Abstract
The initial microbial colonization of tooth surfaces is a repeatable and selective process, with certain bacterial species predominating in the nascent biofilm. Characterization of the initial microflora is the first step in understanding interactions among community members that shape ensuing biofilm development. Using molecular methods and a retrievable enamel chip model, we characterized the microbial diversity of early dental biofilms in three subjects. A total of 531 16S rRNA gene sequences were analyzed, and 97 distinct phylotypes were identified. Microbial community composition was shown to be statistically different among subjects. In all subjects, however, 4-h and 8-h communities were dominated by Streptococcus spp. belonging to the Streptococcus oralis/Streptococcus mitis group. Other frequently observed genera (comprising at least 5% of clone sequences in at least one of the six clone libraries) were Actinomyces, Gemella, Granulicatella, Neisseria, Prevotella, Rothia, and Veillonella. Fluorescence in situ hybridization (FISH) confirmed that the proportion of Streptococcus sp. sequences in the clone libraries coincided with the proportion of streptococcus probe-positive organisms on the chip. FISH also revealed that, in the undisturbed plaque, not only Streptococcus spp. but also the rarer Prevotella spp. were usually seen in small multigeneric clusters of cells. This study shows that the initial dental plaque community of each subject is unique in terms of diversity and composition. Repetitive and distinctive community composition within subjects suggests that the spatiotemporal interactions and ecological shifts that accompany biofilm maturation also occur in a subject-dependent manner.
More than 700 different bacterial phylotypes are found in the oral cavity, as determined by 16S rRNA gene sequencing (1). The initial colonizers of tooth surfaces are a specific subset of the oral microflora (24). Of these bacteria, those that colonize the clean enamel surface independently of other bacteria possess mechanisms for attachment to the acquired salivary pellicle covering the enamel (32) and the ability to metabolize salivary components as the sole nutritional source (27). Alternatively, some bacterial species participate in consortia that are able to attach to enamel and establish as an initial community requiring metabolic interactions among their members (18).
Several studies have identified streptococci as the predominant colonizers of early enamel biofilms (17, 24). Nyvad and Kilian (24) characterized the cultivable microflora colonizing enamel pieces exposed to the oral cavity. Streptococci were shown to compose about 63% (mean value of samples from four individuals) of bacteria isolated after 4 h of plaque formation and 86% of bacteria isolated after 8 h. A variety of other bacteria such as veillonellae and actinomyces were also reported to be present. However, this study was performed at a time when rapid PCR-based taxonomic characterization of bacterial communities was not available. As a consequence, the microflora did not include uncultivated organisms, and many of the rarer isolates were not assigned an identity but rather were placed into broad groups such as gram-negative cocci. Another study reporting the molecular characterization of the initial microflora employed the “checkerboard” DNA-DNA hybridization technique (38) using DNA probes for 40 cultured bacterial species to analyze the early supragingival plaque of 15 healthy individuals (17). That study also identified Streptococcus spp., in particular Streptococcus mitis and Streptococcus oralis, as the predominant early colonizers but was, at its outset, limited in range to 40 culturable organisms. Despite these efforts to characterize initial communities, the broad-based molecular taxonomic information that would eliminate limits imposed by culturability is not available. Furthermore, the application of fluorescence in situ hybridization (FISH) to obtain simultaneous taxonomic and positional information on in situ early biofilms is necessary; cell-to-cell interactions between species might determine subsequent biofilm characteristics.
This study used 16S rRNA gene sequencing and a retrievable enamel chip model (28) to characterize the microflora present at 4 h and 8 h of plaque development in three individuals. FISH was used to visualize the biofilm architecture of the mixed-species communities while confirming the dominance in situ of streptococci in initial dental plaque bacterial populations.
MATERIALS AND METHODS
In vivo biofilm development on enamel chips.
Details on fabrication of enamel chips and use in healthy human subjects have been published previously (28). Briefly, enamel pieces (1.5 mm wide by 1.5 mm long by 1 mm deep) were cut from extracted human third molars, cleaned in an ultrasonic bath (model 1510; Branson, Danbury, CT), sterilized with ethylene oxide, and affixed in custom-fabricated acrylic stents with dental wax. Two bilateral mandibular stents (spanning the posterior buccal surface from the first premolar to first molar), each of which contained two chips, were carried intraorally by each of three subjects. One stent was placed for 4 h and the other was placed for 8 h. No intake of food or liquids (other than water) was allowed except during a lunch period when stents were removed and stored in a humid denture cup at 37°C. Subjects (ages, 21, 29, and 47 years) were recruited through protocol 98-D-0116 at the National Institutes of Health. Criteria for enrollment included systemic health, absence of periodontitis or other pathology of the oral tissues, nonsmoker status, and no record of antibiotic usage in the 3 months prior to enrollment.
PCR amplification, cloning, and sequencing of 16S rRNA genes.
After retrieval from the mouth, each enamel chip was placed in 500 μl of sterile phosphate-buffered saline (PBS) and agitated at 225 rpm for 30 s to release the unattached bacteria present in the coat of saliva surrounding the chip. After washing, each chip was placed in 20 μl sterile water and biofilm cells were released by 20-min mild sonication (model 1510 bath; Branson Ultrasonics Corporation, Danbury, CT). Five microliters of water containing released cells was mixed with 5 μl of Microlysis solution (The Gel Company, San Francisco, CA), followed by rapid heating and cooling of the sample according to the manufacturer's instructions. This template solution was then subjected to 10 min of heating at 100°C, as preliminary experiments revealed that in the absence of this step Actinomyces spp. were incompletely lysed. Specifically, when a mixture containing equal numbers of Streptococcus gordonii DL1 and Actinomyces naeslundii T14V cells was used as a template for PCR, only S. gordonii 16S rRNA gene sequences were obtained unless the sample was heated at 100°C for 10 min, after which S. gordonii and A. naeslundii sequences were retrieved at the same frequency (data not shown).
The entire 10 μl of lysed cells was used as template in a 50-μl PCR mixture containing Ex Taq buffer (TaKaRa Bio, Inc., Shiga, Japan), 400 μM each deoxynucleoside triphosphate, 2.5 U Ex Taq Hot Start DNA polymerase (TaKaRa Bio, Inc.) and 0.6 μM each primer. Universal primers D88 and E94 (30) were used to amplify approximately 1,500 bp of the 16S rRNA gene. PCRs were initially denatured at 94°C for 3 min, followed by 30 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. PCR products were gel purified (gel purification kit; QIAGEN, Valencia, CA) and ligated into pCR 2.1 (Invitrogen, Carlsbad, CA) which was transformed into OneShot Top10 chemically competent Escherichia coli (Invitrogen). After selection (ampicillin, 100 μg/ml) and blue/white screening, 150 transformants were taken from the samples of subject 1, and 100 transformants were taken from the samples of subjects 2 and 3. EcoRI digestion of plasmid DNA was used to determine colonies containing inserts of the right size (∼1,500 bp). Purified plasmid DNA was sequenced in a 3730 DNA analyzer (Applied Biosystems, Foster City, CA). Primers D88 and F15 (30) were used to sequence approximately 500 bp near the 5′ end of the 16S rRNA gene and, when required, the primers B34, E94, F16, F17, F18, F20, and F22 (30) were additionally used to sequence the complete 16S rRNA gene.
All manipulations were carried out in a Plexiglas-enclosed PCR/UV work station (Coy Laboratory Products, Grass Lake, Mich.). Negative controls included a PCR with no added template (water control) and a PCR using template DNA released from an enamel chip fixed on a stent but not placed intraorally (enamel chip control). For the negative controls, the PCR cycle was modified to include 40 cycles of denaturation, annealing, and extension. We sequenced a total of 30 clones obtained for each water and enamel chip control for a total of 60 sequences of environmental contaminants. These were used to determine environmental contaminants in our samples from each subject.
Phylogenetic analysis of insert sequences.
Sequences were assembled using Seqman II from the Lasergene 6 package (DNASTAR, Inc., Madison, WI). BLAST (2) and FASTA (31) searches were performed for initial identification, and possible chimeric sequences were checked using the RDP-II chimera check tool (6). A sequence was assigned a species identity when it shared >98% identity with a published sequence of a cultured organism in the database. Phylogenetic placement of each sequence was confirmed by construction of phylogenetic trees using closely related reference sequences obtained from EMBL and GenBank. Prior to the construction of phylogenetic trees, unambiguous sequences were aligned using CLUSTAL_X (40). Partial 16S rRNA gene sequences of 440 nucleotides were used to construct phylogenetic trees by means of neighbor-joining and maximum-likelihood methods using TREECON (42) and DNAML from the Phylip package (11). All reference sequences used in the tree are from validated bacterial species: each has been cultured, characterized, deposited with at least two recognized culture collections, and named under the procedure described in the Bacteriological Code, 1990 revision (37). Trees were viewed using TREEVIEW (version 1.66) and text was edited in CorelDraw 12. Although 100 or 150 colonies were analyzed at each time point from each subject, the data reflect only clones with a positive insert and, of these, only those sequences that were not chimeric or environmental contaminants.
Phylotype determination.
A square similarity matrix was produced for each library of sequences and for all libraries compiled with DNAdist from the Phylip package (11) and the Kimura two-parameter model of nucleotide substitution. Sequences were grouped into phylotypes using the software DOTUR (for distance-based operational taxonomic unit and richness) (33) and the furthest-neighbor assignment algorithm. Other studies that have sequenced greater portions of the 16S rRNA gene have used a cutoff of 99% identity among sequences to define a phylotype (8, 30). We used a more conservative phylotype definition of 98% because the sequenced region constituted the most variable one-third of the 16S rRNA gene. The partial 16S rRNA gene sequences that possessed <98% identity to 16S rRNA gene sequences in EMBL and GenBank databases were completely sequenced, reanalyzed, and compared to database sequences using BLAST and FASTA search algorithms. Complete 16S rRNA gene sequences that still possessed <98% identity to any known database sequences were described as novel phylotypes.
Estimation of phylotype diversity.
Good's coverage estimation was calculated as [1 − (n/N)] × 100, where n is the number of singleton sequences and N is the total number of sequences for the clone library analyzed. Estimates of phylotype richness were calculated according to the abundance-based coverage estimator (ACE) (5) and the bias-corrected Chao1 estimator (4). Collector's curves of observed and estimated richness (plots of the number of observed and estimated phylotypes as a function of the number of clones sequenced) were calculated in DOTUR (33). The Shannon-Weaver diversity index (21) was also calculated in DOTUR.
Statistical comparisons of 16S rRNA gene libraries.
∫-LIBSHUFF (34, 35) was used to determine if DNA libraries differed between subjects and between time points. A square similarity matrix constructed in DNAdist (11) using the Jukes and Cantor model of nucleotide substitution was used as the input file. Critical P values for individual comparisons were determined based on the correction for multiple comparisons provided by ∫-LIBSHUFF to attain an experiment-wide type I error rate of 5%. P values for each library comparison were estimated by 10,000 random permutations of sequences between libraries.
∫-LIBSHUFF was also used to evaluate interexperiment (days T0 and T21) and intrasubject (chip 1 versus chip 2) variability. Four-hour biofilms from subject 1 were used for these analyses. To evaluate variability between libraries obtained from chips carried simultaneously but amplified independently (intrasubject; chip 1 versus chip 2), we compared 25 sequences obtained from each chip at the genus level by conducting a statistical analysis of the difference between libraries with ∫-LIBSHUFF. To evaluate interexperiment variability, a 4-h biofilm from subject 1 was compared to that sampled 3 weeks later using 40 sequences from each time point. No significant differences were found among libraries; therefore, sequences were pooled with new sequences obtained from subject 1 and used in our overall analysis.
FISH.
Enamel chips were removed from the stent and fixed at 4°C for 3 h with 4% paraformaldehyde in phosphate-buffered saline. Chips were washed in 50% ethanol. For permeabilization, each chip was attached to glass slides with paraffin wax and treated with 25 μl lysozyme (70,000 U ml−1 in 100 mM Tris/HCl [pH 7.5], 5 mM EDTA; Sigma, St. Louis, MO) for 10 min at 37°C in a humid atmosphere. Chips were dehydrated in a series of ethanol washes (50, 80, and 100%; 3 min each wash) and exposed to 10 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris/HCl [pH 7.5], 0.01% sodium dodecyl sulfate, 25% formamide) containing 100 ng of probe. The slides were incubated at 46°C for 90 min in a humid atmosphere. After hybridization, chips were washed first in buffer (20 mM Tris/HCl [pH 7.5], 5 mM EDTA, 0.01% sodium dodecyl sulfate, and 159 mM NaCl) for 15 min at 48°C and then in ice-cold water and stained with 1-μg/ml acridine orange. The oligonucleotide probe STR405 (29, 41) was labeled with Alexa 546 and used to identify all Streptococcus spp. The specificity of the probe was tested against Streptococcus vestibularis ATCC 49124, Streptococcus parasanguinis ATCC 15912, Streptococcus mitis ATCC 49456, Streptococcus mutans ATCC 700610, Streptococcus oralis ATCC 10557, Streptococcus sanguinis ATCC 10556, Streptococcus gordonii ATCC 51656, Streptococcus gordonii ATCC 49818, Streptococcus gordonii ATCC 10558, Streptococcus salivarius ATCC 25975, Actinomyces naeslundii T14V, Fusobacterium nucleatum ATCC 10953, Prevotella intermedia ATCC 15032, Prevotella oralis ATCC 33269, Prevotella melaninogenica ATCC 25845, Veillonella atypica ATCC 17744, Veillonella dispar ATCC 17748, Veillonella parvula ATCC 10790, Granulicatella elegans ATCC 700633, Granulicatella adiacens ATCC 49175, and Abiotrophia defectiva ATCC 49176. All Streptococcus spp. had a positive reaction, whereas species of other genera had a negative reaction, with the exception of Abiotrophia defectiva, which showed a weakly positive signal. The oligonucleotide probe PRV392 (5′-GCACGCTACTTGGCTGG-3) was designed with ARB software (19) and labeled with Alexa 546. The specificity of the probe was tested against Streptococcus parasanguinis ATCC 15912, Streptococcus mitis ATCC 49456, Streptococcus mutans ATCC 700610, Streptococcus oralis ATCC 10557, Streptococcus sanguinis ATCC 10556, Streptococcus gordonii ATCC 49818, Streptococcus salivarius ATCC 25975, Actinomyces naeslundii T14V, Fusobacterium nucleatum ATCC 10953, Prevotella intermedia ATCC 15032, Prevotella oralis ATCC 33269, Prevotella melaninogenica ATCC 25845, and Porphyromonas gingivalis W50 ATCC 53978. All Prevotella spp. had a positive reaction, whereas other genera had a negative reaction. The probe EUB338 (3) was labeled with Alexa 633 and used as a positive control. All labeled probes were purchased from Operon Biotechnologies, Inc., Huntsville, AL.
Microscopy.
Chips were examined using a 63×/0.9 numerical aperture (NA) water-immersible lens on a Leica TCS/SP2 confocal microscope (Leica Microsystems, Malvern, PA). All images presented are maximum projections of the entire confocal image stack, which was collected simultaneously into three channels. Nine 238-μm by 238-μm images from different fields were collected at 63× (low magnification) and nine 79.4-μm by 79.4-μm images were collected at 3× electronic zoom (high magnification). Image stacks were acquired with axial spacing of 0.5 μm or 1.0 μm.
Image analysis and determination of Streptococcus proportions on enamel chips.
Low-magnification images were analyzed by using IMAQ ImageBuilder (National Instruments, Austin, TX), while high-magnification images were counted manually. For software analysis, red-green-blue (RGB) color maximum projections of the image stacks were manually processed in Photoshop to remove debris and enamel autofluorescence and then converted to grayscale. The grayscale images were manually thresholded, and particle analysis results were filtered to remove particles of ≤0.77 μm2 in area (an area slightly less than that of a 1-μm-diameter coccus). The total area of particles in each channel was calculated, and the percentage of STR405-positive area relative to the total EUB338-positive area or acridine orange-stained area was determined.
Nucleotide sequence accession numbers.
Partial and complete 16S rRNA gene sequences were deposited in GenBank under accession numbers DQ016669 to DQ016967 and DQ346213 to DQ346444.
RESULTS
Reproducibility of 16S rRNA amplification results.
Sequences obtained from 4-h biofilms from subject 1 were used to evaluate variability between libraries obtained from chips carried simultaneously but sampled independently. Limited chip-to-chip variability was seen in the proportion of Streptococcus spp. (60% of sequences were identified as Streptococcus spp. on chip 1 and 65% on chip 2) and in the proportion of Veillonella spp. (9% of sequences were identified as Veillonella spp. on chip 1 and 10% on chip 2), while the variability in the proportions of another major non-Streptococcus genus, Prevotella spp., was greater (11% of sequences were identified as Prevotella spp. on chip 1 and 1% on chip 2). ∫-LIBSHUFF analysis indicated that the two libraries (composed of 25 sequences each) were not different [P(X versus Y) = 0.2859; P(Y versus X) = 0.1792; X and Y represent libraries as shown below (see Fig. 3)].
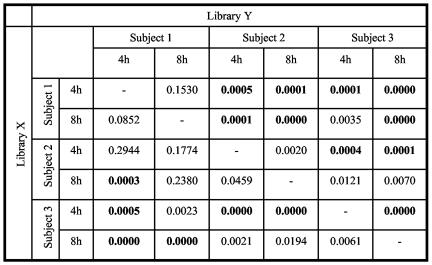
FIG. 3.
∫-LIBSHUFF comparisons of 16S rRNA gene sequence libraries from 4-h and 8-h biofilms in subjects 1, 2, and 3. Significant P values after correction for an experiment-wide type 1 error rate of 5% appear in boldface type and correspond to those values of <0.0017. Libraries are distinct if both X versus Y and Y versus X are statistically significant. If X versus Y is significant but Y versus X is not, then Y is a subset of X. If X versus Y is not significant but Y versus X is significant, then X is a subset of Y. Comparisons of the pooled combination of 4-h and 8-h samples of each subject with the pooled combination of each other subject are not presented; they resulted in significant values (P = 0.0000) for all comparisons (subject 1 versus subject 2, subject 2 versus subject 3, and subject 1 versus subject 3).
To evaluate the repetitive nature of initial colonization in the enamel chip model over time, a total of 40 sequences from an enamel chip carried for 4 h by subject 1 was compared to the same number of sequences obtained from an enamel chip carried for 4 h by the same subject 3 weeks later. The percentage of Streptococcus sequences was 60% at the first sampling and 68% at the later sampling. Veillonella sequences made up 7% (first sampling) and 14% (later sampling), while Prevotella spp. comprised 2% (first sampling) and 5% (later sampling). Furthermore, ∫-LIBSHUFF analysis found no difference between the two libraries [P(X versus Y) = 0.1285; P(Y versus X) = 0.10734). These observations suggested that the composition of initial plaque within an individual is reestablished and consistent over short periods of time (3 weeks).
Environmental contaminants in PCR.
Clones obtained from the amplification of the water control and enamel chip control were sequenced and used to assess contamination within the subject libraries. Contaminant or transient sequences from both sources were similar: an uncultured α-proteobacterium, Pseudomonas spp., Sphingomonas spp., Caulobacter spp., Brevundimonas vesicularis, an uncultured β-proteobacterium, Comamonas spp., and Delftia acidovorans. Neither control yielded sequences identifiable as belonging to resident oral organisms.
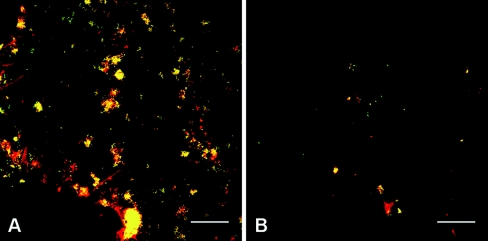
We sequenced 150 colonies from 4-h and 8-h libraries of subject 1. At 4 h, we found that 17 sequences (13%) corresponded to environmental contaminants and 4 sequences (3%) were chimeric sequences. At 8 h, 10 sequences (7%) were of environmental contaminants, and 6 sequences (4%) were chimeric sequences. One hundred colonies were sequenced from subject 2 libraries. At 4 h, 49 sequences (49%) were of environmental contaminants, and 5 sequences (5%) were chimeric sequences. At 8 h, we found 23 environmental contaminants (23%) and 4 (4%) chimeric sequences. In subject 3, 100 colonies were sequenced at each time point, with 18% of sequences representing environmental contaminants and 5% of sequences being chimeric at 4 h of sampling. At 8 h, the proportion of environmental contaminants was 22%, and the proportion of chimeric sequences was 6%. It is evident that the proportion of sequences derived from environmental contaminants varied among subjects and was greatest in subject 2, in particular, in the 4-h library. These results correlated with those of FISH experiments in which biomass also varied greatly among subjects (Fig. 1). In subject 2, very few bacteria were found on the surface of the enamel chip at 4 h of colonization (data not shown), whereas at 8 h, small clusters of cells (two to four cells) were evident (Fig. 1B). In contrast, biofilms from subject 1 typically showed abundant colonization by small to medium (<10 cells) clusters of cells at 4 h (data not shown) and a broad distribution of larger clusters of cells at 8 h (Fig. 1A). Therefore, it is likely that the proportion of environmental contaminants in each library correlated inversely with the amount of template available for PCR amplification.
FIG. 1.
Confocal micrographs (238 by 238 μm) of 8-h plaque development on enamel chips in subject 1 (A) and subject 2 (B). Cells were simultaneously labeled with the kingdom-level bacterial probe EUB338 (red) and the genus-level Streptococcus probe STR405 (green). Panels show the overlay of the two images. Notice the difference in biomass between the two samples but the similar proportion of Streptococcus spp. (yellow, colocalization of the two probes). Bar, 40 μm.
Coverage of 16S rRNA gene libraries and microflora diversity at 4 h and 8 h of enamel colonization.
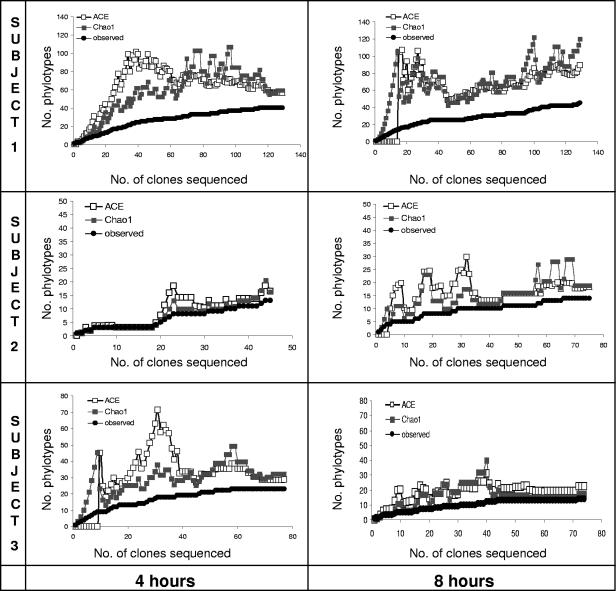
The microflora present at 4 h and 8 h of colonization on enamel chips carried intraorally by three subjects yielded 531 nonchimeric sequences and 97 distinct phylotypes. DOTUR analyses of pooled 4-h and 8-h libraries from each subject showed the following results: 66 distinct phylotypes in subject 1, 23 phylotypes in subject 2, and 32 phylotypes in subject 3. Table 1 presents the number of phylotypes obtained from each subject at each time point. The Good's coverage estimates reflect coverage of roughly 90% in five of the six clone libraries. Coverage in the 8-h library from subject 1 was 80%. The community was more diverse in subject 1 than in subjects 2 and 3, as demonstrated by the number of phylotypes and the Shannon-Weaver diversity index. Although the sampling effort was higher for subject 1, the coverage (in particular at 8 h) was the lowest and was consistent with the higher diversity of the sample. Collector's curves (Fig. 2) for each clone library depict the differences in phylotype richness among libraries and the different results in coverage. The number of unseen phylotypes is represented by the gap between the observed phylotypes and the number of phylotypes estimated by Chao1 and ACE. In most cases, this gap decreased towards the end of sampling. However, for the 8-h library from subject 1, sampling stopped while the gap was still increasing. We estimated that the genetic distance had to be relaxed to 4% (from 2%) difference to obtain a stable ACE and Chao1 richness estimates in the 8-h library of subject 1. Despite the low sampling effort for the libraries of subjects 2 and 3, high coverage and stable richness estimates were obtained because the libraries exhibited low diversity.
TABLE 1.
Phylotype richness and calculated coverage and diversity of each library
| Subject | Clone library (h) | No. clones in library | No. of phylotypes observed | Good's coverage (%) | Shannon-Weaver diversity indexa |
|---|---|---|---|---|---|
| 1 | 4 | 129 | 40 | 87 | 3.28 ± 0.17 |
| 8 | 134 | 45 | 80 | 3.39 ± 0.17 | |
| 2 | 4 | 46 | 13 | 89 | 2.10 ± 0.30 |
| 8 | 73 | 14 | 93 | 2.08 ± 0.23 | |
| 3 | 4 | 77 | 23 | 88 | 2.61 ± 0.26 |
| 8 | 72 | 14 | 92 | 1.83 ± 0.29 |
A higher Shannon-Weaver diversity index is associated with higher diversity. Each value is given as the mean ± the upper and lower bound to the 95% confidence interval (33).
FIG. 2.
Collector's curves of observed and estimated (ACE and Chao1) phylotype richness as a function of the number of clones. These curves depict the change in richness values as the number of clones in the library increased and are used to assess the sufficiency of the sampling effort for each specific library. A sufficient number of clones was sequenced in all libraries except in the 8-h library from subject 1, in which the gap between the observed and estimated phylotype curves had not closed when sampling stopped.
Predominant genera present at 4 h and 8 h of colonization and differences in microflora composition among subjects.
Consistent with the results of cultivable-flora studies (12, 24), the present results report streptococci as constituting 66% to 80% of the microflora at 4 h and 8 h of biofilm development (Table 2) and S. mitis/S. oralis as the predominant streptococci in the three subjects (Table 3). However, interindividual variation was seen in the prevalence and proportions of other genera, with the most prominent differences seen in the genera Abiotrophia, present only in subject 1; Corynebacterium, present only in subject 2; Neisseria, present at higher levels in subject 2; Rothia, absent in subject 2 and present at higher levels in subject 3 than in subject 1; Prevotella, absent in subject 2 and present at higher levels in subject 1 than in subject 3; and uncultured species from the class Clostridia, found only in subject 1. Furthermore, ∫-LIBSHUFF pairwise comparisons of pooled libraries (4 h and 8 h) from each subject revealed that all the libraries were statistically different from one another (P < 0.0001). When we made pairwise comparisons among libraries without pooling 4-h and 8-h samples together (Fig. 3), we found that the 4-h library from subject 3 differed from all the other 4-h libraries. On the contrary, the 4-h library from subject 2 was found to be a subset of the 4-h library of subject 1, a result that indicates the phylogenies in the subject 2 library are contained in the library from subject 1, but the libraries differed, probably because of the greater diversity found in subject 1. At 8 h, the same results were observed in that the 8-h library from subject 2 was a subset of the 8-h library from subject 1. The comparison of the 8-h library from subject 1 to that of subject 3 resulted again in a statistically significant difference, as was observed at 4 h. However, the 8-h library from subject 3 was similar to the 8-h library from subject 2.
TABLE 2.
Microflora present at 4 and 8 h of enamel colonization
| % of sequences ata:
|
|||||||
|---|---|---|---|---|---|---|---|
| Phylum | Genus | Subject 1
|
Subject 2
|
Subject 3
|
|||
| 4 h (n = 129) | 8 h (n = 134) | 4 h (n = 46) | 8 h (n = 73) | 4 h (n = 77) | 8 h (n = 72) | ||
| Firmicutes | Streptococcus | 65.9 | 67.9 | 67.4 | 79.5 | 66.2 | 73.6 |
| Veillonella | 8.5 | 8.2 | 2.2 | None | 5.2 | 2.8 | |
| Gemella | 3.9 | 3.0 | None | 4.1 | 2.6 | 15.3 | |
| Abiotrophia | 1.6 | 0.7 | None | None | None | None | |
| Granulicatella | None | 1.5 | 6.5 | 4.1 | None | None | |
| Peptostreptococcus | 0.8 | None | None | None | None | None | |
| Mogibacterium | None | 0.7 | None | None | None | None | |
| Megasphaera | None | 0.7 | None | None | None | None | |
| Selenomonas | 0.8 | 0.7 | None | None | None | None | |
| Oribacterium | 0.8 | None | None | None | None | None | |
| Unculturedb | 4.7 | 2.2 | None | None | None | None | |
| Actinobacteria | Actinomyces | None | 1.5 | None | 1.4 | 7.8 | None |
| Corynebacterium | None | None | None | 4.1 | None | None | |
| Rothia | 0.8 | 1.5 | None | None | 13.0 | None | |
| Atopobium | 0.8 | None | None | None | None | None | |
| Proteobacteria | Neisseria | 2.3 | 1.5 | 15.2 | 5.5 | 1.3 | 1.4 |
| “Candidatus Burkholderia” | 1.6 | 0.7 | 4.3 | None | None | None | |
| Haemophilus | None | None | 4.3 | 1.4 | None | 1.4 | |
| Bacteroidetes | Prevotella | 7.0 | 7.5 | None | None | 2.6 | None |
| Porphyromonas | 0.8 | 1.5 | None | None | None | 1.4 | |
| Fusobacterium | None | None | None | None | 1.3 | None | |
| Bacteroides | None | None | None | None | None | 1.4 | |
| Unculturedc | None | None | None | None | None | 2.8 | |
Data represent percentages of sequences that clustered within each genus after phylogenetic analysis. n, number of clones sequenced from each sample.
Sequences of uncultured Firmicutes were all placed within the Clostridia class after phylogenetic analysis.
Sequences of uncultured Bacteroidetes were placed under the class Flavobacteria after phylogenetic analysis.
TABLE 3.
Species of Streptoccocus present at 4 and 8 h of enamel colonization
| % of sequences for indicated subject ata:
|
||||||
|---|---|---|---|---|---|---|
| Species | Subject 1
|
Subject 2
|
Subject 3
|
|||
| 4 h | 8 h | 4 h | 8 h | 4 h | 8 h | |
| S. oralis/S. mitis | 26.7 | 23.1 | 70.0 | 100.0 | 54.9 | 84.9 |
| S. salivarius/S. vestibularis/ S. thermophilus | 4.7 | 14.3 | None | None | None | None |
| S. infantis | 14.0 | 11.0 | 6.7 | None | 17.7 | 9.4 |
| S. sanguinis | 15.1 | 6.6 | 16.7 | None | 7.8 | None |
| S. parasanguinis | 3.5 | 13.2 | None | None | None | None |
| S. gordonii | 1.2 | 6.6 | None | None | 7.8 | None |
| S. anginosus | 3.5 | 6.6 | None | None | None | None |
| S. bovis | 1.2 | None | None | None | None | None |
| S. cristatus | None | None | None | None | None | 3.8 |
| Other | 30.2 | 18.7 | 6.7 | None | 11.8 | 1.9 |
Data represent the percentages of sequences from samples that after phylogenetic analysis were >98% similar to the sequence of a validated Streptococcus reference strain. Sequences with <98% similarity to any known Streptococcus spp. were left unclassified and appear in the table as “other.”
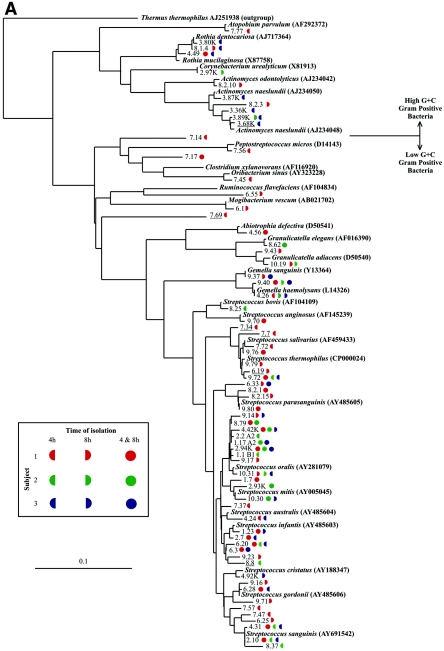
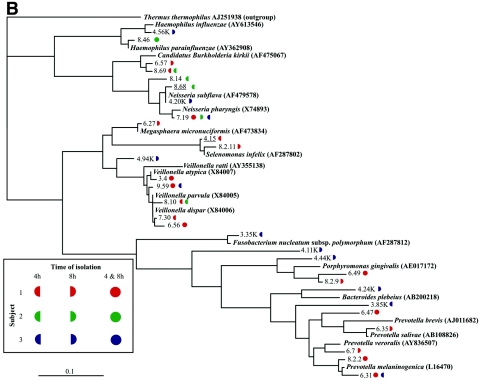
The 531 partial 16S rRNA gene sequences were compared to each other, and those that possessed >98% identity to one another were grouped and classified as phylotypes. Ninety-seven phylotypes were derived, and their phylogenetic relatedness to 16S rRNA gene sequences of previously described cultured organisms was established by the generation of maximum-likelihood phylogenetic trees (Fig. 4). Sixty-eight phylotypes possessed sequences most closely related to 16S rRNA gene sequences from gram-positive species (Fig. 4A) and 29 phylotypes were most closely related to 16S rRNA gene sequences from gram-negative species (Fig. 4B). Only 10 phylotypes were found in all three subjects, while 19 phylotypes were present in two of the three subjects. The remaining 68 phylotypes were unique to a subject. Subject 1 had 44 unique phylotypes, subject 2 had 11 unique phylotypes, and subject 3 had 13 unique phylotypes, resulting in 34 to 63% of phylotypes comprising the initial community found in a specific subject that was unique to that subject. This result highlights the distinct and subject-specific initial microflora composition.
FIG. 4.
Maximum-likelihood phylogenetic trees of the 97 phylotypes and related database sequences of 16S rRNA genes (shown in boldface type with associated EMBL accession numbers). Novel phylotypes are underlined. Symbol colors and shapes, associated with each phylotype, represent the subject (red represents subject 1, green represents subject 2, and blue represents subject 3) and the time period at which the sequence was isolated (left half circle, isolated at 4 h; right half circle, isolated at 8 h; full circle, isolated at 4 h and 8 h). (A) The 69 phylotypes that share close identity with published gram-positive species in the EMBL database library. (B) The 29 phylotypes that share close identity with published gram-negative species in the EMBL database library. The scale bar represents one substitution for every 10 nucleotides. The species used as the outgroup was Thermus thermophilus (EMBL accession number AJ251938).
Table 3 shows the subject-specific abundance of Streptococcus spp. present on the enamel chips carried by the three subjects at 4 h and 8 h of colonization. It was not possible to distinguish by 16S rRNA gene sequencing between S. mitis and S. oralis, or among S. salivarius, S. vestibularis, and Streptococcus thermophilus, as their sequences did not diverge sufficiently. Nevertheless, interindividual variation was seen in the prevalence and proportions of these Streptococcus groups. For example, the proportion of S. mitis/S. oralis sequences obtained from subject 1 was 27%, while in subjects 2 and 3 the sequences in the S. mitis/S. oralis group constituted a much higher percentage (70 to 100% in subject 2 and 55 to 85% in subject 3). Moreover, subject 1, with a more diverse microflora, also harbored a higher number of different Streptococcus spp. than, for example, subject 2.
Interindividual differences were also seen in the presence and proportions of the phylotypes belonging to the predominant genera (those comprising at least 5% of clone sequences in at least one clone library). The majority of Veillonella cloned sequences were closely related to V. parvula/V. dispar, which cannot be differentiated by their 16S rRNA gene sequences. Few clones of Veillonella spp. were identified as V. atypica, and they were present only in subject 1. Only two phylotypes identified as Gemella were observed, the most abundant being Gemella hemolysans, identified in all the subjects. Gemella sanguinis was obtained from two subjects and at lower frequency than G. hemolysans. Phylotypes belonging to the Granulicatella genus were observed in subjects 1 and 2 and were closely related to G. elegans and G. adiacens. Rothia was observed in two subjects and the phylotypes were closely related to those of Rothia mucilaginosa and Rothia dentocariosa. Some divergence was seen in Actinomyces phylotypes closely related to A. naeslundii, present in all subjects, while Actinomyces odontolyticus was present only in subject 1. Difficult to classify by 16S rRNA-based taxonomy (36), Neisseria spp. of the Neisseria pharyngis/Neisseria flava/Neisseria mucosa/Neisseria sicca group were found in all subjects. Subjects 2 and 3 had phylotypes closely related to N. subflava, but only subject 2 had phylotypes of the Neisseria meningitidis/Neisseria cinerea group (for example, sequence 8.14 in Fig. 4B). Subject 1 possessed the highest number of clones of Prevotella, some of which were closely related to P. melaninogenica, Prevotella veroralis, or Prevotella salivae. However, some Prevotella sequences obtained could not be assigned any species identity and belong to uncultured Prevotella spp. An overall tree depicting a representative sequence of each phylotype obtained is presented in Fig. 4.
Population shifts occurring from 4 h to 8 h in each subject.
∫-LIBSHUFF pairwise comparisons of 4-h and 8-h libraries (Fig. 3) revealed that the 4-h and 8-h libraries of subject 1 were similar. The 4-h and 8-h libraries of subject 2 were also similar, while the 8-h library in subject 3 was a subset of the 4-h library. These results suggest that only some species were retained and flourished in the biofilm in subject 3, which correlated with the decreased diversity seen at 8 h with this subject (Table 1).
Uncultured early colonizers and new phylotypes.
We identified 67 sequences that could not be assigned to a species by the criteria utilized in this study (≥98% identity to a named organism in the database). These sequences were therefore classified as sequences of uncultured microorganisms. Among these sequences we found eight novel phylotypes (sequences with <98% identity to any sequence in the database, including sequences originated from clones). Most of the uncultured organisms were found in subject 1, and libraries of subject 2 yielded the smallest number of uncultured bacteria. These observations correlate with the difference in diversity seen between the two subjects.
FISH for quantification of Streptococcus spp. at 4 h of colonization of enamel chips.
To validate the PCR results, we used a genus-level probe to determine the percentage of streptococci on enamel chips at 4 h of colonization in subject 1. Manual counting of nine high-magnification images, covering 1% of the enamel chip surface, indicated that Streptococcus cells were 73% ± 18% of the number of acridine orange-stained cells and 80% ± 19% of the EUB338-positive cells. Automated counting of nine low-magnification images, covering 23% of the area of an enamel chip, indicated that the area of STR405-positive particles was 72% ± 17% of the acridine orange-stained biomass and 73% ± 19% of the EUB338-positive biomass. A comparison of these results with the percentage of Streptococcus clones obtained at 4 h from subject 1 (Table 2) suggests that the PCR-based methodology offered an accurate representation of the relative abundance of streptococci.
Intergeneric juxtaposition in undisturbed plaque.
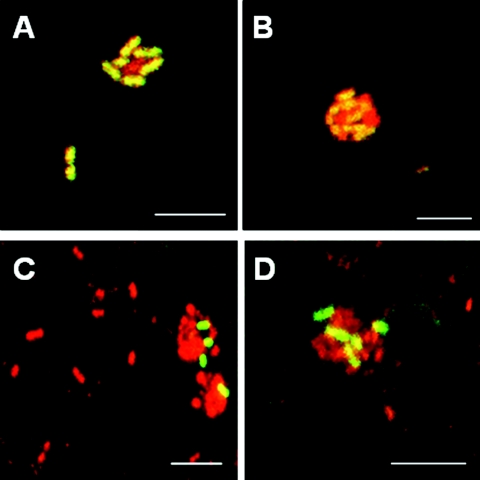
FISH also provided visualization of intact biofilm architecture in subject 1. At 4 h, the enamel surface was dotted with small clusters that typically contained Streptococcus spp. in contact with bacteria not reactive with the Streptococccus probe (Fig. 5A and B). At 8 h, surface coverage increased, and multigeneric clusters were prominent. Examples of such multigeneric clusters containing Prevotella spp. in intimate contact with unidentified bacteria are presented in Fig. 5C and D. The detection of numerically small bacterial constituents of supragingival plaque, such as Prevotella spp., underscores the power of FISH for revealing intergeneric associations.
FIG. 5.
Confocal micrographs of typical multigeneric clusters of cells found on enamel chips at 4 h (A and B) and 8 h (C and D) of plaque development. Cells were simultaneously labeled with all-bacterium-specific EUB338 probe (red) and either the Streptococcus-specific STR405 probe (A and B) (green) or the Prevotella-specific PRV392 probe (C and D) (green). (A and B) Unidentified bacterial cells (EUB338 reactive; red) juxtaposed with Streptococcus cells (EUB338 and STR405 reactive; red + green = yellow). (C and D) Unidentified bacterial cells (EUB338 reactive; red) in association with Prevotella cells (EUB338 and PRV392 reactive; red +green = yellow). Scale bar for all images, 5 μm.
DISCUSSION
The aim of this study was to use molecular techniques to characterize oral bacterial communities after 4 h and 8 h of enamel colonization. We found that the early dental plaque microflora varies on a subject-specific basis. More than two-thirds of the observed phylotypes were unique to a specific subject. Subject-specific variation is commonly overlooked, because studies typically pool samples from different subjects (17, 20, 24). However, a recent extensive characterization of the human intestinal microflora reported major differences in molecular community composition and diversity among three individuals (8). Perhaps interindividual variation in microflora of the digestive tract, including the oral cavity, could be attributed to differences in host factors that modulate colonization of an individual by a specific set of species. We postulate that members of a specific community have adapted to each other and to the host, thus creating interrelationships among community participants that ensure spatiotemporal repeatability and stability of the microbial community composition.
We found 10 phylotypes were common to all three subjects. The closest validated sequences to these phylotypes are Neisseria pharyngis, Gemella hemolysans (two phylotypes), Streptococcus thermophilus, Streptococcus oralis (three phylotypes), Streptococcus infantis, and Streptococcus sanguinis (two phylotypes) (Fig. 4). These phylotypes might constitute a core group of strains, highly adaptable to different human hosts, and constant members of initial biofilm communities. Not surprisingly, the aerobic neisseriae and aerotolerant streptococci and gemellae form this proposed core group, which might provide a favorable biofilm niche for subsequent or concomitant colonization of facultative and obligate anaerobes, which follows the aerobe-to-anaerobe transition for oral bacterial climax community development (22).
Small time-dependent (4-h versus 8-h) shifts in the microflora within subjects were observed, although these changes were not as striking as the differences between the subjects. For example, the diversity in subject 3, as measured by the Shannon-Weaver diversity index (Table 1), decreases from 4 h to 8 h. This change is consistent with a ∫-LIBSHUFF result indicating that the 8-h library of subject 3 was a subset of the 4-h library. This change can be explained by lack of retention in the biofilm of Rothia and Actinomyces spp., while the proportion of Gemella spp. increased at 8 h (Table 2), due solely to a single phylotype, G. hemolysans (data not shown). Shifts in Streptococcus spp. (Table 3) also contributed to the decreased diversity at 8 h in subject 3. The proportion of S. oralis/S. mitis clones increased at 8 h, while other Streptococcus such as S. sanguinis and S. gordonii were not retained. Another example of population shifts from 4 h to 8 h is seen in subject 2, in which an increase in the proportion of streptococci at 8 h (Table 2) coincides with a shift from a mixed Streptococcus population to one composed solely of S. mitis/S. oralis clones (Table 3). On the contrary, subject 1 retained similar species of streptococci at 4 h and 8 h. These results suggest that in some subject-specific communities, of all the phylotypes that attach initially to enamel, only a subset of that population can successfully establish and predominate at later colonization times. The success of a certain bacterial species within the biofilm might be conditioned to interactions with other bacteria present within the same community. Our results indicate that subject-specific communities follow distinct patterns of development. Strain-level 4-h to 8-h shifts within each species might also occur, but the methods used here would not detect such changes. For example, the present results show stable proportions of veillonellae between 4 and 8 h in subject 1 (Table 2), whereas a recent study in our laboratory revealed that, during this time, shifts occur within the veillonella population at the strain level in this subject in such a way that specific veillonella strains seen in moderate proportions at 4 h flourish at 8 h, while others are not retained (25). These changes exemplify the rapid dynamics of early community development and the need for the utilization of multiple approaches to characterize bacteria at different taxonomic levels when studying community changes.
The majority of the sequences reported here corresponded to previously cultivated microorganisms, and only 13% of the sequences and 31% of the phylotypes corresponded to uncultured microorganisms, a proportion lower than that found in subgingival plaque, where approximately 50% of phylotypes/species have not yet been cultured (16, 30). This result is not surprising, however, because the greatest proportion of early colonizers are streptococci, which are highly amenable to cultivation. The coaggregation properties of streptococci favor them as initial colonizers, compared to other oral bacteria. Streptococci are the only oral bacteria that exhibit extensive intrageneric coaggregation (15). In addition, streptococci possess numerous adhesins that recognize receptors in the acquired pellicle that coats the enamel (7, 9, 32, 39). These properties of adhesion likely offer an advantage to streptococci that confers their dominance as the initial colonizing bacteria. Once bound to enamel, the streptococci act as a nascent surface for recognition and binding by other streptococci as well as by species of other genera, such as Actinomyces, Fusobacterium, Haemophilus, Prevotella, and Veillonella (14), which leads to generation of multigeneric communities.
The present study also reports the use of FISH in conjunction with a retrievable enamel chip model. The intersection of these techniques allows taxonomic identification while visualizing architecture and spatial relationships among community members. The occurrence of small clusters of genetically distinct cells in close contact seems to be the most common form of colonization during initial plaque formation (26). Close proximity among early colonizing cells suggests that coaggregation plays a role in vivo in establishing multigeneric microcommunities where metabolic interactions among members might determine community development. Coaggregation and metabolic interactions occurring among some of the species seen as part of the early microflora of subjects in this study have been previously observed in a series of in vitro and in vivo experiments conducted in our laboratory (10, 14, 26, 27). Considering that streptococci are the principal group of the early colonizing bacteria, some of the less predominant genera might be retained by physical or metabolic interactions with streptococci. One example is Veillonella spp., which are known to coaggregate with streptococci (13) and establish a food chain where veillonellae obtain energy from the metabolism of lactic acid produced by the streptococci as an end product of carbohydrate fermentation (23) and where veillonellae signal the streptococci to increase expression of amylase (10). Actinomyces are also known to interact with streptococci in initial microcommunities in vivo (26), and results from in vitro studies indicate that a mutualistic relationship exists between these two species (27). One of the interesting features of the composition of these early communities is the presence of cells with very diverse metabolic requirements. For example, the presence of aerobic organisms such as Neisseria spp. and Rothia spp. contrasts with the presence of anaerobic species such as Porphyromonas spp. and Prevotella spp. The predominance of aerobic and facultative bacteria in early colonizing biofilms is consistent with the long-held hypothesis that initial colonization occurs by aerobic and aerotolerant bacteria, which are replaced by anaerobic and less aerotolerant species (22). The present study found, however, that anaerobic microorganisms such as prevotellae and porphyromonads can be detected with aerobic and facultative species such as gemellae, neisseriae, and streptococci. The prevotellae and porphyromonads might inhabit initial communities in low proportions until biofilm development ensures a proper anaerobic environment for their proliferation. A question for future studies is how bacterial cells interact metabolically within a cluster, as such interactions might drive reestablishment of the community between daily oral hygiene procedures. We propose that repetitive spatiotemporal interactions and ecological shifts in subject-specific communities are integral to the organization of stable natural plaque biofilms.
Acknowledgments
This research was supported by the Intramural Research Program of NIDCR, NIH.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 5.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 8.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egland, P. G., L. D. Du, and P. E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, C. V., R. N. Andersen, and P. E. Kolenbrander. 1992. Characterization of Veillonella atypica PK1910 adhesin-mediated coaggregation with oral Streptococcus spp. Infect. Immun. 60:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ. Microbiol. 56:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97:1311-1318. [DOI] [PubMed] [Google Scholar]
- 18.Liljemark, W. F., C. G. Bloomquist, B. E. Reilly, C. J. Bernards, D. W. Townsend, A. T. Pennock, and J. L. LeMoine. 1997. Growth dynamics in a natural biofilm and its impact on oral disease management. Adv. Dent. Res. 11:14-23. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 21.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 22.Marsh, P., and M. V. Martin. 1999. Oral microbiology, 4th ed. Wright, Oxford, United Kingdom.
- 23.Mikx, F. H., and J. S. van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20:407-410. [DOI] [PubMed] [Google Scholar]
- 24.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. Unpublished data.
- 26.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer, R. J., Jr., R. Wu, S. Gordon, C. G. Bloomquist, W. F. Liljemark, M. Kilian, and P. E. Kolenbrander. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337:393-403. [DOI] [PubMed] [Google Scholar]
- 29.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 30.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scannapieco, F. A. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 5:203-248. [DOI] [PubMed] [Google Scholar]
- 33.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, N. H., E. C. Holmes, G. M. Donovan, G. A. Carpenter, and B. G. Spratt. 1999. Networks and groups within the genus Neisseria: analysis of argF, recA, rho, and 16S rRNA sequences from human Neisseria species. Mol. Biol. Evol. 16:773-783. [DOI] [PubMed] [Google Scholar]
- 37.Sneath, P. H. A. 1992. International code of nomenclature of bacteria: bacteriological code, 1990 revision. ASM Press, Washington, D.C. [PubMed]
- 38.Socransky, S. S., C. Smith, L. Martin, B. J. Paster, F. E. Dewhirst, and A. E. Levin. 1994. “Checkerboard” DNA-DNA hybridization. BioTechniques 17:788-792. [PubMed] [Google Scholar]
- 39.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurnheer, T., R. Gmur, E. Giertsen, and B. Guggenheim. 2001. Automated fluorescent in situ hybridization for the specific detection and quantification of oral streptococci in dental plaque. J. Microbiol. Methods 44:39-47. [DOI] [PubMed] [Google Scholar]
- 42.van de Peer, Y., and R. de Wachter. 1993. TREECON: a software package for the construction and drawing of evolutionary trees. Comput. Appl. Biosci. 9:177-182. [DOI] [PubMed] [Google Scholar]