PhlyA were used to infect C2Bbe1 cells, and the levels of intracellular bioluminescence were monitored and the numbers of intracellular bacterial cells were determined (Fig. 4). Notably, EGD-e::pPL2lux and EGD-e::pPL2lux-PhlyA grown in BHI medium were used to infect the C2Bbe1 cells, as these cells exhibited no detectable bioluminescence (see above). Moreover, no bioluminescence was detected at any time in the wells containing C2Bbe1 cells and control strain EGD-e::pPL2lux, which confirmed the absence of any detectable background bioluminescence (data not shown). Furthermore, no bioluminescence was detected in the gentamicin-treated washes (see Materials and Methods). Similarly, no bioluminescence was detected immediately after infection with EGD-e::pPL2lux-PhlyA, whereas 30 min after infection relatively low absolute levels of intracellular bioluminescence were detected and the absolute levels increased as time progressed. The bacterial counts for EGD-e::pPL2lux-PhlyA increased concomitant with the levels of bioluminescence, indicating that relative hemolysin expression was relatively constant during the course of the experiment (0.60 to 1.30 photons s−1 CFU−1). Moreover, the numbers of intracellular bacterial cells demonstrated that at each time similar numbers of bacteria were recovered from wells infected with EGD-e::pPL2lux and EGD-e::pPL2lux-PhlyA, indicating that luciferase expression does not influence the invasive potential of L. monocytogenes (data not shown). Overall, these experiments established the effectiveness of pPL2lux as a real-time reporter system to study listerial gene expression in an intact monolayer of C2Bbe1 cells.
FIG. 4.
Hemolysin expression during cell invasion. EGDe::pPL2lux-PhlyA was used to infect monolayers of C2Bbe1 cells, and bioluminescence was monitored 0, 30, 60, and 120 min postinfection. The images were obtained using an IVIS 100 system with 5 min of exposure and a binning value of 8. BLC, bioluminescence counts (in photons s−1). The color bar indicates bioluminescence signal intensity (in photons s−1 cm−2). ND, not detectable (bioluminescence counts, <1 × 104 photons s−1). The values for bioluminescence counts and CFU are the means ± standard deviations from triplicate wells. The data are representative of the data from three independent experiments.
Hemolysin expression in the organs of mice.
Mice are used as an in vivo model for L. monocytogenes infections. Therefore, the suitability of the pPL2lux reporter system to monitor listerial gene expression in the organs of mice following infection was investigated. Two groups of two mice were infected with control strain EGD-e::pPL2lux or EGD-e::pPL2lux-PhlyA. Three days postinfection all mice had developed overt symptoms of listeriosis, including piloerection and reduced activity (data not shown). The mice were sacrificed, and the levels of bioluminescence in the intact organs were determined and correlated with the numbers of bacterial cells (Fig. 5). Notably, the numbers of organ-specific CFU recovered from the mice infected with EGD-e::pPL2lux and EGD-e::pPL2lux-PhlyA were very similar, suggesting that luciferase expression does not influence the persistence of L. monocytogenes. No bioluminescence was detected in any of the organs of the two mice in the control group (Fig. 5). In contrast, the livers, spleens, and kidneys obtained from the two mice infected with EGD-e::pPL2lux-PhlyA displayed relatively high levels of bioluminescence. Furthermore, lower levels of bioluminescence were detected in the hearts and lungs of both mice infected with EGD-e::pPL2lux-PhlyA (data not shown). These lower levels of bioluminescence most likely reflected the fact that lower numbers of bacteria were recovered from these organs. Nevertheless, the data clearly indicate that hemolysin was expressed in all tested organs of mice 3 days postinfection and demonstrate that the pPL2lux reporter system can be used to monitor listerial gene expression in intact murine organs.
FIG. 5.
In vivo analysis of L. monocytogenes hemolysin expression and organ-specific colonization in BALB/c mice. Two mice were inoculated with 2 × 106 bacteria via the intraperitoneal route using either EGD-e::pPL2lux (Control) or EGD-e::pPL2lux-PhlyA (hlyA). Three days postinfection, the mice were euthanized, and bioluminescent imaging was performed for 5 min at a binning value of 4 using an IVIS 100 system. BLC, bioluminescence counts (in photons s−1). The color bar indicates the bioluminescence signal intensity (in photons s−1 cm−2). Subsequently, the numbers of bacterial CFU were determined for the individual organs. Results for the left and right kidneys were combined to obtain total BLC and CFU values. ND, not detectable (bioluminescence counts, <1 × 104 photons s−1). The data are representative of the data from three independent experiments.
DISCUSSION
Here we describe construction of a luciferase-based reporter system, pPL2lux, and successful use of this system to quantitatively study gene expression in L. monocytogenes in three model systems (growth in broth, cellular invasion, and murine infection). In all three model systems the luciferase-expressing L. monocytogenes strains displayed characteristics similar to those of control strains that did not express luciferase, as judged by the growth rates and invasive potential. Naturally occurring background levels of bioluminescence have been reported to be extremely low (48), and no background bioluminescence was observed during any of the experiments described here. This feature results in higher sensitivity for luciferase-based reporter systems than for many other reporter systems, including fluorescence-based systems, for which naturally occurring background levels are relatively high (48). One potential limitation of the system described here is the fact that the luciferase reaction requires oxygen; therefore, it remains to be seen if the system is functional in model systems with low oxygen pressures (for instance, anaerobic growth in broth or the colon). The pPL2lux vector is a site-specific integration vector that was designed for construction of exact translational fusions of promoters to the luciferase-encoding genes, mimicking transcription and translation initiation as it occurs at the native chromosomal location of the promoters under investigation. Therefore, it is not surprising that the levels of bioluminescence and luxA transcription closely resembled the actual levels of hlyA transcription found at the native chromosomal location. Moreover, the pPL2lux system allows direct, quantitative determination of luciferase activity and the corresponding promoter strength, intrinsically providing the ability to monitor promoter strength in a real-time fashion. This characteristic provides a major advantage compared with many of the reporter systems available to date for gram-positive bacteria, which typically require disruption of bacterial cells and/or addition of a substrate, followed by spectophotometric measurement to quantitatively determine enzyme and corresponding promoter activities (1, 7, 28, 30, 32).
A previous study in which β-glucuronidase was used as a reporter system in L. monocytogenes revealed that hemolysin expression could be conveniently detected in LB broth buffered at pH 7.4 1 h into the stationary phase (5). Similar conditions were used here for the luciferase reporter system, and our results corroborated the previous findings. In addition, our experiments revealed that hemolysin expression is initiated during the mid-log phase in LB broth buffered at pH 7.4 and is still detectable after prolonged incubation in the stationary phase. Previously, the levels of bioluminescence from luxABCDE systems have been reported to decrease in the stationary phase in several gram-positive bacteria, including Streptococcus pneumoniae (12), Lactobacillus casei (28) and L. monocytogenes (18). To our knowledge, the hemolysin-coupled luciferase expression profiles described here are the first examples of relatively high levels of luciferase expression in a gram-positive bacterium in the stationary phase and thus overcome a final technical hurdle for the use of luciferase reporter systems to monitor bacterial gene expression in broth.
A luxAB-based luciferase reporter system (28) and (quantitative) RT-PCR strategies (4, 8, 28, 38, 40) have been employed previously to monitor in situ gene expression in bacteria during residence in their hosts. Both approaches have been applied only to the murine gut and require extensive homogenization of this organ prior to analysis. In contrast, hemolysin expression profiles could be obtained for intact organs using the luciferase reporter system. Hence, the system described here has great potential for direct comparison of different bacterial promoter activities in intact organs. However, organ-specific variation in quenching of bioluminescence signals does not allow quantitative interorgan comparison for each specific promoter element. Such an analysis would require homogenization of the organs prior to measurement of bioluminescence. Although the quenching differences mentioned above prevent direct correlation of the in vitro and in vivo hlyA promoter strengths observed in our experiments, it is remarkable that the highest relative levels of luciferase found per CFU in vitro are more than 100-fold lower than the levels found in C2 cells and the organs of mice (data not shown). Therefore, our findings further enhance previous experimental suggestions that hemolysin expression might be dramatically elevated in vivo (37).
Overall, the luciferase reporter system described here is a valuable addition to the genetic toolbox available for L. monocytogenes, as it can be employed for in situ real-time investigation of listerial promoter activities after infection of mice. Currently, the luciferase reporter system is being exploited in our laboratory to obtain and compare quantitative, organ-specific expression profiles for the majority of L. monocytogenes virulence genes and regulators, including cwhA, prfA, yycF, inlA, and actA. In parallel, the expression profiles of several household genes, such as atpI and ftsA, are being obtained, which allows normalization of the expression profiles found for the virulence genes (I. R. Monk, P. A. Bron, P. G. Casey, C. G. M. Gahan, and C. Hill, unpublished data). The wealth of in situ information about spatial and temporal gene expression that will become available from this type of animal experiment is expected to contribute significantly to our understanding of the molecular mechanisms involved in the in situ behavior of L. monocytogenes during infection.
Acknowledgments
We thank Pat Casey for conducting the initial animal infection studies. We are indebted to Phil Hill (University of Nottingham), who provided pSB2025.
This research was funded by the Irish Government under the national development plan (2000-2006) and by Science Foundation Ireland.
REFERENCES
- 1.Achen, M. G., B. E. Davidson, and A. J. Hillier. 1986. Construction of plasmid vectors for the detection of streptococcal promoters. Gene 45:45-49. [DOI] [PubMed] [Google Scholar]
- 2.Allen, K. J., and M. W. Griffiths. 2001. Use of luminescent Campylobacter jejuni ATCC 33291 to assess eggshell colonization and penetration in fresh and retail eggs. J. Food Prot. 64:2058-2062. [DOI] [PubMed] [Google Scholar]
- 3.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley, M., R. D. Sleator, C. G. Gahan, and C. Hill. 2005. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73:894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkin, S., D. R. Smulski, A. C. Vollmer, T. K. Van Dyk, and R. A. LaRossa. 1996. Oxidative stress detection with Escherichia coli harboring a katG′::lux fusion. Appl. Environ. Microbiol. 62:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bron, P. A., S. M. Hoffer, I. I. Van Swam, W. M. De Vos, and M. Kleerebezem. 2004. Selection and characterization of conditionally active promoters in Lactobacillus plantarum, using alanine racemase as a promoter probe. Appl. Environ. Microbiol. 70:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 10.Ermolaeva, S., S. Novella, Y. Vega, M. T. Ripio, M. Scortti, and J. A. Vazquez-Boland. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol. Microbiol. 52:601-611. [DOI] [PubMed] [Google Scholar]
- 11.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, K. P., J. Yu, C. Bellinger-Kawahara, D. Joh, M. J. Hawkinson, G. Xiao, T. F. Purchio, M. G. Caparon, M. Lipsitch, and P. R. Contag. 2001. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gahan, C. G., and C. Hill. 2005. Gastrointestinal phase of Listeria monocytogenes infection. J. Appl. Microbiol. 98:1345-1353. [DOI] [PubMed] [Google Scholar]
- 14.Gahan, C. G., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1987. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Guzzo, A., C. Diorio, and M. S. DuBow. 1991. Transcription of the Escherichia coli fliC gene is regulated by metal ions. Appl. Environ. Microbiol. 57:2255-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 19.Israelsen, H., S. M. Madsen, E. Johansen, A. Vrang, and E. B. Hansen. 1995. Environmentally regulated promoters in lactococci. Dev. Biol. Stand. 85:443-448. [PubMed] [Google Scholar]
- 20.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, H., K. J. Boor, and H. Marquis. 2005. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043-1056. [DOI] [PubMed] [Google Scholar]
- 24.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 27.Moors, M. A., B. Levitt, P. Youngman, and D. A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oozeer, R., J. P. Furet, N. Goupil-Feuillerat, J. Anba, J. Mengaud, and G. Corthier. 2005. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 71:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 30.Phillips-Jones, M. K. 2000. Use of a lux reporter system for monitoring rapid changes in alpha-toxin gene expression in Clostridium perfringens during growth. FEMS Microbiol. Lett. 188:29-33. [DOI] [PubMed] [Google Scholar]
- 31.Pieterse, B., R. H. Jellema, and M. J. van der Werf. 2006. Quenching of microbial samples for increased reliability of microarray data. J. Microbiol. Methods 64:207-216. [DOI] [PubMed] [Google Scholar]
- 32.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purdy, D., and S. F. Park. 1993. Heterologous gene expression in Campylobacter coli: the use of bacterial luciferase in a promoter probe vector. FEMS Microbiol. Lett. 111:233-237. [DOI] [PubMed] [Google Scholar]
- 34.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qazi, S. N., C. E. Rees, K. H. Mellits, and P. J. Hill. 2001. Development of gfp vectors for expression in Listeria monocytogenes and other low G+C Gram positive bacteria. Microb. Ecol. 41:301-309. [DOI] [PubMed] [Google Scholar]
- 36.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripio, M. T., G. Dominguez-Bernal, M. Suarez, K. Brehm, P. Berche, and J. A. Vazquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 38.Rokbi, B., D. Seguin, B. Guy, V. Mazarin, E. Vidor, F. Mion, M. Cadoz, and M. J. Quentin-Millet. 2001. Assessment of Helicobacter pylori gene expression within mouse and human gastric mucosae by real-time reverse transcriptase PCR. Infect. Immun. 69:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shelburne, C. E., R. M. Gleason, G. R. Germaine, L. F. Wolff, B. H. Mullally, W. A. Coulter, and D. E. Lopatin. 2002. Quantitative reverse transcription polymerase chain reaction analysis of Porphyromonas gingivalis gene expression in vivo. J. Microbiol. Methods 49:147-156. [DOI] [PubMed] [Google Scholar]
- 41.Sleator, R. D., H. H. Wemekamp-Kamphuis, C. G. Gahan, T. Abee, and C. Hill. 2005. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55:1183-1195. [DOI] [PubMed] [Google Scholar]
- 42.van der Vossen, J. M., J. Kok, and G. Venema. 1985. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl. Environ. Microbiol. 50:540-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Dyk, T. K., D. R. Smulski, T. R. Reed, S. Belkin, A. C. Vollmer, and R. A. LaRossa. 1995. Responses to toxicants of an Escherichia coli strain carrying a uspA′::lux genetic fusion and an E. coli strain carrying a grpE′::lux fusion are similar. Appl. Environ. Microbiol. 61:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vollmer, A. C., S. Belkin, D. R. Smulski, T. K. Van Dyk, and R. A. LaRossa. 1997. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl. Environ. Microbiol. 63:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell. Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 47.Williams, T., S. Bauer, D. Beier, and M. Kuhn. 2005. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 73:3152-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]