Abstract
Gordonia sp. strain P8219, a strain able to decompose di-2-ethylhexyl phthalate, was isolated from machine oil-contaminated soil. Mono-2-ethylhexyl phthalate hydrolase was purified from cell extracts of this strain. This enzyme was a 32,164-Da homodimeric protein, and it effectively hydrolyzed monophthalate esters, such as monoethyl, monobutyl, monohexyl, and mono-2-ethylhexyl phthalate. The Km and Vmax values for mono-2-ethylhexyl phthalate were 26.9 ± 4.3 μM and 18.1 ± 0.9 μmol/min · mg protein, respectively. The deduced amino acid sequence of the enzyme exhibited less than 30% homology with those of meta-cleavage hydrolases which are serine hydrolases but exhibited no significant homology with the sequences of serine esterases. The pentapeptide motif GXSXG, which is conserved in serine hydrolases, was present in the sequence. The enzymatic properties and features of the primary structure suggested that this enzyme is a novel enzyme belonging to an independent group of serine hydrolases.
Phthalate diesters (PAEs), such as di-2-ethylhexyl phthalate (DEHP) and diisononyl phthalate, are used as plasticizers on a massive scale in the industrial production of plastics. The annual amount of PAEs produced was estimated to be several hundred kilotons for Japan alone in 1995 and is estimated to have been increasing. These compounds arise from domestic and industrial wastewater because PAEs easily leak from plastic goods, such as tableware, and might be discharged from plastic-producing plants. PAEs are recalcitrant compounds and have accumulated as environmental contaminants, and they have been detected in the water and sediments of rivers, seas, and lakes at many points around the world (10, 28, 35). PAEs are suspected to be endocrine-disrupting chemicals (5, 13, 16, 32, 37). Reports of developmental toxicity and teratogenic effects of these compounds on animals have been increasing (4, 17, 36).
Because PAEs are considered to be difficult to decompose, removal of these compounds from wastewater has become an issue. It was shown first that biodegradation of PAEs occurs in river water and in acclimated activated sludge (31). After this, microorganisms that could grow using PAEs as a sole carbon source were isolated from soils and enriched wastewater samples. The microorganisms were identified and were reported to be Rhodococcus erythropolis, Pseudomonas species, and members of the genus Corynebacterium (3, 21).
It was reported previously that phthalate esters are hydrolyzed by specific and/or nonspecific esterases in many tissues of mammals and plants and in microorganisms (6, 18, 24, 29, 33). Phthalate diesters are metabolized to monoesters in many tissues of mice (18) and in human blood plasma (33). The purified specific esterases from mouse hepatic microsomes and human saliva have activity that hydrolyzes phthalate diesters to the corresponding monoesters (18, 29). Purified wheat plant esterase exhibits activity that hydrolyzes DEHP to mono-2-ethylhexyl phthalate (MEHP) (20). Higher organisms, such as mammals and plants, metabolize phthalate diesters to the corresponding monoesters. No enzyme that hydrolyzes the monoesters from higher organisms has been reported yet. In microorganisms, an extracellar DEHP hydrolase isolated from the culture broth of a strain of R. erythropolis, strain S-1, has been well characterized and shown to be able to degrade DEHP and other PAEs to phthalic acid (21, 22). However, a two-step reaction in which n-butyl phthalate diesters are decomposed via the corresponding monoesters to phthalic acid has also been reported (2, 8). Recently, an enzyme that hydrolyzes lower monoalkyl phthalates, such as monobutyl phthalate, monopentyl phthalate, and monopropyl phthalate, was purified from Micrococcus sp. strain YGJ1 and characterized (27). This hydrolase has a molecular mass of 60 kDa and is composed of 27-kDa subunits, and its N terminus is homologous to that of the putative phthalate ester hydrolase deduced from pheA of Arthrobacter keyseri 12B (7).
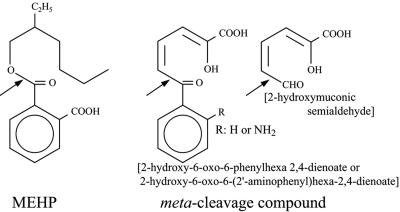
We isolated a new microorganism that dissimilates DEHP and is a member of the genus Gordonia. In this paper we present evidence that this strain has an intracellular hydrolase that catalyzes the decomposition of MEHP to phthalic acid, and we describe purification and characterization of this enzyme. Furthermore, we present evidence that the hydrolase is related phylogenetically to serine hydrolases that hydrolyze the meta-cleavage compounds that are intermediates in the metabolism of biphenyl, carbazole, and catechol in several bacteria (12, 14, 19, 30) (Fig. 1).
FIG. 1.
MEHP and meta-cleavage compound. If R is hydrogen, the compound is 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid metabolized from biphenyl. If R is NH2, the compound is 2-hydroxy-6-oxo-6-(2′-aminophenyl)hexa-2,4-dienoic acid metabolized from carbazole. 2-Hydroxymuconic semialdehyde is a metabolite from catechol. The arrows indicate the cleavage sites of MEHP and meta-cleavage compounds by the corresponding hydrolases.
MATERIALS AND METHODS
Chemicals.
DEHP was purchased from Sigma-Aldrich Japan K.K. (Tokyo, Japan). MEHP and other monoalkyl phthalates were obtained from Tokyo Kasei (Tokyo, Japan). KOD Plus DNA polymerase was obtained from Toyobo Biochemicals, Ltd. (Osaka, Japan). The other reagents were obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Isolation of DEHP-assimilating microorganisms.
A mineral agar medium without a carbon source (M9 agar medium) was prepared. This medium contained 6 g/liter disodium hydrogen phosphate, 3 g/liter potassium dihydrogen phosphate, 1 g/liter ammonium chloride, 0.5 g/liter sodium chloride, 0.011 g/liter calcium chloride, 0.246 g/liter magnesium sulfate hexahydrate, 5 mg/liter ferrous sulfate heptahydrate, 2.86 mg/liter boric acid, 0.22 mg/liter zinc sulfate heptahydrate, 0.08 mg/liter copper sulfate pentahydrate, 0.0021 mg/liter sodium molybdate, and 2% agar. Appropriate numbers of drops of DEHP were put on the agar media on which the supernatants of various soil suspensions had been spread. The agar plates were incubated at 25°C until colonies grew. The colonies were spread onto new mineral agar plates for purification and incubated as described above. The strain isolated has been deposited in the International Patent Organism Depositary (Tsukuba, Ibaraki, Japan) as strain FERM P-18102.
Determination of morphological and physiological properties.
Cell morphology was determined by differential interference microscopy, following growth on NB agar containing 2% glucose (9) at 25°C. Catalase activity was determined by bubble formation in a 3% hydrogen peroxide solution. Oxidase activity was determined by oxidation of 1% p-aminodimethylaniline oxalate. Nitrate reduction, acid fastness, and decomposition of urea were examined by using the methods described by Lanyi (25).
Extraction of DNA from the strain isolated.
The strain that was isolated was cultivated in NB agar containing 2% glucose at 25°C for 1 day and was harvested by centrifugation of 10,000 × g. DNA was extracted with an Isoplant 2 kit (Wako Pure Chemical Industries, Ltd.) used as recommended by the manufacturer.
Amplification of 16S rRNA gene and determination of the DNA sequence.
Two primers designed based on the 16S rRNA gene sequences conserved in bacteria, 16S-5′ (5′-TGGCGGCGTGGTTTAGGC-3′) and 16S-3′ (5′-GGTTACCTTGTTACGACT-3′), were used to amplify the 16S rRNA gene by PCR with extracted DNA and KOD Plus DNA polymerase. An ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA) was used for DNA sequencing according to the manufacturer's directions.
Cultivation of isolated DEHP-dissimilating microorganisms in a liquid medium to determine the ability to use alkyl phthalate esters.
M9 liquid medium was used to cultivate the newly isolated DEHP-decomposing organism. A loopful of cells was inoculated into 10 ml of M9 medium with an appropriate amount of DEHP in a 100-ml Erlenmeyer flask and incubated at 25°C on a rotary shaker (amplitude, 7 cm; 180 rpm) for 3 days. Then 2% of the culture was transferred to 100 ml of medium containing 10 ml of alkyl phthalate ester in a 300-ml flask for determination of growth.
Analyses for DEHP, phthalic acid, and number of cells in the culture broth.
To 100 ml of culture broth, 20 ml of 6 M HCl was added, and then the residual DEHP and newly generated phthalic acid were extracted with an equal volume of ethyl acetate. The ethyl acetate phase was evaporated, and the extracts obtained were resolved in a small amount of methanol and analyzed by reversed-phase high-performance liquid chromatography (HPLC). A C18 column (Wako handy ODS; Wako Pure Chemical Industries, Ltd.) was used. Elution was performed first with 20% methanol containing 10 mM phosphate for 5 min and then with a linear gradient of 20% methanol containing 0.1% (vol/vol) trifluoroacetic acid and methanol for 5 min, and then the elution solution was kept in methanol. Phthalate esters and phthalic acid were photometrically detected at 254 nm. The number of cells in the culture was determined by determining the amount of protein in the cells because the cells attached to DEHP oil drops and formed a heterogeneous suspension in the liquid medium. The cells harvested with oil drops from 100 ml culture broth were suspended with 20 ml of 1 M NaOH, boiled for 10 min, and neutralized with 20 ml of 1 M HCl. The suspension was centrifuged at 3,000 × g for 10 min. The concentration of protein in the resulting supernatant was determined.
Preparation of culture filtrate, cell extracts, and purification of MEHP hydrolase.
The cells were first cultivated in 200 ml of NB medium containing glucose in a 500-ml Erlenmeyer flask for about 70 h at 25°C, collected, and washed twice with an appropriate amount of sterile water, and then they were transferred into the same volume of M9 medium with 0.3 ml of DEHP to activate MEHP hydrolase and incubated at 25°C for 15 h. Culture filtrate and cells were obtained by centrifugation of 5 liters of culture broth at 15,000 × g for 15 min. The cells were suspended in an appropriate amount of 0.1 M potassium phosphate buffer, pH 7.5. The cells were disrupted with glass beads and a Dyno-mill (Willy A. Bachofen AG Machinenfabric, Basel, Switzerland) and centrifuged at 15,000 × g for 15 min to remove the cell debris and undisrupted cells. The supernatant was then centrifuged at 160,000 × g for 1 h to prepare cell extracts, which were transferred a DE-52 column (1.8 by 30 cm; Whatman International Ltd., Kent, England) equilibrated with 50 mM potassium phosphate buffer at pH 7.2, and the gel was washed with the same buffer. MEHP hydrolase was eluted with a 0 to 0.5 M NaCl linear gradient in the washing buffer. The fractions that had relatively strong MEHP hydrolase activity were collected and applied to a TOYOPEARL BUTYL-650 M column (1.6 by 20 cm; TOSOH Corp., Tokyo, Japan) equilibrated with 50 mM potassium phosphate buffer (pH 7.2) containing a 20% saturating concentration of ammonium sulfate. The enzyme was eluted with a linear gradient consisting of decreasing concentrations of ammonium sulfate. The fractions containing MEHP hydrolase were collected and concentrated by ultrafiltration with a YM10 membrane (Millipore Corp., Bedford, MA), and a 20% saturating concentration of ammonium sulfate was added. Then the solution was loaded on top of a gel filtration column (Superdex 200 pg; 2.0 by 50 cm; Amersham Biosciences Co., Piscataway, NJ) and eluted with 50 mM potassium phosphate buffer (pH 7.2) containing 0.5 M KCl. The fractions that had relatively high levels of activity were pooled and applied to a column of hydroxylapatite (Bio-Gel HTP; 1.5 by 8.5 cm; Bio-Rad Laboratories, Inc., Hercules, CA). The column was washed with an appropriate amount of 50 mM potassium phosphate buffer (pH 7.2). The enzyme that adsorbed to the gel was eluted with increasingly higher phosphate concentrations in the buffer.
Determination of the activity of MEHP hydrolase.
The standard reaction mixture (1 ml) for measuring the activity of MEHP hydrolase consisted of 100 mM potassium phosphate buffer (pH 7.2), an appropriate amount of enzyme solution, and 0.01 ml of a 20 mM MEHP-methanol solution. The reaction was performed in a tube incubated at 45°C for 5 or 15 min. The reaction rate was determined by two different methods using HPLC or a spectrophotometer. In the case of the HPLC method, the reaction was stopped by addition of 1 ml of 6 M HCl. The residual substrate and phthalic acid produced were extracted with 2 ml of ethyl acetate, which was evaporated. The extracts obtained were resolved in a small amount of methanol and analyzed by reversed-phase HPLC as described above. The activity of the enzyme was calculated from the rate of formation of phthalic acid. When the spectrophotometer was used, the decrease in absorbance at 242 nm of the reaction mixture was monitored. The initial reaction rates were calculated with an extinction coefficient between MEHP and phthalic acid at 242 nm of 1.83 mM−1 cm−1.
Identification of reaction products.
The phthalic acid formed in the reaction mixture was extracted and isolated by HPLC as described above. The eluate containing the phthalic acid produced was collected and lyophilized. About 50 μg of the lyophilized substance was dissolved in 10 μl methanol, and then an aliquot (1 μl) of the solution was mixed with 1 μl of glycerol and subjected to analysis by fast atom bombardment (FAB) mass spectrometry (MS) (JMS-AX505W; JEOL Ltd., Tokyo, Japan).
Electrophoresis of purified protein.
The purified protein samples were loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) performed on a 10% slab gel according to the method of Laemmli (23). Following electrophoresis, the gels were stained for protein with Coomassie brilliant blue G250.
Amino acid sequencing.
The purified MEHP hydrolase solution was dialyzed against ultrapure water and lyophilized. Dried MEHP hydrolase was digested with cyanogen bromide using the method of Gross (11). The newly generated polypeptides were isolated by reversed-phase HPLC with a C4 column (YMC Co., Kyoto, Japan) and were sequenced using an amino acid sequencer (model 610 A; Applied Biosystems, Foster City, CA).
Analysis of protein concentrations.
Protein concentrations were determined with a BCA protein assay kit (Pierce, Rockford, IL), using bovine serum albumin as the standard (34).
Procedures for sequencing the MEHP hydrolase gene.
Two kinds of degenerate oligonucleotides, 5′-AARTTYCAYCANGTNGA-3′ and 5′-CKYTCRTARAARTCCAT-3′ (where K = G or T, N = A or G or C or T, R = A or G, and Y = C or T), were synthesized on the basis of the sequences of the N terminus and one polypeptide generated by BrCN decomposition of MEHP hydrolase. We amplified a desired nucleotide by PCR using the oligonucleotides as primers and chromosomal DNA as the template. The amplified DNA was sequenced directly. An inverse PCR was performed with an alternative primer pair designed on the basis of the sequence and circular DNA formed by self-ligation of the fragments of chromosomal DNA that had been generated by digestion with an appropriate restriction enzyme. The entire sequence of the open reading frame (ORF) encoding MEHP hydrolase was determined by sequentially repeating inverse PCR procedures and was confirmed by sequencing the DNA amplified from the chromosomal DNA with primers designed based on the 5′ and 3′ flanking sequences of the ORF.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene of strain P8219 and the MEHP hydrolase gene have been deposited in the DDBJ nucleotide database under accession numbers AB054838 and AB214635, respectively.
RESULTS AND DISCUSSION
Isolation and characterization of a bacterium able to degrade DEHP.
A strain, which we designated P8219, was isolated from machine oil-contaminated soil in Kumamoto, and this strain had the ability to utilize DEHP as a sole source of carbon. Colonies of P8219 were glossy, light beige, smooth, and slimy in the early phase of growth and gradually became orange, rough, and dry after the DEHP disappeared. P8219 cells were gram positive, not acid fast, nonmotile, rod shaped or coccoid, and strictly aerobic. In a 9.5-h culture in NB medium, the dimensions of the rods were 0.5 to 0.9 μm by 3 to 5 μm. Cells occurred in pairs and very often in a typical coryneform V shape. In a 48-h culture, the cells were shorter, and the rods were 0.5 to 0.9 μm by 0.8 to 1.5 μm. In stationary cultures, the rods were fragmented into coccoid cells. P8219 could grow in the presence of 7% (wt/vol) NaCl. Acids were not produced from glucose, ribose, xylose, maltose, lactose, sucrose, maltose, or mannitol. We amplified the 16S rRNA gene of P8219 with a bacterium-specific primer pair, 16S-5′ and 16S-3′. The 1,457-bp amplified sequence was determined and compared with DNA sequences in databases. We found that P8219 shares 99.8% of its 16S rRNA gene nucleotides with Gordonia polyisoprenivorans (accession no. Y18310) (26); there are only four different nucleotides in the entire sequence. The morphological features and the 16S rRNA gene sequence indicated that P8219 belongs to the genus Gordonia.
P8219 was cultivated in M9 liquid medium supplemented with 1.5 ml of DEHP per liter. A typical time course for cultivation (Fig. 2) shows that the DEHP in the culture was rapidly degraded with the growth of cells and disappeared in 60 h. Floating drops of DEHP were observed in the first stage of cultivation on the surface of the culture medium, but they disappeared in a short time. White cloudy suspended substances appeared early in the culture period and remained, and the suspension was homogenous at the end. The presumed reaction product, phthalic acid, could not always be detected in the culture medium. Other phthalate diesters, including diethylphthalate, dinonylphthalate, and dihexylphthalate, were also tested for availability as sole carbon sources. All of these compounds except diethylphthalate were consumed with prolonged cultivation of P8219 under the same conditions that were used for DEHP.
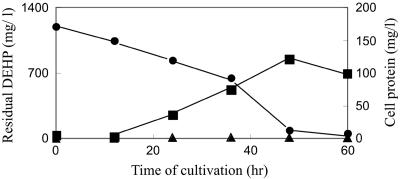
FIG. 2.
Consumption of DEHP and accumulation of phthalic acid along with growth of P8219. The strain was cultivated in M9 liquid medium containing DEHP. Residual DEHP (•) and newly generated phthalic acid (▴) were extracted with ethyl acetate from the acidified culture broth and analyzed by HPLC with a C18 column. The amount of cells (▪) was determined based on the protein extracted from the cells with 1 M NaOH.
Isolation and characterization of MEHP hydrolase.
The ability of the culture filtrate and cell extracts to degrade DEHP to phthalic acid was analyzed by HPLC. The culture filtrate had no such activity, whereas the cell extracts were able to hydrolyze DEHP to MEHP and MEHP to phthalic acid. These results suggested that DEHP was hydrolyzed via MEHP to phthalic acid by intracellular enzymes after uptake by the cell. The activity of MEHP hydrolase was enhanced 100-fold in the presence of DEHP in M9 medium as a carbon source (data not shown). Several rounds of chromatography were carried out to isolate MEHP hydrolase in cell extracts prepared from the cells grown in M9 medium with DEHP as the sole carbon source. The final eluate from Bio-Gel HTP had a specific activity of 17.0 U/mg protein after about 55-fold purification compared with the cell extracts and exhibited only one band on an SDS-PAGE gel. About 17 U of the purified enzyme (ca. 1 mg of protein) was obtained from a 5-liter culture (Table 1). In gel filtration (Superdex 200 pg) column chromatography, the enzyme eluted in the fraction corresponding to a molecular mass of 62 kDa. The SDS-PAGE gel showed that the protein has a molecular mass of 31 kDa. These results made it clear that MEHP hydrolase was a homodimeric protein.
TABLE 1.
Summary of purification of MEHP hydrolasea
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|
| Cell extract | 1,540 | 473 | 0.31 | 100 | 1 |
| DE52 | 20.9 | 154 | 7.37 | 32.6 | 23.8 |
| TOYOPEARL | 11.3 | 101 | 8.94 | 21.4 | 28.8 |
| Superdex | 3.55 | 37.0 | 10.4 | 7.82 | 33.5 |
| Bio-Gel HTP | 0.980 | 16.7 | 17.0 | 3.53 | 54.8 |
One unit was defined as the amount of protein that converted 1 μmol of MEHP per min to phthalic acid. Five liters of culture was used for purification. The reactions were conducted at 45°C for 5 min. HPLC was used for analysis of the activity.
Characterization of the purified enzyme.
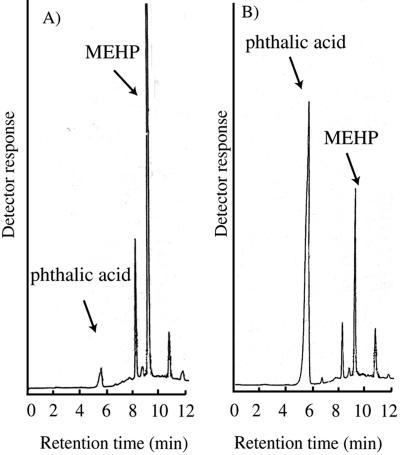
Figure 3 shows typical elution profiles obtained in HPLC analyses for hydrolysis of MEHP by the purified enzyme. A reaction product and MEHP eluted at about 5.3 min and 8.7 min, respectively (Fig. 3B). The reaction product was confirmed to be phthalic acid (molecular weight, 166) by mass spectrometry, as shown in Fig. 4. The molecule ion peak and dehydrate ion peak of phthalic acid were observed in the negative mode and in the positive mode of FAB-MS spectra, respectively. A comparison of the profile with the reaction (Fig. 3B) to the profile without the reaction (Fig. 3A) demonstrated that the amount of phthalic acid increased and the amount of MEHP decreased after the reaction. The molar ratios of the decreased area of MEHP to the increased area of phthalic acid were 1.01 ± 0.23 (mean ± standard error; n = 12). Thus, the stoichiometry of the reaction could be to be assumed as follows:
FIG. 3.
Typical HPLC elution profile. The purified enzyme (1.6 μg) was used for the reaction. MEHP was added to the reaction mixture at a concentration of 500 μM. The reaction conditions are described in the text. (A) Standard reaction mixture (1 ml) supplemented with 1 ml of 6 M HCl before initiation of the reaction. The relative areas of MEHP and phthalic acid were 9.16 × 105 and 4.36 × 104. (B) Standard reaction mixture incubated at 45°C for 15 min. The relative areas of MEHP and phthalic acid were 2.15 × 105 and 7.49 × 105. A C18 column was used. Elution was performed first with 20% methanol containing 10 mM phosphate for 5 min and then with a linear gradient of 20% methanol containing 10 mM phosphate and methanol alone for 5 min, and the elution solution was methanol by the end of the experiment. Phthalate esters and phthalic acid were photometrically detected at 254 nm.
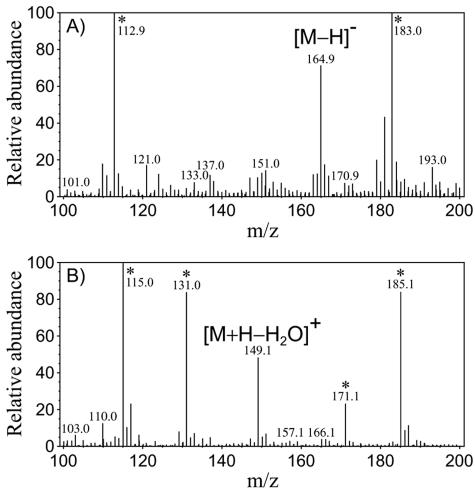
FIG. 4.
FAB-MS spectra of reaction product eluted by HPLC. (A) Negative mode. (B) Positive mode. One microliter of sample-methanol solution (∼5 mg/ml) was mixed with 1 μl glycerol and subjected to analysis. Xenon atoms were used to bombard the sample. The accelerating voltage was 10 kV. The FAB gun was operated at 3 kV. A fragment ion peak was not observed under the analysis conditions used. *, peaks are derived from glycerol.
MEHP + H2O → phthalic acid + 2-ethylhexanol
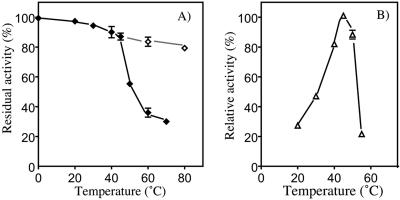
The enzyme was kept in 150 mM potassium phosphate buffer and Tris-HCl buffer at various pH values at 45°C for 10 min to study the effect of pH on the activity of the enzyme. The enzyme was stable between pH 6.0 and pH 9.0. The reaction was performed at various pH values. The relative activities were 50% at pH 6.0, 75% at pH 6.5, 95% at pH 7.0, 100% at pH 7.5, 85% at pH 8.0, 73% at pH 8.5, and 62% at pH 9.0. Thus, the optimum pH was 7.5. Furthermore, the enzyme was incubated at various temperatures for 10 min at pH 7.5. The enzyme was hardly inactivated at temperatures below 45°C and exhibited 85% of the original activity at 80°C if it was allowed to stand at 20°C for 30 min after heat inactivation (Fig. 5A). These results indicated that MEHP hydrolase might be easily and gradually refolded from a heat-denatured state. The activity of the purified enzyme was maximal at 45°C at pH 7.5 (Fig. 5B).
FIG. 5.
Effect of temperature on MEHP hydrolase stability and activity. (A) The enzyme was preincubated for 10 min at 0, 20, 30, 40, 45, 50, 60, and 70°C and immediately quenched on ice for 30 s (⧫) or incubated at 20°C for 30 min after quenching (⋄). Then the enzyme was added to the standard reaction mixture described in the text, which was incubated at 45°C to assay the activity. (B) Activity assayed at 20, 30, 40, 45, 50, and 55°C. The data are means and ranges (error bars) of values obtained from three measurements.
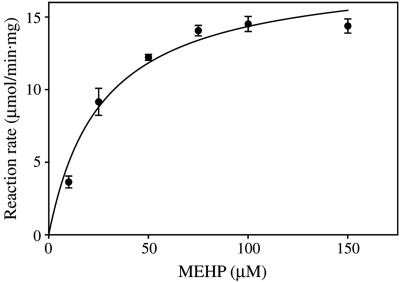
The specificity of the enzyme for various substrates was tested. Monoesters of benzoate, such as 2-ethylhexyl, n-hexyl, n-butyl, and ethyl benzoates, were not hydrolyzed by the purified enzyme. The enzyme could not utilize dialkyl phthalates, including DEHP, as substrates. On the other hand, all monophthalate esters listed in Table 2 were hydrolyzed. Figure 6 shows the regression curve calculated from the plots of reaction rates versus concentrations of MEHP. The Km and Vmax values for the monoesters calculated from regression curves are shown in Table 2. The Km values were similar. However, the Vmax decreased as the length of the alkyl chain decreased. The kcat/Km of the purified enzyme for monoalkyl phthalates was calculated by using the Km, Vmax, and a molecular weight of 64,000. The values were as follows: 3.09 × 105 ± 0.51 × 105 M−1 s−1, 4.08 × 105 ± 0.23 × 105 M−1 s−1, 4.95 × 105 ± 0.69 × 105 M−1 s−1, and 7.19 × 105 ± 0.85 × 105 M−1 s−1 for monoethyl, monobutyl, monohexyl, and mono-2-ethylhexyl phthalates, respectively. These values are comparable to those of the meta-cleavage compound hydrolase from Rhodococcus sp. for the metabolites of biphenyl, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid and 2-hydroxy-6-oxohepta-2,4-dienoic acid (1.08 × 106 M−1 s−1 and 1.11 × 106 M−1 s−1) that were calculated with the Km, Vmax, and molecular weight reported in another paper (14), and they were much higher than those of monoalkyl phthalate esterase from Micrococcus sp. for monobutyl phthalate (1.83 × 104 M−1 s−1 and 2.57 × 104 M−1s−1) (27).
TABLE 2.
Kinetic constants of the purified MEHP hydrolasea
| Phthalate ester | Km (μM) | Vmax (μmol/min · mg) |
|---|---|---|
| Mono-2-ethylhexyl | 26.9 ± 4.3 | 18.1 ± 0.9 |
| Mono-n-hexyl | 30.2 ± 5.8 | 14.0 ± 0.8 |
| Mono-n-butyl | 31.2 ± 2.4 | 12.0 ± 0.3 |
| Mono-n-ethyl | 27.6 ± 6.1 | 8.0 ± 0.5 |
The activities of purified MEHP hydrolase were calculated with the data from the spectrophtometorically detected formation of phthalic acid by using 6.24 μg of the enzyme as described in the text. The reactions were conducted at 45°C for 5 min. The average apparent Km and Vmax values were determined from curves fitted to the data by a nonlinear curve-fitting program, GraphPad Prism 4 (GraphPad Software, San Diego, CA), based on the Michaelis-Menten equation.
FIG. 6.
Reaction rates for the MEHP hydrolase with different concentrations of MEHP. The reaction conditions were the conditions described in the text except that several concentrations of MEHP were used for a substrate. The data are means and ranges (standard errors) calculated by using five individual measurements at each MEHP concentration. The Km and Vmax values were calculated by using a nonlinear curve-fitting program, GraphPad Prism 4 (GraphPad Software, San Diego, CA), based on the Michaelis-Menten equation.
The behavior of the enzyme toward various inhibitors was examined. A metal-chelating agent, such as EDTA (1 mM), did not inhibit the activity. Phenylmethylsulfonyl fluoride (1 mM) reduced the activity by 30%. Diisopropyl fluorophosphate (1 mM) inhibited the activity more than 97%. These typical inhibitors of serine hydrolase had inhibitory effects on the MEHP hydrolase. p-Chloromercuribenzoate (1 mM) reduced the activity by 43%. Fe2+, Ca2+, Co2+, and Ni2+ (2 μM) had no effect on the enzyme. However, 2 μM Hg2+, Cu2+, and Zn2+ inhibited the enzyme (residual activities, 39%, 72%, and 61%, respectively). Although these substances inhibited the MEHP hydrolase, no cysteine residue was present in the amino acid sequence, as described below. These substances might interact with histidine residues in the catalytic triad and/or the pentapeptide motif of serine hydrolase (1, 15). Diethylpyrocarbonate, a specific modification reagent of imidazole, was examined to determine its effect on MEHP hydrolase. The enzyme (3 μg/ml) incubated with 1 mM diethylpyrocarbonate lost 95% of its activity. The results of the experiments with inhibitors indicate that serine and histidine residues are involved in the reaction, and they seem to conform to the features expected if MEHP hydrolase is a serine hydrolase.
Determination of the primary structure of MEHP hydrolase.
We determined the ORF encoding MEHP hydrolase by the repeated inverse procedure, as described in Materials and Methods.
The ORF coded for a 296-residue polypeptide whose N-terminal sequence was consistent with that of MEHP hydrolase, and the molecular weight was calculated to be 32,164. As expected, the deduced amino acid sequence contained three peptide sequences generated by cleavage of MEHP hydrolase by BrCN. The primary structure of the enzyme was compared with those in the Swiss-Prot, PDB, PIR and PRF protein databases by using the BLAST search procedure. The MEHP hydrolase exhibited less than 30% amino acid sequence identity with 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase from Bacillus sp. strain JF8 (accession no. BAC79225) (27%), 2-hydroxymuconic semialdehyde hydrolase from Delftia tsuruhatensis AD9 (AAX47253) (25%), the meta-cleavage compound hydrolase from Pseudomonas resinovorans (BAC41548) (28%), the meta-cleavage compound hydrolase from Janthinobacterium sp. strain J3 (BAC56745) (28%), and the meta-cleavage compound hydrolase from Sphingomonas sp. strain GTIN11 (AAL37979) (28%). These meta-cleavage compound hydrolases are classified as serine hydrolases.
The serine hydrolases include a large number of enzymes, which catalyze the following reactions: hydrolysis of the C—N bond in a number of proteinases, the C—O bond in a number of esterases and lipases, and the C—C bond by hydrolases such as meta-cleavage compound hydrolases (1). Although the MEHP hydrolase hydrolyzes the ester bond of monoalkyl phthalates, the amino acid sequence of the enzyme exhibited homology with meta-cleavage compound hydrolases (Fig. 1) rather than the serine esterases.
An alignment of the amino acid sequences of the MEHP hydrolase and meta-cleavage compound hydrolases was performed with the GENETYX program (GENETYX Co., Tokyo, Japan). The meta-cleavage compound hydrolases, as well as the other serine hydrolases, have a consensus pentapeptide motif, GXSXG (X indicates an unconserved amino acid residue), and the D-S-H catalytic triad (1, 15). A conserved candidate for a catalytic triad, D82-S125-H50, was found in the sequence, although we had no evidence that it actually functions. A candidate of the conserved pentapeptide sequence, G123-H124-S125-R126-G127, was also present in the MEHP hydrolase. This motif of the MEHP hydrolase was, however, distinctive among serine hydrolases. The first X in the pentapeptide motif is N or H (N is major, and H is minor) and the second X is a nonpolar residue (M, F, or L) in meta-cleavage compound hydrolases and other serine hydrolases, whereas both of the residues were basic residues (the first was H124, and the second was R126) in MEHP hydrolase. Maruyama et al. predicted the presence of a positively charged residue in monoalkyl phthalate esterase to neutralize the negative charge of a carboxylate anion because the anion near the ester bond in the substrate might repulse the catalytic nucleophile, inhibiting it from attaching to the bond (27). The basic residue H124 and/or R126 in MEHP hydrolase might neutralize the negative charge of the carboxylate anion in monoalkyl phthalate.
The enzymatic properties and primary structure of the MEHP hydrolase indicated that it was a novel enzyme that seemed to belong to an independent hydrolase group phylogenetically related to the meta-cleavage compound hydrolase group.
REFERENCES
- 1.Ahmad, D., J. Fraser, M. Sylvestre, A. Larose, A. Khan, J. Bergeron, J. M. Juteau, and M. Sondossi. 1995. Sequence of the bphD gene encoding 2-hydroxy-6-oxo-(phenyl/chlorophenyl)hexa-2,4-dienoic acid (HOP/cPDA) hydrolase involved in the biphenyl/polychlorinated biphenyl degradation pathway in Comamonas testosteroni: evidence suggesting involvement of Ser112 in catalytic activity. Gene 156:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Akita, K., C. Naitou, and K. Maruyama. 2001. Purification and characterization of an esterase from Micrococcus sp. YGJ1 hydrolyzing phthalate esters. Biosci. Biotechnol. Biochem. 65:1680-1683. [DOI] [PubMed] [Google Scholar]
- 3.Aleshchenkova, Z. M., A. S. Samsonova, S. V. Baikova, and T. A. Kukulianskaia. 1996. The degradation of plasticizers by Rhodococcus erythropolis 40F. Mikrobiol. Z. 58:34-38. [PubMed] [Google Scholar]
- 4.Barber, E. D., and D. C. Topping. 1995. Subchronic 90-day oral toxicology of di(2-ethylhexyl) terephthalate in the rat. Food Chem. Toxicol. 33:971-978. [DOI] [PubMed] [Google Scholar]
- 5.Blom, A., E. Ekman, A. Johannisson, L. Norrgren, and M. Pesonen. 1998. Effects of xenoestrogenic environmental pollutants on the proliferation of a human breast cancer cell line (MCF-7). Arch. Environ. Contam. Toxicol. 34:306-310. [DOI] [PubMed] [Google Scholar]
- 6.Britt, A. J., N. C. Bruce, and C. R. Lowe. 1992. Identification of a cocaine esterase in a strain of Pseudomonas maltophilia. J. Bacteriol. 174:2087-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhardt, G., and P. R. Wallnofer. 1987. Metabolism of di- and mono-n-butyl phthalate by soil bacteria. Appl. Environ. Microbiol. 35:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii, T., T. Ogawa, and H. Fukuda. 1987. Isobutene production by Rhodotorula minuta. Appl. Microbiol. Biotechnol. 25:430-433. [Google Scholar]
- 10.Giam, C., H. Chan, G. Neff, and E. Atlas. 1978. Phthalate ester plasticizers: a new class of marine pollutant. Science 199:419-420. [PubMed] [Google Scholar]
- 11.Gross, E. 1967. The cyanogens bromide reaction. Methods Enzymol. 11:238-255. [Google Scholar]
- 12.Habe, H., K. Morii, S. Fushinobu, J. W. Nam, Y. Ayabe, T. Yoshida, T. Wakagi, H. Yamane, H. Nojiri, and T. Omori. 2003. Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme). Biochem. Biophys. Res. Commun. 303:631-639. [DOI] [PubMed] [Google Scholar]
- 13.Harris, C. A., P. Henttu, M. G. Parker, and J. P. Sumpter. 1997. The estrogenic activity of phthalate esters in vitro. Environ. Health Perspect. 105:802-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatta, T., T. Shimada, T. Yoshihara, A. Yamada, E. Masai, M. Fukuda, and H. Kiyohara. 1998. Meta-fission product hydrolases from a strong PCB degrader Rhodococcus sp. RHA1. J. Ferment. Bioeng. 85:174-179. [Google Scholar]
- 15.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 16.Jobling, S., T. Reynolds, R. White, M. G. Parker, and J. P. Sumpter. 1995. A variety of environmentally persistent chemicals, including some phthalate plasticizers, are weakly estrogenic. Environ. Health Perspect. 103:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasahara, E., E. F. Sato, M. Miyoshi, R. Konaka, K. Hiramoto, J. Sasaki, M. Tokuda, Y. Nakano, and M. Inoue. 2002. Role of oxidative stress in germ cell apoptosis induced by di(2-ethylhexyl)phthalate. Biochem. J. 365:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayano, Y., K. Watanabe, T. Matsunaga, I. Yamamoto, and H. Yoshimura. 1997. Involvement of a novel mouse hepatic microsomal esterase, ES46.5K, in the hydrolysis of phthalate esters. Biol. Pharm. Bull. 20:749-751. [DOI] [PubMed] [Google Scholar]
- 19.Kilbane, J. J. II, A. Daram, J. Abbasian, and K. J. Kayser. 2002. Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum. Biochem. Biophys. Res. Commun. 297:242-248. [DOI] [PubMed] [Google Scholar]
- 20.Krell, H. W., and H. Sandermann, Jr. 1984. Plant biochemistry of xenobiotics. Purification and properties of a wheat esterase hydrolyzing the plasticizer chemical, bis(2-ethylhexyl)phthalate. Eur. J. Biochem. 143:57-62. [DOI] [PubMed] [Google Scholar]
- 21.Kurane, R. 1997. Microbial degradation and treatment of polycyclic aromatic hydrocarbons and plasticizers. Ann. N. Y. Acad. Sci. 829:118-134. [DOI] [PubMed] [Google Scholar]
- 22.Kurane, R., T. Suzuki, and S. Fukuoka. 1984. Purification and some properties of a phthalate ester hydrolyzing enzyme from Nocardia erythropolis. Appl. Microbiol. Biotechnol. 20:378-383. [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lake, B. G., J. C. Phillips, J. C. Linnell, and S. D. Gangolli. 1977. The in vitro hydrolysis of some phthalate diesters by hepatic and intestinal preparations from various species. Toxicol. Appl. Pharmacol. 39:239-248. [DOI] [PubMed] [Google Scholar]
- 25.Lanyi, B. 1987. Classical and rapid identification methods for medically important bacteria. Methods Microbiol. 19:1-67. [Google Scholar]
- 26.Linos, A., A. Steinbuchel, C. Sproer, and R. M. Kroppenstedt. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tyre. Int. J. Syst. Bacteriol. 49:1785-1791. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama, K., K. Akita, C. Naitou, M. Yoshida, and T. Kitamura. 2005. Purification and characterization of an esterase hydrolyzing monoalkyl phthalates from Micrococcus sp. YGJ1. J. Biochem. (Tokyo) 137:27-32. [DOI] [PubMed] [Google Scholar]
- 28.Mayer, F. L., D. L. Stalling, and J. L. Johnson. 1972. Phthalate esters as environmental contaminants. Nature 238:411-413. [DOI] [PubMed] [Google Scholar]
- 29.Niino, T., T. Ishibashi, H. Ishiwata, K. Takeda, and S. Onodera. 2003. Characterization of human salivary esterase in enzymatic hydrolysis of phthalate esters. J. Health Sci. 49:76-81. [Google Scholar]
- 30.Omori, T., K. Sugiyama, H. Ishigooka, and Y. Minoda. 1986. Purification and some properties of a 2-hydroxy-6-oxo-6phenylhexa-2, 4-dienoic acid hydrolyzing enzyme from Pseudomonas cruciviae S93 B1 involved in the degradation of biphenyl. Agric. Biol. Chem. 50:931-937. [Google Scholar]
- 31.Saeger, V. W., and E. S. Tucker. 1976. Biodegradation of phthalic acid esters in river water and activated sludge. Appl. Environ. Microbiol. 31:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharpe, R. M., J. S. Fisher, M. M. Millar, S. Jobling, and J. P. Sumpter. 1995. Gestational and lactational exposure of rats to xenoestrogens results in reduced testicular size and sperm production. Environ. Health Perspect. 103:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sjoberg, P., and U. Bondesson. 1985. Determination of di(2-ethylhexyl) phthalate and four of its metabolites in blood plasma by gas chromatography-mass spectrometry. J. Chromatogr. 344:167-175. [DOI] [PubMed] [Google Scholar]
- 34.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, T., K. Yamaguchi, S. Suzuki, and T. Suga. 2001. Monitoring of phthalic acid monoesters in river water by solid-phase extraction and GC-MS determination. Environ. Sci. Technol. 18:3757-3763. [DOI] [PubMed] [Google Scholar]
- 36.Willhite, C. C. 2001. Weight-of-evidence versus strength-of-evidence in toxicologic hazard identification: di(2-ethylhexyl)phthalate (DEHP). Toxicology 160:219-226. [DOI] [PubMed] [Google Scholar]
- 37.Zacharewski, T. R., M. D. Meek, J. H. Clemons, Z. F. Wu, M. R. Fielden, and J. B. Matthews. 1998. Examination of the in vitro and in vivo estrogenic activities of eight commercial phthalate esters. Toxicol. Sci. 46:282-293. [DOI] [PubMed] [Google Scholar]