Abstract
The soil bacterium Sinorhizobium meliloti establishes nitrogen-fixing symbiosis with its leguminous host plant, alfalfa, following a series of continuous signal exchanges. The complexity of the changes of alfalfa root structures during symbiosis and the amount of S. meliloti genes with unknown functions raised the possibility that more S. meliloti genes may be required for early stages of the symbiosis. A positive functional screen of the entire S. meliloti genome for symbiotic genes was carried out using a modified in vivo expression technology. A group of genes and putative genes were found to be expressed in early stages of the symbiosis, and 23 of them were alfalfa root exudate inducible. These 23 genes were further separated into two groups based on their responses to apigenin, a known nodulation (nod) gene inducer. The group of six genes not inducible by apigenin included the lsrA gene, which is essential for the symbiosis, and the dgkA gene, which is involved in the synthesis of cyclic β-1,2-glucan required for the S. meliloti-alfalfa symbiosis. In the group of 17 apigenin-inducible genes, most have not been previously characterized in S. meliloti, and none of them belongs to the nod gene family. The identification of this large group of alfalfa root exudate-inducible S. meliloti genes suggests that the interactions in the early stages of the S. meliloti and alfalfa symbiosis could be complex and that further characterization of these genes will lead to a better understanding of the symbiosis.
The soil bacterium Sinorhizobium meliloti establishes nitrogen-fixing symbiosis with its legume plant partner, alfalfa (Medicago sativa), through a series of continuous signal exchanges that results in structural changes of alfalfa roots (16, 21, 38, 57). The first clear structural change is the formation of tightly curled alfalfa root hairs with S. meliloti cells trapped in the middle (21). The presence of bacterial cells inside curled root hairs leads to the formation of tube-like infection threads (23, 35, 55). These infection threads extend toward the bases of root hairs, branch into multiple infection threads, pass layers of root cortical cells, and reach the developing nodule primordia (21, 35, 55). Nodule primordia continue to generate new plant cells and form elongated indeterminate alfalfa nodules (56). S. meliloti cells are released from infection threads into the cytoplasm of new nodule cells and surrounded by plant cell membrane in the small nodule invasion zone (30). These S. meliloti cells differentiate into bacteroids and convert atmospheric nitrogen into ammonia (7, 45, 46).
This complex and interdependent nodule development is reflected in the complexity of the S. meliloti genome (24). Among the 6,204 possible open reading frames of S. meliloti, only 59% have been assigned a possible function (24), some of which are required for symbiosis. Most of the known symbiotic genes were identified through genetic screening as well as genomic approaches. The identification and analyses of genes required for S. meliloti-alfalfa symbiosis have contributed to a better understanding of the nitrogen-fixing symbiosis between bacteria and their host plants.
The identification of nodulation (nod) genes (32) and exopolysaccharide (exo) genes (37) revealed the initial interactions between the two organisms during symbiosis (21). The presence of Nod factor, which is synthesized by the nod gene products in response to alfalfa flavonoids, elicits the formation of curled root hairs, infection threads inside the root hairs, and nodule primordia in the root cortex (47, 51, 54, 58). The formation of infection threads also requires the presence of succinoglycan, an S. meliloti exopolysaccharide (9). Succinoglycan is synthesized by a group of 22 S. meliloti exo genes, with the exoY gene product carrying out the first step of synthesis (3, 27, 42, 53). The nodulation of alfalfa by an exoY null mutant is blocked at the formation of infection threads (9). The production of succinoglycan is regulated by signal transduction pathways that include the ExoR protein and the ExoS-ChvI two-component signal transduction system, which also control flagellum production (10, 15, 50). The sensory protein ExoS has a periplasmic sensing domain, which raises the possibility that the production of succinoglycan is regulated in response to plant signals during the formation of infection threads (10). A number of other S. meliloti genes, such as syrM and syrA (43), and products, such as EPSII (galactoglucan) (26, 28, 46), capsular polysaccharide (8, 18, 33, 46), lipid A (4, 19), and cyclic β-1,2-glucan (6, 13, 17), have been found to be required during the early stages of symbiosis. The identification of additional genes is needed for a better understanding of the interactions between S. meliloti and its plant host, alfalfa.
Loss-of-function mutagenesis has been one of the major approaches used to identify most of the S. meliloti symbiotic genes. This approach is most effective for identifying genes that function as single and essential factors for symbiosis. However, as shown in our recent very limited success in identifying just two new symbiotic genes after screening a group of 90 putative LysR-type transcriptional factors, loss-of-function mutagenesis has become a less effective approach in identifying new symbiotic genes (39).
Positive screening approaches that identify symbiotic genes based on their expression during symbiosis could be more effective in identifying symbiotic genes that may have multiple homologs and that are expressed only transiently during a particular stage of symbiosis. Microarray and proteomic approaches have been used in recent years to identify symbiotic genes based on their expression in bacteroids in nodule cells (1, 2, 14). These approaches, however, could be less effective in identifying genes expressed only in the invasion zones of nodules, which account for a very small portion of nodules. The genes that are only expressed in bacterial cells before the formation of nodules are even more difficult to track using microarray or proteomic approaches because of the difficulties in collecting materials.
Given our understanding and interest in identifying S. meliloti genes that function in the early stages of nodulation, in vivo expression technology (IVET), which has been used to identify S. meliloti genes that function in nodule development (44), seemed to be a suitable approach. This technology was originally developed to identify genes of pathogenic bacteria required for invading their host cells by identifying promoters that are activated during invasion (40, 48). The key to the success of this approach is the use of reporter genes to screen for promoters that are activated at a particular time. The S. meliloti exoY gene is required for the formation of infection threads, so it can be used as a perfect reporter gene to screen for promoters that are active during early stages of symbiosis.
In this paper, we report our efforts in identifying a group of 23 S. meliloti genes that are expressed in the early stages of the S. meliloti-alfalfa symbiosis, using an improved IVET approach.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used for this study are listed in Table 1. Escherichia coli strains used in this study were grown in Luria-Bertani (LB) medium at 37°C, and Sinorhizobium meliloti was grown at 30°C in LB medium with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LB/MC) and plated on LB/MC with 0.02% calcofluor to examine succinoglycan production (37). The following antibiotics were used at the concentrations indicated: ampicillin, 100 μg/μl; chloramphenicol, 10 μg/ml; gentamicin, 50 μg/ml; kanamycin, 25 μg/ml; neomycin, 200 μg/ml; spectinomycin, 100 μg/ml; streptomycin, 500 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | General purpose strain | 29 |
| MT616 | Cmr, MT607, pRK600 | 20 |
| S. meliloti strains | ||
| Rm1021 | Smr, wild type | 37 |
| RmAR9007 | Gmr, Rm1021 exoY-lacZ | 34 |
| RmHC10 | Nmr, Rm1021 dgkA135 | This work |
| Plasmids | ||
| pBBR1MCS3 | Tcr, broad host range | 36 |
| pRK600 | Cmr, helper plasmid | 20 |
| pMCS5 | Apr, cloning vector | 31 |
| pTR102 | Tcr, carries parDE locus | 60 |
| pHC77 | Spr, carries gfp gene | 11 |
| pHC123 | Tcr, promoter trap vector | This work |
| pHC153 | Tcr, pHC123 with exoY native promoter | This work |
| pHC169 | Tcr, pHC123 carrying fusion with SMb21651 promoter | This work |
| pHC170 | Tcr, pHC123 with cyaH open reading frame | This work |
| pHC172 | Spr, pMB393 with nodABCP-gfp fusion | This work |
Construction of promoter trap vector.
The promoter trap vector pHC123 is a derivative of the plasmid pBBR1MCS3 (36) with an additional stable region, the exoY and gfp genes, and the site for cloning S. meliloti genomic fragments. The plasmid pBBR1MCS3 belongs to the pBBR1MCS family of plasmids, which cannot be stably maintained in S. meliloti cells without antibiotic selection (22). The presence of antibiotics, however, will interfere with the growth of alfalfa plants. A DNA fragment containing the E. coli parD and parE genes was amplified by PCR, using plasmid pTR102 (60) as a template, and cloned between the XbaI and SacII sites of pBBR1MCS3. The presence of the parD and parE genes allows the plasmid to be maintained in S. meliloti cells without antibiotic selection (9). The transcriptional fusion of the exoY and gfp genes was first assembled in plasmid pMCS5 (31) and then cloned into pBBR1MCS3 (36). The 0.77-kb XhoI/NheI DNA fragment, containing the exoY gene with its Shine-Dalgarno sequence and the intergenic region between exoY and its downstream exoF gene, was generated by a PCR using S. meliloti genomic DNA as the template. The primers used in this PCR were the XhoI primer 5′ GCC TCG AGT AAC GTA ACG TAA ATG GAG TCA CCT 3′ and the NheI primer 5′ GCG CTA GCC ATT TTT GGA TAT TTC CGT TC 3′. The XhoI primer also contains stop codons in three different reading frames. A 0.86-kb NheI/SpeI DNA fragment containing the gfp open reading frame was generated using pHC77 (11) as the template and cloned into the NheI/SpeI site of pMCS5 (31). The gfp gene used here contains the S65T mutation and is also known as the super gfp gene. The NheI/SpeI DNA region containing the exoY and gfp genes was subcloned from the pMCS5 plasmid into the pBBR1MCS3 plasmid with the stability region to make the promoter trap vector pHC123.
Construction of promoter library.
To construct a promoter fusion library, S. meliloti Rm1021 genomic DNA was partially digested with Sau3AI, and fragments were separated based on size using agarose gel electrophoresis. DNA fragments in the 0.5- to 1-kb range were recovered from the agarose gel, cloned into the promoter trap vector pHC123, using the partial fill-in technique to increase the cloning efficiency (44), and electroporated into E. coli DH5α following procedures recommended by the manufacturer. Plasmid DNAs were purified separately from 50 randomly selected transformants and analyzed by digestion to determine the average size of S. meliloti genome DNA fragments in the promoter trap vector. A total of 70,000 independent transformants were collected and pooled to ensure that the promoter library has 95% coverage of the S. meliloti genome. The promoter library was transferred into S. meliloti strain RmAR9007 (34), which is the exoY null mutant, through conjugation. A total of 300,000 colonies were collected and pooled for RmAR9007 containing the promoter library to ensure the complete transfer of the library.
Targeted mutagenesis of S. meliloti gene.
The S. meliloti dgkA gene was mutagenized by plasmid insertion using a suicide plasmid through homologous recombination, as previously described (39). Specifically, a 0.3-kb DNA fragment containing the middle part of the dgkA gene was obtained by PCR. The fragment was cloned into the unique HindIII/BsrGI sites in the suicide plasmid pK19mob2ΩHMB. The constructed plasmid was electroporated into E. coli DH5α first and then moved into the wild-type strain S. meliloti Rm1021 using a helper strain, E. coli MT616(pRK600), in a triparental mating. The dgkA plasmid insertion mutant was selected on LB/MC agar containing streptomycin and neomycin. The mutagenized dgkA gene was transduced into the wild-type Rm1021 background, using S. meliloti phage φM12, before further examination.
Recording of GFP fluorescence of bacterial colonies.
Bacterial colonies were examined using a Zeiss Bio2 stereofluorescence microscope with a filter set for green fluorescent protein (GFP) fluorescence. Images of bacterial colonies were recorded using an Optronics Magnifire S99802 digital camera (Goleta, CA).
Removal of cells carrying housekeeping gene promoters by FACS.
Fluorescence-activated cell sorting (FACS) was used to separate cells based on promoter activities of the S. meliloti genomic DNA fragments in the promoter trap vector in free-living cells. Cells of strain RmAR9007 carrying the promoter library were collected from fresh LB/MC liquid culture, washed with phosphate-buffered saline (pH 7.4) (PBS) at 4°C, and resuspended in 1 ml ice-cold PBS. Before being sorted, cell suspensions were diluted to a concentration of 2 × 106 cells/ml in ice-cold PBS. Cells were separated into three groups by FACS (Moflo-MLS; Cytomation, Fort Collins, CO) at Albert Einstein College of Medicine, based on GFP fluorescence intensities. The intensity of GFP fluorescence of strain RmAR9007(pHC153), which expresses both the exoY and gfp genes from the native exoY promoter, was used as the standard to separate RmAR9007 cells carrying the promoter library. The cells of RmAR9007 carrying the promoter library and showing GFP fluorescence intensities similar to those of the cells of RmAR9007(pHC153) were collected as the dim sublibrary. Those showing high or low levels of GFP fluorescence intensity were collected as bright or dark sublibraries, respectively. The cells of each sublibrary were amplified with one overnight growth at 30°C and stored at −80°C for future screening.
Plant growth conditions.
Alfalfa plants growing in a hydroponic system with nitrogen-free Jensen medium were used to screen S. meliloti RmAR9007 cells carrying the dark sublibrary. Alfalfa (Medicago sativa) seeds were surface sterilized in 50% Clorox bleach and washed with sterilized distilled water as previously described (37). The surface-sterilized seeds were spread on wet cheesecloth suspended with an aluminum screen in 11-mm by 11-mm by 9-mm containers with 200 ml nitrogen-free Jensen liquid medium. Two ends of the cheesecloth were submerged in liquid medium to ensure that the cheesecloth remained wet. The containers with seeds were kept in the dark at 26°C for 40 h for seed germination and then exposed to light. The liquid medium was inoculated with 1 ml of bacterial suspension at an optical density at 600 nm (OD600) of 0.3. The plants were checked after 4 weeks of inoculation for pink nitrogen-fixing nodules.
Alfalfa plants growing in petri dishes were used to determine the ability of individual bacterial strains to establish symbiosis with alfalfa, as previously described (37). Briefly, alfalfa seeds were surface sterilized and allowed to germinate on 0.7% agar plates in the dark for 40 h. The seedlings were transferred to 9-mm by 9-mm square petri dishes containing Jensen agar medium at 6 seedlings/plate, inoculated with 1 ml (OD600, 0.03) S. meliloti cell suspension, and incubated with light at 26°C for 4 weeks. The alfalfa plants in the petri dishes were examined for height, color, and numbers of white and pink nodules.
Recovery of bacterial cells from root nodules.
Bacterial cells were recovered from alfalfa root nodules as previously described (37). Nodules were removed from roots, surface sterilized with a 2-minute incubation in 50% Clorox bleach, washed several times in sterilized water, and crushed inside the wells of 96-well plates in LB/MC liquid medium with 5.4% glucose. The suspension was then diluted 1:100 in the same medium and plated on LB/MC agar plates for bacterial colonies.
Detection of succinoglycan biosynthesis.
The production of succinoglycan by S. meliloti strains was determined as previously described (37). Briefly, bacterial cells were plated on LB/MC/calcofluor agar medium, incubated for 2 days, and examined under UV light for fluorescence.
Plant root exudate preparation and induction assay.
To prepare exudates, alfalfa seedlings were grown in a hydroponic system with liquid Jensen medium as described above and then inoculated with wild-type S. meliloti Rm1021. The liquid Jensen medium, containing bacterial cells and root exudate, was removed from containers on day 5 after inoculation. Bacterial cells and insoluble materials were removed from the liquid medium by centrifugation (13,000 rpm, 15 min). The supernatants were further purified by passage through a sterilization filter with a pore size of 0.22 μm and used as crude root exudates.
To determine the induction effect of root exudates, bacterial cells were collected from fresh LB/MC culture, washed with sterilized water, and resuspended in the alfalfa root exudates to an OD600 of 0.1. After an 18-hour incubation at 30°C, the specific GFP fluorescence intensities of bacterial cells in the root exudates were determined. The known alfalfa flavonoid apigenin was diluted to 2.5 μg/ml in liquid Jensen medium as a control.
Evaluation of specific GFP fluorescence intensity.
The specific GFP fluorescence intensity of a given bacterial cell suspension was determined by normalizing the intensity of its GFP fluorescence to its cell density. Each S. meliloti cell suspension was transferred to wells of a transparent 96-well plate and a black 96-well plate in equal amounts. The cell suspensions in the transparent 96-well plate were used to determine the cell density, using an absorbance microplate reader (Spectra Max 340PC; Molecular Device, Sunnyvale, CA), and cultures in the black 96-well plate were used to determine the intensities of GFP fluorescence, using a fluorescence microplate reader (Spectra Max Gemini XS; Molecular Device, Sunnyvale, CA). The GFP fluorescence intensity of each cell suspension was normalized to its cell density to generate the average specific GFP expression level. All experiments were replicated twice.
Sequence analysis.
Plasmid DNA was isolated from S. meliloti cells by using a QIAprep Spin miniprep protocol (QIAGEN) following the manufacturer's instructions and then electroporated into E. coli DH5α. Plasmid DNA was purified from E. coli DH5α using a QIAprep miniprep kit and sequenced at the University of Chicago Cancer Research Center DNA sequencing facility using the designed primer 5′-CGAACTGGCCGAGCGCGTC-3′. The sequence results were analyzed with BLASTN at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) and the Sinorhizobium meliloti strain 1021 genome project website (http://bioinfo.genopole-toulouse.prd.fr).
RESULTS
Construction of a promoter library covering the entire S. meliloti genome.
To construct a promoter library for the functional screen (Fig. 1), a promoter trap vector that can be stably maintained in S. meliloti cells without antibiotic selection was first constructed. This promoter trap vector, pHC123, contains a promoterless S. meliloti exoY gene followed by the gfp gene replacing the native exoF open reading frame. Both the exoY and gfp genes will be transcribed in one transcript and translated into two independent proteins. The levels of specific GFP fluorescence of bacterial cells, which reflect the amounts of GFP and the ExoY protein in cells, can be used to monitor the levels of exoY gene expression.
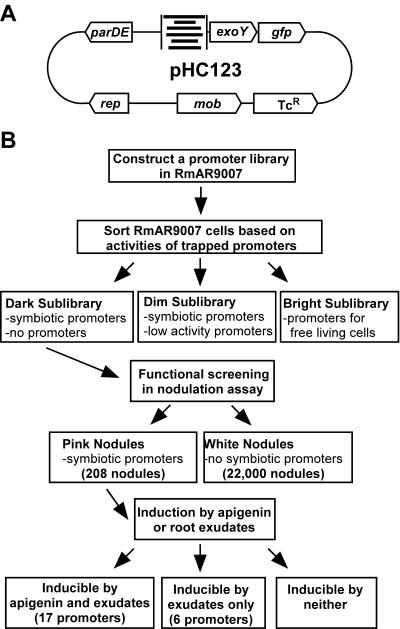
FIG. 1.
(A) Schematic representation of promoter trap vector showing relative positions of key genes, including exoY, gfp, and parDE, and the Rm1021 genomic DNA in front of the exoY gene. (B) Schematic representation of the overall strategy used in functional screening for S. meliloti genes that function in the early stages of S. meliloti-alfalfa symbiosis. The numbers in parentheses indicate either the numbers of nodules screened or the numbers of symbiotic promoters isolated.
To test the effectiveness of the promoter trap vector, the native exoY promoter region was cloned into the vector in front of the exoY gene to form plasmid pHC153 and tested in S. meliloti RmAR9007, a nonpolar exoY null mutant which is also defective in the production of galactoglucan II (EPS II). Strain RmAR9007(pHC153) produced succinoglycan at levels similar to those in the wild-type strain Rm1021 (Fig. 2) and produced high levels of GFP fluorescence (data not shown). More importantly, RmAR9007(pHC153) was able to establish a nitrogen-fixing symbiosis with alfalfa (see Fig. 4). The control strain RmAR9007(pHC123) did not establish symbiosis (data not shown). This confirms that the expression of the S. meliloti exoY gene from a plasmid can suppress the symbiosis deficiency of RmAR9007. This suggests that plasmid pHC123 can be used to trap promoters that can express the exoY gene during early stages of the symbiosis.
FIG. 2.
S. meliloti strains showing different intensities of calcofluor fluorescence, which reflect the levels of succinoglycan production by the strains. The average fluorescence intensities for the strains were determined using Kodak 1D analysis software. They are 105.5 ± 16.5 for Rm1021, 83.0 ± 5.8 for RmAR9007, 146.6 ± 33.8 for RmAR9007(pHC153), 193.7 ± 26.8 for RmAR9007(pHC169), and 80.0 ± 6.2 for RmAR9007(pHC170).
FIG. 4.
Four-week-old alfalfa plants inoculated with or without different S. meliloti strains. (A) No bacteria (control); (B) Rm1021; (C) RmAR9007(pHC170); (D) RmAR9007(pHC169).
To construct a promoter library for more effective promoter screening, a collection of 0.5- to 1-kb S. meliloti genomic DNA fragments was cloned in front of the exoY gene in pHC123 and transformed into E. coli DH5α. A total of 7 × 104 independent E. coli DH5α transformants were collected to ensure that the library has 95% coverage of the 6.7-Mb S. meliloti genome. The promoter library was transferred from E. coli DH5α into S. meliloti RmAR9007 through conjugation. A total of 3 × 105 independent conjugants were collected to ensure the complete transfer of the promoter library.
RmAR9007 cells carrying the promoter library formed colonies with different GFP fluorescence intensities (Fig. 3). About 10% of the colonies exhibited high levels of GFP fluorescence, and 90% displayed low-level or no GFP fluorescence. Plasmids pHC169 and pHC170 were isolated from two randomly picked colonies with high-level and no GFP fluorescence, respectively. The S. meliloti genomic regions of both plasmids were sequenced and identified by comparing them to the S. meliloti genome sequence. The plasmid pHC169 contains the promoter region of a putative open reading frame, SMb21651. The plasmid pHC170 contains a region from the middle of the S. meliloti cyaH gene. In addition, RmAR9007(pHC169) was able to produce succinoglycan, while RmAR9007(pHC170) did not (Fig. 2). In nodulation assays, plants inoculated with the wild-type strain were tall and green, with 4 to 12 pink nodules per plant, as expected, after 4 weeks (Fig. 4, and data not shown). The plants inoculated with RmAR9007 and RmAR9007(pHC170) were yellow and short, with 8 to 25 white nodules per plant (Fig. 4 and data not shown). The plants inoculated with RmAR9007(pHC153) (data not shown) and RmAR9007(pHC169) were tall and green with pink nodules, like those inoculated with the wild type (Fig. 4 and data not shown). These findings suggest that the presence of symbiotic promoters such as the exoY promoter in the promoter trap vector is essential for complementing the genomic exoY null mutation and establishing symbiosis. These results also showed that promoters that are highly active in free-living cells, such as the one in plasmid pHC169, could remain active during early stages of nodulation. These findings demonstrated that a nodulation assay could be used to screen the promoter library for the promoters that are active during symbiosis.
FIG. 3.
Colonies of strain RmAR9007 carrying the unsorted promoter library showing different intensities of GFP fluorescence. Colony C was formed by cells carrying plasmid pHC169, and colony A was formed by cells carrying plasmid pHC170. Colony B was formed by cells showing a low level of GFP fluorescence, which was not further analyzed.
Taken together, these findings suggest that a promoter library was constructed and that it can be used to identify symbiotic promoters based on their ability to express the exoY gene and suppress the symbiotic defect of the exoY null mutant.
Removal of promoters that are active in free-living cells from the promoter library.
The promoter library contains S. meliloti promoters for housekeeping genes that are expressed in both free-living and symbiotic cells. Some of the housekeeping promoters, like the one in plasmid pHC169, could also express the exoY gene and create false-positive nodule isolates. Cells carrying these promoters must therefore be removed from the promoter library.
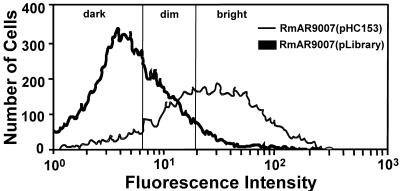
RmAR9007 cells carrying the promoter library were passed through a FACS instrument and separated into three populations, namely, bright, dim, and dark (Fig. 5). The native promoter activity of the exoY gene was used as a standard. RmAR9007 cells carrying the promoter library that had the same GFP fluorescence intensity as RmAR9007(pHC153) cells were collected as a dim sublibrary, which accounted for 25% of the total population. RmAR9007 cells carrying the promoter library which had higher GFP fluorescence intensities than RmAR9007(pHC153) cells were collected as a bright sublibrary, which accounted for 4% of the total population. RmAR9007 cells carrying the promoter library that showed no or lower GFP fluorescence intensity than RmAR9007(pHC153) cells were collected as a dark sublibrary, which accounted for 71% of the population.
FIG. 5.
FACS profiles of GFP fluorescence intensities of RmAR9007(pHC153) and RmAR9007 carrying the entire promoter library. The activities of the exoY promoter in free-living RmAR9007(pHC153) cells, which are reflected in the levels of GFP fluorescence of the cells, were used as a guide to determine the levels of GFP fluorescence to separate the cells of RmAR9007 carrying the promoter library into dark, dim, and bright populations. Cells with fluorescence intensities of <6.31 were collected as the dark sublibrary; cells with fluorescence intensities between 6.31 and 20.62 were collected as the dim sublibrary; and cells with fluorescence intensities of about 20.62 were collected as the bright sublibrary.
The effectiveness of FACS was further examined by plating cells on solid medium because the activity of the same promoter can vary in different cells (Fig. 5). The GFP fluorescence intensity of any given bacterial colony reflects the average activity of the trapped promoter. Colonies formed by cells from the dark sublibrary were almost all dark under a fluorescence microscope, which suggests that most cells in the dark sublibrary did not carry promoters that are active in free-living cells. A small number of colonies formed by cells from the dim sublibrary, however, exhibited high levels of GFP fluorescence, suggesting that the dim sublibrary contained promoters that are highly active in free-living S. meliloti cells. In an effort to remove these nonsymbiotic promoters, the entire dim sublibrary was plated on solid medium for single colonies. The colonies with bright GFP fluorescence were removed, while the dim and dark colonies were collected, pooled, and plated on solid medium for single colonies a second time. Unfortunately, a similar percentage of colonies in the dim sublibrary again showed high levels of GFP fluorescence. This suggests that the dim sublibrary contains housekeeping promoters that cannot be easily removed, which made it too difficult to use in the screening for symbiotic promoters.
Isolation of S. meliloti symbiotic promoters by functional complementation.
Using a special hydroponic system, a total of 2,400 alfalfa plants were used to screen the dark sublibrary. Most of the alfalfa plants were yellow and short, while only a few were slightly green 4 weeks later (data not shown). When nodules were examined, some of the plants had one or two pink nodules, and few plants had more than two pink nodules. The alfalfa plants that had one or a few nodules did appear to be slightly greener than those that did not have any pink nodules. After screening of 22,000 root nodules, a total of 208 pink nodules were identified, collected, surface sterilized, and crushed to recover bacterial cells from inside these nodules. Each of the 208 nodule isolates was purified by streaking to single colonies.
Each of the 208 nodule isolates was used to inoculate six alfalfa plants, all of which became green and tall, with 55 to 100% pink nodules, 4 weeks later (data not shown). Alfalfa plants inoculated with the negative control, RmAR9007(pHC123), were almost dead, with only white nodules. Alfalfa plants inoculated with the positive controls, Rm1021 and RmAR9007(pHC153), were tall and green, with 100% pink nodules. These results suggested that the 208 nodule isolates did indeed carry promoters that are active during early stages of the symbiosis.
The activities of the promoters in free-living cells were checked by determining the specific GFP fluorescence intensities of the 208 nodule isolates. The promoter activities in 25 nodule isolates were much higher than that of the exoY promoter in free-living cells, so the promoters were not symbiotic. The fact that such a small number of the nodule isolates contained nonsymbiotic promoters suggests that the prescreen using the gfp gene really worked. Altogether, these findings suggest that 183 nodule isolates most likely had promoters that were active during early stages of nodulation.
To quickly estimate how many of these 183 nodule isolates carried promoters of previously characterized genes and to determine their levels of duplication, plasmids were purified from 10 randomly selected nodule isolates. The DNA sequence of cloned genomic DNA showed that these 10 nodule isolates carried upstream regions of 10 different genes and putative genes of S. meliloti (Table 2). These genes and putative genes are fliF (SMc03014), SMc02737 (opuC), SMc03927 (nodN2), SMc03807 (amtB), SMc00880, SMc03746, SMa0538, SMb20600, SMb20399, and SMb20156. One of these 10 genes has been previously characterized, and three others were predicted based on homology. The rate of duplication might have been low within the 183 nodule isolates. These results suggested that further analysis of this set of nodule isolates would most likely lead to the discovery of more S. meliloti symbiotic genes.
TABLE 2.
Genes expressed during early stages of nodulation
| Gene or ORFa | Putative function |
|---|---|
| Genes or ORFs induced by alfalfa root exudates only | |
| lsrA (SMc00037) | Transcription regulator |
| dgkA (SMc04213) | Diacylglycerol kinase involved in recycling l-diacylglycerol generated during cyclic β-1,2-glucan synthesis |
| SMb20195 (ppe) | Pentose phosphate epimerase |
| SMc02171 | ABC transporter periplasmic binding protein, involved in cell processes and transport of small molecules |
| SMc02772R | Hypothetical protein |
| SMc03205R | Hypothetical protein |
| Genes or ORFs induced by both exudates and apigenin | |
| SMc00234 (ppiD) | Peptidyl-prolyl cis-trans isomerase |
| SMc00695 (aroK) | Shikimate kinase I |
| SMc01512 (hmuT) | Putative hemin binding and hemin transport |
| SMc02612 (glxD) | Oxidoreductase |
| SMc02653 (lepB) | Probable signal peptidase I transmembrane protein |
| SMb20162 | Putative LuxR family transcription factor |
| SMb20399 | Hypothetical protein |
| SMb20498 | Conserved hypothetical protein |
| SMb21400 | Conserved hypothetical protein |
| SMb21333 | Hypothetical protein |
| SMc00856 | Transmembrane conserved hypothetical protein |
| SMc02426 | Conserved hypothetical protein |
| SMc03130 | Conserved hypothetical protein |
| SMc04194 | Putative transmembrane protein |
| SMa1523R | Hypothetical protein |
| SMb20276R | Hypothetical protein |
| SMb20714R | Hypothetical protein |
| Partial list of genes not induced by exudates or apigeninb | |
| fliF (SMc03014) | FliF flagellar M-ring transmembrane protein |
| SMc02737 (opuC) | OpuC ABC transporter, glycine betaine |
| SMc03807 (amtB) | Ammonium transporter involved in nitrogen metabolism |
| SMc03927 (nodN2) | NodN2 putative nodulation protein |
| SMa0538 | Hypothetical protein |
| SMb20156 | ABC transporter ATP-binding protein |
| SMb20600 | Conserved hypothetical protein |
| SMc00880 | Oxidoreductase |
| SMc03746 | Hypothetical protein |
For previously characterized genes, the names of the genes are listed with the serial numbers of the genes designated by the sequencing project listed in the parentheses. For previously unknown genes, the serial numbers of the genes are listed with the names of the homologs proposed by genome sequencing projects listed in parentheses.
This group of genes was discovered when DNA sequences of trapped promoters from 10 randomly selected nodule isolates were determined. None of this group is inducible by root exudates or apigenin.
Selection of S. meliloti symbiotic genes that are activated during the early stage of symbiosis.
The expression of the S. meliloti genes that function only in the early stage of symbiosis could be activated by plant signals, which would most likely come from alfalfa root exudates. The specific GFP fluorescence intensities of each of the 183 nodule isolates in the presence and absence of alfalfa root exudates were determined (data not shown). Among the 183 nodule isolates, 37 isolates showed elevated promoter activity in the presence of alfalfa root exudates. These 37 nodule isolates may have genes from the nod gene family. To remove possible nod genes, the expression of the promoters in the 37 nodule isolates was further characterized in the presence or absence of apigenin, which is a known alfalfa root flavonoid that induces nod gene expression. Among the 37 nodule isolates, the activities of promoters in 27 nodule isolates were activated by apigenin. They were further characterized to identify the promoters and genes, as described below. The promoter activities in 10 nodule isolates were not activated by root apigenin (Fig. 6; for statistical analysis of the data included in Fig. 6, see the supplemental material). This raised the possibility that characterization of the promoters in these 10 nodule isolates could lead to the identification of previously unknown symbiotic genes.
FIG. 6.
Activities of promoters in the absence or presence of alfalfa root exudates or apigenin, as measured by specific GFP fluorescence intensities. (A) nodABC promoter; (B) exoY promoter; (C) SMb21651 promoter on plasmid pHC169; (D) genomic DNA containing no promoter from plasmid pHC170 (negative control); (E to J) promoters of the lsrA, dgkA, ppe, SMc02171, SMc02773R, and SMc03205R open reading frames, respectively.
Identification of S. meliloti genes induced by alfalfa root exudates but not apigenin.
To identify the promoters and genes carried by the 10 nodule isolates, the promoter trap vector was extracted from the nodule isolates. The genomic DNA fragments in the promoter trap vector were sequenced using a primer that anneals to the exoY open reading frame. The upstream ends of the fragments were estimated based on the sizes of the fragments. DNA sequencing showed that the genomic DNAs in the 10 nodule isolates originated from six different regions of the S. meliloti genome (Fig. 7; Table 2), which included three different putative promoters, two putative open reading frames, and one noncoding region.
FIG. 7.
Schematic representations of symbiotic promoters activated by alfalfa root exudates. The genomic locations of the trapped promoters are indicated by arrows and numbers above the diagrams of the open reading frames. The numbers reflect the genomic positions assigned by the genome sequencing project. *, positions of upstream ends of the promoter fragments were estimated based on the sizes of the inserts in the promoter trap vector. The names of the genes or putative open reading frames are indicated in the open arrows.
The S. meliloti genomic DNA fragment carried by the promoter trap vector in the nodule isolate J052 covers the entire promoter region of the open reading frame SMc00037 on the chromosome, which codes for the lsrA gene (Fig. 7). One putative promoter was identified using prokaryotic promoter predicting software (http://www.fruitfly.org). The putative promoter scored 0.4, with a score of 1.0 indicating an ideal promoter. The lsrA gene is predicted to encode a LysR-type transcriptional regulatory protein and has recently been identified as a symbiotic gene by our lab in a loss-of-function mutagenesis screen (39). The presence of the lsrA gene is essential for the early stage of nodule development (39). The fact that the promoter of the lsrA gene was isolated in our current promoter trap screen suggests that our screen is quite effective, and more importantly, the current results suggest that the expression of the lsrA gene is regulated by unknown factors in alfalfa root exudates. This provides a new avenue for analyzing the regulatory function of the LsrA protein.
Three nodule isolates, J003, J046, and J204, carry the identical S. meliloti genomic DNA fragments covering the promoter region of the dgkA gene (Fig. 7). One putative promoter scoring 0.9 was identified using prokaryotic promoter predicting software (http://www.fruitfly.org). This gene codes for a diacylglycerol kinase required for recycling l-diacylglycerol generated during cyclic β-1,2-glucan synthesis (41). The presence of cyclic β-1,2-glucan is required for the successful nodulation of alfalfa by S. meliloti (5). This is consistent with the finding that the expression of the gene is induced by alfalfa root exudates. The possible roles of the dgkA gene in symbiosis were further explored through the construction of a dgkA plasmid insertion mutant, which is described below.
The nodule isolate J072 carries S. meliloti genomic DNA that covers the promoter region of the putative open reading frame SMc02171 (Fig. 7). Two promoters were identified using prokaryotic promoter predicting software (http://www.fruitfly.org). The putative upstream promoter scored 0.9, while the putative downstream promoter scored 0.7. SMc02171 is predicted to encode a putative periplasmic binding protein of an ABC transporter, which is typically involved in the transport of small molecules. SMc02171 shares high levels of homology with the Agrobacterium tumefaciens C58 frcB gene, encoding 85% identical amino acids, and the Mesorhizobium loti MAFF303099 mlr7582 open reading frame, encoding 80% identical amino acids. The finding of these two close homologs further increases the chance that the SMc02171 open reading frame codes for a protein that plays important roles in symbiosis.
The genomic DNA in the promoter trap vector in nodule isolate J076 comes from the putative cbbFPTALSX (Calvin-Benson-Bassham) operon, encoding enzymes in the Calvin cycle (Fig. 7) (25). It is not known whether the gene products of this operon play any role in symbiosis. The genomic fragment in the promoter trap vector covers the second half of the putative cbbS (SMb20197) gene and the first half of the cbbX (SMb20196) gene, which is the last gene in the operon. Immediately following the cbbX gene, which stops at position 204392, is the putative ppe gene, a pentose phosphate epimerase gene, which starts at position 204392 in the same direction as the cbbFPTALSX operon. The ppe gene could be transcribed together with the cbb operon, but it might also be transcribed separately from the two promoters overlapping the cbbS and cbbX genes (24) (Fig. 7). These two promoters were identified using prokaryotic promoter predicting software (http://www.fruitfly.org). The putative upstream promoter scored 0.9, while the putative downstream promoter scored 0.8. A score of 1.0 is an ideal promoter. These findings increase the possibility that the ppe gene is expressed by these two promoters.
The promoter trap vectors in three nodule isolates, J132, J180, and J199, carry the exact same genomic fragment, covering most of the SMc02772 open reading frame (Fig. 7) (24). But the direction of the fragment showing promoter activities is opposite to that of the SMc02772 and SMc02773 open reading frames, which have been predicted to code for an ABC transporter for sugar (24). Two putative promoters were identified in this region opposite to that of the SMc02772 and SMc02773 open reading frames, using prokaryotic promoter predicting software (http://www.fruitfly.org). Both promoters scored 0.97. There are three possible open reading frames, SMc02773R1, SMc02773R2, and SMc02773R3, in the direction of the putative promoter (Fig. 7). The putative Smc02773R1 (436 amino acids [aa]) protein shares 28% identical amino acids with the Kineococcus radiotolerans SRS30216 TrkA-C protein, 26% identical amino acids with the Chlorobium phaeobacteroides BS1 HslU heat shock protein, and 26% to 32% identical amino acids with more than 10 hypothetical proteins from different bacteria, including Mesorhizobium loti, Burkholderia cenocepacia, Azotobacter vinelandii, Kineococcus radiotolerans, and Chlorobium phaeobacteroides. These findings raised the possibility that SMc02773R1 might encode a functional protein for symbiosis.
The nodule isolate J175 carries the promoter trap vector with an S. meliloti genomic DNA fragment covering the second half of the purU1 (SMc03205) gene and the intergenic region between purU1 and the open reading frame ipdA3 (SMc03204) (Fig. 7) (24). The PurU1 protein is a putative formyltetrahydrofolate deformylase which is involved in small-molecule metabolism and nucleotide biosynthesis (12). The direction of the trapped promoter, however, is opposite to that of the purU1 and ipdA3 (SMc03204) genes. Three putative promoters were identified in this region, using prokaryotic promoter predicting software (http://www.fruitfly.org). The middle promoter scored 0.9, while both the upstream and downstream promoters scored 0.6. There are three possible open reading frames overlapping the purU1 gene in the opposite direction. The putative Smc03205R1 (220 aa) protein shares 38 to 43% identities with the four hypothetical proteins of Kineococcus radiotolerans SRS30216, raising the possibility that SMc03205R1 might encode a functional protein. The putative Smc03205R2 (241 aa) protein shares 38 to 42% identical amino acids with four hypothetical proteins from Kineococcus radiotolerans SRS30216 and 29% identical amino acids with a putative Archaeoglobus fulgidus DSM 4304 acetylglutamate kinase. The putative Smc03205R3 (267 aa) protein shares 25% identical amino acids with the archaebacterium Haloarcula japonica TR-1 cell division protein FtsZ1, 38 to 42% identical amino acids with four hypothetical proteins of Kineococcus radiotolerans SRS30216, and 29% identical amino acids with a putative Archaeoglobus fulgidus DSM 4304 acetylglutamate kinase.
Altogether, four genes and two putative symbiotic genes have been identified by screening the promoter library. Further characterization of these genes will provide a better understanding of the roles of S. meliloti in the early stage of symbiosis.
The dgkA gene is not essential for symbiosis.
A loss-of-function mutant of the dgkA gene was constructed by targeted insertion of a suicide plasmid in the middle of the dgkA open reading frame using a previously described method (39). In the nodulation assay, the plants inoculated with the dgkA mutant were as green and healthy as those inoculated with the wild-type strain. The numbers of pink nodules on plants inoculated with the dgkA mutant were about the same as those on plants inoculated with the wild-type control (data not shown). This suggests that the presence of the dgkA gene is not essential for symbiosis and, presumably, that the function of the DgkA protein is replaced by other proteins.
Identification of genes induced by both root exudates and apigenin.
Further analysis of the 27 nodule isolates that carry root exudate- and apigenin-inducible promoters yielded a total of 17 different genes and putative genes (Table 2). Surprisingly, no nod gene was among them, even though apigenin is a known inducer of nod genes. Five of the genes, lepB, glxD, ppiD, aroK, and hmuT, have been named based on their homology to previously characterized genes in other bacteria. Little is known about their roles in S. meliloti-alfalfa symbiosis. All five genes are located on the chromosome but appear to be unrelated. Two of these, lepB and glxD, were found to have elevated levels of expression in bacteroids inside root nodules in a previous microarray analysis (1). The ppiD gene was expressed at lower levels in bacteroids (1). The last two of these genes, aroK and hmuT, were not detected in the microarray study (1). For the remaining 12 putative genes, the expression of 3 of them, SMb21333, SMb21400, and SMc04194, was found to be increased in bacteroids, the expression of 1 of them, SMc03130, was decreased, and the expression of the other 8 was not detected in previous microarray studies (1). These findings of additional apigenin-inducible S. meliloti genes raised the possibility that the interactions between S. meliloti and alfalfa at the early stage of symbiosis are much more complicated than previously thought.
DISCUSSION
The complexity of the S. meliloti-plant symbiosis suggests that more S. meliloti genes could be involved in symbiosis than is currently understood. Some of the symbiotic genes function as single and essential factors, which could have already been identified through saturation loss-of-function mutagenesis. Some of the symbiotic genes could be activated and remain highly expressed throughout the symbiosis. Those genes should be identified in microarray and proteomic analyses, which have identified at least 350 symbiotic genes that are highly expressed in bacteroids (1). Some of the symbiotic genes, however, may have multiple functional homologs and may be expressed only very briefly during the early stages of the symbiosis. Since the symbiosis is typically started by a few bacterial cells colonizing the right spot on alfalfa root hairs, it would be difficult to collect enough material for microarray or proteomic analyses for a genome-wide screen. To carry out a thorough genome-wide search for genes that are expressed in the early stages of the symbiosis, we have modified the IVET approach (40). The key modification we made was to construct a transcriptional fusion of the gfp gene and the reporter gene exoY. The use of the gfp gene made it possible to prescreen the library for nonsymbiotic genes. This modification made it possible to work with a much larger promoter library to screen the genome more effectively.
To ensure that most S. meliloti promoters were screened for their symbiotic activities, short genomic DNA fragments were used in the construction of the promoter library, which caused an increase in the size of the promoter trap library. The larger size of the promoter library also dramatically increased the number of clones carrying S. meliloti housekeeping promoters that would interfere with the screening for symbiotic promoters. These housekeeping promoters had to be removed from the promoter library before the screening. Anticipating this difficult task, we included the gfp gene in the same transcript following the exoY gene to monitor the promoter activities from the cloned genomic DNA fragments in front of the exoY gene. This is different from the method of differential fluorescence induction, which uses the expression of the gfp gene alone to select for promoters activated under conditions of interest (48). We first removed the promoters that were active in free-living cells and then carried out functional screening for promoters that were active during the early stage of symbiosis.
To remove the promoters of housekeeping genes, a particular level of gene expression has to be determined. One also has to take into consideration that some of the symbiotic genes might be expressed at low levels in free-living cells and at high levels during symbiosis. The exoY gene is expressed at low levels in free-living cells, which results in the production of succinoglycan (52). The expression of the exoY gene is regulated by the S. meliloti ExoR protein and the ExoS/ChvI two-component regulatory system, and this gene is highly expressed in the exoR95 and exoS96 mutant background (11, 49, 50). These findings raise the possibility that isolating promoters that have low activities, like the exoY promoter, could also lead to the isolation of symbiotic promoters that are up-regulated during symbiosis.
Cells of the S. meliloti exoY deletion mutant RmAR9007 carrying the promoter library were separated into three populations, bright, dim, and dark, based on the intensity of GFP fluorescence of individual cells using FACS. The bright population consisted of RmAR9007 cells carrying trapped S. meliloti promoters that are highly expressed in free-living cells, which formed the bright sublibrary. The bright sublibrary could include promoters that are highly active in both free-living and symbiotic cells, but they cannot be separated through the nodulation assay. The dim population consisted of RmAR9007 cells carrying trapped S. meliloti promoters that are expressed at low levels in free-living cells, which formed the dim sublibrary. The dim sublibrary could include symbiotic promoters that are expressed at low levels in free-living cells and high levels during symbiosis. This dim library, however, was contaminated with cells carrying highly active S. meliloti promoters, which made it impossible to screen for symbiotic promoters using the nodulation assay. This was perhaps the result of the variation of expression of the same promoters in different cells of the same strain (48). The dark population consisted of RmAR9007 cells carrying trapped S. meliloti promoters that are not active or are less active than the exoY promoter, which formed the dark sublibrary. Fortunately, the dark sublibrary did not produce noticeable numbers of bright colonies when examined under a fluorescence microscope, which allowed it to be used to screen for symbiotic promoters using the nodulation assay.
Root nodules are typically colonized by bacterial cells derived from the one or few bacterial cells that initiate root hair invasion, so a large number of nodules had to be screened to ensure that all trapped S. meliloti promoters in the dark sublibrary were screened. A total of 208 nodule isolates were collected after screening 22,000 alfalfa root nodules, which is five times the minimum requirement, 4,000 nodules, to cover the dark sublibrary. Further analyses of these 208 nodules isolates led to a group of six S. meliloti symbiotic genes or putative genes that function in the early stages of symbiosis.
Rediscovery of the lsrA gene.
The S. meliloti lsrA gene is predicted to encode a LysR-like transcription regulator, based on its DNA sequence (24). It was discovered in our recent study that LsrA plays an essential role in the development of nitrogen-fixing pink nodules (39). Alfalfa inoculated with the lsrA null mutant developed only white nodules, which suggests that LsrA functions in the early stage of nodule development. The identification of the lsrA gene in this screen not only further validated the effectiveness of the approach used in this study but also provided new leads in characterizing the symbiotic function of the LsrA protein. The fact that the lsrA promoter can be used to express the exoY gene effectively during nodulation suggests that the lsrA gene is likely expressed very early during symbiosis. The expression of the lsrA gene will be further analyzed for its possible links to the succinoglycan production genes. The finding of the induction of lsrA gene expression by root exudates provides an extra handle on identifying the mechanism or genes regulating the expression of the lsrA gene during symbiosis.
Discovery of the dgkA gene as a symbiotic gene.
The S. meliloti dgkA gene has long been linked to symbiosis through its role in the biosynthesis of cyclic β-glucan (5). The symbiotic phenotypes of any dgkA null mutants have not been reported. If dgkA is an essential symbiotic gene, then the loss of this gene would block symbiosis. Our finding that the dgkA null mutant remains symbiotically active would suggest that the dgkA gene is not essential and that the function of the DgkA protein might have been provided by other proteins in the dgkA mutant. Although no other dgkA homolog is found in the S. meliloti Rm1021 genome, SMc02382 encodes a conserved hypothetical protein which also contains a kinase catalytic domain. This protein may replace DgkA function. This is perhaps the reason why the dgkA gene has not been previously identified as a symbiotic gene in loss-of-function screening. The induction of the dgkA gene by root exudates provides a new avenue for studying the role of the dgkA gene and cyclic β-glucan in symbiosis. This also supports the method of finding symbiosis genes using IVET.
None of the 17 apigenin-inducible genes is a nod gene.
The results of the apigenin induction assay suggested that the trapped promoters in 27 of 37 nodule isolates could be related to nod genes. The trapped promoters from all 27 nodule isolates were sequenced. This led to the discovery of 17 apigenin-inducible S. meliloti genes, none of which was a known nod gene. This suggests that expression of the nod genes is regulated differently from that of the exo genes; the nod promoter cannot be used to express the exoY gene at the correct stage of symbiosis. More interestingly, the discovery of these unexpected apigenin-inducible S. meliloti genes suggests that the successful nodulation of alfalfa by S. meliloti might be more complex than previously proposed and that there are more S. meliloti genes involved in the early stage of the symbiosis.
Promoters and genes contained in the other 146 isolates.
The remaining 146 nodule isolates should carry promoters that are active during the early stages of symbiosis because most of them are not active or are expressed at very low levels in free-living cells. They could include promoters that are activated by as yet unknown plant or symbiosis signals or special microenvironments during symbiosis. Additional assays and conditions are being developed to help identify symbiosis-specific promoters from this group of promoters.
IVET is effective for genome-wide screening for symbiotic genes.
While 5 of 23 inducible S. meliloti symbiotic genes identified in this study have been previously identified as possible symbiotic genes by microarray analysis of S. meliloti genes expressed in bacteroids (1), the other 18 were not detected in the same microarray analysis. None of the 23 genes identified in this study have been described in proteomic studies of gene expression in bacteroids (14). S. meliloti genes that function in the early stages of symbiosis should be highly expressed in bacterial cells in infection threads and in the invasion zone of the nodules. The one or few root hairs containing infection threads, which lead to the development of nodules, typically degenerate during the early stage of nodule development, making it extremely difficult to collect enough RNA for microarray analysis (59). The invasion zone remains a small area inside indeterminate alfalfa nodules, which could account for a small portion of the total RNA extracted from the nodules. When total RNA is extracted from mature nodules, it could include a small amount of RNA from the bacterial cells in the invasion zone, but it might not include detectable amounts of RNA from bacterial cells inside infection threads. This could account for the fact that lsrA, dgkA, and other genes were isolated based on their function but not detected in other published microarray analyses of symbiotic genes. The genes identified in both this functional analysis and microarray analysis may function for an extended period of time during nodule development.
In summary, a group of previously unknown symbiotic genes has been identified through the use of a modified IVET approach, which suggests that more S. meliloti genes are involved in the early stages of the symbiosis than was previously known. Further characterization of these symbiotic genes will lead to a better understanding of the roles of S. meliloti in symbiosis. Our success also showed that this approach can be applied to studies of other symbiotic or pathogenic bacterium-host interactions.
Supplementary Material
Acknowledgments
We thank Hilda Ye of Albert Einstein College of Medicine for helping us carry out bacterial cell sorting using a fluorescence-activated cell sorter.
This work was supported by grants PSC-CUNY 652170034 and 662580035 and by a grant from NIH (5S06GM08225) to H.-P. Cheng.
Footnotes
Supplemental material for this article may be found at http://www.aem.asm.org/.
REFERENCES
- 1.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, A., H. Berges, E. Krol, C. Bruand, S. Ruberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Kuster, C. Liebe, A. Puhler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 3.Becker, A., A. Kleickmann, M. Keller, W. Arnold, and A. Puhler. 1993. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol. Gen. Genet. 241:367-379. [DOI] [PubMed] [Google Scholar]
- 4.Bhat, U. R., H. Mayer, A. Yokota, R. I. Hollingsworth, and R. W. Carlson. 1991. Occurrence of lipid A variants with 27-hydroxyoctacosanoic acid in lipopolysaccharides from members of the family Rhizobiaceae. J. Bacteriol. 173:2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breedveld, M. W., and K. J. Miller. 1994. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58:145-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breedveld, M. W., and K. J. Miller. 1995. Synthesis of glycerophosphorylated cyclic (1,2)-beta-glucans in Rhizobium meliloti strain 1021 after osmotic shock. Microbiology 141:583-588. [DOI] [PubMed] [Google Scholar]
- 7.Broughton, W. J., and X. Perret. 1999. Genealogy of legume-Rhizobium symbioses. Curr. Opin. Plant Biol. 2:305-311. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, G. R., L. A. Sharypova, H. Scheidle, K. M. Jones, K. Niehaus, A. Becker, and G. C. Walker. 2003. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, H. P., and S. Y. Yao. 2004. The key Sinorhizobium meliloti succinoglycan biosynthesis gene exoY is expressed from two promoters. FEMS Microbiol. Lett. 231:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiribau, C. B., C. Sandu, G. L. Igloi, and R. Brandsch. 2005. Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans. J. Bacteriol. 187:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickstein, R., T. Bisseling, V. N. Reinhold, and F. M. Ausubel. 1988. Expression of nodule-specific genes in alfalfa root nodules blocked at an early stage of development. Genes Dev. 2:677-687. [DOI] [PubMed] [Google Scholar]
- 14.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 15.Doherty, D., J. A. Leigh, J. Glazebrook, and G. C. Walker. 1988. Rhizobium meliloti mutants that overproduce the Rhizobium meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170:4249-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downie, J. A., and S. A. Walker. 1999. Plant responses to nodulation factors. Curr. Opin. Plant Biol. 2:483-489. [DOI] [PubMed] [Google Scholar]
- 17.Dylan, T., P. Nagpal, D. R. Helinski, and G. S. Ditta. 1990. Symbiotic pseudorevertants of Rhizobium meliloti ndv mutants. J. Bacteriol. 172:1409-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epple, G., K. M. van der Drift, J. E. Thomas-Oates, and O. Geiger. 1998. Characterization of a novel acyl carrier protein, RkpF, encoded by an operon involved in capsular polysaccharide biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 180:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson, G. P., A. Datta, R. W. Carlson, and G. C. Walker. 2005. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 56:68-80. [DOI] [PubMed] [Google Scholar]
- 20.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage, D. J., and W. Margolin. 2000. Hanging by a thread: invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 3:613-617. [DOI] [PubMed] [Google Scholar]
- 24.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, J. L., and F. R. Tabita. 1997. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bacteriol. 179:663-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 27.Glucksmann, M. A., T. L. Reuber, and G. C. Walker. 1993. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J. Bacteriol. 175:7033-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 93:8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch, A. M., M. Bang, and F. M. Ausubel. 1983. Ultrastructural analysis of ineffective alfalfa nodules formed by nif::Tn5 mutants of Rhizobium meliloti. J. Bacteriol. 155:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoheisel, J. D. 1994. Reaction of a rare cutter multiple cloning site. BioTechniques 17:456-459. [PubMed] [Google Scholar]
- 32.Jacobs, T. W., T. T. Egelhoff, and S. R. Long. 1985. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J. Bacteriol. 162:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlin, S., M. J. Barnett, A. M. Campbell, R. F. Fisher, and J. Mrazek. 2003. Predicting gene expression levels from codon biases in alpha-proteobacterial genomes. Proc. Natl. Acad. Sci. USA 100:7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller, M., A. Roxlau, W. M. Weng, M. Schmidt, J. Quandt, N. Karsten, D. Jording, W. Arnold, and A. Pühler. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant-Microbe Interact. 8:267-277. [DOI] [PubMed] [Google Scholar]
- 35.Kijne, J. W. 1992. The Rhizobium infection process, p. 349-398. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long, S. R. 1989. Rhizobium-legume nodulation: life together in the underground. Cell 56:203-214. [DOI] [PubMed] [Google Scholar]
- 39.Luo, L., S. Y. Yao, A. Becker, S. Ruberg, G. Q. Yu, J. B. Zhu, and H. P. Cheng. 2005. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J. Bacteriol. 187:4562-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 41.Miller, K. J., M. W. McKinstry, W. P. Hunt, and B. T. Nixon. 1992. Identification of the diacylglycerol kinase structural gene of Rhizobium meliloti 1021. Mol. Plant-Microbe Interact. 5:363-371. [DOI] [PubMed] [Google Scholar]
- 42.Muller, P., M. Keller, W. M. Weng, J. Quandt, W. Arnold, and A. Puhler. 1993. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol. Plant-Microbe Interact. 6:55-65. [DOI] [PubMed] [Google Scholar]
- 43.Mulligan, J. T., and S. R. Long. 1989. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics 122:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 45.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2:641-646. [DOI] [PubMed] [Google Scholar]
- 46.Pellock, B. J., H. P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters, N. K., J. W. Frost, and S. R. Long. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977-980. [DOI] [PubMed] [Google Scholar]
- 48.Rediers, H., P. B. Rainey, J. Vanderleyden, and R. De Mot. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed, J. W., M. Capage, and G. C. Walker. 1991. Rhizobium meliloti exoG and exoJ mutations affect the exoX-exoY system for modulation of exopolysaccharide production. J. Bacteriol. 173:3776-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed, J. W., J. Glazebrook, and G. C. Walker. 1991. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 173:3789-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Relic, B., X. Perret, M. T. Estrada-Garcia, J. Kopcinska, W. Golinowski, H. B. Krishnan, S. G. Pueppke, and W. J. Broughton. 1994. Nod factors of Rhizobium are a key to the legume door. Mol. Microbiol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 52.Reuber, T. L., S. Long, and G. C. Walker. 1991. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta examined using TnphoA fusions. J. Bacteriol. 173:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 54.Roche, P., P. Lerouge, C. Ponthus, and J. C. Prome. 1991. Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J. Biol. Chem. 266:10933-10940. [PubMed] [Google Scholar]
- 55.Timmers, A. C., M. C. Auriac, and G. Truchet. 1999. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126:3617-3628. [DOI] [PubMed] [Google Scholar]
- 56.van Brussel, A. A., T. Tak, K. J. Boot, and J. W. Kijne. 2002. Autoregulation of root nodule formation: signals of both symbiotic partners studied in a split-root system of Vicia sativa subsp. nigra. Mol. Plant-Microbe Interact. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 57.van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker, S. A., and J. A. Downie. 2000. Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant-Microbe Interact. 13:754-762. [DOI] [PubMed] [Google Scholar]
- 59.Wan, J., M. Torres, A. Ganapathy, J. Thelen, B. B. DaGue, B. Mooney, D. Xu, and G. Stacey. 2005. Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 18:458-467. [DOI] [PubMed] [Google Scholar]
- 60.Weinstein, M., R. C. Roberts, and D. R. Helinski. 1992. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa root nodules. J. Bacteriol. 174:7486-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.