Abstract
Microbial communities play important roles in the functioning of coral reef communities. However, extensive autofluorescence of coral tissues and endosymbionts limits the application of standard fluorescence in situ hybridization (FISH) techniques for the identification of the coral-associated bacterial communities. This study overcomes these limitations by combining FISH and spectral imaging.
Microbial communities of corals play substantial roles in normal and perturbed coral physiological states. Recently, studies using phylogenetic analysis have demonstrated high microbial diversity associated with coral tissues (8, 16, 29, 32, 33, 41) and coral mucus (29). Coral microbial communities appear highly complex; they are distinct from those in the water column, are coral species specific, are spatially and temporally stable, and have roles in the physiological function of the coral holobiont (16, 29, 32, 33). Putative roles include nitrogen fixation, carbon fixation, nutrient accumulation, antibiotic production, pathogen protection, and prevention of fouling and colonization (13, 33, 36, 41). Yet to better understand the complexity of the coral microbial community, we need to know the architecture of the coral-microbe association. Understanding how microorganisms interact with corals is important, given that they are increasingly implicated in coral disease and reef degradation. Fluorescence in situ hybridization (FISH) is widely applied in diverse and complex microbial systems (2) where traditional microbial studies are limited. It is estimated that as few as 0.1% of environmental microbes can be isolated in pure culture; in situ studies overcome this limitation (1, 18, 39).
Extensive autofluorescence has been the major limitation to the application of FISH in coral studies. Coral tissues are highly fluorescent due to the presence of pocilloporins (green fluorescent protein-like molecules) (14, 15, 34) and high densities of endosymbiotic dinoflagellates containing chlorophyll (22, 42). This extensive autofluorescence confounds coral microbial FISH studies, as image analysis is unable to resolve the sources of fluorescence (12). Bythell et al. (9) conducted the first study of FISH using enzyme amplification, previously reported to provide a 10- to 20-fold increase in signal intensity (26, 39). The study of Bythell et al. (9) was able to distinguish coral-associated bacterial communities; however, autofluorescence and nonspecific probe binding confounded image quality. Subsequently, Wegley et al. (40) used peptide nucleic acid probes for visualizing Archaea within coral tissue slurries. While the technique appeared effective for enumeration of probe binding, cellular morphology and image clarity were low. While the authors examined the binding potential of the probes with microbial cultures, controlling for sources of fluorescence emission such as coral tissue fragments and nonspecific probe binding were not considered. Amann et al. (2) suggested that improvements with peptide nucleic acid probes are confounded by nonspecific binding; therefore, the reliability of this method is questionable when it is applied to complex coral tissue slurries where confirmation of cell type and sources of fluorescence are not available. Rowher et al. (33) noted that there is yet to be a clear picture of the ecological roles of the diverse coral microbial associates. The present study describes a method that can address these questions by characterizing the architecture of the coral-bacteria association.
Recently, spectral imaging has been applied to plant research where tissue autofluorescence is a limiting factor (5, 6). Spectral profile images enhance the ability to interpret a fluorescent image by analyzing the spectral emission curve data of each pixel, thus providing an added dimension for image analysis (6, 43). Spectral imaging provides a means of distinguishing sources of fluorescence emission, allowing coral autofluorescence and FISH probe-fluorochrome emissions to be distinguished. Our study applied conventional FISH techniques and spectral imaging to accurately visualize the architecture of bacterial communities associated with reef-building corals.
Branching (Acropora aspera, Acropora formosa, and Acropora nana) and tabulate (Acropora cytherea, Acropora clatharata, and Acropora hyacinthus) acroporids, as well as nonacroporids (Seriatopora hysterix and Stylophora pistillata), were collected from Heron Island on the Southern Great Barrier Reef (23°44′17"S, 151°91′25"E), Australia. Corals were collected from March through September 2004 (at a depth of 1 to 8 m), transported to the Heron Island Research Station, and held in flowthrough seawater aquaria prior to fixation in 4% (wt/vol) paraformaldehyde in sterile phosphate-buffered saline (1) for 12 h. Samples were embedded in 1.5% (wt/vol) agarose (9) prior to decalcification with 20% (wt/vol) EDTA in phosphate-buffered saline (37). Tissues were processed sequentially through 70%, 80%, 95%, and 100% ethanol and three xylene and three paraffin washes, each for 40 min, prior to paraffin embedding. Serial tissue sections (4 μm) were collected onto Superfrost Plus slides (Menzel, Germany). Visualization of bacterial communities associated with coral tissues and lesions was conducted with the bacterial probe suite EUBmix (3, 11) using a conventional FISH protocol (3, 4, 18). Sequential adjacent tissue sections were probed with bacterial group probes (Table 1). All oligonucleotides were labeled with Cy3 (Thermo Electron Corp.). The hybridization was conducted in buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 0.01 M Tris-HCl, pH 7.2) for 1.5 h at 46°C, followed by a 10-min wash in prewarmed buffer (0.08 M NaCl, 0.01% sodium dodecyl sulfate, 0.01 M Tris-HCl, 0.05 M EDTA). Image analysis was conducted on a Meta 510 confocal scanning laser microscope (Zeiss, Germany) with the Zeiss Image Browser software. Several coral tissue sections treated with the FISH protocol without the application of probe were used for spectral profiling of autofluorescence without confounding sources of fluorochrome fluorescence. This was repeated for coral species to determine variability in fluorescence profiles. Coral tissue sections washed with 4% paraformaldehyde-fixed cultures of Escherichia coli were used as positive controls for the combination FISH and spectral imaging procedure.
TABLE 1.
Bacterial group probes used in FISH for identification of bacterial populations
| Probe | Target group | Oligonucleotide sequence | % Formamide | Reference |
|---|---|---|---|---|
| ALF969 | α-Proteobacteria | 5-′TGGTAAGGTTCTGCGCGT-3′ | 20 | 23 |
| BET42a | β-Proteobacteria | 5′-GCCTTCCCACATAGTTT-3′ | 35 | 23 |
| GAM42a | γ-Proteobacteria | 5′-GCCTTCCCACATCGTTT-3′ | 35 | 23 |
| CF319 | Cytophaga-Flavobacterium | 5′-TGGTCCGTGTCTCAGTAC-3′ | 35 | 24 |
| HCG69a | Actinobacteria | 5′-TATAGTTACCACCGCCGT-3′ | 25 | 30 |
| LGC354 | Firmicutes | 5′-TGGAAGATTCCCTACTGC-3′ | 35 | 25 |
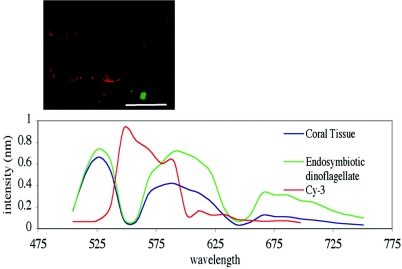
The application of spectral imaging successfully separated coral and endosymbiont autofluorescence from the probe fluorochrome, thereby overcoming previous autofluorescence problems that limited in situ analysis. The success of spectral imaging of the autofluorescence was evident by distinguishing the fluorescence conferred by EUBmix-Cy3-labeled oligonucleotides hybridized to individual E. coli cells. Individual E. coli cells coating coral tissues sections could be easily distinguished from the background fluorescence (Fig. 1).
FIG. 1.
Spectral imaging of A. aspera coral tissue, endosymbiotic dinoflagellate autofluoresence, and EUBmix-Cy3-labeled E. coli. Red, bacteria; blue, coral tissue; green, endosymbiotic dinoflagellates. Scale bar, 50 μm.
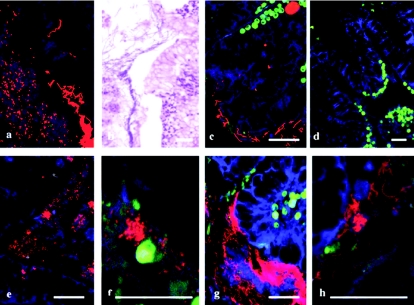
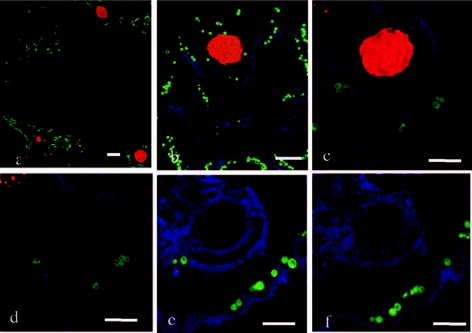
The FISH technique and spectral imaging were successfully applied to visualize and identify bacterial communities associated with predation lesions of corals. Individual bacterial cells and communities of bacteria associated with the lesion created by predation of A. formosa by the snail Drupella sp. were identified. The mixed bacterial community formed a mat that penetrated the coral gastroderm and tissue layers and coated the epithelium (Fig. 2). Two morphologically distinct bacterial communities (filamentous and coccoid) were associated with the predation lesion and identified as belonging to the γ-proteobacteria and Cytophaga-Flavobacterium (Fig. 2). A small population consisting of α-proteobacteria was identified associated with the surface of the coral tissue coating a region of epithelial tissue away from the lesion area. Nonspecific binding was not evident within this study, with no binding of the HGC69a, LGC354, or β-proteobacteria (BET42a) probes on sequential sections. This study also identified bacterial aggregates associated with coral tissues. EUBmix universal bacterial probes marked large ovoid inclusions associated with the gastroderm and mesenterial filaments of the coral tissues, and subsequently these were consistently identified as γ-proteobacteria (Fig. 3). Previously, bacterial aggregates within coral tissues have been described using standard histopathological techniques as large basophilic bodies of up to 40 μm or more associated with tissue layers (27, 28, 35). Interestingly, using 16S rRNA gene sequencing, Rowher et al. (33) consistently found a γ-proteobacterium that they designated PA1 in over 50 Porites asteroides colonies sampled and speculated that this bacterium was the one observed as ovoid aggregates by Santavy and Peters (35). The lack of available and reliable technology previously prevented detailed analysis of these aggregates within the context of the coral holobiont, but this problem has been overcome with this technique. This study's consistent identification of γ-proteobacterial aggregates in branching corals supports the speculation of Rowher et al. (33) and indicates the wider presence of γ-proteobacteria across scleractinian corals.
FIG. 2.
Bacterial communities associated with the lesion created by predation of A. formosa by Drupella sp. FISH identified two bacterial morphologies using EUBmix-Cy3 (a) which were also evident in adjacent sections using hematoxylin and eosin staining (b) and coating the surface of adjacent coral tissues (c). Nonspecific staining of the FISH protocol was not evident (d), and the two bacterial morphologies were identified as a coccoid γ-proteobacterium using the probes GAM42a (e and f) and as a filamentous Cytophaga-Flavobacterium using the probe CF319 (g and h). Red, bacteria; blue, coral tissue; green, endosymbiotic dinoflagellates. Scale bar, 50 μm.
FIG. 3.
FISH using universal bacterial probe EUBmix of large bacterial aggregates associated with coral tissues S. pistillata and A. formosa (a and b) and identification as γ-proteobacteria using GAM42a (c), with no binding of CF319 (d), HGC69a (e), or LGC 354 (f) probes. Bacterial aggregates, red; coral tissue, blue; endosymbiotic dinoflagellates, green. Scale bar, 50 μm.
FISH technology allows for spatial and temporal analysis of bacterial community variability (7), and its application to coral microbial ecology can begin to address questions on the stability and role of bacterial communities. Jones et al. (19) conducted FISH, limiting excitation wavelengths to the red/far-red regions of the spectrum, with coral tissue slurries for confirmation of 16S rRNA sequence analysis and quantification of coral disease-associated microbial communities but did not conduct image analysis, nor did the investigators address the structural arrangement of the microbial communities and interaction with coral tissues. Importantly, the authors noted a difficulty in differentiating disease-causing organisms from secondary invaders (19), a problem that could be addressed by investigating the structural environment of the disease lesions and adjacent regions. The bacterial populations identified within this study are some of the most abundant oceanic bacterial groups and are involved in a wide range of functions (10, 20, 21). Yet investigating the spatial arrangement of microbial communities around lesions and tissues can allow the differentiation of pathogens, lesion colonizers, and persistent microbial communities.
Previous studies utilizing FISH for investigating coral microbial ecology have been limited in determining community interactions by autofluorescence, poor image quality, and probe specificity. This study demonstrates that FISH combined with spectral imaging is a rapid and powerful tool to assess the microbial architecture of reef-building corals. With increasing disease and stress of scleractinian corals (17, 31, 38), the timely description of this technique allows for localization and identification of coral-associated bacterial community changes, suggesting a central role in the future analysis of microbial communities.
Acknowledgments
We are grateful for support provided by the GEF Coral Reef Targeted Research Program (www.gefcoral.org), the Australian Research Council Centre of Excellence for Coral Reef Studies, and the Centre for Advanced Light Microscopy at the University of Queensland.
REFERENCES
- 1.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Scleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of micro-organisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, R. H. 2004. Evaluation of spectral imaging for plant cell analysis. J. Microsc. 214:174-181. [DOI] [PubMed] [Google Scholar]
- 6.Bezanilla, M., A. Pan, and R. S. Quatrano. 2003. RNA interference in the moss Physcomitrella patens. Plant Physiol. 133:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridisation (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., R. Bhagooli, S. Griffin, I. Johnston, and F. Rowher. 2005. Microbial communities associated with skeletal tumors on Porites compressa. FEMS Microbiol. Lett. 243:431-436. [DOI] [PubMed] [Google Scholar]
- 9.Bythell, J. C., M. R. Barer, R. P. Cooney, J. R. Guest, A. G. O'Donnell, O. Pantos, and M. D. A. Le Tissier. 2002. Histopathological methods for the investigation of microbial communities associated with disease lesions in reef corals. Lett. Appl. Microbiol. 34:359-364. [DOI] [PubMed] [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, M. E., G. Bearman, S. Tille, R. Lansford, and S. E. Fraser. 2001. Multi-spectral imaging and linear unmixing add whole new dimensions to laser scanning fluorescence microscopy. BioTechniques 31:1272-1278. [DOI] [PubMed] [Google Scholar]
- 13.Dobretsov, S., and P.-Y. Qien. 2004. The role of epibiotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 299:35-50. [Google Scholar]
- 14.Dove, S. G., M. Takabayashi, and O. Hoegh-Guldberg. 1995. Isolation and partial characterisation of the pink and blue pigments of pocilloporid and acroporid corals. Biol. Bull. 189:288-297. [DOI] [PubMed] [Google Scholar]
- 15.Dove, S. G., O. Hoegh-Guldberg, and S. Rangananthan. 2001. Major colour patterns of reef-building corals are due to a family of GFP-like proteins. Coral Reefs 19:197-204. [Google Scholar]
- 16.Frias-lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surface. Appl. Env. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvell, C. D., C. E. Mitchell, J. R. Ward, S. Altizer, A. P. Dobson, R. S. Ostfeld, and M. D. Samuel. 2002. Climate warming and disease risks for terrestrial and marine biota. Science 296:2158-2162. [DOI] [PubMed] [Google Scholar]
- 18.Hugenholtz, P., B. M. Goebel, and M. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, R. J., J. Bowyer, O. Hoegh-Guldberg, and L. L. Blackall. 2004. Dynamics of a temperature-related coral disease outbreak. Mar. Ecol. Prog. Ser. 281:63-77. [Google Scholar]
- 20.Jorquera, M. A., M. Lody, Y. Leyton, and C. Riquelme. 2004. Bacteria of subclass γ-proteobacteria associated with commercial Argopecten purpuratus (Lamark, 1819) hatcheries in Chile. Aquaculture 236:37-51. [Google Scholar]
- 21.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Micro. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 22.Lesser, M. P. 2004. Experimental biology of coral reef ecosystems. J. Exp. Mar. Bio. Ecol. 300:217-252. [Google Scholar]
- 23.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 24.Manz, W., G. Arp, G. Schumann-Kindel, U. Szemzyke, and J. Reitner. 2000. Widefield deconvolution epifluorescence microscopy combined with fluorescence in situ hybridisation reveals the spatial arrangement of bacteria in sponge tissue. J. Microbiol. Methods 40:125-134. [DOI] [PubMed] [Google Scholar]
- 25.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 26.Moter, A., and U. B. Gobel. 2000. Fluorescence in situ hybridisation (FISH) for direct visualisation of microorganisms. J. Micro. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 27.Peters, E. C., J. J. Oprandy, and P. P. Yevich. 1983. Possible causal agent of “white band disease” in Caribbean acroporids corals. J. Invertebr. Pathol. 41:394-396. [Google Scholar]
- 28.Peters, E. C. 1984. A survey of cellular reactions to environmental stress and disease in Caribbean scleractinian corals. Helgol. Meeresunters. 37:113-137. [Google Scholar]
- 29.Ritchie, K. B., and G. W. Smith. 1997. Physiological comparison of bacterial communities from various species of scleractinian corals. Proc. 8th Int. Coral Reef Symp. 1:521-526. [Google Scholar]
- 30.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram positive bacteria with high G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg, E., and Y. Ben-Haim. 2002. Microbial diseases of corals and global warming. Environ. Microbiol. 4:318-326. [DOI] [PubMed] [Google Scholar]
- 32.Rowher, F., M. Breitbart, J. Jara, F. Azam, and N. Knowlton. 2001. Diversity of bacteria associated with the Caribbean coral Montastrea franksi. Coral Reefs 20:85-95. [Google Scholar]
- 33.Rowher, F., V. Seguritan, F. Azam, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 34.Salih, A., A. Larkum, G. Cox, M. Kuhl, and O. Hoegh-Guldberg. 2000. Fluorescent pigments in corals are photoprotective. Nature 408:850-853. [DOI] [PubMed] [Google Scholar]
- 35.Santavy, D. L., and E. C. Peters. 1997. Microbial pests: coral disease in the western Atlantic. Proc. 8th Int. Coral Reef Symp. 1:607-612. [Google Scholar]
- 36.Shashar, N., A. T. Banaszak, M. P. Lesser, and D. Amrami. 1997. Coral endolithic algae: life in a protected environment. Pac. Sci. 51:167-173. [Google Scholar]
- 37.St. John, J. A., K. T. Tisay, I. W. Caras, and B. Key. 2000. Expression of EphA5 during development of the olfactory nerve pathway in rat. J. Comp. Neuorol. 416:540-550. [DOI] [PubMed] [Google Scholar]
- 38.Sutherland, K. P., J. W. Porter, and C. Torres. 2004. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266:273-302. [Google Scholar]
- 39.Wagner, M., M. Horn, and H. Daims. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6:302-309. [DOI] [PubMed] [Google Scholar]
- 40.Wegley, L., Y. Yu, M. Breitbart, V. Casas, D. I. Kline, and F. Rohwer. 2004. Coral-associated Archaea. Mar. Ecol. Prog. Ser. 273:89-96. [Google Scholar]
- 41.Williams, W. M., A. B. Viner, and W. J. Broughton. 1987. Nitrogen fixation (acetylene reduction) associated with the living coral Acropora variabilis. Mar. Biol. 94:531-535. [Google Scholar]
- 42.Yokouchi, H., H. Takeyama, H. Miyashita, T. Maruyama, and T. Matsunaga. 2003. In situ identification of symbiotic dinoflagellates, the genus Symbiodinium with fluorescence-labelled rRNA-targeted oligonucleotide probes. J. Microbiol. Methods 53:327-334. [DOI] [PubMed] [Google Scholar]
- 43.Zimmerman, T., L. Rietdorf, and R. Pepperkork. 2003. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 546:87-92. [DOI] [PubMed] [Google Scholar]