Abstract
N2O reductase activity in soybean nodules formed with Bradyrhizobium japonicum was evaluated from N2O uptake and conversion of 15N-N2O into 15N-N2. Free-living cells of USDA110 showed N2O reductase activity, whereas a nosZ mutant did not. Complementation of the nosZ mutant with two cosmids containing the nosRZDFYLX genes of B. japonicum USDA110 restored the N2O reductase activity. When detached soybean nodules formed with USDA110 were fed with 15N-N2O, they rapidly emitted 15N-N2 outside the nodules at a ratio of 98.5% of 15N-N2O uptake, but nodules inoculated with the nosZ mutant did not. Surprisingly, N2O uptake by soybean roots nodulated with USDA110 was observed even in ambient air containing a low concentration of N2O (0.34 ppm). These results indicate that the conversion of N2O to N2 depends exclusively on the respiratory N2O reductase and that soybean roots nodulated with B. japonicum carrying the nos genes are able to remove very low concentrations of N2O.
Nitrous oxide (N2O) is a key atmospheric greenhouse gas that contributes to global climate change through radiative warming and depletion of stratospheric ozone (1, 24). Agricultural land is a major source through denitrification and nitrification (14, 35) and contributes significantly to the net increase in atmospheric N2O (1, 34). Several attempts have been made to reduce the emission of N2O from agricultural systems (34, 35).
The complete denitrification of nitrate by bacteria to dinitrogen (N2) is generally an anaerobic respiratory process, where the last step is mediated by N2O reductase (54). The corresponding structural gene is nosZ and is assembled in the nosRZDFYL gene operon (54). Several species capable of denitrification are also nitrogen-fixing bacteria, including rhizobia such as Bradyrhizobium japonicum (4, 6, 44, 49) and Sinorhizobium meliloti (7, 20). Indeed, genes responsible for denitrification have been found in rhizobial genomes (15, 23).
Earlier studies (19, 37) reported the evolution of 15N-N2 from 15N-N2O from sliced or detached soybean nodules. Recently, Velasco et al. (50) reported that nosZ and nosR insertion mutants of B. japonicum USDA110 accumulate N2O when cultured microaerobically in the presence of nitrate. The nosZ gene was also expressed in soybean nodules (29). However, it has not yet been fully proved that N2 evolution from N2O by soybean nodules is mediated by N2O reductase encoded by the nosZ gene in B. japonicum (4, 9, 38). The aims of this work were to confirm whether the nos gene cluster of B. japonicum is responsible for respiratory N2O reduction to N2 in nodules and to evaluate the capability of the nodulated roots to transform N2O into N2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used are listed in Table 1. Bradyrhizobium cells were grown at 30°C in HM salt medium (8) supplemented with 0.1% arabinose and 0.025% (wt/vol) yeast extract (Difco, Detroit, MI), which is termed HM medium here. HM medium was further supplemented with 0.55 μM Na2MoO4 · 2H2O, 1 μM FeCl3, and 1 μM CuSO4 · 5H2O (HMM medium) for the denitrification assay (44). Escherichia coli cells were grown at 37°C in Luria-Bertani medium (42). Antibiotics were added to the media at the following concentrations: for B. japonicum, 100 μg of tetracycline (Tc)/ml, 100 μg of spectinomycin (Sp)/ml, 100 μg of streptomycin (Sm)/ml, 100 μg of kanamycin (Km)/ml, and 50 μg of polymyxin B/ml; for E. coli, 15 μg of Tc/ml, 50 μg of Sp/ml, 50 μg of Sm/ml, 50 μg of Km/ml, and 100 μg of ampicillin/ml.
TABLE 1.
Bacterial strains and cosmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Bradyrhizobium japonicum | ||
| USDA110 | Wild type; nosZ+ | 23 |
| T7 | Field isolate in Tokachi, Hokkaido, Japan; nosZ natural mutant | 32, 43 |
| USDA110ΔnosZ | USDA110 nosZ::del/ins Ω cassette; Smr Spr | This study |
| Escherichia coli | ||
| DH5α | recA; cloning strain | Toyobo Inc. |
| JM109 | recA; cloning strain | Toyobo Inc. |
| HB101 | recA hsdR hsdM pro Smr | 5 |
| S17-1 | hsdR RP4-2(kan::Tn7)(tet::Mu); Smr Spr | 47 |
| Plasmids | ||
| pRK2013 | ColE1 replicon carrying RK2 transfer genes; Kmr | 13 |
| pTZ18R | Cloning vector; pMB1ori; Apr | 46 |
| pTZ18RnosZ | pTZ18R carrying 4.0-kb nosZ fragment; Apr | This study |
| pTZ18rRΔSacI-nosZ | SacI-deleted pTZ18R carrying 4.0-kb nosZ fragment; Apr | This study |
| pHP45Ω | Plasmid carrying 2.1-kb Ω cassette; Spr Smr Apr | 40 |
| pK18mob | Cloning vector; pMB1ori oriT; Kmr | 46 |
| pK18mob-nosZ::Ω | pK18mob carrying 4.0-kb nosZ fragment; Kmr Smr Spr | This study |
| Cosmids | ||
| pKS800 | Derivative of broad-host-range cosmid pLAFR1; IncP Tcr | 17, 23 |
| brc02856 | pKS800 carrying nos gene clusterb; Tcr | 23 |
| brc01733 | pKS800 carrying nos gene clusterb; Tcr | 23 |
Apr, ampicillin resistant; Tcr, tetracycline resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant; Spr, spectinomycin resistant.
See Fig. 1.
DNA manipulations.
Isolation of plasmids, DNA ligation, and transformation of E. coli were performed as described by Sambrook et al. (42). DNA preparation and Southern hybridization were carried out as described previously (22, 30, 43).
Construction of a B. japonicum USDA110 nosZ mutant.
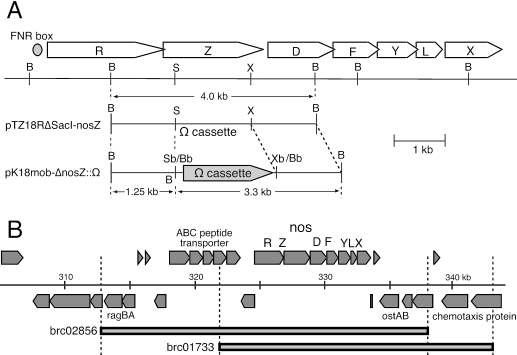
A 4-kb BamHI DNA fragment identified from the genome sequence of B. japonicum USDA110 (23) was excised from BamHI-digested total DNA and inserted into the BamHI site of pTZ18R (Fig. 1A). The plasmid containing the fragment was selected by PCR amplification specific for the nosZ gene of B. japonicum USDA110 with primer 3 (5′-GAGGATCGTTTCGCATGAGCGACAGCGACAACAT-3′) and primer 4 (5′-CGCTCCCGATCAGACCGATTT-3′), in which the underlined nucleotides form the USDA110 nosZ sequence. From the cloned 4-kb DNA fragment, pK18mob-nosZ::Ω was constructed (Fig. 1A). Biparental mating was conducted on HM agar plates using E. coli S17-1 (47). Double crossover was verified by Southern hybridization with the 4-kb nosRZD fragment as a probe (Fig. 1A).
FIG. 1.
Construction of a nosZ mutant (A) and complementation cosmid inserts (B) of Bradyrhizobium japonicum USDA110. (A) Cloned fragments in pTZ18RΔSacI-nosZ and pK18mob-ΔnosZ::Ω are shown alongside the physical map of the nos gene cluster of B. japonicum USDA110. B, BamHI; S, SacI; X, XhoI; Sb, SacI blunt end; Bb, BamHI blunt end; Xb, XhoI blunt end. During blunt-end ligation, the Sb/Bb site produced a BamHI site in pK18mob-ΔnosZ::Ω. Upstream of nosR lies a consensus sequence of an FNR box (TTGAT-N4-ATCAA) (50). The cloned 4-kb DNA fragment containing the nosZ gene in pTZ18R-nosZ was reconnected to pTZ18RΔSacI, which was produced by blunting of the SacI site of pTZ18R, and pTZ18RΔSacI-nosZ was generated. The Ω cassette, which was excised from pHP45Ω (40), digested with BamHI, and blunt ended, was inserted into the blunt-ended SacI and XhoI sites of pTZ18RΔSacI-nosZ to generate pTZ18RΔSacI-nosZ::Ω. Finally, the fragment containing the nosZ and Ω cassette fusion was inserted between the SmaI and XbaI sites of pK18mob to generate pK18mob-nosZ::Ω. (B) Positions covered by two cosmid inserts, brc02856 and brc01733 (Table 1), in the genome of B. japonicum USDA110 (http://www.kazusa.or.jp/rhizobase/) (23).
Genetic complementation by nos genes.
Two cosmids, brc02856 and brc01733, carrying the nos gene cluster (nosRZDFYLX) were selected from the pKS800 cosmid library, which was constructed for the sequencing of B. japonicum USDA110 (23). The cosmids were introduced into B. japonicum strains by triparental mating via pRK2013 (39, 47, 52).
Gas chromatography.
To determine N2O concentrations, we used two 63Ni electron capture detector gas chromatographs. In the quick N2O analysis, 0.2 ml of sample gas was injected into a Shimadzu (Kyoto, Japan) GC-17A gas chromatograph with a CP-PoraBOND Q capillary column (internal diameter, 0.32 mm, length, 25 m; Varian, Palo Alto, CA). The temperatures of injection, the column, and the detector were 250, 65, and 300°C, respectively. In the sensitive N2O analysis, 0.8 ml of sample gas was injected into a Shimadzu GC-14BPsE gas chromatograph equipped with tandem packed columns of a Porapak N (80/100 mesh; diameter, 0.3 mm; length, 1 m) and a Porapak Q (80/100 mesh; diameter, 0.3 mm; length, 3 m). The temperatures of injection, the column, and the detector were 80, 80, and 340°C, respectively. The carrier gas used for both N2O analyses was composed of 5% (vol/vol) CH4 in Ar. 15N-N2 was determined with a thermal conductivity detector gas chromatograph as described previously (44). For the acetylene reduction assay, ethylene was measured by a flame ionization detection gas chromatograph, as described previously (12).
N2O uptake by free-living cells.
B. japonicum cells were washed with HMM medium by centrifugation (5,000 × g, 15 min, 4°C), and the cell density was adjusted to 6 × 109 cells/ml (33). The cell suspension (4.6 ml, 6 × 109 cells/ml) was introduced into a 123-ml airtight vial (V-100; Nichiden-Rika Glass, Kobe, Japan). After the gas phase was replaced with N2, N2O gas was introduced into the vial at a final concentration of approximately 0.1% (vol/vol).
15N-N2 emission from 15N-labeled N2O by free-living cells.
B. japonicum cells (7 ml; 6 × 109 cells/ml) were introduced into a 14-ml airtight specimen vial. After the N2 gas replacement, 15N-N2O (15N, >98 atom%; Cambridge Isotope Laboratories Inc., Andover, MA) was introduced. After incubation at 30°C for 67 h, the concentration of 15N-N2 in the gas phase was determined by TCD gas chromatography (44).
Real-time PCR.
Beacon Designer software (Premier Biosoft International, Palo Alto, CA) was used to design the nosZ primer set (nosZ2F, 5′-GTCGTCATCTTCAACCTCAAG; nosZ2R, 5′-TATTCATGCCATGCGGATTG). Total RNA was prepared as described previously (53). Quantitative reverse transcription-PCR analysis was carried out by the i-Cycler optical system (Bio-Rad Laboratories, Inc., Tokyo, Japan) as described previously (53).
Plant cultivation and inoculation.
The surface-sterilized soybean seeds (Glycine max cv. Enrei) were germinated and transplanted to a Leonard jar (25, 48), which contained sterile vermiculite and nitrogen-free sterilized nutrient solution (31, 39). The cell suspension of B. japonicum (1 ml) was used for inoculation at 1 × 107 cells per seed. For N2O-feeding experiments, plants were grown in a growth chamber (LH200; Nippon Medical & Chemical Industries, Tokyo, Japan). For the nitrogen fixation experiment, soybeans were grown in pots (1 liter) filled with vermiculite in a phytotron (Koitotron type KC; Koito Industries, Tokyo, Japan). A nitrogen-free sterilized nutrient solution was periodically supplied to the pots (31, 39).
N2O uptake, 15N-N2 emission, and 15N incorporation by nodules.
After the nodulated roots of 45-day-old soybean plants were washed with water, the nodules were detached from the roots and weighed. The nodules were introduced into a 15-ml airtight vial. After the vials were sealed, 15N-N2O was injected. After the gas measurements, the nodules were dried at 80°C for 3 days and then powdered with a mortar and pestle. The nodule samples were subjected to tracer 15N and total-N analyses by an automatic gas chromatograph-mass spectrometer (EA1110-DELTAplus Advantage ConFloIII system, Thermo Electron Co., Bremen, Germany).
The whole root system of 32-day-old soybean plants was inserted into a 51-ml test tube (19.4 mm in diameter by 176 mm in height) containing 20 ml nitrogen-free nutrient solution (31, 39). The test tube was sealed with a silicon rubber stopper with a gas sampling port and a hole in the center. This system enclosed the whole root system of intact soybeans in the test tube, leaving the aerial parts in air. Then N2O gas was injected into the closed test tube.
Nitrogen fixation and plant growth.
The nitrogen-fixing activity of nodules was examined by acetylene reduction assays in an incubation jar (300-ml capacity) containing the excised root system. Acetylene gas was injected into the jar at 15% (vol/vol), and the roots were incubated for 60 min at 25°C. Separated shoot, roots, and nodules were weighed.
Bacteroid antibiotic test.
Nodules were removed from soybeans 18 days after inoculation with T7 or T7 with brc01733. Homogenates of surface-sterilized nodules were plated on HM agar medium with and without tetracycline.
RESULTS
Construction of the nosZ deletion mutant.
To examine whether the nos gene cluster of B. japonicum was responsible for N2O reductase activity, we constructed a nosZ deletion mutant of B. japonicum USDA110 (Table 1). After deletion of most of the nosZ gene from the cloned DNA fragment containing nosRZD, the Ω cassette was inserted into the position (Fig. 1). When free-living cells of USDA110 and USDA110ΔnosZ were anaerobically incubated in gas containing 0.1% (vol/vol) N2O, the parent strain consumed N2O, whereas the mutant did not show N2O uptake (Fig. 2A). To further examine the N2O reductase reaction, 15N-N2O was supplied to cultures of B. japonicum USDA110 and USDA110ΔnosZ. Wild-type USDA110 stoichiometrically produced 15N-N2 from 15N-N2O, whereas USDA110ΔnosZ did not (data not shown). These results suggest that nosZ of B. japonicum USDA110 is responsible for the N2O reductase activity that converts N2O to N2.
FIG. 2.
N2O uptake by free-living cells of the nosZ mutant (A) and nos complementation of USDA110ΔnosZ (B) and T7 (C). pKS800 is the cosmid vector used for complementation (17). Two cosmid clones, brc02856 and brc01733, have inserts containing nos genes of USDA110 (Table 1; Fig. 1). N2O reductase activities (means ± SDs) estimated from maximum rates of N2O uptake were 4.6 ± 0.2 [USDA110(pKS800)], 22.9 ± 0.7 [USDA110ΔnosZ(brc02856)], and 23.6 ± 1.9 N2O [USDA110ΔnosZ(brc01733)] μmol h−1 107 cells−1.
Complementation of USDA110ΔnosZ with nos genes.
To further examine the validity of nosZ-dependent N2O reductase activity, two cosmids containing the nos gene cluster were introduced into USDA110ΔnosZ (Fig. 1B). The transconjugants were anaerobically incubated in the presence of 0.1% (vol/vol) N2O and tetracycline, and USDA110 and USDA110ΔnosZ carrying the pKS800 vector were used as controls (Fig. 2B). USDA110 and USDA110ΔnosZ carrying pKS800 (Fig. 2B) showed N2O uptake profiles similar to those of wild-type USDA110 and USDA110ΔnosZ, respectively (Fig. 2A). However, the two cosmids in Fig. 1B restored strong N2O uptake (Fig. 2B). The amino acid sequences of B. japonicum nosRZDFYLX (23, 50) have a high degree of similarity to those of other N2O reductases from Pseudomonas stutzeri (21). Both B. japonicum cosmids contain nosRZDFYLX and adjacent genes (Fig. 1B). Their addition to USDA110ΔnosZ produced similar activities of N2O reductase, as indicated by N2O uptake (Fig. 2B). Moreover, the introduction of a cosmid (brc02856 or brc01733) into USDA110ΔnosZ restored the strain's ability to convert 15N-N2O into 15N-N2 (data not shown). Thus, the complementation experiments confirmed that the nos gene cluster per se is responsible for N2O reductase activity in B. japonicum USDA110.
Expression of nosZ.
Strain USDA110ΔnosZ with the two cosmids containing the nos cluster rapidly and strongly absorbed N2O, much more than the parent strain USDA110 containing the pKS800 vector (Fig. 2B). When the N2O reductase activity was estimated from the maximum rate of N2O uptake in Fig. 2B, the activities of USDA110ΔnosZ containing brc02856 and brc01733 (22.9 ± 0.7 and 23.6 ± 1.9 N2O μmol h−1 107 cells−1) were five times that of wild-type USDA110 (4.6 ± 0.2 N2O μmol h−1 107 cells−1). In addition, the lag time for the induction of N2O reductase activity in the wild-type USDA110(pKS800) disappeared in the cosmid-complemented USDA110ΔnosZ.
Because we used pKS800, a derivative of cosmid pLAFR1 (RK2), as a vector for the nos complementation (17), it is possible that the cosmid introduction increases the copy number and thus results in higher expression of nos genes. USDA110ΔnosZ carrying brc01733 expressed the nosZ gene at 5-times-higher levels on the basis of total RNA than USDA110ΔnosZ carrying the pKS800 vector 1 h after anaerobic incubation in the presence of 0.1% (vol/vol) N2O. This result suggests that the higher expression of the nos genes enhanced N2O reductase activity and shortened the lag time for induction in USDA110ΔnosZ with the nos cosmids.
N2O uptake and N2 emission by nodules.
Denitrification genes of B. japonicum are expressed in a microaerobic state in both legume nodules and free-living bacteria via the fixLJ and fixK2 regulatory cascades (3, 28). Mesa et al. (29) reported that the nir, nor, and nos denitrification genes in B. japonicum were expressed in soybean root nodules. We thus examined the nodulation phenotypes of the nosZ mutant. The nodule weight and nitrogen-fixing activity of USDAΔnosZ showed no significant differences from those of USDA110 (Table 2), indicating that nosZ mutation does not alter symbiotic nitrogen fixation. This agrees with the results of a report on the nodulation kinetics (29) of a nosZ insertion mutant of B. japonicum (50).
TABLE 2.
Growth and N2-fixing activity of soybean plants inoculated with Bradyrhizobium japonicum USDA110 or the nosZ mutanta
| Strain | Growth (g [fresh wt] plant−1)
|
N2-fixing activity (C2H2-reducing activity) (μmol h−1 g of nodule [fresh wt]−1) | ||
|---|---|---|---|---|
| Shoot | Root | Nodule | ||
| USDA110 | 10.8 ± 0.7 | 4.8 ± 0.4 | 1.0 ± 0.1 | 10.3 ± 1.3 |
| USDA110ΔnosZ | 10.4 ± 0.9 | 4.5 ± 0.4 | 0.9 ± 0.1 | 9.0 ± 2.0 |
| Uninoculated | 6.9 ± 0.2 | 5.5 ± 0.2 | 0 | 0 |
Values for growth and N2-fixing activity are shown as means ± SDs, with six replications.
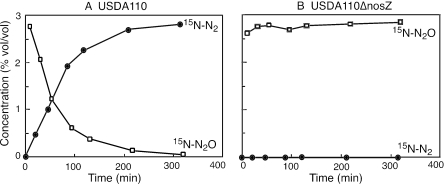
To evaluate the extent of N2O uptake and N2 emission by N2O reductase in soybean nodules, 15N-N2O was supplied to detached nodules formed with the nosZ mutant or the parent strain (Fig. 3). Soybean nodules formed with USDA110 rapidly absorbed 15N-N2O and emitted 15N-N2 outside the nodules, whereas nodules formed with the nosZ mutant did not mediate the conversion of 15N-N2O to 15N-N2 at all (Fig. 3).
FIG. 3.
15N-N2O uptake and 15N-N2 emission by detached soybean nodules formed with Bradyrhizobium japonicum USDA110 (A) and the nosZ mutant USDA110ΔnosZ (B). 15N-labeled N2O was supplied to vials (15 ml) containing soybean nodules (1.6 to 1.8 g). Five independent experiments gave similar results.
Because the transformation of 15N-N2O into 15N-N2 was likely stoichiometric (Fig. 3), we determined the amount of fixed 15N in the nodules and made a 15N balance sheet for 15N-N2O exposure. 15N atoms of 15N-N2O were distributed 98.5% ± 0.1% (mean ± standard deviation [SD]) outside the nodules as 15N-N2 and 1.5% ± 0.1% (mean ± SD) fixed in the nodules. Thus, most N2 molecules produced by N2O reductase in bacteroids were released outside the nodules, and nitrogenase was not able to utilize most of them as a substrate for nitrogen fixation. This is probably due to the diffusion of N2 into various compartments for N2O reductase and nitrogenase. The apparent rate of N2O uptake by USA110 nodules under about 3% N2O (Fig. 3) was estimated to be 7.8 μmol h−1 per g (fresh weight) of nodule; this was comparable with the acetylene-reducing activity (Table 2). These results indicate that the conversion of N2O into N2 in soybean nodules depends on the function of the nos gene cluster in B. japonicum.
N2O uptake by intact soybeans nodulated with USDA110.
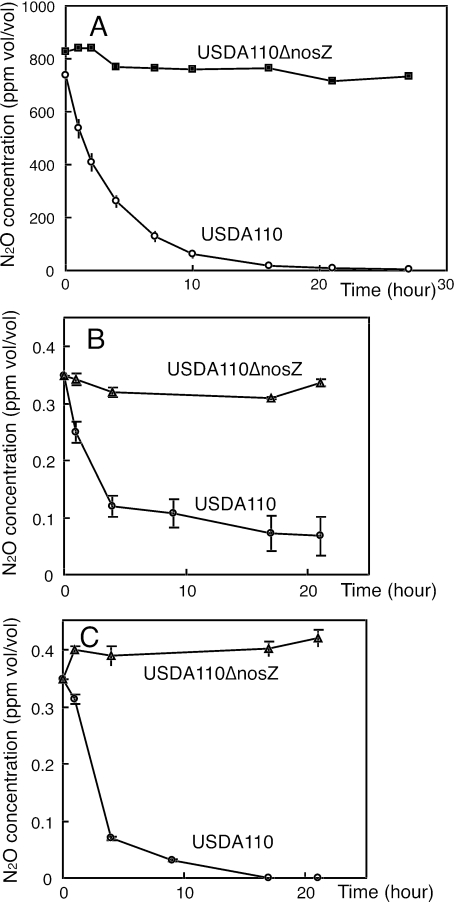
To examine the extent of N2O concentrations that the nodulated roots can take up, the whole root systems of intact plants nodulated by USDA110 or the nosZ mutant were enclosed. After the introduction of N2O gas, N2O concentrations around the whole root system were monitored (Fig. 4A and B). At an initial N2O concentration of 800 ppm (Fig. 4A), the root systems nodulated by USDA110 absorbed N2O, whereas the nosZ mutant-inoculated roots did not take up N2O at all. After a 21-h incubation, the N2O concentration reached 4 to 8 ppm around the roots nodulated with USDA110 (Fig. 4A). Thus, the initial concentrations of N2O were gradually decreased to 50, 5, 1, and 0.34 ppm. Surprisingly, the root systems nodulated by USDA110 were able to take up 0.34 ppm N2O, reducing the concentration to 0.11 ± 0.02 ppm (mean ± SD) 22 h after the start of incubation (Fig. 4B). When a shoot-decapitated root system was completely enclosed in the test tube with a rubber stopper to avoid gas leakage, the N2O concentration around the root system nodulated by USDA110 decreased to below the detection limit (<0.02 ppm) by 17 h (Fig. 4C).
FIG. 4.
N2O uptake from root systems of intact plants (A and B) and decapitated root systems (C) inoculated with the wild-type strain or the nosZ mutant of Bradyrhizobium japonicum USDA110. Initial gas phases were adjusted to approximately 800 ppm (A) (intact plant) and 0.34 ppm (B [intact plant] and C [decapitated root system]) of N2O in air. Soybean plants that harbored 0.26 to 0.30 g (A), 0.15 to 0.22 g (B), or 0.18 to 0.36 g (C) of nodules (fresh weight per plant) were examined by a test tube system (see the text). Triplicate determinations were carried out, except that data for USDA110ΔnosZ inoculation in panel A show means of duplicate determinations. Error bars, SDs.
Generally, the N2O concentration in soils (0.36 ppm to 8,300 ppm) is higher than that in ambient air (0.31 ppm) (1, 26, 34, 45). Therefore, these results strongly suggest that soybean nodules formed with B. japonicum carrying nos genes potentially scavenge N2O gas in soil.
Introduction of nos genes into a naturally occurring nosZ-deficient strain.
Strains of B. japonicum can apparently be classified into three denitrification types: full denitrifiers (up to N2), truncated denitrifiers (up to N2O), and nondenitrifiers (44, 49). Their incidence depends on the field site and the phylogeny (44). We tested whether introduction of the nos gene cluster into the naturally occurring nosRZD-deficient strain T7 would induce N2O reductase activity. Introduction of brc02856 or brc01733 into strain T7 induced N2O uptake, but T7 carrying the cosmid vector did not show such activity (Fig. 2C).
To test whether strain T7 carrying brc01733 exhibited N2O reductase activity in nodules, this strain was inoculated into soybean plants. However, the instability of cosmid brc01733 interfered with this experiment: Bacteroid cells from the nodules lost cosmid brc01733 during nodule development, as suggested by plate counts of tetracycline-resistant colonies (<10−6). Because similar instability of pLAFR1 derivatives has been reported (10), other strategies are needed for the symbiotic testing of strain T7 carrying nos genes.
DISCUSSION
The nosZ mutant of USDA110 was not able to convert 15N-N2O to 15N-N2 in nodules (Fig. 3) or as free-living cells (Fig. 2) and continued to produce nitrogen-fixing nodules (Table 2). Thus, the evolution of 15N-N2 from 15N-N2O in soybean nodules is due exclusively to N2O reductase, which is encoded by nosZ.
An important finding of this work is that N2O reductase expressed in B. japonicum can take up very low concentrations of N2O from outside of the nodules, equivalent to the natural concentration of N2O in air (approximately 0.31 ppm). Probably this indicates that N2O reductase has a high affinity for N2O as a substrate (54), and the gas movement through the tissues of the nodules does not severely interfere with the enzyme property.
Agricultural soil is a major source of the N2O emissions that contribute to global warming (1, 34). The N2O concentration in the soil gas phase fluctuates with water, organic matter, and nitrogen fertilizer but is generally higher than that of ambient air (1, 14, 18, 24, 26, 45). The results of N2O uptake by soybean nodules (Fig. 3 and 4) suggest that soybean nodules formed with B. japonicum carrying nos genes scavenge N2O in soil, thus lessening N2O emission to the atmosphere from soybean fields.
However, several groups have reported that cultivation of legume crops often enhances N2O emission from fields of alfalfa (11), soybean (51), white clover (36), and Bengal gram (16). Thus, we want to discuss the N2O paradox of legume root nodules for those results and ours. One possible explanation for the N2O paradox is that the nitrate absorbed via legume roots and nodule surfaces from the soil solution might be transported in nodules (27, 41), which would emit N2O from nitrate in bacteroids that lack nos genes. Another explanation is that N2O is produced by rhizosphere organisms from degrading nodules (51), where fixed nitrogen would become a source of N2O emission irrespective of the nos genes in the bacteroids. If either assumption is correct, the presence of nos genes in rhizobia ought still to decrease N2O levels in the rhizosphere and N2O emission from field soils.
Rhizobial inoculation technology focuses on increasing legume yields. The nos gene-dependent transformation of N2O into N2 in soybean nodules prompted us to enhance this activity in bradyrhizobial inoculants—a technology that could be environmentally friendly. The introduction of cosmids carrying nos genes enhanced nosZ transcription and N2O reductase activity (Fig. 2B), suggesting that up-regulation of nos gene expression would result in the enhancement of N2O reductase activity in B. japonicum.
Natural populations of B. japonicum that lacks N2O reductase activity were often dominant in soybean fields (44). To generate bradyrhizobial inoculants with high N2O reductase activity in soybean nodules, one can adopt several strategies: (i) introduction of nos genes by using stable vectors or chromosome integration with strong promoters, (ii) selection of natural isolates with high N2O reductase activity, and (iii) simple use of USDA110 and other wild-type strains carrying nos genes as inoculants. Denitrification steps, including N2O reduction, are anaerobic respiration processes. It is thus important to examine the allocations of electrons to O2 and N2O in soybean nodules, because energy metabolism in bacteroids is supported by cbb3 terminal oxidase, which has a high affinity for O2 and adapts to a microoxic environment in root nodules (2).
Acknowledgments
This work was supported by grants 13007988-00 to R. Sameshima-Saito and 14360037 and 17380046 to K. Minamisawa from the Ministry of Education, Science, Sports and Culture of Japan.
We are grateful to M. J. Sadowsky (University of Minnesota) for providing B. japonicum strain USDA110; T. Kaneko and S. Tabata (Kazusa DNA Research Institute) for providing the cosmids carrying the nos gene cluster; and T. Ito (Tohoku University) for help in N2O analysis by 63Ni electron capture detector gas chromatography.
REFERENCES
- 1.Banin, A., J. G. Lawless, and R. C. Whitten. 1984. Global N2O cycles—terrestrial emissions, atmospheric accumulation and biospheric effects. Adv. Space Res. 4:207-216. [DOI] [PubMed] [Google Scholar]
- 2.Batut, J. 2004. Genomics of the ccoNOPQ-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 181:89-96. [DOI] [PubMed] [Google Scholar]
- 3.Bedmar, E. J., E. F. Robles, and M. J. Delgado. 2005. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 33:141-144. [DOI] [PubMed] [Google Scholar]
- 4.Bhandari, B., and D. J. D. Nicholas. 1984. Denitrification of nitrate to nitrogen gas by washed cells of Rhizobium japonicum and by bacteroids from Glycine max. Planta 161:81-85. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Breitenbeck, G. A., and J. M. Bremner. 1989. Ability of free-living cells of Bradyrhizobium japonicum to denitrify in soils. Biol. Fertil. Soils 7:219-224. [Google Scholar]
- 7.Chan, Y. K., L. Barran, and E. S. P. Bromfield. 1989. Denitrification activity of phage types representative of two populations of indigenous Rhizobium meliloti. Can. J. Microbiol. 35:737-740. [Google Scholar]
- 8.Cole, M. A., and G. H. Elkan. 1973. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob. Agents Chemother. 4:248-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne, M. S., and D. D. Focht. 1987. Nitrous oxide reduction in nodules—denitrification or N2 fixation? Appl. Environ. Microbiol. 53:1167-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in Gram-negative bacteria. Mol. Plant-Microbe. Interact. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 11.Duxbury, J. M., D. R. Bouldin, R. E. Terry, and R. L. Tate. 1982. Emission of nitrous oxide from soils. Nature 298:462-464. [Google Scholar]
- 12.Elbeltagy, A., K. Nishioka, T. Sato, H. Suzuki, B. Ye, T. Hamada, T. Isawa, H. Mitsui, and K. Minamisawa. 2001. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67:5285-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firestone, M. K., R. B. Firestone, and J. M. Tiedje. 1980. Nitrous oxide from soil denitrification: factors controlling its biological production. Science 208:749-751. [DOI] [PubMed] [Google Scholar]
- 15.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, S., D. Majumdar, and M. C. Jain. 2002. Nitrous oxide emissions from kharif and rabi legumes grown on an alluvial soil. Biol. Fertil. Soils 35:473-478. [Google Scholar]
- 17.Hattori, Y., H. Omori, M. Hanyu, N. Kaseda, E. Mishima, T. Kaneko, S. Tabata, and K. Saeki. 2002. Ordered cosmid library of the Mesorhizobium loti MAFF303099 genome for systematic gene disruption and complementation analysis. Plant Cell Physiol. 43:1542-1557. [DOI] [PubMed] [Google Scholar]
- 18.Heincke. M., and M. Kaupenjohann. 1999. Effects of soil solution on the dynamics of N2O concentrations: a review. Nutr. Cycl. Agroecosyst. 55:133-157. [Google Scholar]
- 19.Hoch, G. E., K. C. Schneider, and R. H. Burris. 1960. Hydrogen evolution and exchange, and conversion of N2O to N2 by soybean root nodules. Biochim. Biophys. Acta 37:273-279. [DOI] [PubMed] [Google Scholar]
- 20.Hollway, P., W. McCormick, R. J. Watson, and Y. K. Chan. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J. Bacteriol. 178:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honisch, U., and W. G. Zumft. 2003. Operon structure and regulation of the nos gene region of Pseudomonas stutzeri, encoding an ABC-type ATPase for maturation of nitrous oxide reductase. J. Bacteriol. 185:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isawa, T., R. Sameshima, H. Mitsui, and K. Minamisawa. 1999. IS1631 occurrence in Bradyrhizobium japonicum highly reiterated sequence-possessing strains with high copy numbers of repeated sequences RSα and RSß. Appl. Environ. Microbiol. 65:3493-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 24.Lashof, D. A., and D. R. Ahuja. 1990. Relative contribution of greenhouse gas emission to global warming. Nature 344:529-531. [Google Scholar]
- 25.Leonard, L. T. 1943. A simple assembly for use in the testing of cultures of rhizobia. J. Bacteriol. 45:523-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X., K. Inubushi, and K. Sakamoto. 2002. Nitrous oxide concentration in an Andisol profile and emissions to the atmosphere as influenced by the application of nitrogen fertilizers and manure. Biol. Fertil. Soils 35:108-113. [Google Scholar]
- 27.Lucinski, R., W. Polcyn, and L. Ratajczak. 2002. Nitrate reduction and nitrogen fixation in symbiotic association Rhizobium-legumes. Acta Biochim. Pol. 49:537-546. [PubMed] [Google Scholar]
- 28.Mesa, S., E. J. Bedmar, A. Chanfon, H. Hennecke, and H. Fisher. 2003. Bradyrhizbium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J. Bacteriol. 185:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesa, S., J. de Dios Alche, E. J. Bedmar, and M. J. Delgado. 2004. Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol. Plant. 120:205-211. [DOI] [PubMed] [Google Scholar]
- 30.Minamisawa, K. 1990. Division of rhizobitoxine-producing and hydrogen-uptake positive strains of Bradyrhizobium japonicum by nifDKE sequence. Plant Cell Physiol. 31:81-89. [Google Scholar]
- 31.Minamisawa, K., M. Itakura, M. Suzuki, K. Ichige, T. Isawa, K. Yuhashi, and H. Mitsui. 2002. Horizontal transfer of nodulation genes in soils and microcosms from Bradyrhizobium japonicum to B. elkanii. Microbes Environ. 17:82-90. [Google Scholar]
- 32.Minamisawa, K., Y. Nakatsuka, and T. Isawa. 1999. Diversity and field site variation of indigenous populations of soybean bradyrhizobia in Japan by fingerprints with repeated sequences RSα and RSß. FEMS Microbiol. Ecol. 29:171-178. [Google Scholar]
- 33.Miyamoto, T., M. Kawahara, and K. Minamisawa. 2004. Novel endophytic nitrogen-fixing clostridia from the grass Miscanthus sinensis as revealed by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70:6580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosier, A. R., and C. Kroeze. 2000. Potential impact on the global atmospheric N2O budget of the increased nitrogen input required to meet future global food demands. Chemosphere Global Change Sci. 2:465-473. [Google Scholar]
- 35.Mosier, A. R., C. Kroeze, C. Nevison, O. Oenema, S. Seitzinger, and O. van Cleemput. 1998. Closing the global N2O budget: nitrous oxide emissions through the agricultural nitrogen cycle. Nutr. Cycl. Agroecosys. 52:225-248. [Google Scholar]
- 36.Mori, A., M. Honjito, H. Kondo, H. Matsunami, and D. Scholefield. 2005. Effects of plant species on CH4 and N2O fluxes from a volcanic grassland soil in Nasu, Japan. Soil Sci. Plant Nutr. 51:19-27. [Google Scholar]
- 37.Mozen, M. M., and R. H. Burris. 1954. The incorporation of 15N-labelled nitrous oxide by nitrogen fixing agents. Biochim. Biophys. Acta 14:577-578. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, S. G., and G. H. Elkan. 1965. Nitrogen metabolism of some strains of R. japonicum having different nodulating capabilities. Can. J. Microbiol. 11:1039-1041. [DOI] [PubMed] [Google Scholar]
- 39.Okazaki, S., M. Sugawara, and K. Minamisawa. 2004. Bradyrhizobium elkanii rtxC gene is required for expression of symbiotic phenotypes in the final step of rhizobitoxine biosynthesis. Appl. Environ. Microbiol. 70:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 41.Rigaud, J., F. J. Bergersen, G. L. Turner, and R. M. Daniel. 1973. Nitrate dependent anaerobic acetylene-reduction and nitrogen-fixation by soybean bacteroids. J. Gen. Microbiol. 77:137-144. [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sameshima, R., T. Isawa, M. J. Sadowsky, T. Hamada, H. Kosai, A. Shutsrirung, H. Mitsui, and K. Minamisawa. 2003. Phylogeny and distribution of extra-slow-growing Bradyrhizobium japonicum harboring high copy numbers of RSα, RSß and IS1631. FEMS Microbiol. Ecol. 44:191-202. [DOI] [PubMed] [Google Scholar]
- 44.Sameshima-Saito, R., K. Chiba, and K. Minamisawa. 2004. New method of denitrification analysis of Bradyrhizobium field isolates by gas chromatographic determination of 15N-labeled N2. Appl. Environ. Microbiol. 70:2886-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawamoto, T., K. Kusa, R. Hu, and R. Hatano. 2002. Dissolved N2O, CH4, and CO2 in pipe drainage, seepage, and stream water in a livestock farm in Hokkaido, Japan. Soil Sci. Plant Nutr. 48:433-439. [Google Scholar]
- 46.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 48.Trung, B. C., and S. Yoshida. 1983. Improvement of Leonard jar assembly for screening of effective Rhizobium. Soil Sci. Plant Nutr. 29:97-100. [Google Scholar]
- 49.van Berkum, P., and H. H. Keyser. 1985. Anaerobic growth and denitrification among different serogroups of soybean rhizobia. Appl. Environ. Microbiol. 49:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velasco, L., S. Mesa, C. Xu, M. J. Delgado, and E. J. Bedmar. 2004. Molecular characterization of nosRZDFLX genes coding for denitrifying nitrous oxide reductase of Bradyrhizobium japonicum. Antonie Leeuwenhoek 85:229-235. [DOI] [PubMed] [Google Scholar]
- 51.Yang, L., and Z. Cai. 2005. The effect of growing soybean (Glycine max. L.) on N2O emission from soil. Soil Biol. Biochem. 37:1205-1209. [Google Scholar]
- 52.Yasuta, T., S. Okazaki, H. Mitsui, K. Yuhashi, H. Ezura, and K. Minamisawa. 2001. DNA sequence and mutational analysis of rhizobitoxine biosynthesis genes in Bradyrhizobium elkanii. Appl. Environ. Microbiol. 67:4999-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You, M., T. Nishiguchi, A. Saito, T. Isawa, H. Mitsui, and K. Minamisawa. 2005. Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light-dark cycle. Appl. Environ. Microbiol. 71:8183-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]