Abstract
The biosynthesis of the β-lactam antibiotic penicillin in the filamentous fungus Aspergillus nidulans is catalyzed by three enzymes that are encoded by the acvA, ipnA, and aatA genes. A variety of cis-acting DNA elements and regulatory factors form a complex regulatory network controlling these β-lactam biosynthesis genes. Regulators involved include the CCAAT-binding complex AnCF and AnBH1. AnBH1 acts as a repressor of the penicillin biosynthesis gene aatA. Until now, however, little information has been available on the signal transduction cascades leading to the transcription factors. Here we show that inhibition of protein kinase C (Pkc) activity in A. nidulans led to cytoplasmic localization of an AnBH1-enhanced green fluorescent protein (EGFP) fusion protein. Computer analysis of the genome and screening of an A. nidulans gene library revealed that the fungus possesses two putative Pkc-encoding genes, which we designated pkcA and pkcB. Only PkcA showed all the characteristic features of fungal Pkc's. Production of pkcA antisense RNA in A. nidulans led to reduced growth and conidiation in Aspergillus minimal medium, while in fermentation medium it led to enhanced expression of an aatAp-lacZ gene fusion, reduced pencillin production, and predominantly cytoplasmic localization of AnBH1. These data agree with the finding that inhibition of Pkc activity prevented nuclear localization of AnBH1-EGFP. As a result, repression of aatA expression was relieved. The involvement of Pkc in penicillin biosynthesis is also interesting in light of the fact that in the yeast Saccharomyces cerevisiae, Pkc plays a major role in maintaining cell integrity.
Penicillins and cephalosporins belong chemically to the group of β-lactam antibiotics. The biosyntheses of both penicillins and cephalosporins have the first two steps in common. They are synthesized from the same three amino acids, l-α-aminoadipic acid (l-α-AAA), l-cysteine, and l-valine (reviewed in reference 9). In the first reaction of the cephalosporin and penicillin biosynthesis pathway, the amino acid precursors are condensed to the tripeptide δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine (ACV). This reaction is catalyzed by a single enzyme, ACV synthetase. ACV synthetase is encoded by a single structural gene designated acvA (pcbAB). In the second step, oxidative ring closure of the linear tripeptide leads to formation of a bicyclic ring, i.e., the four-membered β-lactam ring fused to the five-membered thiazolidine ring which is characteristic of all penicillins. This reaction is catalyzed by isopenicillin N synthase (IPN synthase), encoded by the ipnA (pcbC) gene. IPN is the branch point of penicillin and cephalosporin biosyntheses. In the third and final step of penicillin biosynthesis, the hydrophilic l-α-AAA side chain of IPN is exchanged for a hydrophobic acyl group catalyzed by acyl coenzyme A (acyl-CoA):isopenicillin N acyltransferase (IAT). The corresponding gene was designated aatA (penDE) (reviewed in references 10 and 13). It is generally believed that the activated forms of the side chains consist of their CoA-thioesters, but the mechanism behind this activation is still not fully elucidated (19). The formation of hydrophobic penicillins has been reported in fungi only, notably Penicillium chrysogenum and Aspergillus nidulans, whereas the hydrophilic cephalosporins are produced by both fungi and bacteria, e.g., Acremonium chrysogenum and Streptomyces clavuligerus, respectively (reviewed in reference 13).
Within the last few years, several studies have indicated that the β-lactam biosynthesis genes of fungi are controlled by a complex regulatory network, and a comparison with known regulators and DNA elements involved in the regulation of genes of primary metabolism is of interest. A variety of cis-acting DNA elements and regulatory factors are involved. In A. nidulans, among others, these regulators include AnCF and AnBH1. AnCF was shown to bind to a CCAAT box (I) in the intergenic region between the acvA and ipnA genes and to a CCAAT box (II) present in the promoter of the A. nidulans aatA gene. AnCF consists of the subunits HapB, HapC, and HapE (27, 45). AnBH1 belongs to the family of basic-region helix-loop-helix (bHLH) transcription factors. AnBH1 binds in vitro as a homodimer to an asymmetric E-box (not previously described) within the aatA promoter which overlaps with the AnCF binding site (CCAAT box II). Since deletion of anbH1 appeared to be lethal, the anbH1 gene was replaced by a regulatable alcAp-anbH1 gene fusion. Analysis of aatAp-lacZ expression in such a strain indicated that AnBH1 acts as a repressor of aatA gene expression and therefore counteracts the positive action of AnCF (15). Until now, however, little information has been available on the signal transduction cascades leading to the transcription factors. Based on the amino acid sequence of AnBH1 and because all bHLH transcription factors were shown to be phosphorylated (25), it was likely that AnBH1 was phosphorylated. Therefore, analysis of phosphorylation and putative kinases involved might give hints on the signal transduction cascades controlling the biosynthesis of the secondary metabolite penicillin. Here we provide evidence that the protein kinase C (Pkc) PkcA is involved directly or indirectly in the regulation of AnBH1 and penicillin biosynthesis.
MATERIALS AND METHODS
Strains and plasmids.
Fungal and bacterial strains and plasmids used in this study are listed in Table 1. Vectors and plasmids were propagated in Escherichia coli TOP10F′. For generation of A. nidulans strain C2.3, strain R21 was crossed with strain LOGOAnBH1, and one of the resulting progeny was crossed with strain OLAB1URA (Table 1). Ascospores were plated on Aspergillus minimal medium (AMM) (see below) agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 μg ml−1) and glucose as the carbon source. Uracil auxotrophic colonies staining the agar blue, which is indicative of the presence of the aatAp-lacZ gene fusion, were used. One of the resulting progeny with the desired genotype was designated C2.3 (Table 1).
TABLE 1.
Fungal strains, bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| Aspergillus nidulans | ||
| R21 | pabaA1 yA2 | 20 |
| LOGOAnBH1 | pyrG89 biA1 fwA1 bga0 argB2::pOLAB1 (aatAp-lacZ) pRG1 pLOGO-AnBH1 (anbH1-egfp) ArgB+ PyrG+ | 29 |
| OLAB1URA | pyrG89 biA1 fwA1 bga0 argB2::pOLAB1 (aatAp-lacZ) ArgB+ | 15 |
| AXB4A | bga0 biA1 argB2::pAXB4A (acvAp-uidA ipnAp-lacZ) ArgB+ | 11 |
| HapB-eGFP | pyrG89 pabaA1 fwA1 bga0 argB2::pAXB4A (acvAp-uidA ipnAp-lacZ) pHapB-GFP (hapB-egfp) ArgB+ PyrG+ | 44 |
| Palp32.3 | pyrG89 pabaA1 fwA1 bga0 argB2::pAXB4A (acvAp-uidA, ipnAp-lacZ) ΔsuAprgA1 paba1p32 ArgB+ PyrG+ PabaA+ | A. Gehrke, unpublished data |
| R21OLAB7 | pabaA1 yA2 argB2::pOLAB1 (aatAp-lacZ) ArgB+; progeny from a cross of strains R21 and OLAB1URA | This study |
| C2.3 | pyrG89 biA1 fwA1 bga0 argB2::pOLAB1 (aatAp-lacZ) pLOGO-AnBH1 (anbH1-egfp) ArgB+ | This study |
| PkcAinv1.1, -1.2, -1.3, -1.4, -1.5 | pyrG89 fwA1 bga0 biA1 argB2::pOLAB1 (aatAp-lacZ); pLOGO-AnBH1 (anbH1-egfp) (pAL4-pkcAinv)n ArgB+ PyrG+ | This study |
| Escherichia coli TOP10F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC), φ80d/lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen, The Netherlands |
| Bacillus calidolactis C953 | 11 | |
| Plasmids | ||
| pCR2.1TOPO | Ampr | Invitrogen, The Netherlands |
| pAL4 | Amprpyr-4 alcAp | 52 |
| pAL4-pkcAinv | Amprpyr-4 alcAp-pkcAantisense | This study |
Media and cultivation of strains.
E. coli was grown in LB medium supplemented with ampicillin (50 μg ml−1). AMM was prepared as previously described (12). For growth under repressing conditions, glucose (0.8%, wt vol−1) was used as the carbon source. For growth under inducing conditions, lactose (1.6%, wt vol−1) was used as the carbon source, and 10 mM cyclopentanone was added to the medium. Another alcAp-inducing compound is threonine. However, threonine was not used, because it is consumed by the fungus, and therefore a constant concentration cannot be maintained. In addition, consumption of threonine changes the pH of the culture and consequently influences penicillin production (48). Fermentation experiments with A. nidulans were carried out in fermentation medium (FM) essentially as described previously (49). Experimental cultures were inoculated with 1 ml of freshly harvested and filtered spore suspensions (2 × 108 conidia ml−1) instead of using seed culture suspensions. Experimental cultures (20 ml of FM in 250-ml flasks) contained 4% (wt vol−1) lactose as the carbon source and 10 mM cyclopentanone under inducing conditions and 4% (wt vol−1) glucose under repressing conditions. Cultures were incubated for 72 h. If required, biotin (0.3 μg ml−1) or p-aminobenzoic acid (15 μg ml−1) was added to the medium.
Quantification of sporulation.
A 50-μl volume of spore suspension containing 1 × 105 conidia of freshly harvested and filtered spore suspensions was plated onto AMM agar plates containing either glucose (0.8%, wt vol−1) or lactose (1.6%, wt vol−1) plus 10 mM cyclopentanone. Because of the high number of conidia, the plates were covered by mycelium. Four plates for each strain and each condition were incubated for 3 days, and the conidia produced from each plate were harvested with 10 ml saline solution containing 2% (vol/vol) Tween 80 (Merck, Germany). Spore suspensions were filtered, and the number of conidia was counted using a Thoma chamber.
Penicillin bioassay.
Penicillin bioassays using “Bacillus calidolactis” C953 as an indicator organism were carried out as described previously (11).
Determination of dry weight.
Dry weights of cultures grown in AMM or FM were determined as previously described (11). Cultures were grown under both repressing (glucose) and inducing (lactose plus 10 mM cyclopentanone) conditions. Fermentation cultures were harvested after 24 h, 48 h, and 72 h. AMM cultures were harvested after 12 h, 24 h, 36 h, 48 h, and 60 h.
Genetic techniques.
Sexual crosses and characterization of the resulting progeny were performed according to Pontecorvo et al. (37).
Standard DNA techniques.
For small-scale preparation of A. nidulans chromosomal DNA, the technique of Raeder and Broda (39) was used. Standard techniques in the manipulation of DNA were used as detailed in Sambrook et al. (40). Southern blot analysis was performed essentially as previously described (12). Chromosomal DNA of A. nidulans was digested with KpnI and XbaI. DNA fragments were separated on an agarose gel and blotted onto Hybond N+ nylon membranes (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). Labeling of DNA probes with fluorescein-11-dUTP and detection of hybridized fragments by Southern blot analysis were carried out using the Gene Images random prime labeling module and CDP-Star detection reagent (both from Amersham Pharmacia Biotech, Germany), respectively, according to the manufacturer's instructions.
Generation of recombinant plasmids and probes for Southern blot analyses.
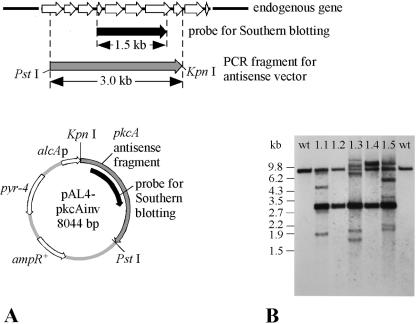
PCR products were cloned using the TOPO cloning kit (Invitrogen, Karlsruhe, Germany). For construction of the pkcA antisense RNA plasmid, a 3.0-kb DNA fragment covering about 80% of the pkcA coding sequence was amplified from genomic DNA using primers PkcAinv_5 and PkcAinv_3, the latter primer encoding a KpnI site (Table 2; Fig. 3A). The PCR fragment was cloned into the pCR2.1TOPO vector (Invitrogen, Karlsruhe, Germany). After restriction digestion with KpnI and PstI, the pkcA fragment was cloned into the KpnI-PstI-digested pAL4 vector (52) (Table 1), which contains the alcA promoter of A. nidulans and the pyr-4 gene of Neurospora crassa as a selectable marker. The resulting plasmid, pAL4-pkcAinv, thus contained a 2.9-kb fragment of the pkcA sequence in the antisense orientation under the control of the alcA promoter (Fig. 3A). Probes for Southern blot analysis were generated by PCR amplification of a 1.5-kb pkcA fragment using oligonucleotides PkcAnar_5 and PkcAnar_3 (Fig. 3A).
TABLE 2.
Oligonucleotide list
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| PkcAinv_5 | CGTCGAATATGCGACAGTCAACCG |
| PkcAinv_3 | GGGGTACCACAGAAGGTGCTTGTA |
| PkcAnar_5 | GGCGCCTCAGCAGGCTATAGCAGC |
| PkcAnar_3 | GGCGCCGAGGTTGAGAAGGAACGG |
| PkcAanti5 | ATCCATACCACATGTTCTCC |
| PkcAanti3 | ATTTTCCAGGTTCTATGCGG |
| PkcAanti33 | ATGGAGTATATCAGTGGTGG |
| PkcAintr7 | ATTTGGCGATACAACTGACG |
| Tef1forw | TTCCCCTCCAGGATGTCTACA |
| Tef1rev | TGCTGGTGGTGCATCTCAAC |
FIG. 3.
(A) Schematic map of the pkcA gene of A. nidulans and of the alcAp-pkcA-encoding antisense plasmid pAL4-pkcAinv. For the endogenous gene, exon (white arrows) and intron (black line) regions are indicated. ampR, ampicillin resistance gene. The pyr-4 gene of N. crassa was used as the selection marker gene in A. nidulans. (B) Southern blot analysis of five selected pkcA antisense transformants (PkcAinv1.1, PkcAinv1.2, PkcAinv1.3, PkcAinv1.4, and PkcAinv1.5, abbreviated as 1.1, 1.2, 1.3, 1.4, and 1.5, respectively) and two wild-type (wt) strains, R21OLAB7 (left) and AXB4A (right). Genomic DNA was digested with XbaI and KpnI. The probe was targeted against both the genomic site of the pkcA gene and the integration sites of the pkcA antisense RNA-encoding vector.
PCR amplification, DNA sequence analysis, and computer programs.
For PCR amplification, sequence-specific oligonucleotides were synthesized (Invitrogen, Karlsruhe, Germany). PCRs were carried out using BIOTAQ Red DNA polymerase (Bioline, Luckenwalde, Germany). The DNA fragments produced were purified using the NucleoSpin purification kit (Macherey-Nagel, Dueren, Germany). The sequences of DNA fragments were determined on both strands by the dideoxy chain termination method (41) using fluorescent dyes analyzed by an Applied Biosystems ABI 310 sequencer. DNA sequence data were edited by using the “Sequence Navigator” and “Auto Assembler” programs (Applied Biosystems, Darmstadt, Germany). Similarities between different amino acid sequences were analyzed using the ClustalW program (16) (http://www.ebi.ac.uk/clustalw).
Preparation of RNA.
Cultures of A. nidulans were incubated in AMM containing either glucose (repressing conditions) or lactose plus 10 mM cyclopentanone (inducing conditions) at 180 rpm and 37°C for 36 h. Mycelia were harvested and broken using liquid nitrogen as previously described (26). Total RNA was prepared using the RNeasy Plant minikit (QIAGEN, Hilden, Germany). The amount of RNA was determined both spectrophotometrically and by agarose gel electrophoresis. DNA remaining in the RNA solutions was digested with RNase-free DNase I (Roche, Mannheim, Germany). DNase I (2 μl; 10 U ml−1) was added to a 20-μl reaction volume containing 10 to 20 μg RNA, 4.4 μl 25 mM MgCl2, and 2 μl 10× TaqMan RT buffer (Applied Biosystems, Darmstadt, Germany). The reaction mixture was incubated at 37°C for 2 h, followed by heat denaturation at 95°C for 5 min. Control PCRs using primers Tef1forw and Tef1rev (Table 2) were carried out to ensure that the RNA extracts did not contain any DNA contamination.
Reverse transcription of RNA.
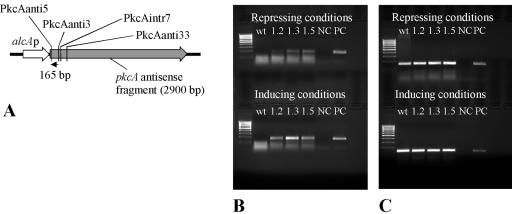
cDNA was created using the TaqMan reverse transcription reagents (Applied Biosystems, Darmstadt, Germany). Twenty-five-microliter reaction volumes contained 2.0 to 5.0 μg RNA, 2.5 μl 10× TaqMan RT buffer, 5.5 μl 25 mM MgCl2, 5 μl deoxynucleoside triphosphate mix, 3.0 μl of each reverse primer (1.2 pmol μl−1), 0.5 μl RNase inhibitor, and 31.25 U MultiScribe reverse transcriptase. A mixture of RNA and oligonucleotides was first incubated at 70°C for 10 min and then cooled to 4°C. After addition of the other reagents listed above, the reaction mixture was incubated at 48°C for 60 min. Finally, the enzyme was heat inactivated at 95°C for 5 min. Reverse transcription of pkcA antisense RNA was carried out using the specific primer PkcAanti3. cDNA of pkcA mRNA was generated using the specific primer PkcAanti5. cDNA of tef1 as an endogenous control was obtained using the specific primer Tef1rev (Table 2; Fig. 5A). Subsequent PCRs with cDNA as the template were carried out using oligonucleotides PkcAanti5 and PkcAanti3, or Tef1forw and Tef1rev. cDNAs obtained from both pkcA mRNA and pkcA antisense RNA were cloned (TA cloning kit; Invitrogen, Karlsruhe, Germany) and sequenced.
FIG. 5.
Detection of pkcA antisense RNA in A. nidulans transformant strains. (A) Scheme showing the strategy to detect antisense RNA by RT-PCR. The predicted antisense RNA was transcribed to cDNA using the specific primer PkcAanti3. The cDNA is indicated by a black arrow. Primers PkcAanti5 and PkcAanti3 were used in subsequent PCRs, yielding a 165-bp PCR product. The binding sites of the reverse primers PkcAintr7 and PkcAanti33, which were used in additional RT-PCR assays (see the text), are also indicated. (B) Detection of pkcA antisense RNA. RT-PCR was performed using primers specific for antisense RNA (primer PkcAanti3). Cultures for RNA extraction were grown either with glucose (repressing conditions) or with lactose plus 10 mM cyclopentanone (inducing conditions). wt, wild-type strain R21OLAB7; 1.2, 1.3, or 1.5, pkcA antisense-encoding transformant strain PkcAinv1.2, PkcAinv1.3, or PkcAinv1.5, respectively; NC, negative control (no template); PC, positive control (chromosomal DNA as the template). (C) Detection of tef1 RNA. RT-PCR was performed using primers specific for the tef1 gene (primer Tef1rev) as a control. Cultures for RNA extraction were grown with either glucose (repressing conditions) or lactose plus 10 mM cyclopentanone (inducing conditions).
Transformation of A. nidulans.
A. nidulans strain C2.3 was transformed to uracil prototrophy according to a method previously described (6). For transformation, plasmid DNA was purified using the E.Z.N.A. Plasmid Miniprep kit (peqlab Biotechnologie GmbH, Erlangen, Germany) according to the manufacturer's instructions.
β-Gal activity assays.
β-Galactosidase (β-Gal) activities were determined in crude extracts from mycelia grown in FM as previously described (49). β-Gal activities at each time point were measured in cell extracts from three simultaneously incubated cultures. Protein concentrations were determined according to Bradford (8) using Roti-Nanoquant (Roth, Karlsruhe, Germany). Specific activities of β-Gal were calculated as previously described (11).
Nucleotide sequence accession numbers.
The nucleotide sequence of A. nidulans pkcA and the deduced amino acid sequence were previously deposited at the NCBI GenBank (http://www.ncbi.nlm.nih.gov) under accession no. AB103459.1. The pkcB sequence was obtained by screening of an A. nidulans gene bank using a probe targeting the catalytic domain of protein kinase C and subsequent cloning and sequencing steps (data not shown). The sequence of the pkcB gene is part of the contigs ANI61C10640, ANI61C2064, ANI60C210894_1, ANI61C7279, and ANI61C9348. The nucleotide sequence of the pkcB mRNA and the deduced amino acid sequence were previously deposited as “Aspergillus nidulans FGSC A4 hypothetical protein” (AN5973.2; REFSEQ accession no. XM 658485). The nucleotide sequence of the A. nidulans tef1 cDNA and the deduced amino acid sequence are accessible at the NCBI GenBank (accession no. XM 408355).
RESULTS
Addition of Pkc inhibitor to culture medium led to localization of an AnBH1-EGFP fusion in the cytoplasm.
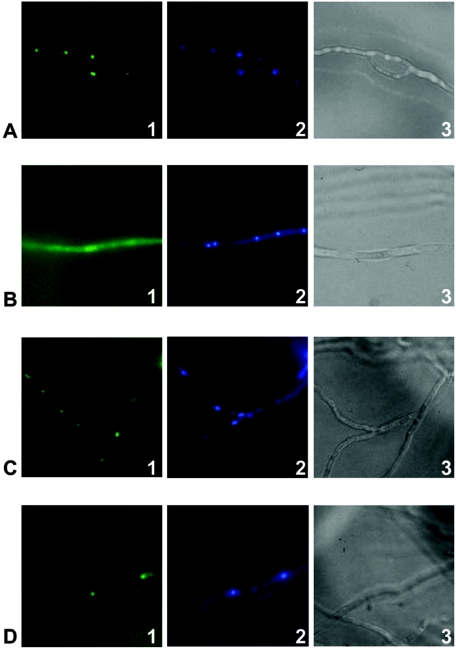
AnBH1 contains several putative phosphorylation motifs, making it likely that Pkc could be involved in the regulation of this transcription factor and consequently in penicillin biosynthesis. Therefore, A. nidulans strain LOGOAnBH1 was analyzed. This strain contains an anbH1-egfp gene fusion integrated into the genome (29). The strain was cultivated in AMM either with or without the Pkc inhibitor calphostin C. Results are shown in Fig. 1. Without the addition of the inhibitor, the AnBH1-enhanced green fluorescent protein (EGFP) fusion protein was found to be located in the nucleus (Fig. 1A). By contrast, addition of the Pkc inhibitor led to a decrease in the fluorescence in the nucleus, and additionally, fluorescence was visible in the cytoplasm, suggesting that AnBH1-EGFP either could not enter the nucleus any more or was actively exported from the nucleus (Fig. 1B). As a control, the nuclear localization of a HapB-EGFP fusion was studied. HapB does not contain an obvious phosphorylation site. As shown in Fig. 1C and D, the HapB-EGFP fusion protein of the A. nidulans strain HapB-eGFP was located in the nucleus irrespective of whether calphostin C was added to the culture medium. It was thus likely that Pkc influenced the nuclear localization of AnBH1.
FIG. 1.
Cellular localization of AnBH1-EGFP and HapB-EGFP fusion proteins. Strains LOGOAnBH1 (containing AnBH1-EGFP) and HapB-eGFP were incubated in AMM either with or without the Pkc inhibitor calphostin C. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (panels 2). Samples were analyzed by fluorescence microscopy (panels 1, showing localization of the fusion proteins, and panels 2, showing nuclei) or light microscopy (panels 3). (A and B) AnBH1-EGFP-derived fluorescence of A. nidulans strain LOGOAnBH1 without calphostin C (A) and after addition of calphostin C (B). (C and D) HapB-EGFP-derived fluorescence of A. nidulans strain HapB-eGFP without calphostin C (C) and after addition of calphostin C (D).
A. nidulans encodes two putative pkc genes.
Analysis of the A. nidulans database (http://www.broad.mit.edu/annotation/fungi/aspergillus/index.html) indicated that there are two putative Pkc-encoding genes, which we designated pkcA and pkcB. The pkcB gene was originally obtained by screening an A. nidulans gene library using a probe targeting the catalytic domain of protein kinase C (see Materials and Methods). Based on computer analysis, the putative pkcB gene consists of 2,224 nucleotides coding for 653 amino acids. The putative pkcA gene comprises 3,680 nucleotides encoding 1,059 amino acids. Most probably, the coding sequences of pkcB and pkcA are interrupted by 4 and 9 introns, respectively.
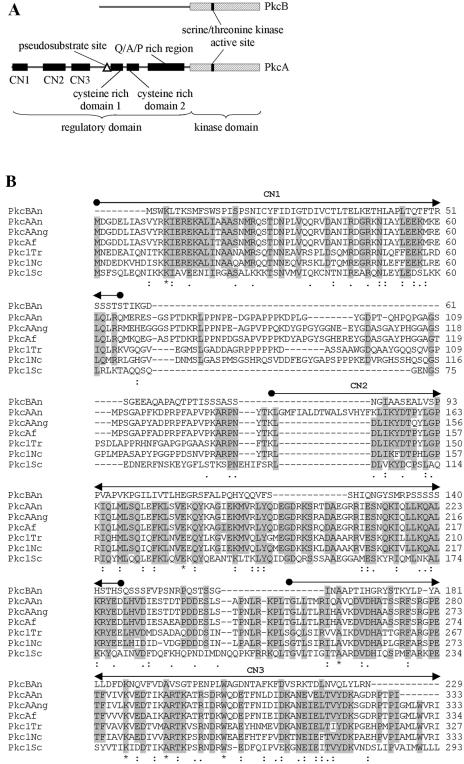
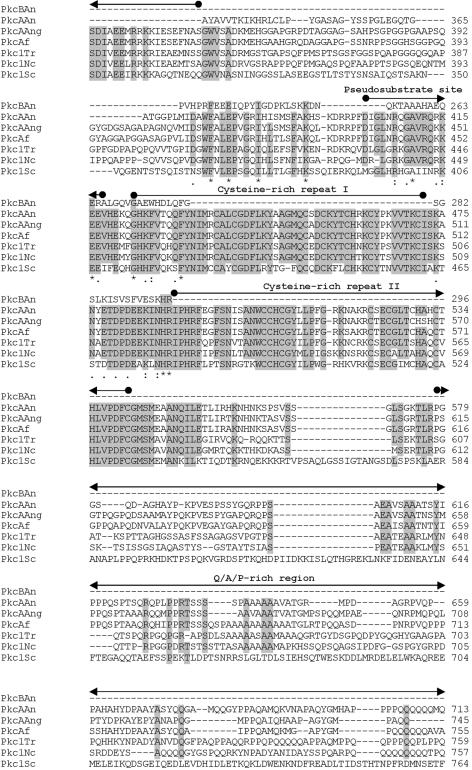
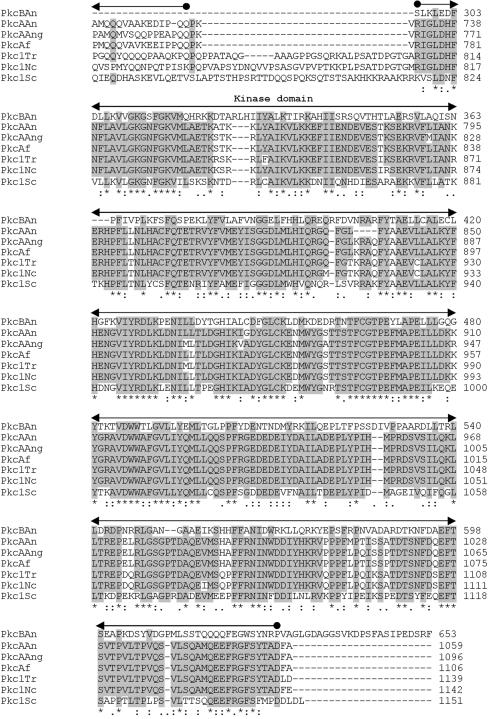
Comparative sequence analysis of the PkcB amino acid sequence revealed that the similarity to other known Pkc's is confined to the amino-terminal region of the protein, which harbors the catalytic region of all Pkc proteins (Fig. 2B). Moreover, PkcB contains several ATP binding sites and putative Pkc phosphorylation regions. However, cysteine-rich regions, usually involved in the binding of diacylglycerol or phorbol esters, as well as a pseudosubstrate sequence involved in the regulation of Pkc activity (5), could not be found (Fig. 2A).
FIG. 2.
(A) Structures of A. nidulans PkcA and PkcB. Domains are based on the overall structure of fungal Pkc as given by Ambra and Macino (2). (B) Comparison of the deduced amino acid sequences of Pkc in A. nidulans (PkcAAn and PkcBAn) with the Pkc sequences of A. niger (PkcAAng; Swiss-Prot accession no. Q00078), Aspergillus fumigatus (PkcAf; REFSEQ accession no. XM 748361.1), T. reesei (Pkc1Tr; Swiss-Prot accession no. Q99014), N. crassa (Pkc1Nc; SWISS-PROT accession no. P87253), and S. cerevisiae (Pkc1Sc; REFSEQ accession no. NC 001134.7). Amino acids that are identical in at least five of the seven different amino acid sequences at the same position are boxed. Numbers on the right indicate the position of the last amino acid shown in each protein. Also indicated are residues identical in all sequences of the alignment (*), conserved substitutions (:), and semiconserved substitutions (.). Conserved features and domains are indicated (2).
The A. nidulans PkcA amino acid sequence greatly resembles those of other fungal Pkc's (Fig. 2B). It contains an amino-terminal-extended regulatory domain, which, so far, has been found in fungal Pkc's only. This region contains the three conserved subdomains CN1, CN2, and CN3. In Trichoderma reesei Pkc1 and Aspergillus niger PkcA, two consecutive hydrophobic helices within CN1, which are also conserved in A. nidulans PkcA, were suggested to interact with cellular membranes (32). The CN3 subdomain shows a high similarity to the Ca2+ binding region of classical Pkc's (38). However, the sequence of this binding region lacks aspartate residues that are important for the binding of Ca2+. This has been observed for all fungal Pkc enzymes characterized so far. Thus, fungal Pkc's have been assigned to the subgroup of novel Pkc's, whose regulation is independent of Ca2+ (2, 38). Moreover, the deduced amino acid sequence of PkcA contains a Q/A/P-rich region, which, so far, has been reported exclusively for fungal Pkc's (17). Taken together, the deduced amino acid sequence of A. nidulans PkcA shows a higher number of features characteristic of typical Pkc's than the PkcB sequence. These findings suggest that PkcA most probably represents a member of the group of novel Pkc's. Consequently, PkcA was more likely to be involved in phosphorylation of AnBH1 than PkcB, because the Pkc inhibitor calphostin C interacts with the regulatory domain, which is conserved in PkcA but not in PkcB (14, 18) (Fig. 2). Therefore, it is very likely that PkcA is involved in the phosphorylation-dependent nuclear localization of AnBH1. Direct phosphorylation of a transcription factor by Pkc has been described for the N. crassa protein WC-1 (4, 22, 28).
Generation of pkcA antisense RNA-producing plasmids.
To study a possible influence of PkcA on AnBH1 in vivo, deletion of the structural gene was attempted. Efforts to replace the pkcA gene by the pabaA1 gene of A. nidulans using a p-aminobenzoic acid-auxotrophic A. nidulans strain for transformation failed (data not shown). Also, the use of the N. crassa pyr-4 gene instead of pabaA1 and the transformation of a uracil-auxotrophic strain as well as of a strain lacking the gene homologous to the human KU70 gene (nkuA), essential for nonhomologous end joining of DNA in double-strand break repair (34), did not result in generation of a knockout mutant (data not shown).
Therefore, an antisense strategy using a regulatable promoter was followed, i.e., the production of a pkcA antisense transcript was controlled by the regulatable alcA promoter. For this purpose, the uracil-auxotrophic A. nidulans strain C2.3 was transformed with plasmid pAL4-pkcAinv, carrying the alcAp-pkcAantisense gene and the pyr-4 gene as the selectable marker gene (Fig. 3A). After transformation, 30 PyrG+ transformants were isolated, and most of them were checked by Southern blot analyses for the presence of the plasmid integrated into the genome (data not shown).
For Southern blot analysis, genomic DNA was digested with KpnI and XbaI. The probe used was targeted against the genomic site of the pkcA gene and the integration sites of the pkcA antisense RNA-encoding vector (Fig. 3A). For further investigations, five transformants, named PkcAinv1.1, PkcAinv1.2, PkcAinv1.3, PkcAinv1.4, and PkcAinv1.5, were selected; these had been shown by Southern blot analysis to carry few (1.2), several (1.1 and 1.4), and many (1.3 and 1.5) copies of the plasmid integrated into the genome at multiple sites (Fig. 3B; Table 1).
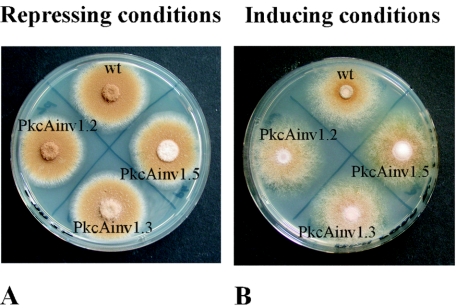
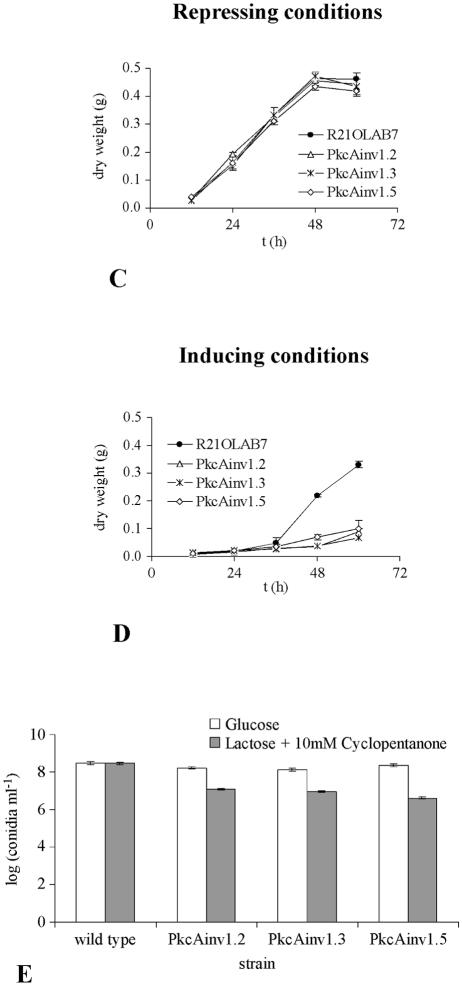
The phenotypes of the different transformants were analyzed. The growth of the transformant strains and that of the untransformed wild-type strain were indistinguishable under alcAp-repressing conditions, i.e., on AMM agar plates with glucose as the sole carbon source (Fig. 4A). Growth of the strains under alcAp-inducing conditions on AMM agar plates containing 1.6% (wt vol−1) lactose plus 10 mM cyclopentanone led to a different picture (Fig. 4B). In contrast to the wild-type A. nidulans strain Palp32.3, which carried the alcAp plasmid pabalp32 integrated into the genome (A. Gehrke, unpublished data), the strains exhibited reduced conidiation and increased formation of aerial hyphae. This finding was further supported when the dry weights of mycelia of the different strains were measured from liquid cultures. Under inducing conditions, the dry weights of transformant strains were lower than that of the wild-type strain R21OLAB7 (Fig. 4D), whereas under repressing conditions, there was no difference between wild-type and transformant strains (Fig. 4C). Measurement of the production of conidia under inducing and repressing conditions revealed that the transformant strains produced about 10-fold fewer conidia under inducing conditions than under repressing conditions, whereas there was no difference in the wild-type strain (Fig. 4E). These data indicated that repression of pkcA affected both growth and conidiation of A. nidulans.
FIG. 4.
Phenotypic characterization of pkcA antisense-encoding strains PkcAinv1.2, PkcAinv1.3, and PkcAinv1.5 and of control strains Palp32.3 and R21OLAB7 (wild type). Repressing conditions, glucose as the carbon source. Inducing conditions, lactose as the carbon source plus 10 mM cyclopentanone. (A and B) Growth and sporulation on AMM agar plates under repressing conditions (A) and inducing conditions (B). Plates were inoculated with 4 μl of conidial suspension and incubated at 37°C for 72 h. Strain Palp32.3, which was derived from transformation of strain DE58 (51) with the alcAp-containing vector pabalp32 (A. Gehrke, unpublished data), was used as the wild-type control (wt). (C and D) Dry weight. Growth of A. nidulans strain R21OLAB7 (wild type) and pkcA antisense transformants PkcAinv1.2, PkcAinv1.3, and PkcAinv1.5 grown in AMM under either repressing (C) or inducing (D) conditions at 37°C and 180 rpm for 60 h was determined. (E) Conidiation after 72 h of incubation at 37°C. Cultures were grown under either repressing or inducing conditions.
pkcA antisense RNA was detectable only in alcAp-pkcAantisense-encoding A. nidulans transformant strains.
To study whether pkcA antisense RNA was present in the transformant strains, the production of pkcA antisense RNA was analyzed by reverse transcription-PCR (RT-PCR). As shown in Fig. 5, antisense RNA was detected only in RNA of transformant strains. pkcA antisense RNA was detectable under both alcAp-repressing and -inducing conditions. However, the amount of antisense RNA produced under alcAp-inducing conditions greatly exceeded that produced under repressing conditions (Fig. 5B). pkcA antisense RNA was not detected in the wild-type strain R21OLAB7 (Fig. 5B). Also, the amount of pkcA antisense RNA correlated with the number of pkcA antisense RNA-encoding plasmids integrated into the genome in the different transformant strains, i.e., the intensity of the cDNA band derived from antisense RNA was strongest for strain PkcAinv1.3, which carried the greatest number of copies of plasmid pAL4-pkcAinv integrated into the genome (Fig. 5B). As a control, tef1 RNA was analyzed. The tef1 gene encodes translational factor TEF1, which is one of the most abundant proteins in eukaryotic cells (43). The tef1 gene has been shown to be highly expressed in filamentous fungi (33, 46). RT-PCR using primers specific for the tef1 gene indicated that tef1 mRNA was present in all samples, i.e., the wild-type strain and the transformant strains, irrespective of whether they were grown under repressing or inducing conditions (Fig. 5C). These data suggest that tef1 was constitutively expressed and therefore could be used as an endogenous control.
To prove the authenticity of the PCR fragments obtained, cDNAs obtained from both pkcA mRNA and pkcA antisense RNA were cloned and sequenced. The sequences agreed with the sequences of pkcA without intron and the deduced antisense RNA (data not shown). To analyze the length of the antisense RNA, reverse primers PkcAanti3, PkcAintr7, and PkcAanti33 (Fig. 5A; Table 2) were used in different RT-PCR experiments. Combining the forward primer PkcAanti5 and either of the reverse primers PkcAanti3 and PkcAintr7 in subsequent PCRs with cDNA as the template yielded a PCR product of the expected size of 165 bp or 186 bp, respectively. However, by using the reverse primer PkcAanti33, a PCR product of the expected size of 280 bp could not be obtained. These results suggest that the antisense RNA had at least 218 but fewer than 312 bases, as measured from the assumed starting point of transcription.
Induction of pkcA antisense RNA led to increased expression of an aatAp-lacZ gene fusion and reduced penicillin titers.
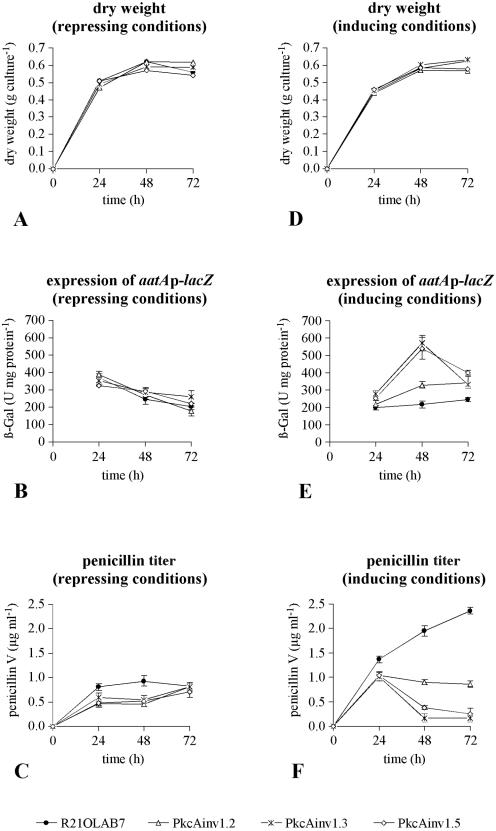
Since we have shown that pkcA antisense RNA was produced, the influence of PkcA on aatA gene expression was studied. Previously, it was shown that AnBH1 acted as a repressor of aatAp-lacZ expression (15). If PkcA negatively affected AnBH1, this should result in increased aatAp-lacZ expression. To test this hypothesis, the expression of an aatAp-lacZ gene fusion was analyzed in A. nidulans strain R21OLAB7 (wild type) and three pkcA antisense-encoding transformants (PkcAinv1.2, PkcAinv1.3, PkcAinv1.5). The strains carry an aatAp-lacZ gene fusion integrated in a single copy at the chromosomal argB gene locus. In addition, the transformant strains contain inducible alcAp-pkcAantisense fusions (Table 1). The strains were grown in FM containing 4% (wt/vol) glucose (repressing conditions) or 4% (wt/vol) lactose plus 10 mM cyclopentanone (inducing conditions) at 27°C and 250 rpm. Mycelia were harvested after 24 h, 48 h, and 72 h for analysis of aatAp-lacZ expression by β-galactosidase assays. Results are shown in Fig. 6. As the determination of the dry weight indicated, in contrast to the results in AMM, in FM there was no difference in growth rate between strains irrespective of the carbon source used, i.e., glucose or lactose plus cyclopentanone (Fig. 6A and D).
FIG. 6.
Fermentation run of A. nidulans R21OLAB7 (wild type) and three pkcA antisense RNA-encoding transformants (PkcAinv1.2, PkcAinv1.3, PkcAinv1.5) with lactose plus cyclopentanone or glucose as the carbon source. Data for each time point are means and standard deviations for three simultaneously harvested flasks (for dry weight, data represent only one flask). (A, B, and C) Glucose was used as the carbon source (repressing conditions). (D, E, and F) Lactose was used as the carbon source, and 10 mM cyclopentanone was added to the medium (inducing conditions). Results were expressed as dry weight (A and D), expression of the aatAp-lacZ fusion gene determined as β-Gal specific activity (B and E), or penicillin titer (C and F).
The results of the analysis of aatAp-lacZ expression are shown in Fig. 6B and E. Under repressing conditions (glucose), expression of the aatAp-lacZ gene fusion was similar in all strains, while under inducing conditions (lactose plus cyclopentanone), the expression patterns were different. After 48 h, aatAp-lacZ expression in the antisense RNA-producing transformants PkcAinv1.3 and PkcAinv1.5 was threefold higher than in the wild-type strain. Interestingly, the increase in aatAp-lacZ expression was less prominent in transformant strain PkcAinv1.2. This finding correlated well with the reduced amounts of antisense RNA-encoding DNA and antisense RNA in this transformant. Also, under alcAp-pkcAantisense-inducing conditions, the amount of penicillin produced correlated negatively with the amount of pkcA antisense RNA produced in the different strains (Fig. 6F). The amount of penicillin produced was highest in the wild-type strain and lowest in the transformant strains PkcAinv1.3 and PkcAinv1.5, which produced the greatest amount of pkcA antisense RNA. By contrast, the wild-type and transformant strains showed similar penicillin production under repressing conditions (glucose) (Fig. 6C), further indicating the dependence of penicillin production on induction of the alcAp-pkcAantisense fusion. Taken together, these results indicated that reduced expression of pkcA resulting from the production of pkcA antisense RNA led to increased aatA expression and reduced penicillin titers. This apparent contradiction is discussed below.
Induction of pkcA antisense RNA production led to localization of an AnBH1-EGFP fusion in the cytoplasm.
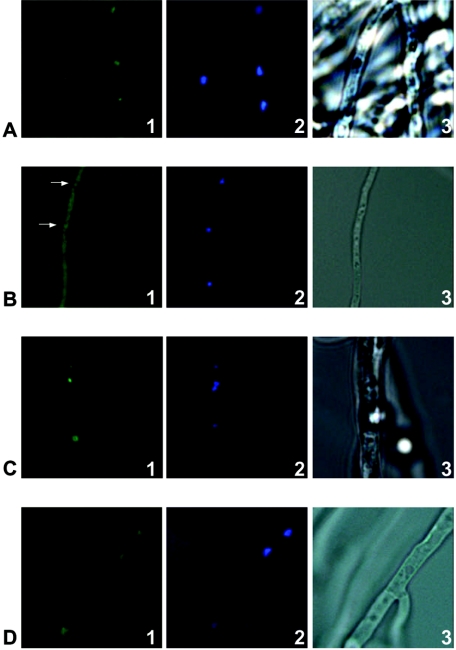
As mentioned above, the increased aatAp-lacZ expression under alcAp-pkcAantisense-inducing conditions suggests a direct relationship of PkcA and AnBH1. Based on the inhibitory studies with calphostin C (Fig. 1), it is conceivable that nuclear localization of AnBH1 is regulated by PkcA. Consequently, if PkcA positively affected nuclear localization of AnBH1, under alcAp-pkcAantisense-inducing conditions an AnBH1-EGFP fusion protein should be located, at least in part, in the cytoplasm. To prove this hypothesis, strain PkcAinv1.3, containing an anbH1-egfp gene fusion and the alcAp-pkcAantisense construct (Table 1), was analyzed. The strain was cultivated in AMM under either alcAp-pkcAantisense-repressing or alcAp-pkcAantisense-inducing conditions. Results are shown in Fig. 7.
FIG. 7.
Cellular localization of the AnBH1-EGFP fusion protein. Strains PkcAinv1.3 and C2.3, respectively, were incubated in AMM under either alcAp-repressing (0.8% [wt vol−1] glucose) or alcAp-inducing (1.6% [wt vol−1] lactose plus 10 mM cyclopentanone) conditions. Nuclei were stained with DAPI (panels 2). Samples were analyzed by fluorescence microscopy (panels 1, showing localization of the AnBH1-GFP fusion protein, and panels 2, showing nuclei) or light microscopy (panels 3). (A and B) AnBH1-EGFP-derived fluorescence of A. nidulans strain PkcAinv1.3 under alcAp-repressing (A) and alcAp-inducing (B) conditions. Arrows in panel B1 indicate nuclei. (C and D) AnBH1-EGFP-derived fluorescence of A. nidulans strain C2.3 under alcAp-repressing (C) and alcAp-inducing (D) conditions.
Under alcAp-pkcAantisense-repressing conditions, the AnBH1-EGFP fusion protein was found to be located in the nucleus (Fig. 7A). By contrast, when pkcAantisense expression was induced by lactose plus cyclopentanone, fluorescence was also visible in the cytoplasm (Fig. 7B). Nevertheless, even under inducing conditions, the nuclei still showed fluorescence (Fig. 7B), indicating that AnBH1-EGFP only partially entered the nucleus when pkcA mRNA was silenced. As a control, the AnBH1-EGFP fusion protein was analyzed in strain C2.3, which does not contain the alcAp-pkcAantisense construct (Table 1). As shown in Fig. 7C and D, the AnBH1-EGFP fusion protein was located in the nucleus irrespective of whether incubation occurred under repressing or inducing conditions. These results indicated that reduced expression of pkcA due to production of pkcA antisense RNA led to reduced localization of AnBH1 in the nucleus and, furthermore, to an accumulation of the protein in the cytoplasm.
DISCUSSION
Until now, little information has been available on the signal transduction cascades involved in β-lactam biosynthesis. Previous work revealed that FadA, the α subunit of a heterotrimeric G protein, is involved in the regulation of penicillin biosynthesis. In an A. nidulans strain containing constitutively activated FadA [fadA(G42R)], an increased steady-state mRNA level of the ipnA gene and concomitantly increased penicillin titers were observed. FadA appears to be a member of a signal transduction cascade activating penicillin biosynthesis in A. nidulans (47). Here we showed that PkcA is involved in penicillin biosynthesis. Members of the family of Pkc's are generally serine/threonine kinases and are found exclusively in eukaryotic cells. Many, if not all, Pkc enzymes are central components of signal transduction chains (reviewed in reference 42). Computer analysis of the A. nidulans genome revealed that the fungus contains two putative Pkc's. Whereas PkcA looks like a typical fungal Pkc, PkcB apparently lacks some of the characteristic features of Pkc's, making it likely that PkcB represents only a Pkc-related protein kinase.
We failed to generate a knockout strain of the pkcA gene by using the usual AMM regeneration agar plates, even when a strain lacking the gene homologous to the human KU70 gene (nkuA), essential for nonhomologous end joining of DNA in double-strand break repair (34), was used (data not shown). Although the reason for this could be that pkcA lies in a less recombinogenic region of the genome, we favor the theory that deletion of pkcA would be lethal for the cell, even in a heterokaryon. This assumption is supported by the observation that in filamentous fungi the deletion of pkc-encoding genes is obviously lethal (35), whereas this is apparently not the case in yeasts. In Saccharomyces cerevisiae, a single gene encoding Pkc1p was identified. Mutants carrying a complete deletion were able to proliferate only in the presence of osmotic stabilizing agents and underwent immediate lysis upon transfer to a medium lacking osmotic stabilizers (24). Within the Schizosaccharomyces pombe genome, two PKC genes (pck1+ and pck2+) were identified (50). Different functions for pck1+ and pck2+ were found. Only the combined deletion of both genes was lethal (3). While the deletion of pck1+ only displayed no apparent phenotype, deletion of pck2+ showed an aberrant cell morphology (50). Deletion of both endogenous copies of the CaPKC1 gene, encoding a Pkc in diploid Candida albicans cells, resulted in an osmotically remedial cell lysis defect of both the budding and the hyphal growth form and morphologically aberrant cells of the budding form (36). By contrast, in filamentous fungi deletion of pkc-encoding genes has not been reported yet, apparently because their deletion appears to be lethal (22). A similar conclusion was drawn by Oeser (35), who failed to delete the Pkc-encoding pkc1 gene of Cochliobolus heterostrophus. Because, as shown here, the deletion of pkcA in A. nidulans is also potentially lethal, an antisense RNA strategy using an inducible promoter was applied. The production of pkcA antisense RNA was an alternative way to study the function of PkcA, which was further investigated in this study because it showed the highest degree of similarity to typical fungal Pkc's. Antisense RNA was successfully applied to filamentous fungi in a number of recent studies (7, 23, 53). As shown here, a good level of pkcA antisense RNA was obtained by using the promoter region of the alcA gene of A. nidulans when transformant strains were grown with lactose as the carbon source plus cyclopentanone. Expression of antisense pkcA led to reduced growth of the fungus in AMM but not in FM, indicating that either the effect of the antisense RNA was less prominent when the fungus grew in FM or, alternatively, the phenotype of reduced Pkc activity is less drastic in FM than in AMM.
The observation that under alcAp-pkcAantisense-inducing conditions, i.e., when the pkcA transcript was silenced, only reduction of growth occurs, and not—as expected for an essential gene—complete growth inhibition, can be explained by the fact that, by using the antisense approach, expression of a given gene usually cannot be completely turned off but can only be reduced (53). The same applies to the observation that under alcAp-pkcAantisense-inducing conditions most, but not all, AnBH1-EGFP molecules were located in the cytoplasm. The effect based on the production of antisense RNA was quantified by Moralejo et al. (31), who measured a pepB mRNA reduction ranging from 10% to 70% when pepBantisense RNA was produced from the A. nidulans gpdA promoter in Aspergillus awamori. Therefore, the level of antisense RNA may not be sufficient to titrate out the respective mRNA completely. Nevertheless, when the gene is essential—which is likely for pkcA—an antisense approach still allows its functional characterization, especially when formation of the antisense RNA is regulatable (53).
In FM, the penicillin titer was reduced to 10% in multicopy transformant strains expressing antisense RNA and to 40% in low-copy-number transformant strains. The amounts of antisense RNA in the different transformants, the increased expression level of the aatA gene, and the copy number of antisense RNA-encoding plasmids integrated into the genome correlated well, i.e., the higher the copy number in the genome, the more antisense RNA was produced, the more aatAp-lacZ expression was observed, and the less penicillin was produced. Also, the initial observation that the addition of Pkc inhibitor to the medium led to a decrease in nuclear localization of AnBH1 accords well with the observation that the production of pkcA antisense RNA led to higher expression of the aatAp-lacZ gene fusion, most likely because the level of AnBH1 present in the nucleus was reduced. These observations suggest that PkcA is involved in the nuclear entry of AnBH1 or in its maintenance in the nucleus, most likely by phosphorylation of AnBH1 either directly or by activating other kinases that phosphorylate AnBH1. This hypothesis is supported by the observation that reduced expression of pkcA resulting from production of pkcA antisense RNA led to reduced maintenance of AnBH1-EGFP in the nucleus and, furthermore, to an accumulation of the protein in the cytoplasm (Fig. 7). Therefore, taken together, these data suggest that PkcA is a direct target of the Pkc inhibitor calphostin C, because the data obtained from Fig. 7 are in accordance with those of Fig. 1. The latter showed that the addition of calphostin C also led to a decrease in AnBH1-EGFP levels in the nucleus and, furthermore, to an increase in levels of this protein in the cytoplasm.
In yeasts, one of the main functions of Pkc's is to maintain cellular integrity (reviewed in reference 42). In filamentous fungi, it is very likely that Pkc's have pleiotropic effects and affect different, in part essential, cellular functions. Recently, Franchi et al. (22) reported that Pkc modulates light responses in N. crassa by regulating the blue light photoreceptor WC-1. To elucidate the function of PkcA, it will also be helpful to identify target proteins by proteome analysis.
An obvious contradiction was the observation that despite higher expression of the aatAp-lacZ gene fusion, the penicillin titer was reduced when the alcAp-pkcAantisense gene fusion was induced. This indicated that higher expression of the third penicillin biosynthesis gene, aatA, leads to reduced penicillin titers. Interestingly, a similar observation supporting this conclusion was made by Fernández-Cañón and Peñalva (21). These authors also reported that overproduction of IAT activity reduced penicillin biosynthesis. They speculated that the negative influence on the synthesis of penicillins exerted by excessive levels of IAT, relative to levels of the other enzymes of the pathway, might be mediated by the penicillin amidase (1) or phenylacetyl-CoA hydrolase (30) activities of IAT. Previously, it was shown that IAT has five enzyme activities (1). The penicillin transacylase activity of IAT exchanges side chains between two hydrophobic penicillin molecules, or between one penicillin molecule and 6-aminopenicillanic acid (6-APA). The penicillin amidase activity of IAT is apparently the reverse of the biosynthetic acyl-CoA:6-APA acyltransferase activity (1). Therefore, it is conceivable that the increase in these enzyme activities by overproduction of IAT leads to drastically less active penicillins or to an increase in 6-APA which also has no significant antibiotic activity. Our observations fully agree with these assumptions. In addition, it is conceivable that PkcA acts directly on other elements of penicillin biosynthesis.
In summary, PkcA is involved in penicillin biosynthesis via regulation of the nuclear localization of the transcription factor AnBH1. Furthermore, PkcA appears to be involved in other cellular processes. Although the exact function of Pkc's in filamentous fungi has not been resolved yet, it is interesting to speculate that if PkcA were also required for maintaining cellular integrity, this would have a direct effect on penicillin biosynthesis, e.g., hydrolysis of the cell wall of A. nidulans by competing bacteria could increase the production of penicillin via activation of PkcA.
Acknowledgments
André Tüncher, Goran Martic, Stefan Steidl and Maria Louise Caruso are gratefully acknowledged for carrying out some initial experiments. We thank Alexander Gehrke for providing A. nidulans strain Palp32.3.
This research was supported by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm 1152).
REFERENCES
- 1.Alvarez, E., B. Meesschaert, E. Montenegro, S. Gutiérrez, B. Diez, J. L. Barredo, and J. F. Martin. 1993. The isopenicillin N-acyltransferase of Penicillium chrysogenum has isopenicillin N-amidohydrolase, 6-aminopenicillanic acid acyltransferase and penicillin amidase activities, all of which are encoded by the single penDE gene. Eur. J. Biochem. 215:323-332. [DOI] [PubMed] [Google Scholar]
- 2.Ambra, R., and G. Macino. 2000. Cloning and characterization of PKC-homologous genes in the truffle species Tuber borchii and Tuber magnatum. FEMS Microbiol. Lett. 189:45-53. [DOI] [PubMed] [Google Scholar]
- 3.Arellano, M., M. H. Valdivieso, T. M. Calonge, P. M. Coll, A. Duran, and P. Perez. 1999. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J. Cell Sci. 112:3569-3578. [DOI] [PubMed] [Google Scholar]
- 4.Arpaia, G., F. Cerri, S. Baima, and G. Macino. 1999. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 262:314-322. [DOI] [PubMed] [Google Scholar]
- 5.Azzi, A., D. Boscoboinik, and C. Hensey. 1992. The protein kinase C family. Eur. J. Biochem. 208:547-557. [DOI] [PubMed] [Google Scholar]
- 6.Ballance, D. J., and G. Turner. 1985. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36:321-331. [DOI] [PubMed] [Google Scholar]
- 7.Bautista, L. F., A. Aleksenko, M. Hentzer, A. Santerre-Henriksen, and J. Nielsen. 2000. Antisense silencing of the creA gene in Aspergillus nidulans. Appl. Environ. Microbiol. 66:4579-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-252. [DOI] [PubMed] [Google Scholar]
- 9.Brakhage, A. A. 1998. Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol. Mol. Biol. Rev. 62:547-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brakhage, A. A. 1999. Biosynthesis of β-lactam compounds in microorganisms, p. 159-193. In D. Barton and K. Nakanishi (ed.), Comprehensive natural products chemistry, vol. 4. Amino acids, peptides, porphyrins and alkaloids. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 11.Brakhage, A. A., P. Browne, and G. Turner. 1992. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J. Bacteriol. 174:3789-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brakhage, A. A., and J. Van den Brulle. 1995. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 177:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brakhage, A. A., Q. Al-Abdallah, A. Tüncher, and P. Spröte. 2005. Evolution of β-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry 66:1200-1210. [DOI] [PubMed] [Google Scholar]
- 14.Bruns, R. F., F. D. Miller, and R. L. Merriman. 1991. Inhibition of protein kinase C by calphostin C is light-dependent. Biochem. Biophys. Res. Commun. 176:288-293. [DOI] [PubMed] [Google Scholar]
- 15.Caruso, M. L., O. Litzka, G. Martic, F. Lottspeich, and A. A. Brakhage. 2002. A novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J. Mol. Biol. 323:425-439. [DOI] [PubMed] [Google Scholar]
- 16.Chenna, R., H. Sugawara, T. Koike, R. Lopez, R. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courey, A. J., and R. Tijan. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 18.Diwu, Z., J. Zimmermann, T. Meyer, and J. W. Lown. 1994. Design, synthesis and investigation of mechanisms of action of novel protein kinase C inhibitors: perylenequinonoid pigments. Biochem. Pharmacol. 47:373-385. [DOI] [PubMed] [Google Scholar]
- 19.Evers, M. E., H. Trip, M. A. van den Berg, R. A. L. Bovenberg, and A. J. M. Driessen. 2004. Compartmentalization and transport in β-lactam antibiotics biosynthesis. Adv. Biochem. Eng. Biotechnol. 88:111-136. [DOI] [PubMed] [Google Scholar]
- 20.Fantes, P. A., and C. F. Roberts. 1973. Beta-galactosidase activity and lactose utilization in Aspergillus nidulans. J. Gen. Microbiol. 77:417-486. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Cañón, J. M., and M. A. Peñalva. 1995. Overexpression of two penicillin structural genes in Aspergillus nidulans. Mol. Gen. Genet. 246:110-118. [DOI] [PubMed] [Google Scholar]
- 22.Franchi, L., V. Fuici, and G. Macino. 2005. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 56:334-345. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann, B., S. K. LaPaglia, E. Kübler, M. Andermann, S. E. Eckert, and G. H. Braus. 2000. Developmental and metabolic regulation of the phosphoglucomutase-encoding gene, pgmB, of Aspergillus nidulans. Mol. Gen. Genet. 262:1001-1011. [DOI] [PubMed] [Google Scholar]
- 24.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Littlewood, T., and G. I. Evan. 1994. Transcription factors 2: helix-loop-helix. Protein Profile 1:635-709. [PubMed] [Google Scholar]
- 26.Litzka, O., K. Then Bergh, and A. A. Brakhage. 1995. Analysis of the regulation of Aspergillus nidulans penicillin biosynthesis gene aat (penDE) encoding acyl coenzyme A:6-aminopenicillanic acid acyltransferase. Mol. Gen. Genet. 249:557-569. [DOI] [PubMed] [Google Scholar]
- 27.Litzka, O., P. Papagiannopolous, M. A. Davis, M. J. Hynes, and A. A. Brakhage. 1998. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur. J. Biochem. 251:758-767. [DOI] [PubMed] [Google Scholar]
- 28.Loros, J. 2005. A kinase for light and time. Mol. Microbiol. 56:299-302. [DOI] [PubMed] [Google Scholar]
- 29.Martic, G. 1999. Diploma thesis. Ludwig-Maximilians-Universität München, Munich, Germany.
- 30.Martín-Villacorta, J., A. Reglero, and J. M. Luengo. 1991. Acyl-CoA:6-APA acyltransferase from Penicillium chrysogenum. Studies on its hydrolytic activity. J. Antibiot. 44:108-110. [DOI] [PubMed] [Google Scholar]
- 31.Moralejo, F. J., R. E. Cardoza, S. Gutierrez, M. Lombraña, F. Fierro, and J. F. Martín. 2002. Silencing of the aspergillopepsin B (pepB) gene of Aspergillus awamori by antisense RNA expression or protease removal by gene disruption results in a large increase in thaumatin production. Appl. Environ. Microbiol. 68:3550-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morawetz, R., T. Lendenfeld, H. Mischak, M. Mühlbauer, F. Gruber, J. Goodnight, L. H. de Graaff, J. Visser, J. F. Mushinski, and C. P. Kubicek. 1996. Cloning and characterisation of genes (pkc1 and pkcA) encoding protein kinase C homologues from Trichoderma reesei and Aspergillus niger. Mol. Gen. Genet. 250:17-28. [DOI] [PubMed] [Google Scholar]
- 33.Nakari, T., E. Alatalo, and M. E. Penttila. 1993. Isolation of Trichoderma reesei genes highly expressed on glucose-containing media: characterization of the tef1 gene encoding translation elongation factor 1α. Gene 136:313-318. [DOI] [PubMed] [Google Scholar]
- 34.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2005. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 35.Oeser, B. 1998. PKC1, encoding a protein kinase C, and FAT1, encoding a fatty acid transporter protein, are neighbours in Cochliobolus heterostrophus. FEMS Microbiol. Lett. 165:273-280. [DOI] [PubMed] [Google Scholar]
- 36.Paravicini, G., A. Mendoza, B. Antonsson, M. Cooper, C. Losberger, and M. A. Payton. 1996. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity but not dimorphism. Yeast 12:741-756. [DOI] [PubMed] [Google Scholar]
- 37.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 38.Ponting, C. P., and P. J. Parker. 1996. Extending the C2 domain family: C2s in PKCs δ, ɛ, η, θ, phospholipases, GAPs, and perforin. Protein Sci. 5:162-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz, H.-P., and J. Heinisch. 2003. Evolution, biochemistry and genetics of protein kinase C in fungi. Curr. Genet. 43:245-254. [DOI] [PubMed] [Google Scholar]
- 43.Slobin, L. I. 1980. The role of eucaryotic factor Tu in protein synthesis. The measurement of the elongation factor Tu content of rabbit reticulocytes and other mammalian cells by a sensitive radioimmunoassay. Eur. J. Biochem. 110:555-563. [DOI] [PubMed] [Google Scholar]
- 44.Steidl, S., A. Tüncher, H. Goda, C. Guder, N. Papadopoulou, T. Kobayashi, N. Tsukagoshi, M. Kato, and A. A. Brakhage. 2004. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 342:515-524. [DOI] [PubMed] [Google Scholar]
- 45.Steidl, S., P. Papagiannopoulos, O. Litzka, A. Andrianopoulos, M. A. Davis, A. A. Brakhage, and M. J. Hynes. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol. Cell. Biol. 19:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner, S., and P. Philippsen. 1994. Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol. Gen. Genet. 242:263-271. [DOI] [PubMed] [Google Scholar]
- 47.Tag, A., J. Hicks, G. Garifullina, C. Ake, Jr., T. D. Phillips, M. Beremand, and N. Keller. 2000. G-protein signalling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38:658-665. [DOI] [PubMed] [Google Scholar]
- 48.Then Bergh, K., and A. A. Brakhage. 1998. Regulation of the Aspergillus nidulans penicillin biosynthesis gene acvA (pcbAB) by amino acids: implication for involvement of transcription factor PACC. Appl. Environ. Microbiol. 64:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Then Bergh, K., O. Litzka, and A. A. Brakhage. 1996. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J. Bacteriol. 178:3908-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toda, T., M. Shimanuki, and M. Yanagida. 1993. Two novel protein kinase C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO J. 12:1987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Brulle, J., S. Steidl, and A. A. Brakhage. 1999. Cloning and characterization of an Aspergillus nidulans gene involved in the regulation of penicillin biosynthesis. Appl. Environ. Microbiol. 65:5222-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 53.Zadra, I., B. Abt, W. Parson, and H. Haas. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 66:4810- 4816. [DOI] [PMC free article] [PubMed] [Google Scholar]