Abstract
As part of an ongoing effort to catalog spore-forming bacterial populations in environments conducive to interplanetary transfer by natural impacts or by human spaceflight activities, spores of Bacillus spp. were isolated and characterized from the interior of near-subsurface granite rock collected from the Santa Catalina Mountains, AZ. Granite was found to contain ∼500 cultivable Bacillus spores and ∼104 total cultivable bacteria per gram. Many of the Bacillus isolates produced a previously unreported diffusible blue fluorescent compound. Two strains of eight tested exhibited increased spore UV resistance relative to a standard Bacillus subtilis UV biodosimetry strain. Fifty-six isolates were identified by repetitive extragenic palindromic PCR (rep-PCR) and 16S rRNA gene analysis as most closely related to B. megaterium (15 isolates), B. simplex (23 isolates), B. drentensis (6 isolates), B. niacini (7 isolates), and, likely, a new species related to B. barbaricus (5 isolates). Granite isolates were very closely related to a limited number of Bacillus spp. previously found to inhabit (i) globally distributed endolithic sites such as biodeteriorated murals, stone tombs, underground caverns, and rock concretions and (ii) extreme environments such as Antarctic soils, deep sea floor sediments, and spacecraft assembly facilities. Thus, it appears that the occurrence of Bacillus spp. in endolithic or extreme environments is not accidental but that these environments create unique niches excluding most Bacillus spp. but to which a limited number of Bacillus spp. are specifically adapted.
Spore-forming bacteria of the species Bacillus are ubiquitous in the environment, and their endospores represent some of the hardiest and longest-lived cells on Earth (reviewed in references 37, 39, and 41). Endolithic Bacillus spp. have been obtained from a wide variety of sites within the lithosphere, such as the interiors of mined amber (6), halite crystals (58), marine manganese nodules (36), rock varnishes from Sonoran, Mohave, and Negev desert rocks (23, 35, 45), deep subsurface boreholes (2, 4), Sonoran desert basalts (3), and Florida limestones from intertidal zones (1). In addition, several Bacillus spp. (e.g., Bacillus brevis, B. licheniformis, B. mycoides, B. megaterium, and B. subtilis) have repeatedly been found associated with the biodegradative activities destroying ancient stone buildings and monuments in Europe (12, 15, 49, 52).
Of particular recent interest to us has been the potential role that endolithic microbes may play in the transfer of life between the terrestrial planets. Because of their extreme resistance properties, bacterial spores have been a model system of choice for testing the theory that viable microbes could be successfully transferred through space between the environments of terrestrial planets such as Earth and Mars, which has variously been dubbed “panspermia,” “lithopanspermia,” and “transpermia” (reviewed in references 7, 11, 19, 33, 41, and 43). Interplanetary transfer of microbes can be envisioned to occur by (i) human spaceflight activities or (ii) natural impacts (11). In both processes, the initial microbial populations for Earth-to-Mars transfer originate in Earth environments; thus, it is of relevance to understand the compositions of populations which are in a position to be transferred and the likelihood that they could survive the journey (11).
Candidates for transfer by human spaceflight.
The possibility that space exploration could lead to the microbial “forward contamination” of pristine planetary environments such as Mars has led to current international Planetary Protection protocols which require that spacecraft destined for Mars be constructed and assembled under conditions as nearly as possible approaching sterility (reviewed in reference 46). To achieve disinfection, robotic spacecraft are assembled in spacecraft assembly facilities (SAFs), clean rooms which are considered extreme selective environments where air circulation is controlled, strict hygienic practices are observed, and a number of sterilants, such as hydrogen peroxide (H2O2) vapor and UV radiation, are used (25, 27). Spores of a number of Bacillus spp. have been identified as dominant inhabitants of SAFs and spacecraft, both on Earth and in space (6a, 26, 55, 56). Recurring isolates from SAFs belong to Bacillus spp., particularly B. pumilus; it has further been demonstrated that spores of several Bacillus spacecraft isolates exhibit hyperresistance to sterilants such as H2O2 and UV (25, 27, 55).
Candidates for natural interplanetary transfer.
The notion that endolithic microbes could naturally be transferred between Earth and Mars has been bolstered in part by the existence on Earth of at least 30 meteorites originating from Mars (14, 32), which represent samples of the martian endolithic environment. Sound theoretical and experimental support exists for a spallation mechanism by which near-surface rocks could be launched into space by the energy released from large impacts (30, 31; reviewed in references 19, 33, and 41). Spores of B. subtilis have been shown to survive the shock, heating, and acceleration forces calculated to prevail during impact-mediated launch into space (5, 9, 21, 29). In addition, B. subtilis spores embedded in artificial meteorites have been demonstrated to survive long-term exposure to the space environment (18, 20) and hypervelocity atmospheric entry from space (10).
Characterization of the collection of martian meteorites and the predictions of spallation theory indicate that the most likely rock types which could serve as putative vehicles of natural interplanetary transfer are igneous rocks situated close to the surface (29-31). On Earth, these rocks consist mainly of basalts and granites. In a previous communication (3), we described the isolation of endolithic Bacillus sp. spores from the interior of near-subsurface basalt. We found exceedingly low numbers of spores in basalt (a few tens of spores per gram of rock), but interestingly, as found in SAFs (26, 56), the major species recovered was B. pumilus; these B. pumilus isolates formed spores which were more resistant to acceleration shock and UV than were spores of the reference B. subtilis lab strain (3). An examination of the occurrence, distribution, and UV resistance properties of endolithic Bacillus spp. from granite would provide us with a more complete catalog of spore formers naturally positioned for space travel. Therefore, in this communication we describe sampling of the interiors of near-subsurface granite rocks collected in the Sonoran desert (Arizona) to obtain, identify, and characterize endolithic spores.
MATERIALS AND METHODS
Bacterial strains, media, and cultivation conditions.
Reference strains from our collection used in this study were B. subtilis strains 168 (trpC2), HA101 (hisA1 metB5) (44), and ATCC 6633 (22) and the B. pumilus type strain, ATCC 7061 (3). The media used were Luria-Bertani (LB) medium (34), either liquid or solidified with 1.5% agar, and Schaeffer sporulation medium (SSM) (47), either liquid or solidified with 1.7% agar. All bacteria were cultivated at 37°C, and liquid cultures were incubated with vigorous aeration.
Sampling locations and methods.
Granite samples were obtained from natural outcrops at six locations in the Santa Catalina mountain range near Tucson, Ariz. At each location, surface material was first removed to a depth of 3 to 5 cm using a rock hammer and chisel, each of which had been field disinfected by soaking in hypochlorite bleach. The chisel was again disinfected, and subsurface granite samples of 200 to 500 g apiece were removed and placed inside zipper-closure plastic bags for transport to the laboratory. Using aseptic technique in a disinfected laminar flow hood, the outer faces of the sample from sampling site 2 were further pared off with a flame-sterilized chisel, and the resulting cores were crushed to 1- to 2-mm-diameter particles using a flame-sterilized hammer. Particles were weighed, transferred to 3-ml aliquots of phosphate-buffered saline (10 mM potassium phosphate, 150 mM NaCl, pH 7.4) (42), suspended by vigorous vortexing, diluted serially 10-fold, and plated on solid LB medium, either before or after heat shock (80°C, 10 min), which selects for heat-resistant spores (42). After incubation at 37°C for several days, colonies were counted. Colonies arising from isolates which survived heat shock were picked at daily intervals, streak purified at least three times on LB plates, grown overnight in liquid LB medium or SSM, Gram stained, and stored as glycerol stocks at −80°C.
Visualization of blue fluorescence.
Colonies or culture supernatants were illuminated with a hand-held medium-wavelength UV lamp (model UVM-57; UVP, Upland, CA), which emits a spectrum of UV wavelengths spanning 280 to 320 nm, with an emission maximum at 302 nm. Photographs were taken with a digital camera using the same UV illumination.
DNA isolation.
Individual strains from frozen glycerol stocks were streaked onto LB plates and incubated overnight at 37°C. Cells were lifted directly from plates, and their chromosomal DNAs were extracted using an Ultraclean Microbial DNA isolation kit (Mo Bio Laboratories, Inc., Solana Beach, CA). The quality and quantity of DNA were verified by electrophoresis through 0.8% agarose gels versus DNA standards.
Bacterial DNA fingerprinting by rep-PCR.
Bacterial isolates were analyzed by repetitive extragenic palindromic PCR (rep-PCR) (57), using the commercial Diversilab microbial typing system for Bacillus spp. (Bacterial Barcodes Inc., Athens, GA) according to the manufacturer's protocols and using the manufacturer's proprietary Bacillus sp. primers and reagents. rep-PCR products were separated on an Agilent 2100 bioanalyzer (Agilent Technologies, Wilmington, DE) and analyzed with Diversilab system, version 1.4, software as directed by the manufacturer.
16S rRNA gene sequencing.
Approximately 10 ng of purified chromosomal DNA from each selected isolate was used as the template for 16S rRNA gene amplification by standard PCR, using the universal bacterial primers B27F (5′-GAGTTTGATCMTGGCTCAG-3′) and B1512R (5′-AAGGAGGTGATCCANCCRCA-3′) (M = A or C; N = A, T, C, or G; and R = A or G) (3). PCRs were performed in a PTC-200 thermal cycler (MJ Research, Waltham, MA), using 35 cycles of denaturation for 1 min at 95°C, annealing for 2 min at 55°C, and elongation for 3 min at 72°C. After a final incubation for 10 min at 72°C, the PCR products were purified using a Qiaquick PCR purification kit (QIAGEN), and the purified products were sequenced at the University of Florida Interdisciplinary Center for Biotechnology Research. The taxonomic identification of organisms was accomplished by comparison to 16S rRNA gene sequences in the Ribosomal Database Project II, release 9.32 (8), available at http://rdp.cme.msu.edu/ and hereafter called the RDP.
Spore preparation.
Strains were streaked onto solid SSM and incubated at 37°C for 48 h, the resulting growth was resuspended in sterile water, and spores were purified by lysozyme treatment followed by a series of buffer and detergent washes, as described previously (42), and then by heat shock (80°C, 10 min). All spore preparations were determined by phase-contrast microscopy to consist of >99% phase-bright spores.
Spore UV resistance.
Purified spores were suspended in water at a concentration of 1 × 106/ml and irradiated with 254-nm UV using a model UVGL-25 low-pressure mercury vapor lamp (UVP). The absorbance at 254 nm (A254) of each spore suspension was determined in an Ultraspec 3000 UV-visible spectrophotometer (Pharmacia). The lamp output at 254 nm was measured with a UVX radiometer fitted with a UVX-25 filter (UVP). UV fluence rates for each experiment were determined by entering the values of the lamp output and the A254 of the spore suspension into UVCalc, a software program for custom UV calculations available from Bolton Photosciences. UV inactivation curves were determined and used to compute the average UV dose lethal to 90% of the spore population (LD90) and the average dose necessary to reduce the spore population by 1 order of magnitude (D value) (3, 38, 40). All UV inactivation experiments were repeated three or four times. Basic statistical parameters and analysis of variance were computed using commercial statistical software (Kaleidagraph, version 3.6.2; Synergy Software, Reading, PA).
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences obtained in this work have been deposited in the GenBank database, and their accession numbers are listed in Table 1.
TABLE 1.
Summary of group I to XII Bacillus granite isolates
| Group | Strain | Blue fluorescence | Sporulation in SSM | GenBank accession no. | Nearest neighbor by 16S rRNA gene sequencinga | % Identity by 16S rRNA gene sequencingb |
|---|---|---|---|---|---|---|
| I | WN603 | + | + | DQ275182 | B. megaterium | 99.2 |
| II | WN611 | + | + | DQ275184 | B. megaterium | 99.6 |
| III | WN585 | + | + | DQ275179 | B. megaterium | 99.4 |
| IV | WN613 | − | − | DQ275185 | B. barbaricus | 87.1 |
| V | WN586 | + | + | DQ275180 | B. megaterium | 100.0 |
| VI | WN606 | + | + | DQ275183 | B. megaterium | 100.0 |
| VII | WN591 | + | + | DQ275181 | B. megaterium | 100.0 |
| VIII | WN570 | + | − | DQ275175 | B. simplex | 99.6 |
| IX | WN579 | ++ | + | DQ275178 | B. simplex | 100.0 |
| X | WN577 | − | − | DQ275177 | B. niacini | 95.1 |
| XI | WN559 | − | + | DQ275174 | B. drentensis | 96.2 |
| XII | WN575 | − | − | DQ275176 | B. drentensis | 98.6 |
Full-length 16S rRNA gene (∼1.5 kb) PCR fragments were sequenced.
Isolates with a percent identity of >97.5% were considered the same species (59).
RESULTS AND DISCUSSION
Cultivable endolithic spore populations from near-subsurface granite.
The interiors of granite rocks were sampled for bacteria, with particular emphasis on characterizing spore-forming Bacillus spp. We previously reported that exceedingly low numbers of spore-forming Bacillus spp. were isolated from Sonoran desert basalt, on the order of 101 CFU/g rock (3). In contrast, sampling of near-subsurface granite yielded much higher numbers of both total cultivable bacteria and heat-resistant spores. Suspensions prepared from granite were plated on solid LB medium before and after heat shock and then incubated at 37°C for 3 days, with colonies being counted each day. The results showed that after 3 days of incubation, approximately 104 total viable bacteria and ∼5 × 102 heat-resistant viable spores were obtained per gram of granite (Fig. 1). Further incubation of the plates did not yield appreciably larger numbers of isolates (data not shown). Colonies from a total of 56 isolates which had survived heat shock were picked randomly as they arose at daily intervals, streak purified several times on solid LB medium, and assigned the identifying codes WN558 to WN613 in our laboratory strain collection. All 56 isolates formed colonies on solid SSM with the appearance typical of Bacillus spp., i.e., flat, tan colonies with rough margins. All 56 isolates were grown overnight in liquid SSM and subjected to Gram staining. All were observed to be gram-positive rods, and many isolates were observed to contain endospores. From these observations, we tentatively concluded that all 56 isolates were Bacillus spp.
FIG. 1.
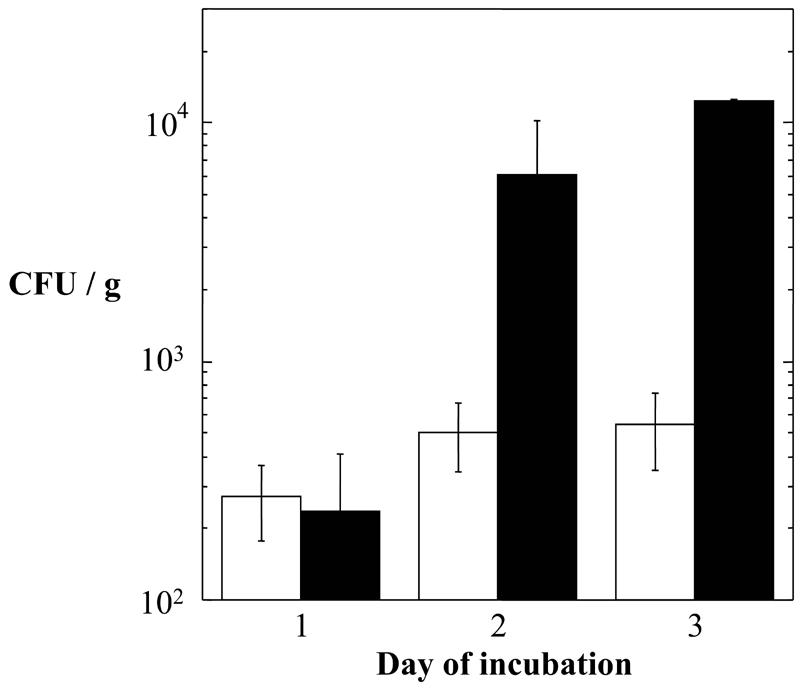
Numbers of total viable bacteria (solid bars) and heat-resistant spores (open bars) per gram of granite, grown on solid LB medium. Numbers are averages ± standard deviations (n = 2 or 3).
Blue fluorescence of endolithic Bacillus isolates.
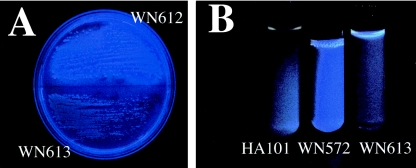
During streak purification of the 56 isolates, it was noticed that under certain lighting conditions, some of the isolates exhibited a slight bluish-green color. Upon further examination, we found that many, but not all, of the 56 isolates fluoresced blue under medium-wavelength UV light (Fig. 2A). To our knowledge, the production of blue fluorescent compounds by Bacillus spp. has not been reported to date. In a search of the literature, we were able to find a publication describing the characterization of chlorxanthomycin, a yellow fluorescent compound, produced by a strain of Bacillus isolated from soil (28). In contrast to the yellow fluorescent compound, which is located intracellularly (28), the blue fluorescent compound from the granite isolates diffused out of the cells and accumulated in the surrounding medium (Fig. 2B). We reasoned that production of the blue fluorescent compound could be used as a further distinguishing characteristic to group the granite isolates (Fig. 3). At this time, the identity of the blue fluorescent compound is unknown; details of its purification and characterization will be reported elsewhere.
FIG. 2.
Blue fluorescence exhibited by Bacillus spp. from granite. (A) Colonies of strains WN612 (fluorescent granite isolate; top) and WN613 (nonfluorescent granite isolate; bottom) after 2 days on solid LB medium. (B) Cell-free culture supernatants of 2-day cultures in liquid LB medium of B. subtilis HA101 (nonfluorescent reference strain; left), strain WN572 (fluorescent granite isolate; center), and strain WN613 (nonfluorescent granite isolate; right). Photographs were taken under medium-wavelength UV illumination.
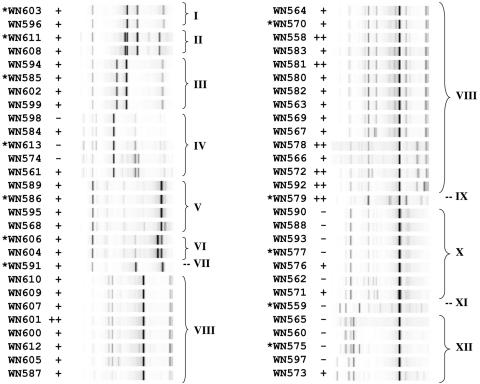
FIG. 3.
rep-PCR analysis of 56 endolithic Bacillus sp. isolates from granite. The rep-PCR gel image is shown to the right of the corresponding strain number. Bracketed groups I to XII denote strains sharing >93% homology in their rep-PCR patterns based on cluster analysis. Strains with asterisks were chosen as group representatives for 16S rRNA gene sequence determination. The production of blue fluorescence after 48 h on LB solid medium is denoted as follows: −, no fluorescence; +, weak to moderate fluorescence; ++, strong fluorescence.
rep-PCR grouping of endolithic granite and basalt isolates.
In order to classify the 56 granite isolates in a rapid and cost-effective manner, we decided to use rep-PCR as a relatively quick, high-throughput method capable of “fingerprinting” bacteria at the subspecies level (16, 57). Chromosomal DNAs from all 56 isolates were analyzed by rep-PCR, using the Diversilab system, and sorted using the proprietary web-based cluster analysis software provided with the system (Fig. 3). Cluster analysis revealed a complex set of relatedness of rep-PCR patterns among the granite isolates, suggesting a rich population diversity of spore-forming Bacillus spp. within the granite sample. We arbitrarily decided to group isolates whose rep-PCR patterns were at least 93% similar, which resulted in division of the 56 isolates into 12 groups, designated groups I through XII (Fig. 3). We attempted to assess the concordance of blue fluorescence with the rep-PCR groupings, with a moderate amount of success. All members of groups I, II, III, V, VI, VII, VIII, and IX displayed blue fluorescence, while groups IV, X, XI, and XII contained mostly nonfluorescent members, with a few exceptions (Fig. 3).
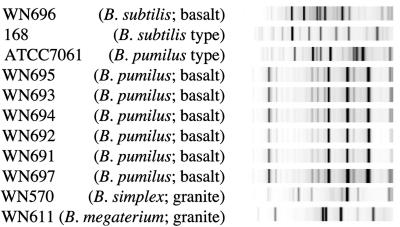
In a previous communication, we reported the isolation of endolithic Bacillus spores from the interior of Sonoran desert basalt; seven total isolates were obtained and characterized using randomly amplified polymorphic DNA PCR (RAPD-PCR) and 16S rRNA gene sequencing (3). Six of the isolates were identical by RAPD-PCR and 16S rRNA gene sequencing and were closely related (99.9%) by 16S rRNA gene analysis to B. pumilus. The seventh isolate, WN696, was shown to be most closely related (99.1%) by 16S rRNA gene analysis to B. subtilis but was distinct by RAPD-PCR and 16S rRNA gene analysis from B. subtilis laboratory strains (3). Because the RAPD-PCR technique used in that experiment resulted in a rather low resolution (only two visible RAPD-PCR bands), we reexamined the basalt isolates using the higher-resolution technique of rep-PCR. Chromosomal DNAs of the seven endolithic Bacillus spp. previously isolated from basalt (3) were also subjected to rep-PCR analysis. All of the basalt isolates exhibited rep-PCR patterns which were completely distinct from those of the granite isolates (Fig. 4). It was observed that six of the isolates from basalt identified by 16S rRNA gene sequencing as B. pumilus (WN691-695 and WN697) exhibited rep-PCR patterns that were visually indistinguishable from each other and were >97% similar by cluster analysis but were readily distinguished from the B. pumilus type strain, ATCC 7061 (Fig. 4). These observations are consistent with the notion that the isolates are likely siblings originating from the same microcolony within the basaltic rock (3). Furthermore, the rep-PCR patterns of the B. pumilus isolates were easily distinguishable from that of the sole B. subtilis isolate from basalt, strain WN696 (Fig. 4). Furthermore, WN696 itself was easily distinguishable from the typical laboratory B. subtilis strain 168 (Fig. 4). Thus, both the absolute numbers and the diversity of Bacillus sp. spores from granite were considerably higher than those from basalt.
FIG. 4.
rep-PCR analysis of seven endolithic Bacillus sp. isolates from basalt (3). Strain WN696 was most closely related to B. subtilis, and strains WN691-695 and WN697 were most closely related to B. pumilus by 16S rRNA gene analysis (3). Isolates WN570 (B. simplex) and WN611 (B. megaterium) from granite and type strains B. subtilis 168 and B. pumilus ATCC 7061 were included for comparison.
Comparison of 16S rRNA gene sequences from granite isolates.
One representative granite isolate from each of groups I to XII was chosen for 16S rRNA gene sequencing, and each 16S rRNA gene sequence obtained was analyzed using the RDP web-based software. In our analysis, we used the criterion that bacteria sharing ≥97.5% identity in their 16S rRNA gene sequences belong to the same species (59). It is well established that rep-PCR distinguishes strains below the species level (16, 57). Because multiple groups were found whose rep-PCR patterns were <93% similar but which were essentially the same species at the level of the 16S rRNA gene sequence (see below), we assumed that the unsequenced members of each group were also of the same species as the sequenced representative. Preliminary classification of 16S rRNA genes confirmed that all 12 sequenced individuals belonged to the genus Bacillus. Using the criterion outlined above, it was determined that the 15 isolates from groups I to III and V to VII were strains of B. megaterium (99.2 to 100.0% identity), the 23 isolates from groups VIII and IX were strains of B. simplex (99.6 to 100.0% identity), and the 5 isolates from group XII were strains of B. drentensis (98.6% identity) (Table 1). In addition, the seven isolates from group X were most closely similar to B. niacini (95.1% identity), and the single isolate from group XI was most similar to B. drentensis (96.2% identity); however, both of these values were slightly below the 97.5% cutoff for assignment to the same species (59). The five isolates from group IV were intriguing, as a search of the RDP revealed that these strains were most closely related to B. barbaricus; however, the degree of identity was low, at 87.1% (Table 1). A search of the RDP with the 16S rRNA gene sequence from the group IV representative did reveal two closely related (97.1 to 97.5%) uncharacterized Bacillus spp. isolated from deep-sea sediments in the Mariana Trench (10,897-m depth) (51). Thus, these three isolates may represent a new Bacillus species.
Relationship of granite isolates to environmental Bacillus spp.
During our search of the RDP for 16S rRNA gene homologies to our 56 granite isolates, we were struck by the repeated number of high-scoring hits to Bacillus spp. deposited in the RDP which have also been isolated from rocks (Table 2). For example, the granite isolates described here for every group except group XI were found to be closely related in 16S rRNA gene sequence to Bacillus sp. isolates from biodeteriorated murals, stone churches, tombs, and experimental wall paintings in Europe (17, 50); an underground cavern in Arizona (L. Ikner et al., unpublished data); and concretions of siderite (i.e., the mineral iron carbonate) from India (48). Furthermore, granite isolates from all groups except groups IV and XI were also closely related in 16S rRNA gene sequence to Bacillus spp. previously isolated from extreme environments such as Antarctic soils (M. Rodriguez-Diaz et al., unpublished data), sediment from the deep-ocean Mariana Trench (51), and ultraclean spacecraft assembly facilities (54; K. Venkateswaran et al., unpublished data) (Table 2).
TABLE 2.
Relationships of granite isolates to environmental Bacillus isolates
| Group(s) | Species | Strain | Accession no. | Environmental origin | % Identity | Reference |
|---|---|---|---|---|---|---|
| I, II, III, V, VI, VII | Bacillus sp. | R-16769 | AJ748259 | Soil, Mars oasis, Antarctica | 99.1-100 | Rodriguez-Diaz et al., unpublished |
| B. megaterium | SAFN-002 | AY167824 | SAF | 98.6 | Venkateswaran, unpublished | |
| SAFR-038 | AY167862 | SAF | 98.6 | |||
| SAFR-011 | AY167865 | SAF | 98.9-99.3 | |||
| SAFR-040 | AY167861 | SAF | 98.7-99.3 | |||
| Bacillus sp. | LMG20240 | AJ316310 | Biodeteriorated mural painting | 98.9-100 | 17 | |
| LMG19497 | AJ315065 | Biodeteriorated mural painting | 98.9-100 | |||
| Bacillus sp. | L2 (Kartchner) | DQ192209 | Kartchner Caverns, AZ | 98.6 | Ikner et al., unpublished | |
| B. megaterium | KL-197 | AY030338 | SAF | 99.1-99.4 | 54 | |
| IV | B. barbaricus | VII-B3-A2 (type strain) | AJ422145 | Experimental wall painting | 87.1 | 50 |
| B. decolorationis | LMG19507 (type strain) | AJ315075 | Biodeteriorated mural painting | 78.9 | Heyrman et al., unpublished | |
| B. arsenicus | con a/3 | AJ606700 | Siderite concretion | 82.3 | 48 | |
| Bacillus spp. | HTA437 | AB002642 | Sea floor sediment, Mariana trench | 97.5 | 51 | |
| HTA506 | AB002643 | Sea floor sediment, Mariana trench | 97.1 | |||
| VIII, IX | B. simplex | LMG19489 | AJ628744 | Biodeteriorated mural painting | 98.5-98.6 | Heyrman et al., unpublished |
| LMG21002 | AJ628745 | Biodeteriorated mural painting | 99.5-99.6 | |||
| LMG17633 | AJ628746 | Biodeteriorated mural painting | 99.0-100 | |||
| LMG17636 | AJ628747 | Biodeteriorated mural painting | 99.0-100 | |||
| Bacillus spp. | LMG19491 | AJ315057 | Biodeteriorated mural painting | 99.2 | 17 | |
| LMG19494 | AJ315062 | Biodeteriorated mural painting | 99.1 | |||
| Bacillus spp. | HI-D3 | DQ196470 | Kartchner Caverns, AZ | 99.7 | Ikner et al., unpublished | |
| MI-qb2 | DQ196481 | 99.2 | ||||
| X | B. niacini | SAFN-019 | AY167811 | SAF | 94.0 | Venkateswaran, unpublished |
| Bacillus spp. | LMG19498 | AJ315066 | Biodeteriorated mural painting | 94.6 | 17 | |
| LMG20241 | AJ316313 | Biodeteriorated mural painting | 94.0 | |||
| XI | Bacillus spp. | Various | Various | Soils, sediments, tap water biofilms | 94.1-97.2 | RDP |
| XII | B. niacini | SAFN-019 | AY167811 | SAF | 94.6 | Venkateswaran, unpublished |
Spore UV resistance of endolithic Bacillus isolates.
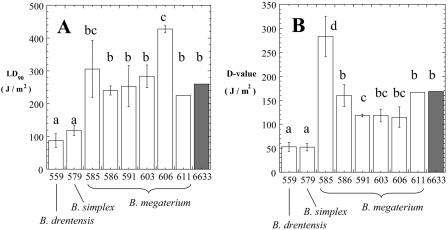
A number of environmental Bacillus spp. have been isolated, both from within rock (3) and from within the extremely clean environments of spacecraft and spacecraft assembly facilities (26, 56) (Table 2), which produce spores exhibiting elevated resistance to sporicidal treatments in comparison to standard spore dosimeters (25, 27, 55). We are particularly interested in extreme spore resistance to UV, as it relates to spore survival in response to both clean room disinfection treatments and spore exposure to the space environment during interplanetary transport (reviewed in references 41 and 43). We therefore undertook the measurement of spore UV resistance from the representative isolates from groups I to XII that had been characterized by 16S rRNA gene sequencing (Table 1). Despite the fact that all 12 representative strains had been originally isolated as heat-resistant spores from granite suspensions, four isolates failed to produce spores under our standard laboratory conditions, i.e., cultivation for several days on SSM plates at either 37°C or room temperature (Table 1). The eight remaining representative strains sporulated on SSM plates, and purified spores of each strain were tested for resistance to 254-nm UV radiation in comparison to spores of the standard UV dosimetry strain B. subtilis ATCC 6633 (22, 40). Spore UV inactivation curves characteristically exhibit a “shoulder” at low UV doses followed by an exponential decline in viability at higher UV doses. To quantify spore resistance properties, two parameters, the LD90 value and the D value, are often used (for extensive discussion of these parameters, see references 3, 27, 38, and 40). The LD90 value is defined as the lethal dose for 90% of the population and mostly reflects the length of the “shoulder” of the curve. The D (decimal reduction) value, defined as the dose producing 1 log10 of inactivation, is derived from the exponential portion of the inactivation curve and reflects its slope (24).
The spore UV resistance of the strains tested appeared to fall into four general categories (Fig. 5), as follows. (i) Spores of granite isolates WN559 (group XI, B. drentensis) and WN579 (group IX, B. simplex) displayed very similar levels of UV resistance, and both strains were significantly more UV sensitive than the reference strain ATCC 6633, using either the LD90 (Fig. 5A) or the D value (Fig. 5B) as a criterion. (ii) Spores of granite isolates WN586 (group V), WN591 (group VII), WN603 (group I), and WN611 (group II), all classified as B. megaterium, exhibited LD90 values similar to that of ATCC 6633 (Fig. 5A) and D values either similar to or marginally lower than that of ATCC 6633 (Fig. 5B). (iii) Granite isolate WN585 (group III), classified as B. megaterium, exhibited a significantly higher D value than ATCC 6633 but had a similar LD90 (Fig. 5). (iv) Granite isolate WN606 (group VI), also classified as B. megaterium, displayed the reverse, having a significantly higher LD90 value than ATCC 6633 but a similar D value (Fig. 5). Therefore, of the eight granite isolates tested, only WN585 and WN606 could justifiably be considered to produce spores with more UV resistance than the reference B. subtilis strain; furthermore, even the most UV-resistant granite isolates did not approach the extreme spore UV resistance exhibited by B. pumilus isolates from basalt (3) or spacecraft assembly facilities (27).
FIG. 5.
Spore UV resistance. The graphs show comparisons of LD90 values (A) and D values (B) of granite isolates (white bars) (numbers denote strain numbers; see Table 1) and the UV dosimetry strain B. subtilis ATCC 6633 (6633; shaded bars). Bars and error bars represent averages and standard deviations, respectively (n = 3 or 4). Strains with P values of <0.05 by analysis of variance were judged as belonging to separate groups (indicated with lowercase letters).
In conclusion, as part of our investigation of endolithic Bacillus spores, we report the isolation and characterization of spores found inside basalt (3) and granite (this study) from the extreme environment of the Sonoran desert. The location of these rocks just below the Earth's surface was deliberately chosen to represent sites potentially available for natural launch into space by hypervelocity impacts according to the spallation model of lithopanspermia theory (7, 11, 19, 33, 41, 43). We describe here the discovery of a likely new taxon represented by group IV isolates, and we report the discovery of a previously unknown blue fluorescent compound produced by granite-inhabiting Bacillus spp. We found that granite appeared to harbor larger numbers and a richer diversity of cultivable Bacillus spp. than basalt. We suggest that this may be due to two factors. First, it has been demonstrated that bacteria can infiltrate porous rock in association with the percolation of groundwater (23a). The lower porosity of basalt (3 to 35% void space) than that of granite (34 to 57% void space) (34a) may limit the infiltration of microorganisms into basalt (39). Second, the Sonoran desert is characterized by extreme solar radiation and heating; the darker color of near-surface basalt may result in stronger diurnal heating cycles, limiting microbial survival.
A question naturally arising from our observations is whether the Bacillus spp. found were actually living (i.e., undergoing metabolism and growing) inside these endolithic environments or were merely accidental inhabitants. Our observation that six B. pumilus spores inhabiting basalt were likely siblings (Fig. 4) (3) suggests either replication within the rock or, alternatively, transport into the rock as a group. Certainly, the current study did not set out to answer this question, as assays of in situ metabolism were not performed. Indeed, we specifically selected heat-resistant (and presumably dormant) spores as part of the isolation procedure. However, prior evidence has been obtained of Bacillus spp. making their living in isolated deep subsurface endolithic environments (4). Furthermore, as of 2003, well over 200 distinct species of aerobic endospore-forming bacteria have been described from a wide variety of environments (13). In contrast, it is striking that only a limited subset of apparently closely related Bacillus spp. has been repeatedly isolated from rock environments distributed worldwide (Table 2). These observations strongly suggest either (i) that these Bacillus spp. are indeed living within the rock matrix itself or (ii) that there exists some unknown mechanism by which only these species become accidentally and preferentially transported into, or survive within, rock interiors. In either case, it appears that the endolithic environment creates niches that exclude all but a few Bacillus spp. Understanding the mechanisms by which this group of microbes infiltrates, colonizes, and lives within rock habitats will yield important insights, with impacts on fields as disparate as astrobiology, geomicrobiology, microbial ecology, and biodeterioration.
Acknowledgments
We thank Heather Maughan and Terry Hurford for technical assistance and Savita Shankar at the UF ICBR for 16S rRNA gene sequencing.
This work was supported by grants (NCC2-1342 and NNA04CI35A) from the NASA Exobiology program to P.F.-C. and W.N.
REFERENCES
- 1.Andrews, M., H. J. Sun, and K. H. Nealson. 2002. Novel bacterial 16S rRNA sequences from a marine endolithic community, p. 333. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, D.C.
- 2.Balkwill, D. L., R. H. Reeves, G. R. Drake, J. Y. Reeves, F. H. Crocker, M. B. King, and D. R. Boone. 1997. Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol. Rev. 20:201-216. [DOI] [PubMed] [Google Scholar]
- 3.Benardini, J. N., J. Sawyer, K. Venkateswaran, and W. L. Nicholson. 2003. Spore UV and acceleration resistance of endolithic Bacillus pumilus and B. subtilis isolates obtained from Sonoran desert basalt: implications for lithopanspermia. Astrobiology 3:709-717. [DOI] [PubMed] [Google Scholar]
- 4.Boone, D. R., Y. Liu, Z.-J. Zhao, D. L. Balkwill, G. R. Drake, T. O. Stevens, and H. C. Aldrich. 1995. Bacillus infernus sp. nov., an Fe(III)- and Mn(IV)-reducing anaerobe from the deep terrestrial subsurface. Int. J. Syst. Bacteriol. 45:441-448. [DOI] [PubMed] [Google Scholar]
- 5.Burchell, M. J., J. R. Mann, and A. W. Bunch. 2004. Survival of bacteria and spores under extreme shock pressures. Mon. Not. R. Astron. Soc. 352:1273-1278. [Google Scholar]
- 6.Cano, R. J., and M. K. Borucki. 1995. Revival and identification of bacterial spores in 25- to 40-million-year-old Domican amber. Science 268:1060-1064. [DOI] [PubMed] [Google Scholar]
- 6a.Castro, V. A., A. N. Thrasher, M. Healy, C. M. Ott, and D. L. Pierson. 2004. Microbial diversity aboard spacecraft: evaluation of the International Space Station. Microb. Ecol. 47:119-126. [DOI] [PubMed] [Google Scholar]
- 7.Clark, B. 2001. Planetary interchange of bioactive material: probability factors and implications. Orig. Life Evol. Biosphere 31:185-197. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fajardo-Cavazos, P., L. Link, H. J. Melosh, and W. L. Nicholson. 2004. Testing lithopanspermia theory: survival of bacterial spores in granite to forces generated during impact-driven launch and high-velocity atmospheric entry, p. 74. In Proceedings of the 10th International Symposium on Microbial Ecology (ISME-10). International Society for Microbial Ecology, Geneva, Switzerland.
- 10.Fajardo-Cavazos, P., L. Link, H. J. Melosh, and W. L. Nicholson. 2005. Bacillus subtilis spores in simulated meteorite survive hypervelocity atmospheric entry: implications for lithopanspermia. Astrobiology 5:726-736. [DOI] [PubMed] [Google Scholar]
- 11.Fajardo-Cavazos, P., A. C. Schuerger, and W. L. Nicholson. Testing interplanetary transfer of bacteria by natural impact phenomena and human spaceflight activities. Acta Astronautica, in press.
- 12.Flores, M., J. Lorenzo, and G. Gomez-Alarcón. 1997. Algae and bacteria on historic monuments at Alcala de Henares, Spain. Int. Biodeterior. Biodegrad. 40:241-246. [Google Scholar]
- 13.Fritze, D. 2004. Taxonomy and systematics of the aerobic endospore forming bacteria: Bacillus and related genera, p. 17-34. In E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spore formers: probiotics and emerging applications. Horizon Scientific Press, Norfolk, United Kingdom.
- 14.Gladman, B. 1997. Destination: Earth. Martian meteorite delivery. Icarus 130:228-246. [Google Scholar]
- 15.Gomez-Alarcón, G., J. Lorenzo, and Y. B. Cilleros. 1995. Weathering factors of granite in the building of the Royal Academy of Pharmacy. Anales Real Acad. Farm. 61:373-389. [Google Scholar]
- 16.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyrman, J., and J. Swings. 2001. 16S rDNA sequence analysis of bacterial isolates from biodeteriorated mural paintings in the Servilia tomb (Necropolis of Carmona), Seville, Spain. Appl. Microbiol. 24:417-422. [DOI] [PubMed] [Google Scholar]
- 18.Horneck, G., H. Bücker, and G. Reitz. 1994. Long-term survival of bacterial spores in space. Adv. Space Res. 14:41-45. [DOI] [PubMed] [Google Scholar]
- 19.Horneck, G., C. Miliekowsky, H. J. Melosh, J. W. Wilson, F. A. Cucinotta, and B. Gladman. 2002. Viable transfer of microorganisms in the solar system and beyond, p. 55-76. In G. Horneck and C. Baumstark-Khan (ed.), Astrobiology: the quest for the conditions of life. Springer, Berlin, Germany.
- 20.Horneck, G., P. Rettberg, G. Reitz, J. Wehner, U. Eschweiler, K. Strauch, C. Panitz, V. Starke, and C. Baumstark-Khan. 2001. Protection of bacterial spores in space, a contribution to the discussion on panspermia. Orig. Life Evol. Biosphere 31:527-547. [DOI] [PubMed] [Google Scholar]
- 21.Horneck, G., D. Stöffler, U. Eschweiler, and U. Hornemann. 2001. Bacterial spores survive simulated meteorite impact. Icarus 149:285-290. [Google Scholar]
- 22.Hoyer, O. 2000. The status of UV technology in Europe. Int. UV Assoc. News 2:22-27. [Google Scholar]
- 23.Hungate, B., A. Danin, N. B. Pellerin, J. Stemmler, P. Kjellander, J. B. Adams, and J. T. Staley. 1987. Characterization of manganese-oxidizing (manganese II to manganese IV) bacteria from Negev Desert (Israel) rock varnish: implication in desert varnish formation. Can. J. Microbiol. 33:939-943. [Google Scholar]
- 23a.Jang, L. K., P. W. Chang, J. E. Findley, and T. F. Yen. 1983. Selection of bacteria with favorable transport properties through porous rock for the application of microbial-enhanced oil recovery. Appl. Environ. Microbiol. 46:1066-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joslyn, L. 1983. Sterilization by heat, p. 3-46. In S. Block (ed.), Disinfection, sterilization, and preservation. Lea & Febiger, Philadelphia, Pa.
- 25.Kempf, M. J., F. Chen, R. Kern, and K. Venkateswaran. 2005. Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5:391-405. [DOI] [PubMed] [Google Scholar]
- 26.La Duc, M. T., W. Nicholson, R. Kern, and K. Venkateswaran. 2003. Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ. Microbiol. 5:977-985. [DOI] [PubMed] [Google Scholar]
- 27.Link, L., J. Sawyer, K. Venkateswaran, and W. L. Nicholson. 2004. Extreme spore UV resistance of Bacillus pumilus isolates obtained from an ultra-clean spacecraft assembly facility. Microb. Ecol. 47:159-163. [DOI] [PubMed] [Google Scholar]
- 28.Magyarosy, A., J. Z. Ho, H. Rapoport, S. Dawson, J. Hancock, and J. D. Keasling. 2002. Chlorxanthomycin, a fluorescent, chlorinated, pentacyclic pyrene from a Bacillus spp. Appl. Environ. Microbiol. 68:4095-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastrapa, R. M. E., H. Glanzberg, J. N. Head, H. J. Melosh, and W. L. Nicholson. 2001. Survival of bacteria exposed to extreme acceleration: implications for panspermia. Earth Planet. Sci. Lett. 189:1-8. [Google Scholar]
- 30.Melosh, H. J. 1988. The rocky road to panspermia. Nature 332:687-688. [DOI] [PubMed] [Google Scholar]
- 31.Melosh, H. J. 1989. Impact cratering: a geologic process. Oxford University Press, New York, N.Y.
- 32.Meyer, C. 2003. Mars meteorite compendium—2003. NASA publication JSC#27672. [Online.] http://www-curator.jsc.nasa.gov/antmet/mmc/index.cfm.
- 33.Mileikowsky, C., F. A. Cucinotta, J. W. Wilson, B. Gladman, G. Horneck, L. Lindegren, H. J. Melosh, H. Rickman, M. Valtonen, and J. Q. Zheng. 2000. Natural transfer of viable microbes in space. 1. From Mars to Earth and Earth to Mars. Icarus 145:391-427. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34a.Morris, D. A., and A. I. Johnson. 1967. Summary of hydrologic and physical properties of rock and soil materials as analyzed by the Hydrologic Laboratory of the U.S. Geological Survey. Water supply paper 1839-D. U.S. Geological Survey, Reston, Va.
- 35.Nagy, B., L. A. Nagy, M. J. Rigaly, W. D. Jones, D. H. Krinsley, and N. A. Sinclair. 1991. Rock varnish in the Sonoran desert: microbiologically mediated accumulation of manganiferous sediments. Sedimentology 38:1153-1171. [Google Scholar]
- 36.Nealson, K. H., and J. Ford. 1980. Surface enhancement of bacterial manganese oxidation: implications for aquatic environments. Geomicrobiol. J. 2:21-37. [Google Scholar]
- 37.Nicholson, W. L. 2002. Roles of Bacillus spores in the environment. Cell. Mol. Life Sci. 59:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson, W. L. 2003. Using thermal inactivation kinetics to determine bacterial spore longevity: implications for paleomicrobiology and lithopanspermia. Orig. Life Evol. Biosphere 33:621-631. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson, W. L. 2004. Ubiquity, longevity, and ecological roles of Bacillus spores, p. 1-15. In E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spore formers: probiotics and emerging applications. Horizon Scientific Press, Norfolk, United Kingdom.
- 40.Nicholson, W. L., and B. Galeano. 2003. UV resistance of Bacillus anthracis spores revisited: validation of Bacillus subtilis spores as UV surrogates for spores of B. anthracis Sterne. Appl. Environ. Microbiol. 69:1327-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of bacterial endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. J. Wiley and Sons, New York, N.Y.
- 43.Nicholson, W. L., A. C. Schuerger, and P. Setlow. 2005. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat. Res. 571:249-264. [DOI] [PubMed] [Google Scholar]
- 44.Okubo, S., and T. Yanagida. 1968. Isolation of a suppressor mutant in Bacillus subtilis. J. Bacteriol. 95:1187-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer, F. E., J. T. Staley, R. G. E. Murray, T. Counsell, and J. B. Adams. 1986. Identification of manganese-oxidizing bacteria from desert varnish. Geomicrobiol. J. 4:343-360. [Google Scholar]
- 46.Rummel, J. D. 2001. Planetary exploration in the time of astrobiology: protecting against biological contamination. Proc. Natl. Acad. Sci. USA 98:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivaji, S., K. Suresh, P. Chaturvedi, S. Dube, and S. Sengupta. 2005. Bacillus arsenicus sp. nov., an arsenic-resistant bacterium isolated from a siderite concretion in West Bengal, India. Int. J. Syst. Evol. Microbiol. 55:1123-1127. [DOI] [PubMed] [Google Scholar]
- 49.Stassi, A., E. Zanardini, F. Cappitelli, A. Schiraldi, and C. Sorlini. 1998. Calorimetric investigations on the metabolism of Bacillus strains isolated from artistic stoneworks. Ann. Microbiol. Enzymol. 48:111-120. [Google Scholar]
- 50.Taeubel, M., P. Kaempfer, S. Buczolitz, W. Lubitz, and H. J. Busse. 2003. Bacillus barbaricus sp. nov., isolated from an experimental wall painting. Int. J. Syst. Evol. Microbiol. 53:725-730. [DOI] [PubMed] [Google Scholar]
- 51.Takami, H., A. Inoue, F. Fuji, and K. Horikoshi. 1997. Microbial flora in the deepest sea mud of Mariana Trench. FEMS Microbiol. Lett. 152:279-285. [DOI] [PubMed] [Google Scholar]
- 52.Turtura, G. C., A. Perfetto, and P. Lorenzelli. 2000. Microbiological investigation on black crusts from open-air stone monuments of Bologna (Italy). New Microbiol. 23:207-228. [PubMed] [Google Scholar]
- 53.Reference deleted.
- 54.Venkateswaran, K., N. Hattori, M. T. La Duc, and R. Kern. 2003. ATP as a biomarker of viable microorganisms in clean-room facilities. J. Microbiol. Methods 52:367-377. [DOI] [PubMed] [Google Scholar]
- 55.Venkateswaran, K., M. Kempf, F. Chen, M. Satomi, W. L. Nicholson, and R. Kern. 2003. Bacillus nealsonii sp. nov., isolated from a spacecraft-assembly facility, whose spores are gamma-radiation resistant. Int. J. Syst. Evol. Microbiol. 53:165-172. [DOI] [PubMed] [Google Scholar]
- 56.Venkateswaran, K., M. Satomi, S. Chung, R. Kern, R. Koukol, C. Basic, and D. White. 2001. Molecular microbial diversity of a spacecraft assembly facility. Syst. Appl. Microbiol. 24:311-320. [DOI] [PubMed] [Google Scholar]
- 57.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vreeland, R. H., W. D. Rosenzweig, and D. W. Powers. 2000. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407:897-900. [DOI] [PubMed] [Google Scholar]
- 59.Wayne, L., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, et al. 1987. International Committee on Systematic Bacteriology: report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]