Abstract
Extensive gene duplication is thought to have occurred in the vertebrate lineage after it diverged from cephalochordates and before the divergence of lobe- and ray-finned fishes, but the exact timing remains obscure. This timing was investigated by analysis of the Dlx gene family of a representative cartilaginous fish, the leopard shark, Triakis semifasciata. Dlx genes encode homeodomain transcription factors and are arranged in mammals as three convergently transcribed bigene clusters. Six Dlx genes were cloned from Triakis and shown to be orthologous to single mammalian Dlx genes. At least four of these are arranged in bigene clusters. Phylogenetic analyses of Dlx genes were used to propose an evolutionary scenario in which two genome duplications led to four Dlx bigene clusters in a common ancestor of jawed vertebrates, one of which was lost prior to the diversification of the group. Dlx genes are known to be involved in jaw development, and changes in Dlx gene number are mapped to the same branch of the vertebrate tree as the origin of jaws.
COMPARISONS of vertebrate and invertebrate gene families have provided evidence for large-scale gene duplication in the vertebrate lineage after it diverged from its extant sister group, the cephalochordates (Holland et al. 1994; Sidow 1996; Spring 1997; Furlong and Holland 2002; Panopoulou et al. 2003). One of the most extensively studied of these gene families is the Hox family of homeodomain transcription factors (Pendleton et al. 1993; Ruddle et al. 1994a; Prince 2002; Wagner et al. 2003). The 39 Hox genes of mammals are arranged as four clusters of closely linked genes, each located on separate chromosomes. In contrast, the cephalochordate amphioxus has 14 Hox genes linked in a single cluster (Ferrier et al. 2000). The existence of a single Hox cluster in more distantly related invertebrate outgroups and the presence of multiple Hox clusters in actinopterygian (ray-finned) fishes suggest that a single Hox cluster gave rise to four prior to the divergence of actinopterygians and sarcopterygians (lobe-finned fishes and tetrapods; Amores et al. 1998; Finnerty and Martindale 1998; Balavoine et al. 2002; Prince 2002; Wagner et al. 2003). Additional Hox cluster duplications are likely to have occurred in the actinopterygian lineage after it diverged from sarcopterygians (Amores et al. 1998, 2004; Prince 2002; Wagner et al. 2003; Chiu et al. 2004).
Less information is available for assessing whether four Hox clusters were present prior to the divergence of osteichthyans (actinopterygians and sarcopterygians) from chondrichthyans (cartilaginous fishes) or the divergence of lampreys and hagfishes from gnathostomes (jawed vertebrates, which include osteichthyans and chondrichthyans). Recent analyses of lamprey Hox genes have suggested that not all of the duplications leading to the four-cluster condition antedated the lamprey-gnathostome divergence and, furthermore, that independent Hox duplications occurred in the lamprey lineage (Force et al. 2002; Irvine et al. 2002; Fried et al. 2003; Wagner et al. 2003). While only two Hox clusters have been described to date from a chondrichthyan (Kim et al. 2000), it has been speculated that the four-cluster condition characterized the ancestral gnathostome (Wagner et al. 2003).
Hox cluster duplications are commonly described as the result of complete genome duplications (Sidow 1996; Furlong and Holland 2002; Larhammar et al. 2002), but this interpretation remains controversial (Hughes et al. 2001; Martin 2001; Horton et al. 2003; Hughes and Friedman 2003). Much attention in this controversy has focused on the linkage of additional families of paralogous genes to the Hox clusters of vertebrates and more specifically on the question of whether their duplication history parallels that of the Hox genes (Lundin 1993; Ruddle et al. 1994b; Bailey et al. 1997; Hughes et al. 2001; Larhammar et al. 2002; Hughes and Friedman 2003; Lundin et al. 2003). As pointed out by Larhammar et al. (2002), many of the analyses of these gene families have not included a sample of vertebrate lineages adequately representing the diversity of the group. A partial exception to this generalization is the Dlx family of homeodomain transcription factors related to the Drosophila gene Distal-less.
Dlx genes have been shown to play a role in the development of a variety of vertebrate structures, including the brain, ears, nose, jaws, teeth, and limbs (Panganiban and Rubenstein 2002). Mammals possess six Dlx genes arranged as three convergently transcribed bigene clusters, with each cluster located on a different Hox-containing chromosome (Panganiban and Rubenstein 2002; Sumiyama et al. 2003). Analyses of the eight described zebrafish Dlx genes (arranged as three bigenes clusters and two unlinked single genes) have suggested a duplication history resembling that of Hox genes (Stock et al. 1996; Amores et al. 1998; Neidert et al. 2001; Sumiyama et al. 2003). Specifically, duplication of an ancestral bigene cluster led to four clusters in a common ancestor of osteichthyans, followed by loss of a cluster before the divergence of actinopterygians and sarcopterygians. These events were followed by independent duplication and losses of Dlx genes in the actinopterygian lineage. As in the Hox gene family, it has been suggested that independent Dlx duplications have occurred in the lamprey lineage (Neidert et al. 2001).
No data have been available to indicate whether four Dlx bigene clusters arose before the divergence of the main lineages of gnathostomes. Such data are of interest for a number of reasons. Jaws, the possession of which gives gnathostomes their name, are believed to be patterned by Dlx genes (Depew et al. 2002) and expression differences exist between lamprey and tetrapod Dlx genes (Neidert et al. 2001; Shigetani et al. 2002). The possibility that Dlx genes were involved in the evolution of gnathostome jaws raises the question of whether gene duplication events were important in addition to changes in expression. The linkage of Hox and Dlx genes in osteichthyans allows interpretation of an identical duplication history of these genes as evidence for duplication of at least large chromosomal regions. In addition, information on Dlx genes of chondrichthyans may aid the interpretation of the incompletely characterized Hox clusters of this group. Finally, if it is assumed that Hox and Dlx duplications are the result of whole-genome duplications, information on the Dlx genes of chondrichthyans would provide insight on the organization of the genome of the common ancestor of gnathostomes.
This study seeks to address the above issues through a characterization of the Dlx gene complement of a representative chondrichthyan, the leopard shark, Triakis semifasciata. Two main alternative hypotheses tested are (1) that the four bigene-cluster-condition postdated the divergence of osteichthyans and chondrichthyans and (2) that the four-cluster condition antedated this divergence. The first scenario predicts that shark genes will be orthologous to multiple osteichthyan genes, while the second predicts that shark genes will be orthologous to single osteichthyan genes. Under either scenario, sharks may have lost orthologs of gnathostome genes or undergone independent gene duplication. The cloning and analyses of Triakis Dlx genes presented herein are interpreted as support for the origin of four bigene clusters and the subsequent loss of one of these prior to the divergence of the main gnathostome lineages. No evidence was found for additional Dlx duplications or losses in the chondrichthyan lineage. The acquisition of the three-cluster condition of sharks and mammals therefore maps to the same branch of the phylogenetic tree of extant vertebrates as the origin of jaws.
MATERIALS AND METHODS
Specimens:
Gravid females of the Leopard shark, T. semifasciata, were provided by fisherman participating in the Pajaro Valley Rod and Gun Club Shark Derby of July 10, 1994. The specimens were captured in Elkhorn Slough, Monterey County, California. Embryos were collected from the females by dissection and were dechorionated, separated from the yolk, and staged according to the series constructed by Ballard et al. (1993) for Scyliorhinus canicula, a member of the same order. The embryos were then frozen on dry ice and stored at −70°.
RNA isolation:
A single embryo 30 mm in total length (approximate stage 30) was ground to a fine powder in liquid N2 using a mortar and pestle. Total cellular RNA was extracted from the powder using the RNAzol B reagent (Biotecx Laboratories, Houston) according to the manufacturer's instructions.
Reverse transcriptase-mediated PCR of Dlx homeoboxes:
Reverse transcription of total RNA was carried out at 42° using an RNase H− Moloney murine leukemia virus reverse transcriptase (Superscript II; Invitrogen, Carlsbad, CA) and random hexamer primers. Single-stranded cDNA from this reaction was used as the template for PCR amplification with degenerate Dlx primers described by Stock et al. (1996). These included the primers 5′-GCCGGGATCCAARCCNMGNACNATHTAYTC-3′ (where the underlined sequence represents a restriction site added to the 5′-end) and 5′-TTYTGRAACCADATYTTNAC-3′, which bind codons near the 5′- and 3′-ends, respectively, of the homeoboxes of all previously known Dlx genes. The additional primers used (5′-GCCGGGATCCGGNAARAARATHMGNAARCC-3′ and 5′-GCCGGGATCCATNGTNAAYGGNAARCCNAA-3′) bind to regions conserved in a subset of vertebrate Dlx genes and located just upstream of the homeobox. The thermocycling profile used was 2 min of denaturation at 94° followed by 40 cycles of 1 min at 94°, 1 min at 45°, and 1 min at 72°.
PCR amplification of 5′ and 3′ cDNA ends:
To obtain complete cDNA sequences corresponding to the reverse transcriptase-mediated PCR (RT-PCR) products described above, the homeobox sequences determined were used to design gene-specific primers for the rapid amplification of cDNA ends (RACE) procedure (Frohman 1990). 3′-RACE was carried out on poly(A)+ RNA using the Marathon RACE method (Chenchik et al. 1996) and 5′-RACE was carried out on total RNA by the SMART RACE method (Chenchik et al. 1998). In both cases, the RNA came from the same individual used for RT-PCR and the protocol followed that provided by the manufacturer (BD CLONTECH, Palo Alto, CA). RACE products corresponding to the gene of interest were identified by Southern blotting with oligonucleotide probes or by restriction enzyme digestion.
PCR amplification of exons and introns from genomic DNA:
The exon/intron structure of Dlx genes has been described for human (Nakamura et al. 1996; Sumiyama et al. 2002), mouse (McGuinness et al. 1996; Nakamura et al. 1996; Liu et al. 1997; Sumiyama et al. 2002), and zebrafish (Ellies et al. 1997). Intron location is conserved in all described cases, with one intron 5′ of the homeobox and a second intron within. To determine whether this genomic structure is also conserved with Dlx genes of Triakis, PCR primers were designed to amplify from genomic DNA the entire coding region of each gene in two overlapping fragments. These two fragments extended from the 5′-UTR to the predicted second exon and from this exon to the 3′-UTR. This strategy identifies all introns located within the coding region of the genes, but cannot exclude the possibility of introns at the extreme ends of the 5′- and 3′-UTRs.
Genomic DNA (extracted from individual, unstaged embryos by overnight proteinase K digestion at 55° followed by phenol/chloroform extraction and isopropanol precipitation) was used as a template for long PCR (Barnes 1994). These PCR reactions employed the Expand DNA polymerase mixture and associated buffer (Roche, Indianapolis) and the TaqStart Antibody (CLONTECH), with the latter allowing a “hot start” (Kellogg et al. 1994). Thermocycling consisted of 2 min of denaturation at 94°, followed by 35 cycles of 30 sec at 94° and 5 min at 68°. PCR products were subcloned and intron positions were determined by sequencing. The sizes of the introns were estimated by a combination of partial sequencing and agarose gel electrophoresis of PCR products.
Genomic long PCR:
The presence of Dlx bigene clusters in Triakis was investigated by long PCR, using conditions as described above (with the exception that the annealing/extension step at 68° was increased to 10 or 15 min). PCR products were characterized by restriction mapping, subcloning, and partial sequencing.
Cloning and sequencing:
PCR products were subcloned into pBluescript II SK+ (Stratagene, La Jolla, CA) or pCR4-TOPO (Invitrogen) by standard procedures (Sambrook et al. 1989). Single- or double-stranded plasmid DNA was subjected to automated sequencing. With the exception of a few nucleotides in the 5′-untranslated regions, each nucleotide of the six Triakis Dlx cDNAs was determined from at least five different clones, representing each strand at least twice. Sequences were manipulated and translated using programs in the DNASTAR package (DNAstar, Madison, WI).
Phylogenetic analyses:
Amino acid sequences of Distal-less-related genes were obtained from the GenBank database with the following accession numbers: nematode (Caenorhabditis elegans) C28A5.4 (Z32680); fruit fly (Drosophila melanogaster) Dll (S47947); acorn worm (Ptychodera flava) Pf-Dlx (AB028221); tunicate (Ciona intestinalis) Dll-A (AJ278696), Dll-B (AJ278697), and Dll-C (AJ278698); amphioxus (Branchiostoma floridae) AmphiDll (U47058); lamprey (Petromyzon marinus) DlxA (AY010116), DlxB (AY010117), DlxC (AY010118), and DlxD (AY010119); lamprey (Lethenteron japonicum) LjDLX1/6 (AB048759); zebrafish (Danio rerio) dlx1 (U67842), dlx2 (U03875), dlx3 (X65060), dlx4 (U03876), dlx5 (U67843), dlx6 (U67844), dlx7 (U67845), and dlx8 (U67846); frog (Xenopus laevis) Xdll (D10259), X-DLL1 (A56570), Xdll-2 (S74210), X-dll2 (L09730), X-dll3 (L09729), and X-dll4 (L09728); newt (Notophthalmus viridescens) NvHBox-4 (X63531) and NvHBox-5 (X63532); chicken (Gallus gallus) DLX3 (AJ243432) and Dlx5 (U25274); mouse (Mus musculus) Dlx1 (NM_010053), Dlx2 (NM_010054), Dlx3 (NM_010055), Dlx5 (U67840), and Dlx7 (AF452637); and human (Homo sapiens) DLX1 (XM_087198), DLX2 (NM_004405), DLX3 (NM_005220), DLX5 (NM_005221), DLX6 (NM_005222), and DLX7 (AF452638). The nomenclature of human, mouse, and zebrafish Dlx genes was adjusted from the above GenBank records where necessary to follow Panganiban and Rubenstein (2002). Related genes outside of the Distal-less family were obtained for use as outgroups (Stock et al. 1996). These sequences were mouse Barx1 (Y07960) and Msx1 (NM_010835) and D. melanogaster Msh (U33319).
Upon inspection of initial alignments, a few of the above sequences were interpreted as containing errors or truncations of their 5′-ends. Adjustments to these sequences were made as follows. The lamprey sequences DlxD (P. marinus) and LjDLX1/6 (L. japonicum) are clearly orthologous on the basis of a high degree of nucleotide identity. However, comparison of these genes with each other and with other vertebrate Dlx genes suggests that DlxD is truncated at its 5′-end, while LjDLX1/6 has a frameshift error in the region 3′ of the homeobox (which itself has a six-nucleotide deletion found in no other animals). A composite lamprey DlxD sequence was therefore constructed with the missing amino terminus of P. marinus DlxD replaced by the corresponding region from L. japonicum LjDLX1/6. The reported human DLX6 sequence similarly appears to have an internal methionine designated as the start codon. Translation of the genomic sequence of a BAC clone (accession AC004774) revealed the likely true start codon in frame and part of the same exon; this region was added to the reported amino acid sequence. X. laevis Xdll-2 was reported to have a truncated 5′-end. The missing nucleotides were added to this sequence from an X. laevis EST that appeared to be derived from the same gene (accession BF613971). Finally, a single amino acid adjustment was made to the homeodomain of zebrafish dlx2a on the basis of unpublished sequence data, which exhibited a better match to all other sequenced animal Distal-less-related genes. After these modifications, the only incomplete coding region remaining among the chordate sequences analyzed was that of P. marinus DlxB (reported as such by Neidert et al. 2001).
The 61-amino-acid homeodomain of all Dll/Dlx genes may be aligned reliably without gaps. However, alignment outside of the homeodomain of the most distantly related genes is problematic (Stock et al. 1996; Neidert et al. 2001). To identify the location of the root of the tree of vertebrate Dlx genes, an initial phylogenetic analysis of homeodomain sequences was attempted. Pairwise distances between amino acid sequences were calculated as percentage of similarities [P-values, as recommended by Zhang and Nei (1996) in their analysis of Hox gene homeodomains] and used to construct neighbor-joining trees with the program MEGA, version 2.1 (Kumar et al. 2001). To reduce the number of identical sequences in the analyses, gnathostome sequences were restricted to those of human, zebrafish, and Triakis.
To provide increased resolution of the phylogeny of vertebrate Dlx sequences, an alignment of complete vertebrate Dlx sequences was constructed using Clustal X (Thompson et al. 1997). The profile alignment feature of this program was used to align sequences in clades identified from the analysis of homeodomain sequences, followed by sequential alignment of these subalignments into a global alignment. Adjustments to alignments at all stages of the process were made either by changing alignment parameters for specific regions or by manual editing. The alignment is available as supplementary material at http://www.genetics.org/supplemental/.
Maximum likelihood and neighbor-joining analyses of complete Dlx protein sequences were conducted with the PROML, PROTDIST, and NEIGHBOR programs of the PHYLIP package (version 3.6; Felsenstein 1989). Both types of analyses employed the Jones-Taylor-Thornton model of amino acid change (Jones et al. 1992) and a gamma distribution of rates among sites (coefficient of variation is 0.707, corresponding to an α-value of 2.0, as recommended by Kumar et al. 2001). For maximum likelihood analysis, six rate categories, 40 random addition sequences, and global rearrangements were used.
RESULTS
Six Dlx genes detected in T. semifasciata:
Sixty-five cloned Dlx homeobox fragments obtained by RT-PCR were sequenced. These fragments arose from two independent reactions performed with each of three different primer combinations (using a common antisense primer). The sequences obtained fell into six classes and were used to design primers for 5′- and 3′-RACE PCR reactions. Cloning and sequencing of the latter PCR products resulted in complete cDNA sequences that were divergent enough to confirm that the six classes represented different loci rather than multiple alleles. The loci were named according to their orthology to other vertebrate Dlx genes (Panganiban and Rubenstein 2002), as inferred from the phylogenetic analyses described below. These loci and the number of homeobox clones obtained by RT-PCR are Dlx1 (5), Dlx2 (2), Dlx3 (35), Dlx4 (10), Dlx5 (5), and Dlx6 (8).
Genomic organization of Triakis Dlx genes:
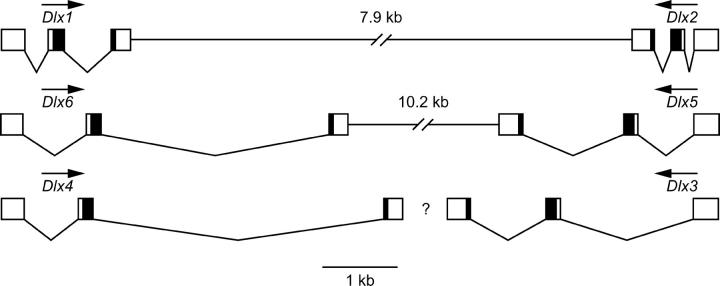
The exon/intron structures and linkage arrangements of the six T. fasciata Dlx genes are shown in Figure 1. All six genes possess two introns and three exons. The second intron occupies an identical location in the homeobox of all genes and is phase 0 (between codons). The first intron is phase 1 (between the first and second nucleotides of a codon) in all genes and is located 50–68 nucleotides 5′ of the homeobox. Intron 1 ranges in size from 128 bp (Dlx2) to ∼1.8 kb (Dlx3), while intron 2 ranges in size from 221 bp (Dlx2) to ∼3.9 kb (Dlx4).
Figure 1.—
Genomic organization of Triakis Dlx genes. Phylogenetically related genes are aligned in columns. Boxes indicate protein-coding portions of exons (homeodomain in black), angled lines indicate introns, and horizontal lines indicate intergenic regions. Transcriptional orientation is denoted by arrows. All distances between the start and stop codons of an individual gene are drawn to scale, while intergenic distances (listed) are not. Question mark indicates lack of data from genomic PCR to support or refute linkage predicted from phylogenetic analyses.
Analysis of genomic long PCR products revealed that Triakis possesses at least two Dlx bigene clusters (Dlx1-Dlx2 and Dlx5-Dlx6), with the genes oriented in the convergently transcribed arrangement characteristic of chordate Dlx genes (Figure 1). Furthermore, the linkage of these specific orthologs (identified by phylogenetic analysis of amino acid sequences—see below) is the same as that described in other gnathostome vertebrates (Panganiban and Rubenstein 2002). The stop codons of Dlx1 and Dlx2 are separated by ∼7.9 kb, while those of Dlx5 and Dlx6 are separated by ∼10.2 kb. Despite attempts with multiple sets of primers, no genomic PCR product was obtained that included Dlx3 and Dlx4, genes whose presumed orthologs form a bigene cluster in other gnathostomes.
Phylogenetic analysis of bilaterian Dlx homeodomain sequences:
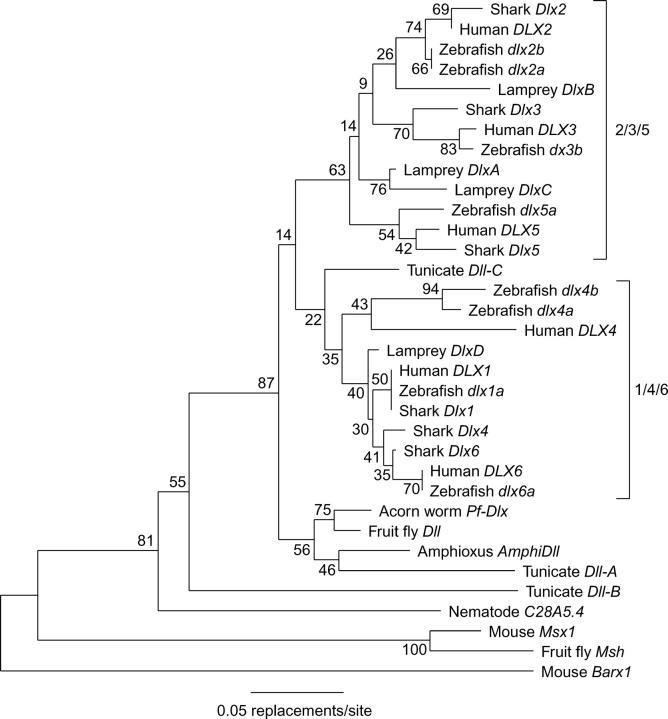
A neighbor-joining tree constructed from homeodomain sequences is shown in Figure 2. Gnathostome Dlx sequences in this tree are found in two main clades. One of these (35% bootstrap support) contains Dlx1, Dlx4, Dlx6, and one lamprey Dlx sequence, while the other contains Dlx2, Dlx3, Dlx5, and three lamprey sequences (63% bootstrap support). One tunicate Dll gene forms the sister group to the Dlx1/Dlx4/Dlx6 clade, while the single reported Dll/Dlx genes of Drosophila, the hemichordate Ptychodera, and the cephalochordate amphioxus, as well as one tunicate Dll gene form a clade outside of both gnathostome-containing Dlx clades. An additional tunicate Dll gene and the single reported nematode (C. elegans) Dll/Dlx gene group outside of other animal Dll/Dlx genes.
Figure 2.—
Neighbor-joining analysis of Dll/Dlx homeodomain sequences. Numbers indicate the percentage of 1000 bootstrap replicates in which the indicated node was present. The tree is rooted at the midpoint of the longest path between two taxa. Brackets indicate the two main clades of vertebrate Dlx genes named after the mammalian members.
This analysis of homeodomain sequences provides little phylogenetic resolution (as judged by low bootstrap values). Nevertheless, it reveals that Triakis possesses three members of each of the main gnathostome Dlx clades and suggests that the root of the vertebrate Dlx tree lies on the internal branch connecting these clades.
Phylogenetic analysis of complete vertebrate Dlx sequences:
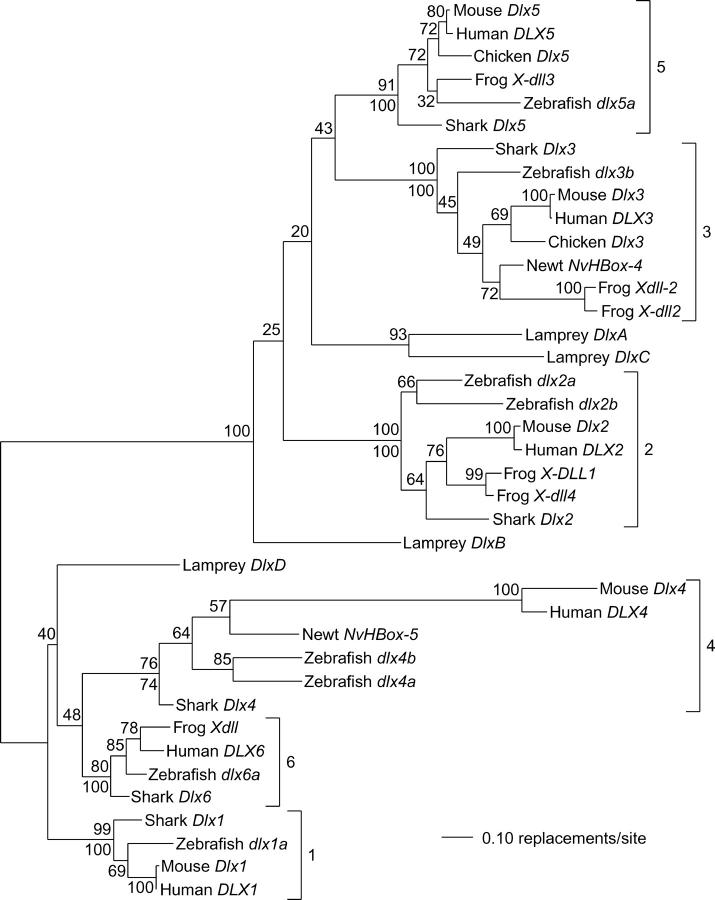
The phylogenetic analyses of Dlx genes reported by Stock et al. (1996) and Neidert et al. (2001) provided evidence for the existence of six clades of osteichythyan Dlx genes, each possessing a single mammalian Dlx gene and one or two zebrafish genes. The results of maximum likelihood and neighbor-joining analysis of complete vertebrate Dlx genes shown in Figure 3 indicate that each of these clades also contains a single Triakis gene. The monophyly of five of these six clades is supported by bootstrap values of at least 99% with one or both methods of analysis. Somewhat lower bootstrap values for the monophyly of the Dlx4 clade (74 and 76%) may be due to the rapid evolution of mammalian Dlx4 apparent in the tree in Figure 3.
Figure 3.—
Maximum likelihood and neighbor-joining analysis of vertebrate Dlx genes. Tree topology and branch lengths are from a maximum likelihood analysis, with midpoint rooting. Nodes with single numbers indicate the results of a bootstrap maximum likelihood analysis with 100 replicates and options as described in materials and methods, except for the use of two random addition sequences for each replicate. Where two numbers appear, the upper is from the bootstrap maximum likelihood analysis and the lower is from a bootstrap neighbor-joining analysis with 1000 replications. Brackets indicate the six clades of jawed vertebrate Dlx genes, each named after and containing a single Triakis gene.
If the six Dlx genes isolated from Triakis are in fact the orthologs of the osteichthyan genes, then they would be expected to exhibit a sister group relationship to all the other members of a given clade. This relationship is found for all clades other than Dlx2 in which the expected relationship is contradicted by an internal branch with relatively weak bootstrap support (64%; Figure 3).
The phylogenetic analysis in Figure 3 reveals 100% bootstrap support for an internal branch separating lamprey DlxA, DlxB, and DlxC from lamprey DlxD. It is likely that this branch also contains the root of the tree (on the basis of midpoint rooting in Figure 3 and outgroup analysis in Figure 2). If this is the true location of the root of the tree, then DlxA, DlxB, and DlxC are members of the vertebrate Dlx2/Dlx3/Dlx5 clade and DlxD is a member of the Dlx1/Dlx4/Dlx6 clade. The analysis shown in Figure 3 provides little support for the phylogenetic position of the lamprey genes within these major clades, with the exception of strong bootstrap support (93%) for a sister group relationship between lamprey DlxA and DlxC.
DISCUSSION
Orthology of the Triakis Dlx genes:
The number of Dlx genes isolated from Triakis is the same as that reported from mammals and all are supported as members of clades containing a single human gene (Figure 3). It has been argued that the six-Dlx-gene condition of the osteichthyan common ancestor arose by the loss of a Dlx bigene cluster linked to the HoxC complex (Stock et al. 1996; Neidert et al. 2001; Sumiyama et al. 2003). Because of the phylogenetic position of chondrichthyans as the sister group of all other gnathostomes in the analyses, it is not possible to rule out the orthology of one or two of the Dlx genes of Triakis to such HoxC-linked Dlx genes. The failure to amplify a PCR product containing Triakis Dlx3 and Dlx4 is in fact consistent with the possibility that one of these genes is orthologous to a Dlx gene lost in the ancestry of gnathostomes. This result does not constitute positive evidence for the nonorthology of one of these genes to osteichthyan Dlx genes, however. Not all primer combinations attempted amplified the other bigene clusters and it is possible that Dlx3 and Dlx4 are too far apart to be reliably amplified by the PCR methods employed. Given the high degree of divergence between osteichthyan Dlx clades relative to that within, orthology of a Triakis Dlx gene to one lost from osteichthyans would be expected to be associated with extensive divergence of the Triakis sequence from its closest relatives in the tree. No clear evidence exists for this (Figure 3), and it is concluded on the basis of parsimony that each Triakis Dlx gene is orthologous to the members of the osteichthyan Dlx clade of the same name.
Conservation of Dlx intron structure:
Comparison of the locations of introns in Triakis Dlx genes with those of zebrafish (dlx1a, dlx2a, dlx3b, dlx5a, and dlx6a; Ellies et al. 1997), mouse (Dlx1, Dlx2, Dlx3, and Dlx4; McGuinness et al. 1996; Sumiyama et al. 2002), and human (DLX3, DLX4, DLX5, and DLX6; Sumiyama et al. 2002; GenBank accession no. AC004774) revealed the exact correspondence of intron location among Dlx orthologs, with the exception of the first intron in zebrafish dlx3b, which was displaced by two amino acids from the corresponding alignment position in shark and mammal orthologs. All vertebrate Dlx genes exhibited an identical location for the second intron (within the homeobox), while the first intron varied in location (but not phase) by up to six alignment positions.
The location of the intron in the homeobox of vertebrate Dlx genes is identical to that of Drosophila. Five additional introns are present in the Drosophila gene (Vachon et al. 1992), however, and in the absence of outgroup information, it is not possible to determine whether the differences in intron number arose through loss in vertebrates or gain in Drosophila. Of the three reported Dll genes in the tunicate C. intestinalis, two have been reported to have a homeobox intron in the same location as that of vertebrates (Di Gregorio et al. 1995). The structure of the region upstream of the homeobox has not been reported for any of the genes, but Ci-DllB has two introns 3′ of the homeobox (Caracciolo et al. 2000). As discussed below, it is likely that Dlx bigene clusters arose by a tandem duplication before the divergence of vertebrates and tunicates. The virtually identical intron/exon structure of members of bigene clusters (more distantly related than orthologous tunicate and vertebrate Dlx genes) suggests that Ci-DllB gained introns after the divergence of vertebrates and tunicates. Such gain of introns over this time scale is not unusual (Lynch and Richardson 2002).
Dlx arrangement as bigene clusters:
A convergently transcribed bigene-cluster arrangement has been described for two of the three Dll genes of the tunicate C. intestinalis (Caracciolo et al. 2000), six of the eight zebrafish Dlx genes (Ellies et al. 1997; Ghanem et al. 2003), four Dlx genes of the pufferfishes Takifugu rubripes and Spheroides nephelus (Ghanem et al. 2003), and the six mammalian Dlx genes (reviewed by Sumiyama et al. 2003). The Dlx1/Dlx2 and Dlx5/Dlx6 bigene clusters identified for Triakis are consistent with the organization of Dlx genes in the other vertebrates examined. Furthermore, the intergenic distances (between stop codons) of 7.9 and 10.2 kb, respectively, fall within the range of those reported for ray-finned fish and mammals (5.3–12.3 kb and ∼3–10.7 kb, respectively; Ghanem et al. 2003; Sumiyama et al. 2003). The bigene cluster for which no evidence was obtained in T. semifasciata, Dlx3/Dlx4, contains an intergenic region of ∼17.6 kb in humans (Sumiyama et al. 2003). Such a distance between these genes in Triakis may explain the failure to obtain an intergenic PCR product. In the absence of evidence of Dlx genomic organization in the lamprey, the existence of Dlx bigene clusters in Triakis provides further support for the hypothesis that this arrangement is primitive for chordates (Stock et al. 1996; Neidert et al. 2001; Sumiyama et al. 2003).
Evolutionary history of vertebrate Dlx genes:
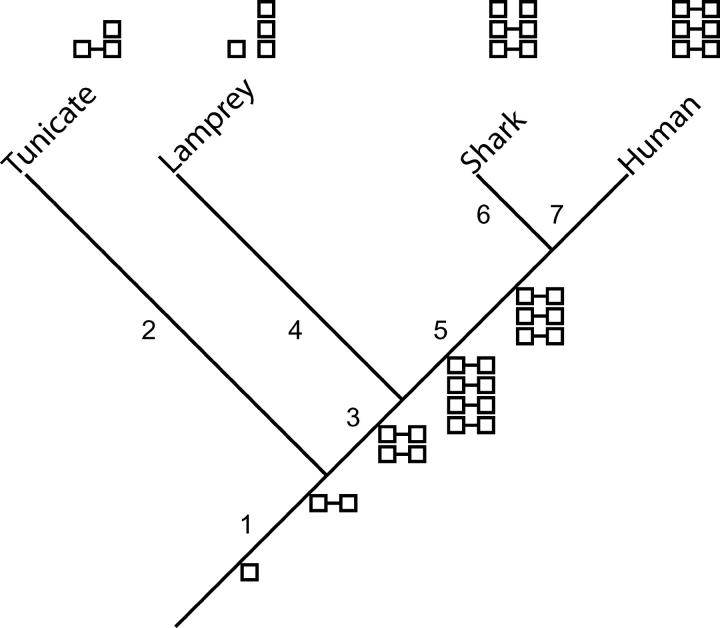
A scenario for the evolution of chordate Dlx genes is shown in Figure 4. The closer relationship of Dlx genes among rather than within bigene clusters provides the evidence for the postulated initial tandem duplication of Dlx genes shown on branch 1. That this event antedates the divergence of vertebrates and tunicates is hypothesized on the basis of the presence of bigene clusters in both groups. It should be noted, however, that phylogenetic analysis of homeodomain sequences (Figure 2) did not yield the predicted clustering of all tunicate Dll genes as members of one or the other of the two main clades of vertebrate Dlx genes. Such a relationship was found for Dll-C, but was weakly supported, while the linked Dll-A and Dll-B genes clustered outside of all vertebrate Dlx genes. Rather than postulating the complex pattern of gain and loss implied by a literal interpretation of the tree in Figure 2, the scenario in Figure 4 follows Stock et al. (1996) in interpreting the bigene clusters of tunicates and vertebrates as unlikely to have evolved independently.
Figure 4.—
Scenario for the evolution of Dlx genes showing selected deuterostome taxa. Boxes indicate individual Dlx genes arranged in columns according to phylogenetic relationship. Horizontal lines joining boxes indicate demonstrated linkage relationships. See text for details of events postulated to have occurred along numbered branches.
Placing the initial tandem duplication on branch 1 of Figure 4 implies that the single Dll gene isolated from amphioxus was ancestrally (if not still) a member of a bigene cluster. The position of the amphioxus sequence in Figure 2 does not support this assertion, but the expected clustering in one of the two major clades of vertebrate Dlx genes is not strongly contradicted. Whether the initial tandem duplication occurred before protostomes and/or hemichordates diverged from tunicates and vertebrates remains equivocal; Drosophila and hemichordate sequences cluster outside of all vertebrate Dlx genes but are members of a clade containing amphioxus and tunicate sequences. A literal interpretation of the tree would require loss of a Dll-like gene from vertebrates and loss of a Dlx-like gene from protostomes, hemichordates, and amphioxus.
The events on branch 2 of Figure 4 that led to three tunicate Dll genes after an initial tandem duplication remain unclear because of the contradiction between the results of phylogenetic analyses and gene linkage described above. The scenario in Figure 4 postulates independent duplications on branches 2 (tunicate lineage) and 3 (vertebrate lineage) on the basis of the common view that duplication of Hox clusters (linked to Dlx genes) all occurred after the divergence of amphioxus from vertebrates (Holland et al. 1994; Sidow 1996; Amores et al. 1998; Finnerty and Martindale 1998; Ferrier et al. 2000; Force et al. 2002; Furlong and Holland 2002; Irvine et al. 2002; Panopoulou et al. 2003; Wagner et al. 2003).
The general pattern of Dlx bigene cluster duplication from one (branch 1) to two (branch 3) to four (branch 5) shown in Figure 4 is based on the hypothesis that the four Hox clusters of mammals arose by two rounds of genome duplication (Sidow 1996; Furlong and Holland 2002; Larhammar et al. 2002). It is important to note that this hypothesis remains controversial (Hughes et al. 2001; Martin 2001; Horton et al. 2003; Hughes and Friedman 2003) and that Dlx genes cannot in themselves refute or support such a means of duplication because only three bigene clusters are found in jawed vertebrates that have not undergone relatively recent genome duplication. The exact placement of genome duplications shown in Figure 4 (one on branch 3 before lampreys diverged from gnathostomes and one on branch 5 afterward) is based on the analyses of Hox genes by Force et al. (2002) and Fried et al. (2003) and remains tentative because of lack of phylogenetic resolution of lamprey Dlx sequences (this study and Neidert et al. 2001). Independent Dlx duplication in the lamprey lineage relative to that in gnathostomes is required by the scenario in Figure 4 and is supported by clustering of lamprey DlxA and DlxC in Figure 3 and in the analysis of Neidert et al. (2001).
The most significant finding of this study bearing on the evolutionary history of vertebrate Dlx genes is evidence that the common ancestor of extant gnathostomes had three Dlx bigene clusters (branch 5 of Figure 4). This is suggested by the analyses described above that identify all six Triakis Dlx genes as orthologous to specific osteichthyan Dlx genes. That this condition evolved by the loss of a bigene cluster is postulated on the basis of the presence of such clusters on only three of the four mammalian Hox-containing chromosomes (Stock et al. 1996; Sumiyama et al. 2003). While the relationship of lamprey Dlx genes shown in Figure 3 differs somewhat from that obtained in the analysis of Neidert et al. (2001), both suggest that the acquisition of four bigene clusters and loss of one occurred after gnathostomes diverged from lampreys.
No evidence was found for independent Dlx gene duplication in the lineage leading to Triakis (branch 6 of Figure 4), in contrast to those leading to the lamprey (branch 4, discussed above), zebrafish, and X. laevis. The latter two species (separate derivatives of branch 7, not shown in Figure 4) are thought to have undergone genome duplications after diverging from the other vertebrates examined (Kobel and Du Pasquier 1986; Amores et al. 1998, 2004; Fried et al. 2003) and these duplications have been postulated to have given rise to additional Dlx genes (Stock et al. 1996; Amores et al. 1998). Because of potential biases in the ability of degenerate PCR primers to amplify multiple Dlx genes and the fact that cDNA-based amplification is limited to genes expressed at the embryonic stage examined, it is not possible to rule out the presence of additional, uncharacterized Dlx in the Triakis genome, however.
Dlx genes and the origin of gnathostome characters:
The earliest divergence among extant jawed vertebrates separates chondrichthyans from osteichthyans (Maisey 1986; Janvier 1996, 2001). As argued above, the presence of three Dlx bigene clusters characterized the common ancestor of both lineages and arose by duplication and loss after the divergence of lampreys and gnathostomes (branch 5 of Figure 4). This same internal branch, the stem gnathostome lineage (Janvier 2001), was undoubtedly the location of the origin of jaws, the defining feature of gnathostomes. Dlx genes have been shown to be involved in patterning the jaws of mice (Depew et al. 2002; Panganiban and Rubenstein 2002), and the expression patterns of Dlx genes differ between the oral regions of lampreys and gnathostomes (Myojin et al. 2001; Neidert et al. 2001; Cohn 2002; Shigetani et al. 2002). In addition to changes in the expression of existing Dlx genes playing a role in the origin of jaws, the scenario for Dlx evolution illustrated in Figure 4 raises the possibility that Dlx gene duplication (and loss) were involved as well.
Janvier (2001) lists 22 characters in addition to jaws that are unique to gnathostomes among living vertebrates and that are likely to be preserved in fossils. Determining whether these originated along the stem gnathostome lineage (and hence may have been related to the acquisition of three Dlx bigene clusters) rather than representing reversals in extant jawless vertebrates depends critically on the relationship of fossil jawless fishes. Recent proposals that many fossil jawless fishes (“ostracoderms”) branch from the stem gnathostome lineage rather than clustering with extant jawless fishes (Janvier 1996, 2001; Donoghue et al. 2000) increase the number of characters that may have arisen after the three Dlx bigene-cluster condition. Those whose development requires Dlx genes include paired nasal openings, three semicircular canals, teeth, and pectoral fins (see Janvier 2001 and Donoghue et al. 2000 for character evolution and Panganiban and Rubenstein 2002 for a review of Dlx function).
The structure of the ancestral gnathostome genome:
Recent proponents of the hypothesis of two rounds of genome duplication in vertebrates have argued that both occurred before the divergence of actinopterygians and sarcopterygians (Amores et al. 1998, 2004; Wagner et al. 2003). The presence of three Dlx bigene clusters in Triakis and mammals is consistent with both having also occurred before the divergence of extant gnathostome taxa. While it has been proposed that the ancestral gnathostome had four Hox clusters (Amores et al. 1998, 2004; Wagner et al. 2003), only two Hox clusters have been reported from chondrichthyans to date (Kim et al. 2000). The linkage of Dlx genes to Hox genes in mammals and zebrafish (Amores et al. 1998; Sumiyama et al. 2003), argued to be a common feature of the bilaterian ancestor (Pollard and Holland 2000), suggests that at least three Hox clusters were present in chondrichthyan ancestors, and the hypothesis of multiple Hox and Dlx clusters arising by whole-genome duplication suggests that there were four. Consistent with this prediction, the two identified chondrichthyan Hox clusters are each orthologous to single osteichthyan clusters (Kim et al. 2000; Wagner et al. 2003).
If Dlx bigene clusters arose by whole-genome duplication, other families of osteichthyan gene duplicates with single orthologs in invertebrates should have multiple orthologs in chondrichthyans. Although chondrichthyan sequences are underrepresented in studies of vertebrate gene and genome duplication, evidence does exist from a number of gene families supporting this prediction. Early studies of gene number by isozyme electrophoresis suggested gene duplication after the divergence of vertebrates and invertebrates and before the divergence of osteichthyans and chondricthyans for the lactate dehydrogenase (LDH), malate dehydrogenase, creatine kinase, and glucose phosphate isomerase gene families (Markert et al. 1975; Fisher et al. 1980). Similar duplication histories have been proposed for several Wnt paralogs (Sidow 1992), proteasome Z subunits (Kasahara et al. 1996), LDH genes [the sequence analysis of Stock et al. (1997) and Stock and Powers (1998) confirmed the results of early isozyme studies], dioxin receptors (Hahn et al. 1997), enolases (Tracy and Hedges 2000), Otx genes (Germot et al. 2001), Tbx genes (Tanaka et al. 2002), Emx genes (Derobert et al. 2002), thyroid hormone receptors (Escriva et al. 2002), dopamine receptors (Le Crom et al. 2003), and neuropeptide Y receptors (Salaneck et al. 2003). The Dlx gene data presented in this study are therefore consistent with data from other gene families in suggesting that the distinctive morphological differences between osteichthyans and chondrichthyans arose in organisms with a similar overall gene content.
Acknowledgments
The author thanks Gregor Caillet, Giacomo Bernardi, Stephanie Clendennen, and anonymous participants in the Elkhorn Slough Shark Derby for assistance in obtaining Triakis specimens; Kenneth Weiss for support during the early stages of this project; R. Travis Merritt and Anne Buchanan for technical assistance; and William Jackman, Jason Pardo, Josh Trapani, Sarah Wise, and two reviewers for helpful comments. Funding was provided by National Science Foundation grants SBR 9408402 to K. Weiss and IBN 0092487 to the author.
References
- Amores, A., A. Force, Y.-L. Yan, L. Joly, C. Amemiya et al., 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Amores, A., T. Suzuki, Y.-L. Yan, J. Pomeroy, A. Singer et al., 2004. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 14: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, W. J., J. Kim, G. P. Wagner and F. H. Ruddle, 1997. Phylogenetic reconstruction of vertebrate Hox cluster duplications. Mol. Biol. Evol. 14: 843–853. [DOI] [PubMed] [Google Scholar]
- Balavoine, G., R. de Rosa and A. Adoutte, 2002. Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 24: 366–373. [DOI] [PubMed] [Google Scholar]
- Ballard, W. W., J. Mellinger and H. Lechenault, 1993. A series of normal stages for development of Scyliorhinus canicula, the Lesser spotted dogfish (Chondrichthyes: Scyliorhinidae). J. Exp. Zool. 267: 318–336. [Google Scholar]
- Barnes, W. M., 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc. Natl. Acad. Sci. USA 91: 2216–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracciolo, A., A. Di Gregorio, F. Aniello, R. Di Lauro and M. Branno, 2000. Identification and developmental expression of three Distal-less homeobox containing genes in the ascidian Ciona intestinalis. Mech. Dev. 99: 173–176. [DOI] [PubMed] [Google Scholar]
- Chenchik, A., F. Moqadam and P. Siebert, 1996 A new method for full-length cDNA cloning by PCR, pp. 273–321 in A Laboratory Guide to RNA: Isolation, Analysis and Synthesis, edited by P. A. Krieg. Wiley-Liss, New York.
- Chenchik, A., Y. Zhu, L. Diatchenko, R. Li, J. Hill et al., 1998 Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR, pp. 305–319 in Gene Cloning and Analysis by RT-PCR, edited by P. Siebert and J. Larrick. BioTechniques Press, Westborough, MA.
- Chiu, C.-h., K. Dewar, G. P. Wagner, K. Takahashi, F. Ruddle et al., 2004. Bichir HoxA cluster sequence reveals surprising trends in ray-finned fish genomic evolution. Genome Res. 14: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, M. J., 2002. Lamprey Hox genes and the origin of jaws. Nature 416: 386–387. [DOI] [PubMed] [Google Scholar]
- Depew, M. J., T. Lufkin and J. L. R. Rubenstein, 2002. Specification of jaw subdivisions by Dlx genes. Science 298: 381–385. [DOI] [PubMed] [Google Scholar]
- Derobert, Y., J. L. Plouhinec, T. Sauka-Spengler, C. Le Mentec, B. Baratte et al., 2002. Structure and expression of three Emx genes in the dogfish Scyliorhinus canicula: functional and evolutionary implications. Dev. Biol. 247: 390–404. [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A., A. Spagnuolo, F. Ristoratore, M. Pischetola, F. Aniello et al., 1995. Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene 156: 253–257. [DOI] [PubMed] [Google Scholar]
- Donoghue, P. C., P. L. Forey and R. J. Aldridge, 2000. Conodont affinity and chordate phylogeny. Biol. Rev. Camb. Philos. Soc. 75: 191–251. [DOI] [PubMed] [Google Scholar]
- Ellies, D. L., D. W. Stock, G. Hatch, G. Giroux, K. M. Weiss et al., 1997. Relationship between the genomic organization and the overlapping embryonic expression patterns of the zebrafish dlx genes. Genomics 45: 580–590. [DOI] [PubMed] [Google Scholar]
- Escriva, H., L. Manzon, J. Youson and V. Laudet, 2002. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol. Biol. Evol. 19: 1440–1450. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1989. PHYLIP—Phylogeny Inference Package, Version 3.2. Cladistics 5: 164–166. [Google Scholar]
- Ferrier, D. E. K., C. Minguillón, P. W. H. Holland and J. Garcia-Fernàndez, 2000. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol. Dev. 2: 284–293. [DOI] [PubMed] [Google Scholar]
- Finnerty, J. R., and M. Q. Martindale, 1998. The evolution of the Hox cluster: insights from outgroups. Curr. Opin. Genet. Dev. 8: 681–687. [DOI] [PubMed] [Google Scholar]
- Fisher, S. E., J. B. Shaklee, S. D. Ferris and G. S. Whitt, 1980. Evolution of five multilocus isozyme systems in the chordates. Genetica 52/53: 73–85. [Google Scholar]
- Force, A., A. Amores and J. H. Postlethwait, 2002. Hox cluster organization in the jawless vertebrate Petromyzon marinus. J. Exp. Zool. (Mol. Dev. Evol.) 294: 30–46. [DOI] [PubMed] [Google Scholar]
- Fried, C., S. J. Prohaska and P. F. Stadler, 2003. Independent Hox-cluster duplications in lampreys. J. Exp. Zool. Mol. Dev. Evol. 299B: 18–25. [DOI] [PubMed] [Google Scholar]
- Frohman, M. A., 1990 RACE: rapid amplification of complementary DNA ends, pp. 28–38 in PCR Protocols: A Guide to Methods and Applications, edited by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White. Academic Press, New York.
- Furlong, R. F., and P. W. H. Holland, 2002. Were vertebrates octoploid? Philos. Trans. R. Soc. Lond. B 357: 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germot, A., G. Lecointre, J.-L. Plouhinec, C. Le Mentec, F. Giradot et al., 2001. Structural evolution of Otx genes in craniates. Mol. Biol. Evol. 18: 1668–1678. [DOI] [PubMed] [Google Scholar]
- Ghanem, N., O. Jarinova, A. Amores, Q. Long, G. Hatch et al., 2003. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 13: 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. E., S. I. Karchner, M. A. Shapiro and S. A. Perera, 1997. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl. Acad. Sci. USA 94: 13743–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P. W., J. Garcia-Fernàndez, N. A. Williams and A. Sidow, 1994 Gene duplications and the origins of vertebrate development. Dev. Suppl., 125–133. [PubMed]
- Horton, A. C., N. R. Mahadevan, I. Ruvinsky and J. J. Gibson-Brown, 2003. Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. J. Exp. Zool. Mol. Dev. Evol. 299B: 41–53. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and R. Friedman, 2003. 2R or not 2R: testing hypotheses of genome duplication in early vertebrates. J. Struct. Funct. Genomics 3: 85–93. [PubMed] [Google Scholar]
- Hughes, A. L., J. da Silva and R. Friedman, 2001. Ancient genome duplications did not structure the human Hox-bearing chromosomes. Genome Res. 11: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, S. Q., J. L. Carr, W. J. Bailey, K. Kawasaki, N. Shimizu et al., 2002. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J. Exp. Zool. Mol. Dev. Evol. 294: 47–62. [DOI] [PubMed] [Google Scholar]
- Janvier, P., 1996 Early Vertebrates. Clarendon Press, Oxford.
- Janvier, P., 2001 Ostracoderms and the shaping of the gnathostome characters, pp. 172–186 in Major Events in Early Vertebrate Evolution: Paleontology, Phylogeny, Genetics and Development, edited by P. E. Ahlberg. Taylor & Francis, New York.
- Jones, D. T., W. R. Taylor and J. M. Thornton, 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., M. Hayashi, K. Tanaka, H. Inoko, K. Sugaya et al., 1996. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc. Natl. Acad. Sci. USA 93: 9096–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, D. E., I. Rybalkin, S. Chen, N. Mukhamedova, T. Vlasik et al., 1994. TaqStart antibody: hot start PCR facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. BioTechniques 16: 1134–1137. [PubMed] [Google Scholar]
- Kim, C.-B., C. Amemiya, W. Bailey, K. Kawasaki, J. Mezey et al., 2000. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl. Acad. Sci. USA 97: 1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel, H. R., and L. Du Pasquier, 1986. Genetics of polyploid Xenopus. Trends Genet. 2: 310–315. [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Larhammar, D., L.-G. Lundin and F. Hallböök, 2002. The human Hox-bearing chromosome regions did arise by block or chromosome (or even genome) duplications. Genome Res. 12: 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Crom, S., M. Kapsimali, P.-O. Barôme and P. Vernier, 2003. Dopamine receptors for every species: gene duplications and functional diversification in Craniates. J. Struct. Funct. Genomics 3: 161–176. [PubMed] [Google Scholar]
- Liu, J. K., I. Ghattas, S. Liu, S. Chen and J. L. R. Rubenstein, 1997. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev. Dyn. 210: 498–512. [DOI] [PubMed] [Google Scholar]
- Lundin, L. G., 1993. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16: 1–19. [DOI] [PubMed] [Google Scholar]
- Lundin, L.-G., D. Larhammar and F. Hallböök, 2003. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of vertebrates. J. Struct. Funct. Genomics 3: 53–63. [PubMed] [Google Scholar]
- Lynch, M., and A. O. Richardson, 2002. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 12: 701–710. [DOI] [PubMed] [Google Scholar]
- Maisey, J. G., 1986. Heads and tails: a chordate phylogeny. Cladistics 2: 201–256. [DOI] [PubMed] [Google Scholar]
- Markert, C. L., J. B. Shaklee and G. S. Whitt, 1975. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science 189: 102–114. [DOI] [PubMed] [Google Scholar]
- Martin, A., 2001. Is tetralogy true? Lack of support for the “one-to-four” rule. Mol. Biol. Evol. 18: 89–93. [DOI] [PubMed] [Google Scholar]
- McGuinness, T., M. H. Porteus, S. Smiga, A. Bulfone, C. Kingsley et al., 1996. Sequence, organization, and transcription of the Dlx-1 and Dlx-2 locus. Genomics 35: 473–485. [DOI] [PubMed] [Google Scholar]
- Myojin, M., T. Ueki, F. Sugahara, Y. Murakami, Y. Shigetani et al., 2001. Isolation of Dlx and Emx gene cognates in an agnathan species, Lampetra japonica, and their expression patterns during embryonic and larval development: conserved and diversified regulatory patterns of homeobox genes in vertebrate head evolution. J. Exp. Zool. Mol. Dev. Evol. 291: 68–84. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., D. W. Stock, K. L. Wydner, J. A. Bollekens, K. Takeshita et al., 1996. Genomic analysis of a new mammalian Distal-less gene: Dlx7. Genomics 38: 314–324. [DOI] [PubMed] [Google Scholar]
- Neidert, A. H., V. Virupannavar, G. W. Hooker and J. A. Langeland, 2001. Lamprey Dlx genes and early vertebrate evolution. Proc. Natl. Acad. Sci. USA 98: 1665–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban, G., and J. L. R. Rubenstein, 2002. Developmental functions of the Distal-less/Dlx homeobox genes. Development 129: 4371–4386. [DOI] [PubMed] [Google Scholar]
- Panopoulou, G., S. Hennig, D. Groth, A. Krause, A. J. Poustka et al., 2003. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 13: 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton, J. W., B. K. Nagai, M. T. Murtha and F. H. Ruddle, 1993. Expansion of the Hox gene family and the evolution of chordates. Proc. Natl. Acad. Sci. USA 90: 6300–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, S. L., and P. W. H. Holland, 2000. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 10: 1059–1062. [DOI] [PubMed] [Google Scholar]
- Prince, V. E., 2002. The Hox paradox: more complex(es) than imagined. Dev. Biol. 249: 1–15. [DOI] [PubMed] [Google Scholar]
- Ruddle, F. H., J. L. Bartels, K. L. Bentley, C. Kappen, M. T. Murtha et al., 1994. a Evolution of Hox genes. Annu. Rev. Genet. 28: 423–442. [DOI] [PubMed] [Google Scholar]
- Ruddle, F. H., K. L. Bentley, M. T. Murtha and N. Risch, 1994b Gene loss and gain in the evolution of vertebrates. Dev. Suppl., 155–161. [PubMed]
- Salaneck, E., D. H. Ardell, E. T. Larson and D. Larhammar, 2003. Three neuropeptide Y receptor genes in the spiny dogfish, Squalus acanthias, support en bloc duplications in early vertebrate evolution. Mol. Biol. Evol. 20: 1271–1280. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shigetani, Y., F. Sugahara, Y. Kawakami, Y. Murakami, S. Hirano et al., 2002. Heterotopic shift of epithelial-mesenchymal interactions in vertebrate jaw evolution. Science 296: 1316–1319. [DOI] [PubMed] [Google Scholar]
- Sidow, A., 1992. Diversification of the Wnt gene family on the ancestral lineage of vertebrates. Proc. Natl. Acad. Sci. USA 89: 5098–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow, A., 1996. Gen(om)e duplications in the evolution of early vertebrates. Curr. Opin. Genet. Dev 6: 715–722. [DOI] [PubMed] [Google Scholar]
- Spring, J., 1997. Vertebrate evolution by interspecific hybridization—Are we polyploid? FEBS Lett. 400: 2–8. [DOI] [PubMed] [Google Scholar]
- Stock, D. W., and D. A. Powers, 1998. A monophyletic origin of heart-predominant lactate dehydrogenase (LDH) isozymes of gnathostome vertebrates: evidence from the cDNA sequence of the spiny dogfish (Squalus acanthias) LDH-B. Mol. Mar. Biol. Biotechnol. 7: 160–164. [PubMed] [Google Scholar]
- Stock, D. W., D. L. Ellies, Z. Zhao, M. Ekker, F. H. Ruddle et al., 1996. The evolution of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 93: 10858–10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock, D. W., J. M. Quattro, G. S. Whitt and D. A. Powers, 1997. Lactate dehydrogenase (LDH) gene duplication during chordate evolution: the cDNA sequence of the LDH of the tunicate Styela plicata. Mol. Biol. Evol. 14: 1273–1284. [DOI] [PubMed] [Google Scholar]
- Sumiyama, K., S. Q. Irvine, D. W. Stock, K. M. Weiss, K. Kawaski et al., 2002. Genomic structure and functional control of the Dlx3–7 bigene cluster. Proc. Natl. Acad. Sci. USA 99: 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama, K., S. Q. Irvine and F. H. Ruddle, 2003. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. J. Struct. Funct. Genomics 3: 151–159. [PubMed] [Google Scholar]
- Tanaka, M., A. Münsterberg, W. G. Anderson, A. R. Prescott, N. Hazon et al., 2002. Fin development in a cartilaginous fish and the origin of vertebrate limbs. Nature 416: 527–531. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy, M. R., and S. B. Hedges, 2000. Evolutionary history of the enolase gene family. Gene 259: 129–138. [DOI] [PubMed] [Google Scholar]
- Vachon, G., B. Cohen, C. Pfeifle, M. E. McGuffin, J. Botas et al., 1992. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71: 437–450. [DOI] [PubMed] [Google Scholar]
- Wagner, G. P., C. Amemiya and F. Ruddle, 2003. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl. Acad. Sci. USA 100: 14603–14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and M. Nei, 1996. Evolution of Antennapedia-class homeobox genes. Genetics 142: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]