Abstract
Tetracyclines are aromatic polyketides biosynthesized by bacterial type II polyketide synthases (PKSs). Understanding the biochemistry of tetracycline PKSs is an important step toward the rational and combinatorial manipulation of tetracycline biosynthesis. To this end, we have sequenced the gene cluster of oxytetracycline (oxy and otc genes) PKS genes from Streptomyces rimosus. Sequence analysis revealed a total of 21 genes between the otrA and otrB resistance genes. We hypothesized that an amidotransferase, OxyD, synthesizes the malonamate starter unit that is a universal building block for tetracycline compounds. In vivo reconstitution using strain CH999 revealed that the minimal PKS and OxyD are necessary and sufficient for the biosynthesis of amidated polyketides. A novel alkaloid (WJ35, or compound 2) was synthesized as the major product when the oxy-encoded minimal PKS, the C-9 ketoreductase (OxyJ), and OxyD were coexpressed in CH999. WJ35 is an isoquinolone compound derived from an amidated decaketide backbone and cyclized with novel regioselectivity. The expression of OxyD with a heterologous minimal PKS did not afford similarly amidated polyketides, suggesting that the oxy-encoded minimal PKS possesses novel starter unit specificity.
Tetracyclines are among the most important antibiotics known to mankind in the last half century (15). The broad-spectrum antimicrobial activities of tetracyclines have resulted in their widespread clinical use against infectious gram-positive and gram-negative bacteria (21). The emergence of microbial resistance to tetracycline has severely limited their effectiveness and has prompted the search for analogs that can overcome the known modes of antibiotic resistance (14, 40, 55). The recently published, elegant total synthesis of 6-deoxytetracyclines by Myers's group has highlighted the importance of being able to access structurally diverse tetracycline derivatives (13).
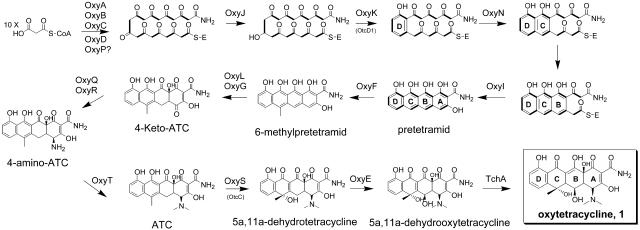
Considering the structural complexity of natural tetracycline products, engineered biosynthesis is an attractive route of generating pharmaceutically important analogs (27). Tetracyclines are aromatic polyketides synthesized by soilborne actinomycetes using type II polyketide synthases (PKSs) (49). The carbon skeleton of an aromatic polyketide is assembled from malonate-derived building blocks through iterative Claisen-like condensations catalyzed by the minimal PKS, which consists of the ketosynthase (KSα), the chain length factor (CLF, or KSβ), and the acyl carrier protein (ACP) (12). Dedicated tailoring enzymes then transform the carbon backbone into fused, richly substituted compounds. Shunt products of the oxytetracycline pathways have been characterized, which enabled deduction of a putative biosynthetic pathway (Fig. 1) (9, 37, 50). The gene clusters responsible for oxytetracycline (compound 1; oxy and otc genes) and chlorotetracycline have been located on the genomes of Streptomyces rimosus (2, 9) and Streptomyces aureofaciens (43), respectively. For the oxy (otc) cluster, gene sequences for the minimal PKS (28), a putative D-ring cyclase (46), and a downstream anhydrotetracycline (ATC)-oxygenase (45) have been deposited in GenBank.
FIG. 1.
Proposed biosynthetic pathway of oxytetracycline (1) in S. rimosus. The biosynthetic pathway was established previously by Hunter and coworkers based on an exhaustive analysis of shunt products. The assignments of the tailoring enzymes are based on their functions obtained through protein-protein BLAST analysis. We confirmed in this report the role of OxyD and the minimal PKS in the biosynthesis of an amidated decaketide. We also confirmed the role of OxyJ as the C-9 ketoreductase.
One of the distinguishing structural features of tetracyclines compared to other aromatic polyketides is the universal presence of an amide unit at one terminus of the polyketide backbone (Fig. 1). Substrate feeding studies have suggested that the amide unit stems from an intact malonate unit (59, 60). The enzymes involved in the biosynthesis of the amide starter unit have not been identified to date. The biosynthesis of the polar amide starter unit is especially interesting from a biocombinatorial perspective, since all other aromatic polyketides are primed by chemically inert aliphatic and aromatic starter units (41). The polar amide unit can serve as a useful reaction handle for orthogonal semisynthetic modifications of polyketides. Elucidating the formation of the amide group will therefore enable the exploration and engineering of tetracycline biosynthesis as well as expand the repertoire of tools that can be used in combinatorial biosynthesis of other polyketides.
In this work, we report the identification of an amidotransferase, OxyD, encoded in the oxy (otc) gene cluster, that is involved in the biosynthesis of the amidated backbone. Using a heterologous host, we show that coexpression of OxyD with the minimal oxy-encoded PKS affords a novel isoquinolone compound derived from an amidated decaketide backbone.
MATERIALS AND METHODS
Bacterial strains and general techniques for DNA manipulation.
Streptomyces coelicolor strain CH999 was used as a host for the transformation of shuttle vectors. Protoplast preparation and polyethylene glycol-assisted transformation were performed as described by Hopwood et al. (23). Escherichia coli XL1-Blue (Stratagene) was used for the manipulation of plasmid DNA. Streptomyces rimosus (ATCC 10970) was obtained from ATCC and was cultured for extraction of genomic DNA. Unmethylated DNA was obtained using the methylase-deficient strain GM2163 (New England Biolabs).
Sequencing of oxy (otc) cluster.
The complete genomic DNA library of S. rimosus was constructed using a pWEB cosmid cloning kit (Epicenter). The cosmid clone pYT264, which harbors the 21.2-kb oxy gene cluster flanked by otrA (18) and otrB, was identified by PCR screening. A combination of shotgun and primer walking techniques was used to obtain the sequence information. Open reading frames (ORFs) were detected and analyzed using Frameplot software (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl), and the putative roles of the proteins were assigned using protein-protein BLAST and Pfam analysis. To simplify naming and analysis of the cluster for subsequent studies, the genes were renamed oxyA to oxyT in a linear fashion. The five previously sequenced genes of the cluster are cross-referenced in Table 1.
TABLE 1.
Genes in the oxytetracycline biosynthetic cluster and deduced roles based on sequence homology
| ORFs
|
Homologs
|
||||||
|---|---|---|---|---|---|---|---|
| Geneb | Start position | Stop position | Predicted size (kDa) | Cts homolog (reference)a | Protein (reference) | Deduced role | % Identity |
| oxyTA1 (otrR) | 134 | 625 | 18 | CAD15553 (52) | Transcription regulation | 40 | |
| oxyA (otcY1-1) | 770 | 2047 | 45 | TcsD (43) | SnoaI (62) | Ketosynthase | 70 |
| oxyB (otcY1-2) | 2044 | 3312 | 44 | TcsE (43) | ChaB (61) | Chain length factor | 66 |
| oxyC (otcY1-3) | 3388 | 3675 | 10 | TcsF (43) | Sim4 (20) | Acyl carrier protein | 57 |
| oxyD (otcY1-4) | 3686 | 5524 | 69 | TcsG (43) | AsnB (8) | Asparagine synthetase | 60 |
| oxyE (otcY1-5) | 5521 | 6777 | 46 | Yes | MtmO1 (32) | Oxygenase | 51 |
| oxyF (otcY2-5) | 7846 | 6809 | 37 | Yes | BAB69170 (44) | Methyltransferase | 61 |
| oxyG (otcY2-4) | 8186 | 7899 | 11 | Yes | MtmOIII (32) | Oxygenase | 45 |
| oxyH (otcY2-3) | 9833 | 8241c | 59 | Yes | MtmL (32) | Acyl-CoA ligase | 47 |
| oxyI (otcY2-2) | 10324 | 9872 | 17 | Yes | MtmX (32) | Cyclase | 49 |
| oxyJ (otcY2-1) | 10593 | 11384 | 27 | Yes | Sim5 (20) | C-9 ketoreductase | 69 |
| oxyK (otcD1)d | 11443 | 12396 | 35 | Yes | ChaF (61) | Aromatase | 54 |
| oxyL (otcD2) | 12386 | 14059 | 60 | Yes | MtmOII (32) | Oxygenase | 47 |
| oxyM (otcD3) | 14077 | 14835 | 25 | No | MtmTI (32) | Ketoreductase | 54 |
| oxyN (otcD4) | 14855 | 15628 | 28 | Yes | MtmY (32) | Cyclase | 68 |
| oxyO (otcD5) | 15734 | 16795 | 38 | No | ZP00569712 | Unknown | 31 |
| oxyP (otcX3) | 17835 | 16813 | 35 | No | AknF (16) | Acyltransferase | 61 |
| oxyQ (otcX2) | 18911 | 17832 | 38 | Yes | AspB2 (44) | Aminotransferase | 56 |
| oxyR (otcX1) | 19336 | 18908 | 16 | Yes | ActVA ORF2 (11) | PNP-oxidase | 50 |
| oxyS (otcC) | 19492 | 21003 | 54 | Yes (17) | MtmOIV (32) | Oxygenase | 46 |
| oxyT (otcZ) | 21064 | 22098 | 37 | Yes (17) | MtmMI (32) | N-Methyltransferase | 45 |
We compared the partially annotated chlorotetracycline PKS cluster (43, 51) from Streptomyces aureofaciens with the oxy c-encoded PKS cluster.
Gene names in parentheses are loci of the oxy (otc) PKS previously identified through mutant complementation studies. Sequence information for these genes is not available publicly, except for that shown in bold. The cluster was renamed in this study to facilitate sequence analysis. References for previously sequenced genes are listed in the text.
No stop codon was detected. The putative protein size was based on alignment with closely related acyl-CoA ligases.
A 60-bp deletion at the 3′ end was found in the deposited otcD1 sequence and has been corrected here.
Construction of shuttle plasmids for biosynthesis.
The following primers were used to amplify the individual genes: for oxyA, 5′-GGTTAATTAAGGAGGAGCCAGCATGTCCAAGATCCATGACGC-3′ (PacI/XbaI) and 5′-GGTCTAGAGGTCATCGCTGCCTCCCCGGCTC-3′; for oxyB, 5′-GGACTAGTGGAGGAGCCAGCATGACCGGCCAGCTCGCCCCC-3′ (SpeI/XbaI) and 5′-GGTCTAGAGGTCAGTCCCGGCCGCTGACCA-3′; for oxyC, 5′-GGTCTAGAGGAGGAGCCCATATGACCCTACTCACCCTCTCC-3′ (XbaI/SpeI) and 5′-GGACTAGTCACTTGTCCCGCGCGGCGC; for oxyD, 5′-AATCTAGAGGAGGAGCCCATATGTGCGGAATCGCCGGCTGGATC-3′ (XbaI/SpeI) and 5′-AAACTAGTCATAGCTCCAGGCTGACGCCGTA; and for oxyP, 5′-GGTCTAGAGGAGGAGCCCATATGACGGCGGACACGAAGGCC-3′ (XbaI/SpeI) and 5′-GGACTAGTCAGGGAATCCGGTACCCCT-3′. The introduced restriction sites are shown in italics, and the restriction enzymes are indicated in parentheses. The optimal ribosome binding site was introduced at the 5′ end of each gene and is underlined. All oxy genes were amplified from pYT264, and multicistronic cassettes were constructed using the compatible XbaI/SpeI cohesive ends for most of the genes, except for oxyA and oxyB, which were cloned as a single PacI/XbaI cassette. Different combinations of genes were introduced into pYT315 (a pRM5-derived vector) to yield the constructs shown in Table 2.
TABLE 2.
Plasmid constructs and resulting polyketide productsa
| Plasmid | Genes | Major product | Approx. yield (mg/liter) |
|---|---|---|---|
| pYT319 | oxyABC | SEK15 | 30 |
| pYT318 | oxyABCJ | RM20b | 30 |
| pWJ35 | oxyABCDJ | Compound 2 | 20 |
| pWJ35a | oxyABCD actIII | Compound 2 | 20 |
| pWJ40 | actI ORFs I-II oxyCDJ | Mutactin | 20 |
| pWJ48 | tcmKL oxyCDJ | RM20b | <5 |
Streptomyces coelicolor strain CH999 was used as the host for polyketide biosynthesis. Each plasmid is derived from pRM5.
Culture conditions and purification of polyketides.
Strains were grown on solid R5 plates with 25 mg/liter thiostrepton at 30°C for 7 to 10 days. For analytical high-performance liquid chromatography (HPLC) analysis, a well-pigmented plate was chopped into fine pieces and extracted with 50 ml of ethyl acetate-methanol-acetic acid (89%-9.8%-1.2%). Extracts were dried over anhydrous Na2SO4. The solvent was removed in vacuo, and the residue was dissolved in 0.5 ml of dimethyl sulfoxide (DMSO). The polyketide products were separated by reverse-phase HPLC and detected at 254 and 280 nm using an analytical C18 column (Varian Pursuit 5u; 250 mm × 4.6 mm) with a linear gradient of 5% acetonitrile (ACN) in water (0.1% trifluoroacetic acid [TFA]) to 95% ACN in water (0.1% TFA) over 30 min with a flow rate of 1 ml/min. HPLC retention times were as follows: for RM20b, 19.3 min; and for WJ35 (compound 2), 15.0 min. For large-scale production and isolation of compound 2, 60 R5 plates (2 liters) streaked with the transformed CH999 strains were incubated at 30°C for 7 to 10 days. The plates were chopped into fine pieces and extracted with 2 liters of ethyl acetate-methanol-acetic acid (89%-9.8%-1.2%). The solvent was removed in vacuo, and the residue was dissolved in 10 ml of H2O-ACN-DMSO (50%-25%-25%) and filtered for injection into a semipreparative reverse-phase HPLC column (Alltech Alltima 5u C18 column; 250 mm × 10 mm). A 10% to 50% acetonitrile and water (0.1% TFA) gradient was used over 45 min with a flow rate of 3 ml/min. The solvent was removed in vacuo from the collected fractions containing the expected biosynthetic product. The residue was dissolved in 2 ml of acetone to be loaded onto two preparative thin-layer chromatography (TLC) plates (20-cm × 20-cm × 0.25-mm silica gel [60F-254]). The preparative TLC plates were developed in ethyl acetate-methanol-acetic acid (94%-5%-1%), and the desired band (Rf = 0.3) was excised from the TLC plate and eluted from silica with ethyl acetate-methanol (90%-10%).
Spectroscopic analysis.
High-resolution mass spectrometry (HRMS) was performed at the UCLA Pasarow Mass Spectrometry Laboratory with IonSpec Ultima 7.0 Telsa electrospray ionization and matrix-assisted laser desorption ionization-Fourier transform mass spectrometry. The HRMS result for compound 2 was m/z = 388.1043 (C19H18NO8; calculated [M + H]+, 388.1027). Nuclear magnetic resonance (NMR) spectra were obtained on Bruker DRX-500 spectrometers at the NMR facility of the Department of Chemistry and Biochemistry at UCLA. 1H and 13C chemical shifts were referenced to the solvent peak (acetone-d6) and were δ 2.05 and 29.9 ppm, respectively. Standard parameters were used for one-dimensional (1D) and 2D NMR experiments, which included 1H, 13C, heteronuclear multiple quantum correlation (HMQC) (1H-13C, 1H-15N), and heteronuclear multiple-bond correlation (HMBC) (1H-13C, 1H-15N) analyses. 15N NMR experiments were performed on a DRX-600 instrument, and formamide (δ = 95; DMSO-d6) was used as an internal reference. The observed 15N NMR signal for compound 2 was δ = 115. For detailed NMR data, see Table 3.
TABLE 3.
Proton and carbon NMR data for WJ35 (compound 2)a
| Positionb | 13C δ (ppm) | 1H δ (ppm) (m, area, JHH [Hz]) | HMBCc |
|---|---|---|---|
| 1 | 165.9 | ||
| 2 | 90.9 | 5.35 (d, 1H, 2.1) | C1, C3, C4 |
| 3 | 172.7 | 11.68 (s, 1H), OH | |
| 4 | 105.0 | 6.09 (d, 1H, 2.0) | C2, C5 |
| 5 | 160.9 | ||
| 6 | 49.2 | 3.73 (s, 2H) | C4, C5, C7 |
| 7 | 204.7 | ||
| 8 | 50.7 | 2.82 (d, 2H, 5.3) | C7, C9, C10 |
| 9 | 68.2 | 4.50 (m, 1H) | |
| 10 | 41.5 | 2.69 (dd, 1H, 8.0, 14.5) | C8, C9, C11, C12, N |
| 2.79 (dd, 1H, 4.4, 14.5) | |||
| 11 | 140.7 | ||
| 12 | 107.8 | 6.28 (s, 1H) | C10, C11, C13, C14, C18, N |
| 13 | 142.8 | ||
| 14 | 102.5 | 6.40 (d, 1H, 2.1) | C12, C15, C16, C18 |
| 15 | 164.9 | 11.16 (s, 1H), OH | |
| 16 | 101.7 | 6.25 (d, 1H, 2.1) | C14, C15, C17, C18 |
| 17 | 165.2 | 12.91 (s, 1H), OH | C16, C17, C18 |
| 18 | 106.2 | ||
| 19 | 167.9 | ||
| NH | 10.18 (s, 1H) |
Spectra were obtained at 500 MHz for protons and 125 MHz for carbon and were recorded in acetone-d6 for compound 2.
Numbering of the carbon backbone is shown in Fig. 4.
Observed 1H-13C and 1H-15N HMBC signals.
Nucleotide sequence accession number.
The sequence of the oxy gene cluster was deposited in GenBank under accession number DQ143963.
RESULTS
Sequencing of oxytetracycline gene cluster.
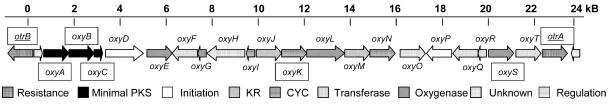
To study the origin of the amide moiety observed among all tetracyclines and to gain additional insight into the biosynthesis of tetracyclines, we sequenced the oxytetracycline (oxy and otc) gene cluster using a combination of shotgun sequencing and cosmid walking techniques. The gene cluster was previously mapped to be between the two resistance genes otrA (18) and otrB (2, 10, 38). A total of 21 ORFs were clustered between otrA and otrB (Fig. 2 and Table 1), including the previously sequenced minimal PKS (otcY1-1, otcY1-2, and otcY1-3) (28), cyclase (otcD1) (46), and ATC oxygenase (otcC) (2, 45) genes. Sequencing information for the remainder of the cluster, which has been studied through mutant complementation experiments (24), was not available from GenBank prior to this study. To simplify designation of the gene cluster, we named the oxy genes oxyA to oxyT from end to end, as graphically represented in Fig. 2. The functions of the proteins were assigned based on sequence similarities to known aromatic PKS enzymes and are listed in Table 1.
FIG. 2.
Organization of oxy (otc) biosynthetic gene cluster. The biosynthetic enzymes are located between the two resistance genes otrA and otrB. Previously sequenced genes are boxed. For details of enzyme function assignments, see Table 1.
A putative oxytetracycline biosynthetic pathway was previously established, largely aided by the identification of metabolic shunt products (Fig. 1) (9, 24). The minimal PKS (OxyA, OxyB, and OxyC) accepts a malonamyl starter unit and condenses eight equivalents of malonate to afford an amidated decaketide backbone. Two distinct nitrogen-inserting enzymes (OxyD and OxyQ) were found to be encoded in the oxy gene cluster. OxyD is an amidotransferase and participates in the biosynthesis of the amide starter unit (see below). OxyQ is homologous to aspartate/tyrosine/aromatic aminotransferases involved in amino acid metabolism (22) and is likely the enzyme that transaminates C-4 of 4-keto-ATC, using pyridoxal 5′-phosphate as a cofactor. OxyR, the putative pyridoxamine 5-phosphate oxidase, is translationally coupled to OxyQ with overlapping stop/start codons. A small set of proteins homologous to OxyR are found in the literature, with the most notable being ActVA-ORF2 (11), an enzyme of unknown function involved in actinorhodin biosynthesis.
Two ORFs (oxyJ and oxyM) encoding NADPH-dependent ketoreductases (KR) are present in the gene cluster. OxyJ catalyzes regiospecific C-9 reduction of the oxytetracycline backbone (see below). The specific role of OxyM in the oxy cluster is unresolved (24) and perhaps reflects redundancy in function. Only one KR gene is present in the recently sequenced chlorotetracycline gene cluster (51). The bifunctional cyclase/dehydratase OxyK (OtcD1) was previously identified by Petkovic and coworkers and was assigned to catalyze formation of the D ring (46). OxyN shows strong sequence similarity to second-ring cyclases (such as DpsY from the daunorubicin cluster [33]) and presumably catalyzes the aldol reaction between the C-5 carbonyl and the acidic C-14 methylene to form the C ring. Subsequent formation of the B ring should be spontaneous, as seen for other aromatic polyketides such as aklanonic acid (33). Formation of the A ring is presumably catalyzed by OxyI, a small protein that is homologous to MtmX. MtmX has been implicated in catalyzing the formation of the final ring during mithramycin biosynthesis (32).
Two S-adenosylmethionine (SAM)-dependent methyltransferases (OxyF and OxyT) are encoded in the gene cluster. OxyT shows high sequence similarity to O-methyltransferases and is cotranscribed with OxyQ, OxyR, and OxyS (OtcC) (37), and it may therefore be associated with these downstream tailoring steps. We putatively assigned OxyT to dimethylation of the free amine present in 4-amino-ATC to yield ATC. OxyF is putatively assigned to methylate C-6 of pretetramid that yields 6-methyl-pretetramid. Four oxygenase genes are present in the oxy gene cluster, including that for the previously identified ATC oxygenase OxyS (OtcC) (2, 45). Interestingly, all four oxygenases are homologous to enzymes encoded in the mithramycin gene cluster in Streptomyces argillaceus (32). OxyE, OxyL, OxyG, and OxyS (OtcC) show strong sequence similarities to MtmOI, MtmOII, MtmOIII, and MtmOIV, respectively, indicating that the oxy and mtm gene clusters are evolutionarily closely related. Prado and coworkers showed that MtmOII is involved in an early hydroxylation step to yield the mithramycin precursor 4-demethylpremithramycinone (47). Thus, OxyL is assigned to catalyze an analogous reaction in the biosynthesis of compound 1, using 6-methyl-pretetramid as a substrate. OxyG is a small (11-kDa) quinone-forming oxygenase and is also homologous to ElmH from the tetracenomycin gene cluster (48). OxyG is therefore possibly involved in the quinone formation of ring A in 4-keto-ATC. OxyE is a flavin adenine dinucleotide-dependent monooxygenase and is putatively assigned to catalyze the C-5 oxidation of 5a,11a-dehydrotetracycline to yield 5a,11a-dehydrooxytetracycline.
The last step in the enzymatic cascade is the reduction of 5a,11a-dehydrooxytetracycline to compound 1. The gene encoding the reductase (TchA) responsible for the same step in chlorotetracycline biosynthesis was mapped outside the chlorotetracycline gene cluster (42). We have used degenerate primers and isolated a homologous ORF in the S. rimosus genome (result not shown).
Enzymes putatively involved in malonamyl starter unit biosynthesis.
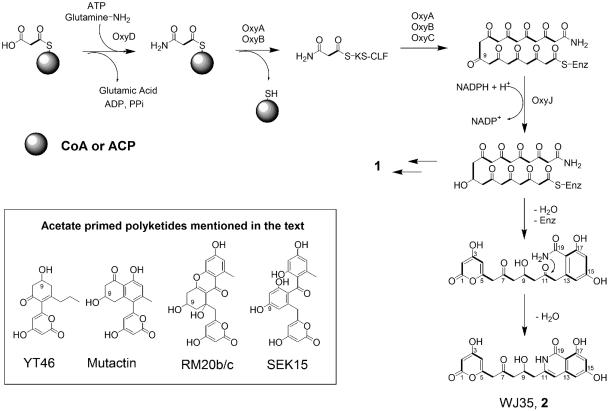
We previously showed that nonacetate-primed aromatic PKSs contain discrete initiation modules that typically consist of (i) a second ACP (ACPp) that primes the minimal PKS with the nonacetate unit (34), (ii) a ketosynthase III that synthesizes the ACPp-bound primer unit (39), and (iii) a potent acetyl-ACP thiolase that hydrolyzes the competing acetyl-ACP species that may otherwise initiate polyketide assembly with acetate (56). An ORF (oxyP) 15 kb downstream of the minimal PKS genes encodes an enzyme of high sequence homology to acetyl-ACP thiolases, such as ZhuC from the R1128 PKS (34) (49% identity) and FrnK from the frenolicin PKS (1) (43% identity). Based on sequence homology, OxyP may play an analogous role in the biosynthesis of compound 1 to ensure correct chain initiation by the malonamyl starter unit.
Surprisingly, no homologs of ketosynthase III and ACPp are present in the oxy cluster, suggesting that the oxy-encoded PKS employs a distinct set of enzymes to initiate polyketide biosynthesis. OxyD, encoded immediately downstream of the minimal PKS genes (oxyABC), is a 613-amino-acid protein that shows high sequence identity (60%) to the type II (also known as the Ntn family) asparagine synthases (31). Asparagine synthase converts aspartic acid to asparagine in an ATP-dependent two-step reaction, using glutamine or ammonia as the amine donor (3-5). OxyD contains the conserved N-terminal nucleophilic cysteine (Cys2) which is involved in the hydrolysis of glutamine (64) as well as the conserved adenylation domain that activates the acid moiety of the amine acceptor (6). We hypothesize that OxyD may therefore amidate either malonyl-coenzyme A (malonyl-CoA) or malonyl-ACP to yield malonamyl-CoA or malonamyl-ACP (see Fig. 4), which then primes the oxy-encoded KS-CLF for chain elongation.
FIG. 4.
Biosynthesis of compound 2 by the extended oxy-encoded minimal PKS.
Biosynthesis of an amidated polyketide by extended minimal oxy-encoded PKS.
To examine the roles of OxyD in the biosynthesis of an amidated polyketide in vivo, a series of Streptomyces coelicolor shuttle vectors derived from pRM5 were constructed (35). The plasmids were transformed into S. coelicolor strain CH999 (35) and were analyzed for polyketide biosynthesis (Table 2).
The minimal oxy-encoded PKS (oxyABC, in pYT319) produced ample amounts of SEK15 (see Fig. 4) (53) (30 mg/liter), while the addition of OxyJ (pYT318) yielded reduced decaketide RM20b as the major product (30 mg/liter), consistent with the data reported by Fu et al. (19). OxyJ is therefore the regiospecific C-9 ketoreductase in the oxy-encoded PKS. Therefore, to reconstitute the steps of oxytetracycline biosynthesis, OxyJ was coexpressed in all subsequent studies.
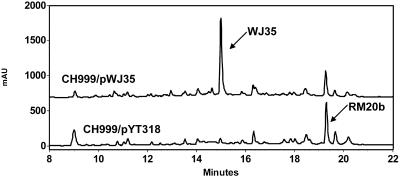
Significantly, a new polyketide product was identified when OxyD was coexpressed with the minimal oxy-encoded PKS and OxyJ. The oxyD gene encoding the amidotransferase was cloned into pYT318 to yield pWJ35. CH999 transformed with pWJ35 produced an intense yellow pigmentation on solid R5 medium that was not observed before. HPLC analysis of the CH999/pWJ35 extract revealed that a new metabolite (WJ35, or compound 2) was synthesized as the major product, with an excellent yield (>20 mg/liter) (Fig. 3). The acetate-primed RM20b product was also synthesized by this strain, although in much lower quantities (<5 mg).
FIG. 3.
HPLC analysis (254 nM) of organic extracts from CH999/pWJ35 and CH999/pYT318. The major biosynthetic products are indicated with arrows. The traces are not drawn to the same scale for comparison purposes.
The mass ([M + H]+) of compound 2 was detected to be 388. LC-MS analysis of the CH999/pYT318 extract with positive ion extraction for m/z 388 showed that no trace of compound 2 was produced in the absence of OxyD, confirming the essential role of OxyD in the biosynthesis of compound 2. A sufficient amount of compound 2 was extracted from 2 liters of CH999/pWJ35 fermentation and was purified to homogeneity using reverse-phase preparative HPLC and normal-phase thin-layer chromatography. HRMS indicated a molecular formula of C19H17NO8 (m/z = 388.1027 [M + H]+; difference of 0.0016), consistent with the molecular composition of an amidated decaketide that has been subjected to one ketoreduction at C-9 and two cyclization/dehydration events. When OxyJ was replaced by the heterologous ActIII KR (pWJ35a; Table 2), no change in yield and selectivity of compound 2 was observed.
The structure of compound 2 was elucidated using a combination of 1D and 2D NMR experiments (Table 3) and is shown in Fig. 4. Data obtained from 13C, distortionless enhancement by polarization transfer (90, 135), and HMQC experiments identified 19 carbon signals, supporting the number of carbon atoms predicted by HRMS. No methyl carbon atom is present, consistent with the lack of an acetate starter unit. The presence of the α-pyrone moiety as a result of O-1/C-5 cyclization is clearly evident from both 1H (δH2 = 5.3 and δH4 = 6.1) and 13C (63) data. The C-6 methylene was assigned using 1H-13C HMBC (Table 3). The linear connectivities among C-6 to C-9 were readily established using correlation spectroscopy and 1H-13C HMBC. The 1H multiplet at δH 4.5 is assigned to H-9 and is nearly identical to the H-9 protons present in mutactin (66) and YT46 (57), with each containing a 9-OH as a result of C-9 ketoreduction (Fig. 4). The assignments of the C-8/C-10 methylenes were readily achieved using correlation spectroscopy, HMBC, and HMQC experiments. The key assignment of the C-7 aliphatic ketone enabled us to eliminate the common C-7/C-12 intramolecular cyclization observed among aromatic polyketides (36). The remaining carbon backbone cyclizes and dehydrates through a C-13/C-18 intramolecular aldol condensation, which is confirmed by the coupling between H-14 and H-16 (JHH = 2.1 Hz). The nucleophilic amide group is thus favorably positioned to attack the electrophilic C-11 to yield, after dehydration, an isoquinolone heterocycle. The sharp proton singlet at δ 12.9, which shows long-range coupling to C-16, C-17, and C-18, is assigned to OH-17. The broad singlet at δ 10.2 is assigned to the NH proton. These assignments are consistent with the isoquinolone protons observed in the natural product fredericamycin (7). The presence of a lactam ring was unambiguously supported by the 1H-15N HMBC experiment, in which both H-10 and H-12 showed long-range coupling to the nitrogen (Table 3). Therefore, WJ35 is a shunt product of the oxy-encoded PKS in the absence of the first-ring cyclase.
OxyD does not interact with heterologous minimal PKSs.
We assayed whether heterologous minimal PKSs that are normally primed by acetate can interact productively with OxyD and OxyC to yield amidate polyketides. The act-encoded KS-CLF (65) and the tcm-encoded KS-CLF (54) were each coexpressed in the presence of Act KR, OxyD, and OxyC in CH999. Heterologous KS-CLFs have been shown to function with the OxyC ACP to produce acetate-primed compounds (26). To our surprise, no amidated polyketides were recovered from these strains (Table 2). We were able to detect only acetate-primed mutactin and RM20b from CH999/pWJ40 and CH999/pWJ48, respectively. This is in sharp contrast to the broad compatibility between the R1128 initiation module and heterologous minimal PKSs (57). Hence, fundamental differences exist between oxy-encoded and other minimal PKSs that allow the former to accept both amide and acetate starter units.
DISCUSSION
The tetracyclines are universally primed with a unique malonamate starter unit not found in any other polyketides. Past work has cloned five oxy (otc) genes and shed light on the minimal PKS. However, the initiation mechanism that leads to the production of the unique malonamate starter unit is not well understood. The lack of this information has hampered efforts to engineer the initiation of tetracycline biosynthesis. Understanding the biosynthesis and incorporation of the novel amide functionality is therefore a top priority in studying tetracycline biosynthesis.
In this work, we sequenced the previously mapped oxy (otc) gene cluster from S. rimosus, which allowed us to propose that OxyD is involved in the formation of the malonamate starter unit.
We reconstituted the minimal oxy-encoded PKS in CH999, as well as an extended minimal PKS including OxyD. Significantly, the coexpression of minimal PKS with the C-9 KR and OxyD produced a unique, amidated polyketide 2 at a high yield (Table 2). Establishing the origin of the amide unit and constructing the complete backbone required for tetracycline biosynthesis are significant steps towards rational bioengineering of this family of compounds. Isolation of this novel isoquinolone 2 revealed several important biochemical properties of the oxy-encoded PKS, as follows.
(i) OxyD is the only enzyme required to biosynthesize and insert an amide starter unit into the polyketide backbone in the heterologous host. CH999/pWJ35 produced predominantly the amidated polyketide, indicating preferential incorporation of a malonamyl unit over acetyl-OxyC by the oxy-encoded minimal PKS, even in the absence of an acetyl-ACP editing enzyme. A homologous enzyme (TcsG) from the chlorotetracycline PKS (43) likely performs the same catalytic function as OxyD during chlorotetracycline biosynthesis.
(ii) The amidated compound 2 is derived from an intact decaketide (10-carbonyl) backbone, indicating that in the heterologous host CH999, the oxy-encoded minimal PKS is able to maintain correct chain length control. We did not find any truncated polyketide products in the extract of CH999/pWJ35, presumably because these truncated products are present at much lower levels in this strain and may have escaped our purification and detection protocols. We did detect a truncated polyketide in a pWJ35-derived construct coexpressing OxyK (OtcD1) (unpublished data), consistent with the conclusion by Petkovic and coworkers that the oxy-encoded PKS also synthesizes truncated polyketides (46).
(iii) The most surprising structural feature of compound 2 is perhaps the lack of C-7/C-12 cyclization. The decaketide backbone must undergo two unique cyclization steps (C-13/C-18 and N-19/C-11) to yield the isoquinolone backbone. This is apparently a dominant mode of cyclization, since no alternatively cyclized amidated polyketides were detected in the fermentation extract. The cyclization regioselectivity of compound 2 was completely unanticipated, considering that C-9-reduced, acetate-primed decaketides cyclize solely between C-7 and C-12 (as observed in RM20b/c) (49). From the crystal structures of act-encoded KR (29) and act-encoded KS-CLF (25), it has been tempting to propose that the C-7/C-12 connectivity is formed within the active site of KS, prior to C-9 reduction by the KR. It is evident from the novel structural features of compound 2 that C-9 ketoreduction must take place independently of C-7/C-12 cyclization. OxyJ must therefore recognize an uncyclized polyketide backbone to yield compound 2.
(iv) Furthermore, formation of the isoquinolone must take place immediately after the complete assembly and release of the polyketide product. Premature C-13/C-18 cyclization will prevent the correct C-7/C-12 cyclization catalyzed by OxyK during the tailoring steps of tetracycline biosynthesis (Fig. 1).
Malonate is a very rare metabolite under normal physiological conditions and is a toxic compound due to its potent inhibition of succinate dehydrogenase in the tricarboxylic acid cycle (30), and hence it is unlikely to serve as a substrate for OxyD. Therefore, we hypothesize that biologically plausible malonyl substrates for OxyD are malonyl-CoA and malonyl-OxyC. Malonyl-OxyC may be preferred over malonyl-CoA for the following reasons. (i) Malonyl-CoA is involved in other essential cellular processes, including fatty acid biosynthesis. Amidation of a significant amount of the intracellular malonyl-CoA pool may therefore be detrimental to host strain viability (we did not observe any difference in the growth characteristics of CH999/pWJ35). Malonyl-OxyC, on the other hand, is dedicated to the oxy-encoded PKS and is not involved in the primary metabolism of the host. The robust growth of CH999/pWJ35 supports the above argument that the malonyl-CoA pool is not depleted. (ii) It is known that a cognate ACP-bound acyl substrate has a micromolar Km towards the KS-CLF, while the same acyl-CoA has Km values exceeding 100 μM (56). The lower Km of acyl-ACP is a result of extensive protein-protein interactions between the acyl-ACP and the KS-CLF. Therefore, under in vivo conditions, a significantly lower concentration of malonamyl-ACP than of malonamyl-CoA may be required to prime the KS-CLF. We are currently investigating the substrate specificities of OxyD in vitro.
It is unknown why heterologous pairing of minimal PKSs with OxyD failed to yield amidated polyketides. One likely possibility is that the exclusively acetate-primed KS-CLFs, including the act- and tcm-encoded PKSs, do not tolerate the presence of a polar starter unit in their active sites. Alternatively, OxyD may interact with the KS-CLF heterodimer to shuttle the malonamyl starter unit to the active site of KS and may require specific residues present on the surfaces of OxyA-OxyB. We have demonstrated that the chain length specificities of KS-CLFs can be drastically altered through rational mutagenesis (58). Therefore, the specificity of oxy-encoded KS-CLF may be similarly engineered to synthesize amidated polyketides of various lengths in coordination with OxyD.
While our in vivo results show that OxyD is a key determinant in the formation of the amide unit in WJ35 and, most logically, oxytetracycline, our results do not completely rule out a late role of OxyD during tetracycline assembly. In this model, the oxy-encoded PKS is primed directly by a malonyl group and proceeds with chain elongation. The acid-primed polyketide can then be amidated by OxyD to yield an amidated polyketide (WJ35) or can undergo spontaneous decarboxylation to yield an acetate-primed compound (RM20b, which will require the oxy-encoded KS to synthesize an 11-carbonyl, C21 backbone). However, it is difficult to hypothesize the exact timing and substrate of the OxyD-catalyzed amidation reaction in this model (e.g., partially elongated, linear, or cyclized). We are currently performing in vitro assays to unequivocally identify the possible malonyl substrates of OxyD, which will provide a definitive mechanism for the formation of the amidated unit.
Acknowledgments
This work was supported by a UCLA faculty research grant and University of California Cancer Research Coordinating Committee funds. The UCLA Pasarow Mass Spectrometry IonSpec 7.0T Ultima FTMS-MALDI and ESI sources are supported by NSF grant CHE0092036. Our NMR instrumentation is partially supported by the NSF under equipment grants CHE9974928 and CHE0116853.
We thank Chaitan Khosla, Christopher Boddy, and Iain Hunter for helpful discussions.
REFERENCES
- 1.Bibb, M. J., D. H. Sherman, S. Omura, and D. A. Hopwood. 1994. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene 142:31-39. [DOI] [PubMed] [Google Scholar]
- 2.Binnie, C., M. Warren, and M. J. Butler. 1989. Cloning and heterologous expression in Streptomyces lividans of Streptomyces rimosus genes involved in oxytetracycline biosynthesis. J. Bacteriol. 171:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehlein, S. K., N. G. Richards, and S. M. Schuster. 1994. Glutamine-dependent nitrogen transfer in Escherichia coli asparagine synthetase B. Searching for the catalytic triad. J. Biol. Chem. 269:7450-7457. [PubMed] [Google Scholar]
- 4.Boehlein, S. K., S. M. Schuster, and N. G. Richards. 1996. Glutamic acid gamma-monohydroxamate and hydroxylamine are alternate substrates for Escherichia coli asparagine synthetase B. Biochemistry 35:3031-3037. [DOI] [PubMed] [Google Scholar]
- 5.Boehlein, S. K., J. D. Stewart, E. S. Walworth, R. Thirumoorthy, N. G. Richards, and S. M. Schuster. 1998. Kinetic mechanism of Escherichia coli asparagine synthetase B. Biochemistry 37:13230-13238. [DOI] [PubMed] [Google Scholar]
- 6.Boehlein, S. K., E. S. Walworth, N. G. Richards, and S. M. Schuster. 1997. Mutagenesis and chemical rescue indicate residues involved in beta-aspartyl-AMP formation by Escherichia coli asparagine synthetase B. J. Biol. Chem. 272:12384-12392. [DOI] [PubMed] [Google Scholar]
- 7.Boger, D. L., O. Huter, K. Mbiya, and M. Zhang. 1995. Total synthesis of natural and ent-fredericamycin A. J. Am. Chem. Soc. 117:11839-11849. [Google Scholar]
- 8.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, M. J., C. Binnie, I. S. Hunter, D. A. Sugden, and M. Warren. 1990. Genetic manipulation of the oxytetracycline biosynthetic pathway genes. Dev. Ind. Microbiol. 31:41-50. [Google Scholar]
- 10.Butler, M. J., E. J. Friend, I. S. Hunter, F. S. Kaczmarek, D. A. Sugden, and M. Warren. 1989. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in the biosynthesis of oxytetracycline. Mol. Gen. Genet. 215:231-238. [DOI] [PubMed] [Google Scholar]
- 11.Caballero, J. L., E. Martinez, F. Malpartida, and D. A. Hopwood. 1991. Organisation and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol. Gen. Genet. 230:401-412. [DOI] [PubMed] [Google Scholar]
- 12.Carreras, C. W., and C. Khosla. 1998. Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry 37:2084-2088. [DOI] [PubMed] [Google Scholar]
- 13.Charest, M. G., C. D. Lerner, J. D. Brubaker, D. R. Siegel, and A. G. Myers. 2005. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science 308:395-398. [DOI] [PubMed] [Google Scholar]
- 14.Chopra, I. 2001. Glycylcyclines: third-generation tetracycline antibiotics. Curr. Opin. Pharmacol. 1:464-469. [DOI] [PubMed] [Google Scholar]
- 15.Chopra, I., P. M. Hawkey, and M. Hinton. 1992. Tetracyclines, molecular and clinical aspects. J. Antimicrob. Chemother. 29:245-277. [DOI] [PubMed] [Google Scholar]
- 16.Chung, J. Y., I. Fujii, S. Harada, U. Sankawa, and Y. Ebizuka. 2002. Expression, purification, and characterization of AknX anthrone oxygenase, which is involved in aklavinone biosynthesis in Streptomyces galilaeus. J. Bacteriol. 184:6115-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dairi, T., T. Nakano, K. Aisaka, R. Katsumata, and M. Hasegawa. 1995. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci. Biotechnol. Biochem. 59:1099-1106. [DOI] [PubMed] [Google Scholar]
- 18.Doyle, D., K. J. McDowall, M. J. Butler, and I. S. Hunter. 1991. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol. Microbiol. 5:2923-2933. [DOI] [PubMed] [Google Scholar]
- 19.Fu, H., S. Ebert-Khosla, D. Hopwood, and C. Khosla. 1994. Relaxed specificity of the oxytetracycline polyketide synthase for an acetate primer in the absence of a malonamyl primer. J. Am. Chem. Soc. 116:6443-6444. [Google Scholar]
- 20.Galm, U., J. Schimana, H. P. Fiedler, J. Schmidt, S. M. Li, and L. Heide. 2002. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch. Microbiol. 178:102-114. [DOI] [PubMed] [Google Scholar]
- 21.Hardman, J. G., and L. E. Limbird. 2001. The pharmacological basis of therapeutics. McGraw-Hill, New York, N.Y.
- 22.Hayashi, H., K. Inoue, T. Nagata, S. Kuramitsu, and H. Kagamiyama. 1993. Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry 32:12229-12239. [DOI] [PubMed] [Google Scholar]
- 23.Hopwood, D., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of streptomyces: a laboratory manual. The John Innes Foundation, Norwich, Conn.
- 24.Hunter, I. S. 2002. Tetracylines, p. 141-166. In J. F. Martin (ed.), Microbial secondary metabolites: biosynthesis, genetics and regulation. Research Signpost, Lucknow, India.
- 25.Keatinge-Clay, A. T., D. A. Maltby, K. F. Medzihradszky, C. Khosla, and R. M. Stroud. 2004. An antibiotic factory caught in action. Nat. Struct. Mol. Biol. 11:888-893. [DOI] [PubMed] [Google Scholar]
- 26.Khosla, C., R. McDaniel, S. Ebert-Khosla, R. Torres, D. H. Sherman, M. J. Bibb, and D. A. Hopwood. 1993. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J. Bacteriol. 175:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khosla, C., and Y. Tang. 2005. Chemistry. A new route to designer antibiotics. Science 308:367-368. [DOI] [PubMed] [Google Scholar]
- 28.Kim, E. S., M. J. Bibb, M. J. Butler, D. A. Hopwood, and D. H. Sherman. 1994. Sequences of the oxytetracycline polyketide synthase-encoding otc genes from Streptomyces rimosus. Gene 141:141-142. [DOI] [PubMed] [Google Scholar]
- 29.Korman, T. P., J. A. Hill, T. N. Vu, and S. C. Tsai. 2004. Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity. Biochemistry 43:14529-14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotlyar, A. B., and A. D. Vinogradov. 1984. Interaction of the membrane-bound succinate dehydrogenase with substrate and competitive inhibitors. Biochim. Biophys. Acta 784:24-34. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, T. M., S. K. Boehlein, S. M. Schuster, N. G. Richards, J. B. Thoden, H. M. Holden, and I. Rayment. 1999. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry 38:16146-16157. [DOI] [PubMed] [Google Scholar]
- 32.Lombo, F., G. Blanco, E. Fernandez, C. Mendez, and J. A. Salas. 1996. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene 172:87-91. [DOI] [PubMed] [Google Scholar]
- 33.Lomovskaya, N., Y. Doi-Katayama, S. Filippini, C. Nastro, L. Fonstein, M. Gallo, A. L. Colombo, and C. R. Hutchinson. 1998. The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J. Bacteriol. 180:2379-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marti, T., Z. H. Hu, N. L. Pohl, A. N. Shah, and C. Khosla. 2000. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist: insights into an unusual priming mechanism. J. Biol. Chem. 275:33443-33448. [DOI] [PubMed] [Google Scholar]
- 35.McDaniel, R., S. Ebert-Khosla, D. A. Hopwood, and C. Khosla. 1993. Engineered biosynthesis of novel polyketides. Science 262:1546-1550. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel, R., S. Ebert-Khosla, D. A. Hopwood, and C. Khosla. 1995. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature 375:549-554. [DOI] [PubMed] [Google Scholar]
- 37.McDowall, K. J., D. Doyle, M. J. Butler, C. Binnie, M. Warren, and I. S. Hunter. 1991. Molecular genetics of oxytetracycline production by Streptomyces rimosus, p. 105-116. In D. Noack, H. Krugel, and S. Baumberg (ed.), Genetics and product formation in Streptomyces. Plenum, New York, N.Y.
- 38.McMurry, L. M., and S. B. Levy. 1998. Revised sequence of OtrB (Tet347) tetracycline efflux protein from Streptomyces rimosus. Antimicrob. Agents Chemother. 42:3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meadows, E. S., and C. Khosla. 2001. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry 40:14855-14861. [DOI] [PubMed] [Google Scholar]
- 40.Mendez, B., C. Tachibana, and S. B. Levy. 1980. Heterogeneity of tetracycline resistance determinants. Plasmid 3:99-108. [DOI] [PubMed] [Google Scholar]
- 41.Moore, B. S., and C. Hertweck. 2002. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19:70-99. [DOI] [PubMed] [Google Scholar]
- 42.Nakano, T., K. Miyake, H. Endo, T. Dairi, T. Mizukami, and R. Katsumata. 2004. Identification and cloning of the gene involved in the final step of chlortetracycline biosynthesis in Streptomyces aureofaciens. Biosci. Biotechnol. Biochem. 68:1345-1352. [DOI] [PubMed] [Google Scholar]
- 43.Nakano, T., K. Miyake, M. Ikeda, T. Mizukami, and R. Katsumata. 2000. Mechanism of the incidental production of a melanin-like pigment during 6-demethylchlortetracycline production in Streptomyces aureofaciens. Appl. Environ. Microbiol. 66:1400-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peric-Concha, N. A., B. Borovicka, P. F. Long, D. Hrnaueli, P. G. Waterman, and I. S. Hunter. 2005. Ablation of the otcC gene encoding a post-polyketide hydroxylase from the oxytetracycline biosynthetic pathway in Streptomyces rimosus results in novel polyketides with altered chain length. J. Biol. Chem. 280:37455-37460. [DOI] [PubMed] [Google Scholar]
- 46.Petkovic, H., A. Thamchaipenet, L. H. Zhou, D. Hranueli, P. Raspor, P. G. Waterman, and I. S. Hunter. 1999. Disruption of an aromatase/cyclase from the oxytetracycline gene cluster of Streptomyces rimosus results in production of novel polyketides with shorter chain lengths. J. Biol. Chem. 274:32829-32834. [PubMed] [Google Scholar]
- 47.Prado, L., F. Lombo, A. F. Brana, C. Mendez, J. Rohr, and J. A. Salas. 1999. Analysis of two chromosomal regions adjacent to genes for a type II polyketide synthase involved in the biosynthesis of the antitumor polyketide mithramycin in Streptomyces argillaceus. Mol. Gen. Genet. 261:216-225. [DOI] [PubMed] [Google Scholar]
- 48.Rafanan, E. R., Jr., L. Le, L. Zhao, H. Decker, and B. Shen. 2001. Cloning, sequencing, and heterologous expression of the elmGHIJ genes involved in the biosynthesis of the polyketide antibiotic elloramycin from Streptomyces olivaceus Tu2353. J. Nat. Prod. 64:444-449. [DOI] [PubMed] [Google Scholar]
- 49.Rawlings, B. J. 1999. Biosynthesis of polyketides (other than actinomycete macrolides). Nat. Prod. Rep. 16:425-484. [DOI] [PubMed] [Google Scholar]
- 50.Rhodes, P. M., N. Winskill, E. J. Friend, and M. Warren. 1981. Biochemical and genetic characterization of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. J. Gen. Microbiol. 124:329-338. [Google Scholar]
- 51.Ryan, M. J., J. A. Lotvin, N. Strathy, and S. F. Fantini. February1999. Cloning of the biosynthetic pathway for chlortetracycline and tetracycline formation and cosmids useful therein. U.S. patent 5,866,410.
- 52.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 53.Shen, B., and C. R. Hutchinson. 1996. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc. Natl. Acad. Sci. USA 93:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, B., and C. R. Hutchinson. 1993. Enzymatic synthesis of a bacterial polyketide from acetyl and malonyl coenzyme A. Science 262:1535-1540. [DOI] [PubMed] [Google Scholar]
- 55.Tamura, N., S. Konishi, and A. Yamaguchi. 2003. Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr. Opin. Chem. Biol. 7:570-579. [DOI] [PubMed] [Google Scholar]
- 56.Tang, Y., A. T. Koppisch, and C. Khosla. 2004. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry 43:9546-9555. [DOI] [PubMed] [Google Scholar]
- 57.Tang, Y., T. S. Lee, and C. Khosla. 2004. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLOS Biol. 2:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang, Y., S. C. Tsai, and C. Khosla. 2003. Polyketide chain length control by chain length factor. J. Am. Chem. Soc. 125:12708-12709. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, R., and D. J. Williams. 1983. Oxytetracycline biosynthesis: mode of incorporation of [1-13C]- and [1,2-13C]-acetate. J. Chem. Soc. Chem. Commun. 1983:128-130. [Google Scholar]
- 60.Thomas, R., and D. J. Williams. 1983. Oxytetracycline biosynthesis: origin of the carboxamide substituent. J. Chem. Soc. Chem. Commun. 1983:677-679. [Google Scholar]
- 61.Xu, Z., K. Jakobi, K. Welzel, and C. Hertweck. 2005. Biosynthesis of the antitumor agent chartreusin involves the oxidative rearrangement of an anthracyclic polyketide. Chem. Biol. 12:579-588. [DOI] [PubMed] [Google Scholar]
- 62.Ylihonko, K., J. Tuikkanen, S. Jussila, L. Cong, and P. Mantsala. 1996. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-block mutations in the anthracycline pathway. Mol. Gen. Genet. 251:113-120. [DOI] [PubMed] [Google Scholar]
- 63.Yu, T. W., Y. Shen, R. McDaniel, H. G. Floss, C. Khosla, D. Hopwood, and B. S. Moore. 1998. Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J. Am. Chem. Soc. 120:7749-7759. [Google Scholar]
- 64.Zalkin, H. 1993. The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 66:203-309. [DOI] [PubMed] [Google Scholar]
- 65.Zawada, R. J., and C. Khosla. 1999. Heterologous expression, purification, reconstitution and kinetic analysis of an extended type II polyketide synthase. Chem. Biol. 6:607-615. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H. L., X. G. He, A. Adefarati, J. Gallucci, S. P. Cole, J. M. Beale, Jr., P. J. Keller, C. J. Chang, and H. G. Floss. 1990. Mutactin, a novel polyketide from Streptomyces coelicolor. Structure and biosynthetic relationship to actinorhodin. J. Org. Chem. 55:1682-1684. [Google Scholar]