Abstract
LuxS is responsible for the production of autoinducer 2 (AI-2), which is involved in the quorum-sensing response of Vibrio harveyi. AI-2 is found in several other gram-negative and gram-positive bacteria and is therefore considered a good candidate for an interspecies communication signal molecule. In order to determine if this system is functional in the gastrointestinal pathogen Listeria monocytogenes EGD-e, an AI-2 bioassay was performed with culture supernatants. The results indicated that this bacterium produces AI-2 like molecules. A potential ortholog of V. harveyi luxS, lmo1288, was found by performing sequence similarity searches and complementation experiments with Escherichia coli DH5α, a luxS null strain. lmo1288 was found to be a functional luxS ortholog involved in AI-2 synthesis. Indeed, interruption of lmo1288 resulted in loss of the AI-2 signal. Although no significant differences were observed between Lux1 and EGD-e with regard to planktonic growth (at 10°C, 15°C, 25°C, and 42°C), swimming motility, and phospholipase and hemolytic activity, biofilm culture experiments showed that under batch conditions between 25% and 58% more Lux1 cells than EGD-e cells were attached to the surface depending on the incubation time. During biofilm growth in continuous conditions after 48 h of culture, Lux1 biofilms were 17 times denser than EGD-e biofilms. Finally, our results showed that Lux1 accumulates more S-adenosyl homocysteine (SAH) and S-ribosyl homocysteine (SRH) in culture supernatant than the parental strain accumulates and that SRH, but not SAH or AI-2, is able to modify the number of attached cells.
Listeria monocytogenes is a human and animal pathogen that is frequently found in feces, soil, and water and on vegetation (33). Due to its ubiquitous nature, its high stress tolerance, and its resistance to cleaning procedures, this bacterium is present in raw materials and in food plant environments, where it can contaminate food products during processing or packaging. Consequently, L. monocytogenes is associated with food-borne disease outbreaks, which are characterized by widespread distribution and high mortality rates.
Most of the time, in the food processing environment, L. monocytogenes forms biofilms on abiotic surfaces in association with other bacteria, such as Pseudomonas spp. (12). Biofilms allow bacteria to better resist environmental stresses, such as dehydration, and antimicrobial and sanitizing agents (19).
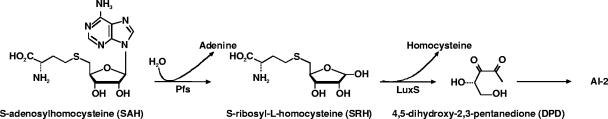
Inside a biofilm, some bacteria are known to communicate using signaling molecules, such as peptides or homoserine lactone derivatives, depending on whether they are gram positive or gram negative. As quorum-sensing mechanisms of the marine bacterium Vibrio harveyi have been deciphered, a new kind of molecule, autoinducer 2 (AI-2), has been added to the list of known signaling compounds (3). Bassler et al. have hypothesized that AI-2 or its derivatives could in fact be a universal signal involved in interspecies communication (1), whereas Joyce et al. suggested that in some species AI-2 serves as a barometer of cellular health (17). Although the biosynthesis pathway of AI-2 is not fully understood, recent data suggest that Pfs and LuxS enzymes catalyze the two-step conversion of S-adenosyl homocysteine (SAH), a very toxic by-product of methyl transfer reactions (28), into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD is a very unstable molecule that can subsequently rearrange into various cyclic compounds, such as furanosyl borate diester (6) or (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (23), which is called AI-2 (Fig. 1). Recent reports have shown that, depending on the bacterium, AI-2 plays a role in motility (11, 34, 35), pathogenicity (36), and biofilm formation (4, 7, 22, 29, 41).
FIG. 1.
Partial pathway for conversion of SAH to AI-2.
In this study, we demonstrated that L. monocytogenes is able to produce AI-2-like molecules via LuxS encoded by lmo1288, a functional ortholog of luxS. We obtained a luxS mutant strain that forms denser biofilms than the parental strain forms in batch or continuous-culture conditions. Finally, the results of addition of in vitro-synthesized SAH, S-ribosyl homocysteine (SRH), and AI-2 suggest that SRH, but not AI-2 or SAH, has an effect on biofilm formation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Biofilm and planktonic cultures of L. monocytogenes EGD-e, which was isolated from a rabbit outbreak (25), and L. monocytogenes Lux1, a mutant derivative of L. monocytogenes EGD-e, were grown in tryptic soy broth (TSB) at 25°C. Escherichia coli DH5α [F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1] (Invitrogen R&D Systems Europe, Oxon, United Kingdom) and E. coli EC101 (E. coli JM101 with repA from pWV01 integrated into the chromosome) (18) were grown aerobically at 37°C in TSB supplemented with 250 μg ml−1 erythromycin and 25 μg ml−1 kanamycin when necessary. E. coli BL21(DE3) (Novagen, VWR International S.A.S., Fontenay-sous-Bois, France) was grown aerobically on LB medium supplemented with 50 μg ml−1 kanamycin.
Vibrio harveyi BB170 (sensor 1−, sensor 2+) (37) and V. harveyi MM30 (autoinducer 1+, autoinducer 2−) (38) were kindly provided by B. Bassler (Princeton University) and were grown aerobically in AB medium at 30°C (2).
Cloning of luxS gene in E. coli DH5α.
A 723-bp DNA fragment was generated by colony PCR from L. monocytogenes EGD-e using primers LUXS1 (5′-TGGCTAGCAATACCCGAAAT-3′) and LUXS2 (5′-CCATAAACCATCACGCCCTT-3′). The fragment containing the coding sequence from lmo1288, along with a 133-bp upstream region and a 122-bp downstream region, was cloned into the pCR2.1 TOPO vector (Invitrogen, Cergy-Pontoise, France) to obtain plasmid pSCB1. This vector was used to electroporate E. coli DH5α.
A 203-bp internal fragment of the coding sequence of luxS was also generated by colony PCR using primers LUXS3 (5′-AATGCCAGCGCTACACTCT-3′) and LUXS4 (5′-CTGTCTTCAAGGACGTACGT-3′) and was cloned into the pCR2.1 TOPO vector to obtain plasmid pSCB2. This vector was transferred into E. coli DH5α and was used as a negative control when necessary.
Construction of an L. monocytogenes luxS-negative strain.
Several steps were necessary to generate the mutant strain. First, plasmid pSCB2 was digested with HindIII and XbaI, and the resulting 315-bp fragment containing an internal part of lmo1288 was ligated into pORI19 (18) to obtain pSCB3. This plasmid was routinely maintained in the recipient strain E. coli EC101 (18). pSCB3 was then transferred into L. monocytogenes EGD-e previously transformed with the thermosensitive helper plasmid pVE6007 (20). In order to select for integration mutants, L. monocytogenes EGD-e(pSCB3, pVE6007) was grown for 28 generations on brain heart infusion (BHI) medium (Difco Laboratories, Elancourt, France). A temperature upshift from 25°C to a nonpermissive temperature, 37°C, resulted in the loss of pVE6007. Plating on BHI agar supplemented with 5 μg erythromycin ml−1 and 25 μg lincomycin ml−1 selected for chromosomal integration of pSCB3 at the point of homology with lmo1288. The mutant selected was designated Lux1.
Complementation of the luxS mutation.
A reversion mutant of L. monocytogenes Lux1 was obtained by excision of pSCB3 from the chromosome. Strain Lux1 was grown for 120 generations on BHI medium without erythromycin at 37°C. A total of 4,000 clones from this culture were tested for loss of the integrated plasmid pSCB3 by plating in duplicate onto BHI medium plates with or without erythromycin. Three clones were selected based on their sensitivity to the antibiotic. Excision of pSCB3 was confirmed by PCR and sequencing of the lmo1288 region. The three revertants were used as controls in order to confirm the phenotype differences between strains EGD-e and Lux1.
Swimming motility, hemolytic, and phospholipase assays.
For swimming motility, tryptone swim plates (1% tryptone, 0.5% NaCl, 0.3% agar) were inoculated with a sterile toothpick and incubated for 16 h at 25°C. Motility was then qualitatively assessed by examining the circular turbid zone formed by the bacterial cells migrating away from the point of inoculation.
Hemolytic and phospholipase activities were examined as previously described (27).
AI-2 bioassay.
Cell-free culture supernatants of V. harveyi BB170, L. monocytogenes EGD-e, L. monocytogenes Lux1, E. coli DH5α, E. coli DH5α(pSCB1), or E. coli DH5α(pSCB2) were obtained by centrifugation (6,000 × g, 10 min, 25°C) and filtration (0.22-μm-pore-size SLGV R25 KS filter; Millipore, Saint Quentin en Yvelines, France) of 1-ml portions of 22-h cultures. AI-2 in the supernatants was detected using the biosensor V. harveyi MM30 as previously described (38), with some modifications. Briefly, V. harveyi MM30 was grown aerobically in AB medium for 16 h at 30°C. The culture was then diluted 1/5,000 (vol/vol) in fresh AB medium, and 90 μl of it was placed into each well of a 96-well microplate (Optiplate-96 white; PerkinElmer Life Sciences, Courtaboeuf, France). Ten microliters of each supernatant to be tested was then added to wells. For each experiment, cell-free culture supernatants of E. coli DH5α and V. harveyi BB170 were used as negative and positive controls, respectively. Luminescence was monitored hourly using a Fusion universal microplate analyzer (PerkinElmer Life Sciences, Courtaboeuf, France) at 30°C until an AI-2 signal was detected (generally 5 h after the beginning of the experiment). Each experiment was repeated four times with three different inocula.
Biofilm formation and quantitation.
Discontinuous biofilm cultures were grown on polystyrene microplates (Nunc; Dominique Dutscher S.A., Brumath, France) as follows. An overnight culture of the bacterium in TSB was used to inoculate (1/100, vol/vol) fresh TSB and was grown at 25°C to an optical density at 600 nm of 0.05. One hundred microliters of the culture to be tested was then transferred to a microtiter plate well, and the plate was tightly sealed and incubated at 25°C for 5 to 72 h.
Cells attached to the well walls were quantified as previously described (9), with some modifications. After incubation, the medium was removed from each well, and the plates were washed twice using a microtiter plate washer (Cellwash; Thermolabsystems, Cergy Pontoise, France) with a 150 mM NaCl solution in order to remove loosely attached cells. The plates were then stained with a 0.05% (wt/vol) aqueous crystal violet solution for 45 min and washed four times. In order to quantitatively assess biofilm production, 100 μl of a destaining solution (96% [vol/vol] ethanol) was added to each well, and the optical density at 595 nm was determined. For each experiment, 40 replicates resulting from five different inocula were analyzed. Each microtiter plate included eight wells with sterile TSB in control wells.
Continuous-flow biofilm cultures were grown on AISI 304 stainless steel chips (Goodfellow SARL, Lille, France) in flow cells inoculated (1/100, vol/vol) with overnight cultures at 25°C. During growth, TSB was pumped through the flow cells at a constant flow rate (14 ml · h−1) by using a peristaltic pump (model 205S; Watson Marlow, Calmouth, Cornwall, England).
ESEM.
For environmental scanning electron microscopy (ESEM) observations, bacterial cells were grown on chemically defined medium MCDB 202 (CryoBiosystem, L'Aigle, France). Biofilms were grown for 36 h on AISI 304 stainless steel chips in MCDB 202 medium. The cells were fixed with glutaraldehyde and osmic vapors as previously described (5) and were observed using a Philips XL30 ESEM.
pET cloning of pfs and luxS genes.
pfs (lmo1494) and luxS (lmo1288) coding sequences were amplified by PCR using primers PFSF3 (5′ TTAATACTCATGACAATTGGTATTATCGG 3′) and PFSR3 (5′ TATTAACTCGAGAATTGTTTTAAGCAATTC 3′) and primers LUXSF3 (5′ TTAATACTCATGATGGCAGAAAAAATGAATG 3′) and LUXSR3 (5′ TATTAACTCGAGTTCACCAAACACATTTTTCC 3′), respectively. In both cases, the primers generated a BspHI site and a XhoI site (underlined) that were used to clone the BspHI/Xho1-digested PCR fragments into NcoI/XhoI-digested pET 28a(+) (Novagen, VWR International S.A.S., Fontenay-sous-Bois, France). The resulting plasmids, pSCB4 and pSCB5, contained the coding sequences of pfs and luxS, respectively, fused in frame with a downstream sequence encoding six histidine residues (His tag). They were controlled by sequencing and used to transform E. coli BL21(DE3) cells for the production of the recombinant proteins.
The recombinant LuxS protein contained an additional N terminus methionine and two additional residues (Leu and Glu) between the C terminus of the mature protein and the His tag, whereas the recombinant Pfs protein contained two additional residues (Leu and Glu) before the His tag.
Production and purification of the LuxS and Pfs proteins.
Both LuxS and Pfs were produced using the same procedure. Briefly, E. coli BL21(DE3) transformed with pSCB4 and pSCB5 was grown aerobically on Luria-Bertani medium supplemented with kanamycin (50 μg/ml) at 37°C to an optical density at 600 nm of 1. The cultures were then cooled to 20°C and incubated aerobically for 15 h in the presence of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG) in order to induce synthesis of the recombinant proteins. The cells were cooled to 5°C and harvested by centrifugation, resuspended in cold 30 mM Tris-HCl (pH 8) binding buffer (BB), and disrupted with a Constant cell disruption system (Cell-D; Constant System Ltd., Roquemaure, France). The crude extract was centrifuged at 26,000 × g for 15 min, and the supernatant was loaded into a 3-ml Ni-nitrilotriacetic acid column previously equilibrated with BB. After 20 column volume washes with BB, the proteins were eluted with BB supplemented with 400 mM imidazole. The eluates were dialyzed against BB and concentrated with an Amicon concentrator. The protein purification steps were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Aliquots of the protein solution were filter sterilized and stored at 4°C.
Protein concentrations were calculated by measuring the absorbance of a 6 M guanidium chloride protein solution at 280 nm using molar extinction coefficients of 11,520 M−1 cm−1 for Pfs and 16,620 M−1 cm−1 for LuxS.
In vitro synthesis of SRH and AI-2.
DPD, the AI-2 precursor, was synthesized in 20 mM KH2PO4 (pH 7.0) buffer containing 1 mM SAH, 14.4 μM purified recombinant Pfs, and 8.6 μM purified recombinant LuxS. The reaction mixture was incubated for 14 h at 30°C. For SRH synthesis, the same procedure was used, except that LuxS was omitted from the reaction mixture. In both cases, after incubation, the enzymes were removed by ultrafiltration, and the reaction products were quantified as follows.
The in vitro reaction was monitored by measuring the amounts of homocysteine and AI-2 released during the reaction using Ellman's reagent (15) and the AI-2 bioassay, respectively. Under these experimental conditions the substrate (SAH) was totally converted into DPD and homocysteine when Pfs and LuxS were used.
AI-2, SAH, and SRH complementation.
Complementation experiments were performed with 42-h-old L. monocytogenes EGD-e and Lux1 biofilms grown on microtiter plates. Biofilm cultures were complemented with 50 μM (final concentration) in vitro-synthesized SAH, SRH, or AI-2 and incubated for another 8 h before cell quantification. The amount of luminescence measured in the AI-2 bioassay in the presence of 50 μM synthetic AI-2 was similar to the amount of luminescence generated by a 50-h-old L. monocytogenes EGD-e biofilm supernatant.
SAH and SRH detection in cell-free biofilm supernatants.
Fifty-hour-old biofilms were used in the experiments for detection of SAH and SRH in cell-free biofilm supernatants and were treated as follows. Biofilm supernatants were filter sterilized, tested for the absence of LDH activity (13), which indicated that no cell lysis occurred, and heat treated (100°C, 30 min) to inactivate AI-2. At the same time, the amount of attached cells was determined by measuring the optical density at 595 nm. To detect the presence of SAH, the supernatants were incubated for 1 h at 37°C with 38 μM Pfs and 56 μM LuxS (final concentrations). For SRH detection, only LuxS was added. After this, supernatants were tested for AI-2 activity using the AI-2 bioassay. The amount of luminescence measured in the bioassay was directly proportional to the amount of SAH or SRH converted into AI-2. The results were expressed in relative light units per optical density unit to take into account the differences in the numbers of attached cells in EGD-e and Lux1 biofilms.
Statistical analysis.
Data were analyzed for statistical significance using the SigmaStat 3.0.1 software (SPSS France S.A., Paris-La-Défense, France) for the t test (Student-Newman-Keuls method).
RESULTS
L. monocytogenes produces an autoinducer 2-like molecule.
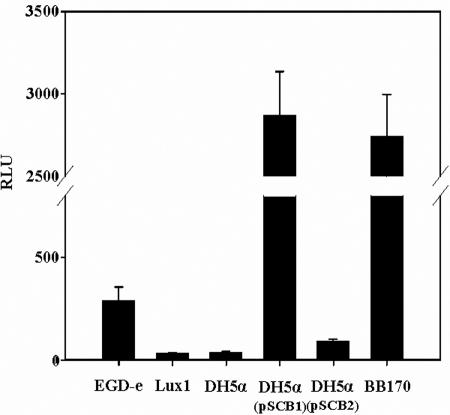
In order to understand cell-to-cell communication mechanisms in L. monocytogenes, we focused on the ability of this bacterium to produce AI-2 molecules. Therefore, cell-free culture supernatants of L. monocytogenes EGD-e were tested for the presence of AI-2 using V. harveyi MM30 as a biosensor. As shown in Fig. 2, cell-free culture supernatants of L. monocytogenes EGD-e induced luminescence of V. harveyi MM30, indicating that AI-2 like molecules were produced by strain EGD-e.
FIG. 2.
Induction of V. harveyi MM30 luminescence by cell-free supernatants of planktonic cultures of L. monocytogenes EGD-e and Lux1, E. coli DH5α, DH5α(pSCB1), and DH5α(pSCB2), and V. harveyi BB170. Each bar indicates the mean from eight experiments, and the error bars indicate standard deviations. RLU, relative light units.
L. monocytogenes EGD-e AI-2 production was growth phase independent, as the same amount of induced luminescence (reported to the biomass) was detected for early-, mid-, and late-log-phase culture supernatants (data not shown).
Identification and isolation of the luxS gene and complementation of E. coli DH5α.
lmo1288, a gene similar to V. harveyi luxS, was found in the genome of L. monocytogenes EGD-e by a BLAST search on the ListiList World Wide Web server (Institut Pasteur) (14, 24). This gene encodes a 155-amino-acid protein that is fully conserved in various Listeria strains (26) and exhibits 41% identity and 59% similarity with the V. harveyi LuxS protein. Sequence comparisons showed that Lmo1288 was closely related to LuxS of other species, such as Staphylococcus aureus Mu50 (78% identity and 87% similarity), Helicobacter pylori 26695 (68% identity and 82% similarity), Clostridium perfringens (51% identity and 68% similarity), and Bacillus subtilis (44% identity and 62% similarity). For example, all the metal ligands (H54, H58, and C126) of LuxS of B. subtilis are conserved in Lmo1288. Moreover the amino acids involved in hydrogen bonding and electrostatic interactions with the substrate or the product of LuxS of B. subtilis (S6, K35, E57, R65, D78, I79, S80, C84, Q125, and G127) are also conserved in Lmo1288 (16, 30, 31, 45). These results strongly suggest that Lmo1288 is a LuxS-like protein.
To confirm the function of lmo1288, we designed a pair of primers, LUXS1 and LUXS2, to amplify the 723-bp region containing the lmo1288 CDS, 133 bases upstream of the initiation codon and 122 bases downstream of the stop codon. The PCR fragment was cloned into the pCR2.1 TOPO vector, and its sequence was confirmed by sequencing. The resulting plasmid, pSCB1, was electroporated into E. coli DH5α, a luxS-negative strain (38), in order to perform complementation experiments. Cell-free culture supernatants of E. coli DH5α (negative control), E. coli DH5α(pSCB2) (negative control), E. coli DH5α(pSCB1), and L. monocytogenes EGD-e were tested for AI-2 activity. As shown in Fig. 2, E. coli DH5α(pSCB1) culture supernatants induced a 77-fold increase in luminescence compared to the luminescence induced by the E. coli DH5α supernatant. This result confirmed that lmo1288 encodes a LuxS-like protein involved in AI-2-like molecule synthesis. Therefore, we refer to the L. monocytogenes protein as LuxSLm below.
Phenotypic characterization of the L. monocytogenes Lux1 mutant.
To confirm the role of LuxSLm in AI-2 synthesis, we generated mutant Lux1 from L. monocytogenes EGD-e by disruption of the luxS gene. This construction was controlled by PCR and Southern blotting (data not shown).
Lux1 cell-free culture supernatants did not generate any luminescence in our bioassay (Fig. 2). Furthermore, complementation of the luxS mutation by excision of pSCB3 from lmo1288 restored the ability to induce luminescence of V. harveyi (data not shown). This result confirmed the involvement of LuxSLm in AI-2 synthesis.
To characterize the mutant phenotype, we compared planktonic growth of L. monocytogenes EGD-e and planktonic growth of Lux1 in TSB at 10°C, 15°C, 25°C, and 42°C. Irrespective of the conditions, the growth rates of the two strains were similar (data not shown), and there was no difference between the two strains in terms of swimming motility and hemolytic and phospholipase activities.
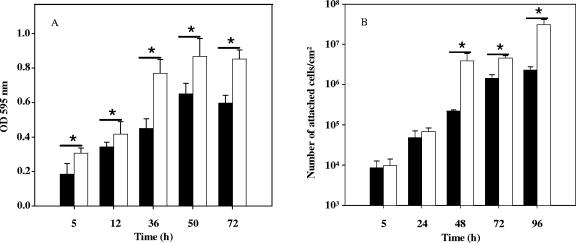
As recent reports have demonstrated the involvement of luxS in biofilm development of various oral pathogens (4, 21, 22, 43), we investigated the abilities of L. monocytogenes EGD-e and Lux1 to form biofilms in batch culture conditions (Fig. 3A). Statistically significant differences between the two strains (P ≤ 0.00021 for 5 h and P ≤ 0.00001 for 12 h, 36 h, 50 h, and 72 h, as determined by t tests) were observed, and interestingly, denser biofilms were obtained with Lux1. The difference was maximal after 36 h of incubation, when 58% more mutant cells than EGD-e cells were attached to the surface. Then the abilities of the two strains to form biofilms on stainless steel in continuous culture conditions were compared. Again, major differences in terms of the number of attached cells were observed. Indeed, the Lux1 strain formed denser biofilms than the EGD-e strain formed, with 10 to 17 times more attached cells. Interestingly, the differences were significant only after 48 h of incubation (Fig. 3B).
FIG. 3.
Biofilm formation by L. monocytogenes EGD-e (solid bars) and L. monocytogenes Lux1 (open bars) in batch conditions (A) or in continuous-culture conditions (B) over time, as described in Materials and Methods. In panel A, each bar indicates the mean from five independent experiments with eight measurements for each point, and in panel B, each bar indicates the mean from three independent experiments with three measurements for each point; the error bars indicate standard deviations. Asterisks indicate statistically significant differences (P < 0.001). OD 595 nm, optical density at 595 nm.
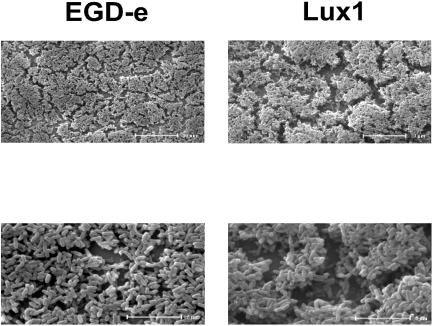
ESEM microscopic observations of Lux1 and EGD-e confirmed these results. As shown in Fig. 4, Lux1 formed thicker biofilms with large spaces separating cell patches, whereas EGD-e biofilms were thinner and more confluent. As reversion of the mutation in Lux1 restored the EGD-e biofilm phenotype (data not shown), we concluded that luxS was important for biofilm formation in L. monocytogenes EGD-e.
FIG. 4.
ESEM images of 36-h-old L. monocytogenes EGD-e and Lux1 biofilms incubated at 25°C on stainless steel in batch conditions.
Complementation of Lux1 biofilms.
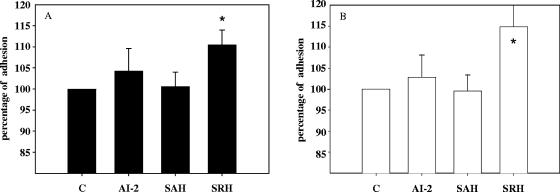
To understand the role of luxS in biofilm formation, synthetic AI-2 was produced from SAH using purified His-tagged PfsLm and LuxSLm proteins and was used to complement supernatants of 42-h-old biofilms. To make sure that the right quantity of AI-2 was added in these experiments, throughout the AI-2 bioassay we measured the amount of luminescence generated by the supernatant of a 42-h-old EGD-e biofilm culture. Then we added to the supernatant of a 42-h-old Lux1 biofilm culture the amount of synthetic AI-2 necessary to generate the same luminescence. Surprisingly, as shown in Fig. 5, addition of AI-2 failed to restore the parental strain biofilm phenotype. As a luxS strain does not produce AI-2 but may accumulate SAH and SRH (Fig. 1), we investigated the effects of these molecules on Lux1 biofilm. As shown in Fig. 5, 50 μM SAH had no effect on Lux1 biofilm formation, whereas on average, addition of 50 μM SRH resulted in a 15% increase in the number of attached cells, irrespective of the strain tested.
FIG. 5.
Effect of addition of 50 μM synthetic AI-2, SAH, or SRH on 42-h-old biofilms. The results are expressed as quantities relative to that for the control experiment (C), in which nothing was added to the supernatant for strain EGD-e (A) and strain Lux1 (B). Asterisks indicate statistically significant differences from the value for the control (P < 0.001).
Presence of SAH and SRH in biofilm culture supernatant.
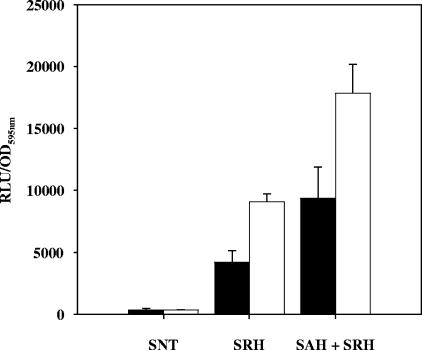
As the effect of SRH on L. monocytogenes biofilms was unexpected, we addressed the following questions. Do EGD-e and Lux1 biofilm culture supernatants contain any SRH? If so, is there any difference between EGD-e and Lux1 in terms of the SRH supernatant concentration, which could explain the Lux1 phenotype? To answer these questions, we used Lux1 and EGD-e supernatants that were heat treated to eliminate AI-2 activity, and we added LuxSLm with or without PfsLm in order to convert SAH and SRH into AI-2; the latter compound was detected using the AI-2 bioassay. At the same time, lactate dehydrogenase activity was monitored in each supernatant to make sure that the presence of the molecules was not the result of cell lysis (data not shown). As shown in Fig. 6, it appeared that both EGD-e and Lux1 supernatants contained SAH and SRH but different quantities of these compounds. There was twice as much SRH in the Lux1 supernatant as in the supernatant of EGD-e. As Lux1 was unable to convert SRH into AI-2 and as SRH increased the number of attached cells, this result could partially explain the Lux1 phenotype.
FIG. 6.
Quantities of SRH and SAH present in heat-treated biofilm supernatants of EGD-e (solid bars) and Lux1 (open bars). SNT, heat-treated supernatants alone; SRH, heat-treated supernatants incubated with recombinant LuxS; SAH + SRH, heat-treated supernatants incubated with recombinant Pfs and LuxS. The amount of luminescence produced is adjusted for the number of cells present in the biofilm, as described in Materials and Methods. RLU, relative light units; OD595nm, optical density at 595 nm.
For SAH, the only way to evaluate the amount in culture supernatants was to subtract the amount of luminescence obtained when only LuxS was added from the amount of luminescence obtained after addition of Pfs and LuxS to the supernatant. Both supernatants contained SAH, but there was almost twice as much SAH present in the Lux1 supernatant as in the supernatant of EGD-e.
Similar results were obtained with planktonic culture supernatants of V. harveyi BB170 and MM30 and E. coli DH5α. In each case, SAH and SRH were detected in supernatants, and proportionally, there was more SAH and SRH in the supernatants of the luxS-negative strains (V. harveyi MM30 and E. coli DH5α).
DISCUSSION
In this study, we demonstrated that L. monocytogenes, like several other gram-positive bacteria, is able to produce an AI-2 or an AI-2-like signal recognized by V. harveyi MM30. We looked for a luxS ortholog in the L. monocytogenes genomic database and found lmo1288, a putative gene encoding a 155-amino-acid protein. As most of the amino acids involved in the substrate binding site and in the metal chelating site of LuxS from B. subtilis perfectly match amino acids in Lmo1288, we postulated that Lmo1288 is an S-ribosylhomocysteinase (16, 31, 45).
Complementation experiments with E. coli DH5α and mutagenesis of lmo1288 from L. monocytogenes EGD-e confirmed the involvement of Lmo1288 in AI-2 biosynthesis. Moreover, overproduction, purification, and an activity assay of the protein clearly demonstrated that it is an S-rybosylhomocysteinase which catalyzes the bioconversion of SRH into homocysteine and DPD. As previously described, the latter compound undergoes chemical modifications to generate AI-2-like signals that are able to induce the luminescence of V. harveyi MM30.
The Lux1 strain does not differ from the parental strain in terms of the growth rates at various temperatures, motility, and phospholipase and hemolytic activities. As a luxS mutation sometimes affects biofilm formation (7, 43), we compared the abilities of the two strains to form biofilms under batch and continuous-culture conditions. Surprisingly, it appeared that Lux1 cells formed denser biofilms than cells of the parental strain formed. For example, in continuous-culture conditions, 17 times more Lux1 cells than EGD-e cells were attached to the support after 96 h of incubation. These results were confirmed by ESEM observations and are in accordance with those obtained by Cole et al. with a luxS mutant of H. pylori (7). In contrast, Wen and Burne, working with Streptococcus mutans, demonstrated that less biofilm was formed by a luxS mutant (41). Nevertheless, irrespective of the phenotype, it seems that luxS plays a key role in the formation of biofilms.
For some bacteria, the pleiotropic effect of a luxS mutation has been demonstrated (8, 35, 44). Considering the differences, we investigated the ability of synthetic AI-2 to restore the phenotype of the parental strain when it was added to Lux1 supernatant. Surprisingly, addition of AI-2 had no effect on L. monocytogenes biofilm formation. Therefore, we hypothesized that perhaps the accumulation of the precursors SAH and SRH may have an effect. Indeed, these molecules were found in biofilm culture supernatants of both strains, but larger quantities were observed for Lux1 than for EGD-e and only SRH was able to increase the number of attached cells. These results raised the question of the SAH detoxification pathway of L. monocytogenes. Indeed, during the activated methyl cycle (AMC) it is accepted that S-adenosylmethionine provides methyl groups for proteins, RNA, and DNA. Depending on the organism, the resulting SAH, a toxic compound, is then converted into homocysteine using a one-step reaction (SAH hydrolase) or a two-step reaction (Pfs and LuxS). The latter reaction, found in L. monocytogenes, generates DPD, which spontaneously rearranges to form AI-2-like molecules (40). Our observations suggest that SAH and SRH can accumulate outside the cell via an unknown mechanism and, moreover, that SRH modulates the ability of L. monocytogenes to form biofilms. The reason why such a mechanism exists is unclear, but it may be necessary to eliminate SAH when the detoxification pathway is overwhelmed or down regulated. Moreover, as there are different AI-2-like compounds (6, 23), it could be useful for the bacteria to be able to detect the presence of other bacteria using SRH, an AMC-linked molecule.
In conclusion, AI-2 has been demonstrated to be a mediator of quorum sensing in V. harveyi, Vibrio cholerae, E. coli, and Salmonella enterica serovar Typhimurium (3, 39, 42). However, in other bacteria, such as Neisseria meningitidis (10, 32), AI-2 does not trigger any global transcriptional response. Similarly, our results suggest that in L. monocytogenes, the physiological role of AI-2 may be limited to the detoxification of SAH and may not play a key role in cell-to-cell signaling. In contrast, SRH, the precursor of AI-2, plays an important role in biofilm formation in L. monocytogenes and may be involved in cell-to-cell communication. The mechanisms of excretion of AI-2, SAH, and SRH must be deciphered in order to better understand the roles of these AMC intermediates in biofilm formation in L. monocytogenes.
Acknowledgments
We thank B. Bassler for providing the V. harveyi strains, D. Valentin for her statistical expertise, and the Centre de Microscopie Appliquée à la Biologie de l'Université de Bourgogne for the ESEM images.
S. Challan Belval holds a doctorate fellowship from INRA and the Conseil Régional de Bourgogne.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 4.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavant, P., B. Martinie, T. Meylheuc, M. N. Bellon-Fontaine, and M. Hebraud. 2002. Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 68:728-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djordjevic, D., M. Wiedmann, and L. A. McLandsborough. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 149:1859-1869. [DOI] [PubMed] [Google Scholar]
- 11.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 12.Fatemi, P., and J. F. Frank. 1999. Inactivation of Listeria monocytogenes/Pseudomonas biofilms by peracid sanitizers. J. Food Prot. 62:761-765. [DOI] [PubMed] [Google Scholar]
- 13.Garmyn, D., T. Ferain, N. Bernard, P. Hols, B. Delplace, and J. Delcour. 1995. Pediococcus acidilactici ldhD gene: cloning, nucleotide sequence, and transcriptional analysis. J. Bacteriol. 177:3427-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Hendricks, C. L., J. R. Ross, E. Pichersky, J. P. Noel, and Z. S. Zhou. 2004. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 326:100-105. [DOI] [PubMed] [Google Scholar]
- 16.Hilgers, M. T., and M. L. Ludwig. 2001. Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proc. Natl. Acad. Sci. USA 98:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce, E. A., A. Kawale, S. Censini, C. C. Kim, A. Covacci, and S. Falkow. 2004. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 72:2964-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 24.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 25.Murray, E. G. D., R. A. Webb, and M. B. R. Swann. 1926. A disease of rabbit characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus: Bacterium monocytogenes (n. sp.). J. Pathol. Bacteriol. 29:407-439. [Google Scholar]
- 26.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 28.Pei, D., and J. Zhu. 2004. Mechanism of action of S-ribosylhomocysteinase (LuxS). Curr. Opin. Chem. Biol. 8:492-497. [DOI] [PubMed] [Google Scholar]
- 29.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajan, R., J. Zhu, X. Hu, D. Pei, and C. E. Bell. 2005. Crystal structure of S-ribosylhomocysteinase (LuxS) in complex with a catalytic 2-ketone intermediate. Biochemistry 44:3745-3753. [DOI] [PubMed] [Google Scholar]
- 31.Ruzheinikov, S. N., S. K. Das, S. E. Sedelnikova, A. Hartley, S. J. Foster, M. J. Horsburgh, A. G. Cox, C. W. McCleod, A. Mekhalfia, G. M. Blackburn, D. W. Rice, and P. J. Baker. 2001. The 1.2 Å structure of a novel quorum-sensing protein, Bacillus subtilis LuxS. J. Mol. Biol. 313:111-122. [DOI] [PubMed] [Google Scholar]
- 32.Schauder, S., L. Penna, A. Ritton, C. Manin, F. Parker, and G. Renauld-Mongenie. 2005. Proteomics analysis by two-dimensional differential gel electrophoresis reveals the lack of a broad response of Neisseria meningitidis to in vitro-produced AI-2. J. Bacteriol. 187:392-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlech, W. F., III, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 34.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stroeher, U. H., A. W. Paton, A. D. Ogunniyi, and J. C. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 40.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 41.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, L., J. D. Hillman, and A. Progulske-Fox. 2005. Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis. Infect. Immun. 73:4146-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, J., E. Dizin, X. Hu, A. S. Wavreille, J. Park, and D. Pei. 2003. S-ribosylhomocysteinase (LuxS) is a mononuclear iron protein. Biochemistry 42:4717-4726. [DOI] [PubMed] [Google Scholar]