Abstract
Bonus, a Drosophila TIF1 homolog, is a nuclear receptor cofactor required for viability, molting, and numerous morphological events. Here we establish a role for Bonus in the modulation of chromatin structure. We show that weak loss-of-function alleles of bonus have a more deleterious effect on males than on females. This male-enhanced lethality is not due to a defect in dosage compensation or somatic sex differentiation, but to the presence of the Y chromosome. Additionally, we show that bonus acts as both an enhancer and a suppressor of position-effect variegation. By immunostaining, we demonstrate that Bonus is associated with both interphase and prophase chromosomes and through chromatin immunoprecipitation show that two of these sites correspond to the histone gene cluster and the Stellate locus.
POSITIONAL information and domains of higher-order chromatin modulate gene expression in eukaryotes. Chromatin can be divided into two types of domains: euchromatic (noncondensed) and heterochromatic (condensed). Euchromatin, which is generally more accessible to the transcriptional machinery and therefore transcriptionally permissive, is composed of mostly single-copy DNA sequences. In contrast, heterochromatin is generally more inaccessible to DNA-binding transcription factors and is predominantly transcriptionally silent (Henikoff 2000; Grewal and Moazed 2003). In Drosophila, large domains of heterochromatin are present at centromeres and telomeres, while smaller domains of heterochromatin are present throughout the genome (Grewal and Elgin 2002). Despite the important role of heterochromatin in chromosomal architecture and gene expression, many of the components underlying its formation and propagation have yet to be identified and characterized.
Much of the information regarding heterochromatin has come from studying its ability to repress transcription in Drosophila (Weiler and Wakimoto 1995). Transcriptional repression occurs when a euchromatic gene is placed near or in regions of heterochromatin through chromosomal rearrangement or transposable-element-mediated insertion. In this new environment, a subset of cells assumes a repressed transcriptional state that is propagated through multiple cell divisions. This mosaic gene expression is termed position-effect variegation (PEV) and is believed to result from the spreading of condensed, higher-order structured heterochromatin into neighboring euchromatin (Demerec and Slizynska 1937; Grigliatti 1992). In Drosophila, genetic screens have identified mutations that either enhance [enhancers of variegation (E(var))] or suppress [suppressors of variegation (Su(var))] the effect of PEV (Reuter and Wolff 1981; Sinclair et al. 1989). E(var) proteins are believed to participate in the formation of active chromatin domains, while Su(var) proteins participate in the formation of repressed chromatin domains. Several of these genes have been cloned and analyzed. Their protein products encode nonhistone components of heterochromatin or proteins that regulate its assembly. The ability of these modifying proteins to suppress or enhance PEV often depends on their dosage and has led to the hypothesis that heterochromatin assembly is regulated by the concentration of available components (Locke et al. 1988; Schotta et al. 2003).
One PEV modifier gene, Su(var)2-5, encodes the heterochromatin-associated protein 1 (HP1; Eissenberg et al. 1990). Su(var)2-5 suppresses PEV when deleted and enhances PEV when duplicated (Eissenberg et al. 1990, 1992). Molecular studies have shown that HP1 is an essential component of heterochromatin and is required for transcriptional regulation, chromosome segregation, and structural integrity of the interphase nucleus (Kellum 2003). Interestingly, the ability of HP1 to efficiently bind and increase heterochromatin assembly depends upon its ability to be phosphorylated (Zhao et al. 2001). Moreover, Piacentini et al. (2003) have recently demonstrated that HP1 is unexpectedly associated with transcriptionally active regions of euchromatin. They showed that HP1 recruited to heat-shock and ecdysone-activated puffs plays a positive regulatory role in the transcription of genes located at these sites as compared to its more studied role in silencing.
Several proteins that either interact with or modify HP1 have been identified (Grewal and Moazed 2003). One of these families of proteins is the TIF1 family. The TIF1 family of proteins, TIF1α (Le Douarin et al. 1995), TIF1β [also called KAP-1 (Friedman et al. 1996) or KRIP-1 (Kim et al. 1996)], and TIF1γ (Venturini et al. 1999), are all structurally and functionally similar. All family members have an N-terminal RING finger, B boxes, and a coiled-coil domain (RBCC) followed by a C-terminal PHD finger and a bromodomain. They also have intrinsic kinase activity and repress transcription when tethered to a promoter (Fraser et al. 1998; Nielsen et al. 1999; Venturini et al. 1999). In addition, TIF1α and TIF1β recruit histone deacetylases (HDAC) to repress transcription (Nielsen et al. 1999). Interestingly, TIF1α and TIF1β have been shown to interact with and phosphorylate vertebrate homologs of HP1 (Le Douarin et al. 1998; Nielsen et al. 1999; Ryan et al. 1999). Thus, the ability of TIF1 family members to recruit HDAC and phosphorylate vertebrate homologs of HP1 suggests that they may have essential roles in the regulation of chromatin. Unfortunately, the in vivo relevance of these biochemical interactions has not been established due to the early lethality associated with mutations in mouse TIF1β and the lack of mutation in vertebrate TIF1α and -γ (Cammas et al. 2000).
Previously, we reported the isolation of the Drosophila homolog of the TIF1 family, Bonus (Bon), and established Drosophila as a valuable model organism for studying the in vivo function of the TIF1 family (Beckstead et al. 2001). Bon is the only Drosophila homolog of the TIF1 family and is expressed throughout development. Mutational analysis revealed that bon is required for numerous events in metamorphosis, including leg elongation, bristle development, and salivary gland cell death. Bon was shown to interact biochemically with several Drosophila nuclear receptors. Specifically, it was demonstrated, both genetically and biochemically, that Bon is able to bind to and inhibit the transcriptional activity of βFTZ-F1, a component of the ecdysone transcriptional cascade, providing the first in vivo evidence for a role of a TIF1 homolog in nuclear receptor signaling.
During our phenotypic characterization of different bon alleles, we observed that partial loss-of-function alleles had a more deleterious effect on males than on females. Here we expand on this observation and report a new allele of bon that is male specific lethal and female viable. We show that the male lethality associated with bon alleles is not due to defects in dosage compensation or sex determination pathways, but rather to the presence of the Y chromosome. We also show that an increase in X-heterochromatin results in a decrease in the viability of bon mutant animals. In addition, loss-of-function bon alleles dominantly suppress the PEV of the y+ gene and dominantly enhance the PEV of the w+ gene, suggesting that Bon plays an important positive and negative regulatory role in transcription. Finally, chromatin immunoprecipitation assays indicate that Bon is associated with the histone cluster and Stellate locus, two loci that display properties of β-heterochromatin.
MATERIALS AND METHODS
Immunohistochemistry and microscopy:
Guinea pig anti-Bon (Beckstead et al. 2001) was used as a primary antibody and fluorescent conjugated goat anti-guinea pig antibody (Molecular Probes, Eugene, OR) was used as the secondary antibody. TOTO-3 iodide (Molecular Probes) was used to visualize DNA. Prepupal brains of 0 hr were dissected in a 0.7% NaCl solution followed by a 10-min incubation in 0.5% sodium citrate solution. Brains were fixed in 45% acetic acid containing 2% formaldehyde for 3 min, squashed between a slide and coverslip, and frozen in liquid nitrogen. After removal of the coverslip, the slide was incubated in 1% Triton X-100 for 10 min prior to staining (Pimpinelli et al. 2000). Alternatively, the slide was incubated in 45% acetic acid for 3 min, squashed, and frozen. The coverslip was removed, and the sample incubated in 1% Triton X-100 for 10 min and then fixed in 2% formaldehyde for 3 min prior to staining. Brain images were captured using a Bio-Rad (Richmond, CA) MRC 600 laser scanning confocal microscope. Animals were photographed using a Zeiss Stemi SV8 and Hamamatsu digital camera. See Table 1 for Drosophila stocks.
TABLE 1.
Drosophila stocks
| yw; bonS024108/TM6B, Tb1 (Beckstead et al. 2001) |
|---|
| yw; bon21B/TM6B, Tb1 (Beckstead et al. 2001) |
| yw; bonS048706/TM6B, Tb1 (Beckstead et al. 2001) |
| w; Df(3R)HB79, e*/TM2 (Wustmann et al. 1989) |
| yw; P{lacw}043420/TM6B, Tb1 (Deak et al. 1997) |
| SxlF1/FM7a (Muller and Zimmering 1960) |
| tra-2B/CyO (Belote and Baker 1987) |
| tra-21/CyO (Belote and Baker 1987) |
| MCdelta3′-10/TM6, Tb1 (Bashaw and Baker 1997) |
| C(1;Y) y1 BS (Bloomington Stock Center; Lindsley et al. 1972) |
| y1 P{y+mDint2 wBR.E.BR=SUPor-P}25-4-3 (Roseman et al. 1995) |
| ln(1)wM4H (Bloomington Stock Center) |
| y1; ry506; γ878, y+ (Le et al. 1995) |
Chromatin immunoprecipitation:
Formaldehyde crosslinked chromatin fragments were prepared by sonication of 8- to 16-hr-old embryos (Orlando et al. 1997). The crosslinked nucleoprotein complexes were isolated by immunoprecipitation using an antibody against Bon (Beckstead et al. 2001) and dimethyl-lysine 9 histone H3 peptide. The DNA-protein complexes were reverse crosslinked for 5 hr at 65°. DNA was purified by phenol-chloroform extraction and recovered by ethanol precipitation. Precipitated DNA was analyzed by non-real-time PCR using primers specific to regions of the his unit (accession no. X14215; Matsuo and Yamazaki 1989) and the Stellate cluster (accession no. X15899.1; Livak 1990). The PCR conditions used for amplification were denaturation at 92° for 50 sec, annealing at 57° for 1 min, and extension at 72° for 2 min 30 sec. This cycle was repeated 25 times for all primer pairs except P6 and P8, which required 28 cycles for efficient detection of the products.
To verify specific enrichment of DNA fragments by the antibodies used, we performed several controls. First, we used an antibody that recognizes the T7-Tag. Drosophila does not contain any proteins with this sequence and therefore we expected no pull-down with the T7 antibody, which proved to be the case. Second, we performed a mock immunoprecipitation reaction using only blocked protein A sepharose beads. This control determines the level of nonspecific DNA interactions with the beads and is essentially negligible in our experiments. Third, to rule out the possibility that any observed enrichment of histone sequences was not a consequence of the multiple gene copies present, we examined another reiterated set of genes located elsewhere in the genome. These were the 5S rRNA cluster (accession no. X06938) and ubiquitin protein gene DROUBIA (accession no. M22428). Sequences corresponding to these loci were not enriched when chromatin was immunoprecipitated with the Bon antibody or the control antibody (Figure 6D). The 5S cluster is reiterated >200 times per haploid genome. However, 5S sequences were significantly enriched when antibodies against acetylated K9 H3 and acetylated H4 were used, as is expected of highly expressed loci (data not shown). Finally, we performed control chromatin immunoprecipitation (ChIP) reactions in parallel using chromatin prepared from Su(var)3-906 null mutant extracts. Su(var)3-9 encodes K9 H3 methyltransferase and is associated with the HIS-C cluster (Ner et al. 2002). Using an antibody raised to Su(var)3-9, we do not detect this protein at HIS-C in the null mutant and, moreover, we observed no K9 H3 methylation at histone sequences compared with immunoprecipitations performed on chromatin from wild-type embryos. See Table 2 for a list of oligonucleotide primers.
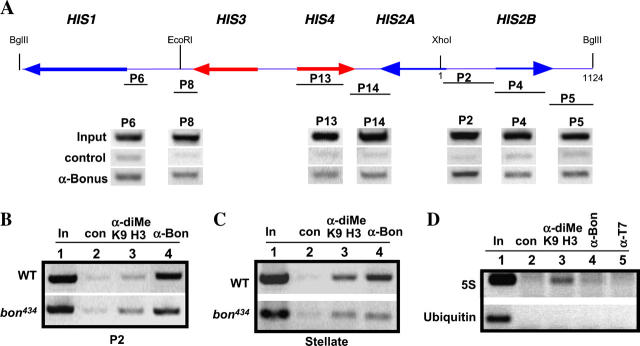
Figure 6.—
Bon is associated with the HIS-C and Stellate sequences. (A) Schematic of the 5-kb his repeat unit isolated as a BglII fragment (Lifton et al. 1978; Samal et al. 1981). The arrows indicate the five histone gene transcription units. The lines below the units labeled P6, P8, P13, P14, P2, P4, and P5 represent the seven regions of the his unit amplified to detect DNA sequences immunoprecipitated by the Bon antibody. The amplified products are shown below the schematic. The control lanes are a mock immunoprecipitation carried out using either no antibody or an antibody specific to the T7-tag (Novagen). All seven regions of the his unit tested were enriched by the Bon antibody whereas the control antibody shows background binding. The intergenic regions (P14, P2, and P5) show a higher degree of enrichment compared with the coding regions (P13 and P4). The association of Bon binding to the his units was tested in a heterozygous bon434 mutant background (B). The data for one region (P2) of the his unit are shown. Bon binding is still present at the histone sequences in the mutant (lane 4). Lane 1 is input DNA, lane 2 is the no-antibody control, and lane 3 is product immunoprecipitated with the di-Me K9 H3 antibody. (C) Stellate sequences are enriched in chromatin immunoprecipitations using the Bon antibody. The enrichment is reduced when chromatin immunoprecipitation analysis is performed on a heterozygous bonS434 mutant (lane 4). Anti-diMe-K9 H3 was used as a control antibody (lane 3). Lane 1 is input DNA and lane 2 is the no-antibody control. (D) DNA sequences from the 5S rDNA and ubiquitin loci are not immunoprecipitated with the Bon antibody. ChIP analysis was performed on a chromatin isolated from wild-type embryos using α-diMe-K9 H3 (lane 3), α-bon (lane 4), and α-T7 tag (lane 5) antibodies. No product is detected for ubiquitin with the three antibodies tested. 5S sequences are amplified with the α-di-Me-K9 H3 antibody whereas a small amount of product is detected with the bonus antibody. Lane 1 is input DNA and lane 2 is the no-antibody control.
TABLE 2.
Oligonucleotide primers
| Primer pair | Position | Sequence |
|---|---|---|
| P6 iH1f | 2065-85 | 5′-ACAATGCTACTGACATCAGTC-3′ |
| P6 iH1Dr | 2380-03 | 5′-TAATAGACGCTTCTTTCAGAAGCC-3′ |
| P8 iH1Bf | 2740-60 | 5′-TTCCGCAACAAAATTAGCCAA-3′ |
| P8 iH3r | 3316-35 | 5′-AGCGCTAGCGTACTCTATAAT-3′ |
| P13 H4R | 4384-02 | 5′-GGTACACAGGATGTACACT-3′ |
| P13 H4F | 4073-92 | 5′-ACTGGTCGTGGTAAAGGAGG-3′ |
| P14 iH4f | 4353-74 | 5′-CCGCACCCTCTACGGATTTGG-3′ |
| P14 iH2Ar | 4852-72 | 5′-CCGAGAAGAAGGCCTAAACGT-3′ |
| P2 iH2Af | 85-105 | 5′-CCGGAGCAAACGGTGAATACG-3′ |
| P2 iH2Br | 506-26 | 5′-GATGGCATAGCTCTCCTTCCT-3′ |
| P4 iH2Bf | 683-702 | 5′-CGGGAGATCCAAACGGCTGT-3′ |
| P4 iH1r | 1081-01 | 5′-TCAGGGCTACAACGTTCCGTT-3′ |
| P5 H2BR | 792-09 | 5′-TGTCCGCATTCGCAGGAG-3′ |
| P5 H2Bf | 419-35 | 5′-CCTCCGAAAACTAGTGGA-3′ |
| Stef | 115-140 | 5′-GGCCATCGAGTCCTCAGCCGA-3′ |
| Ster | 472-497 | 5′-GATCCCGAGGAACCAATCGAT-3′ |
RESULTS
Loss-of-function mutations of bonus are male specific lethal:
Phenotypic analysis of bon loss-of-function mutations suggested that males are more affected by the loss of bon than females (Beckstead et al. 2001). We have identified a mutation, P{lacW}043420 (Deak et al. 1997), that caused male-specific lethality and mapped ∼100 nucleotides into the 5′-UTR of the bon gene. As shown in Figure 1A, complementation tests performed between P{lacW}043420 and other bon alleles demonstrate that this novel mutation fails to complement all bon mutations in regard to the male lethality while it only partially fails to complement the female lethality (allelic series in decreasing strength is Df(3R)HB79 > bon21B > bon487 > bon241; see Beckstead et al. 2001). Thus, P{lacW}043420 is a weak loss-of-function allele of bon. We refer to this allele as bon434. Homozygous bon434 males die as first instar larvae while 86% of mutant female animals eclose (Figure 1, A and B). Most bon434 females appear morphologically normal and are fertile, but a small percentage display a rough-eye phenotype (data not shown). Precise excision of the bon434 P-element reverts both the male lethality and eye phenotype, confirming that the P-element is responsible for the observed phenotypes.
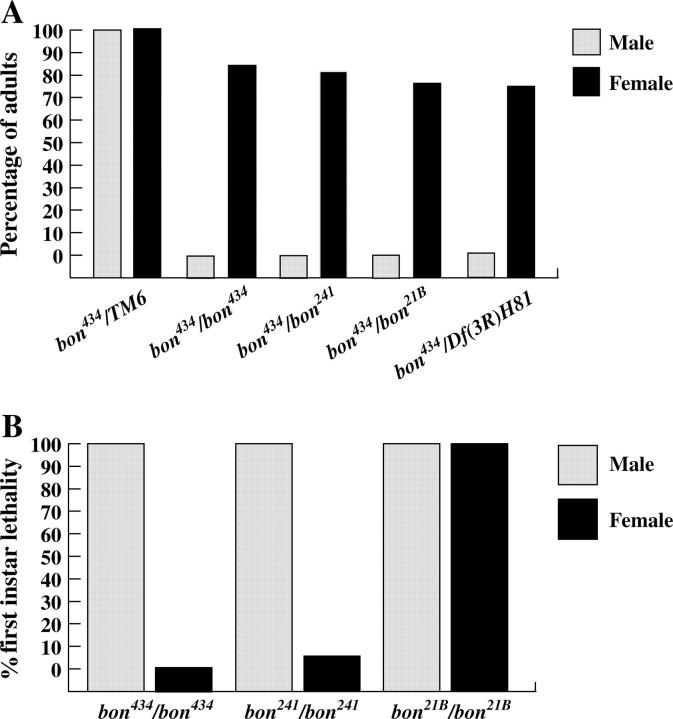
Figure 1.—
Partial loss-of-function alleles of bon have a more deleterious effect on males than on females. (A) The percentage of animals that survived to the adult stage. Percentages for bon434/bon434, bon434/bon241, bon434/bon21B, and bon434/Df(3R)H81 were determined by comparison to the number of bon434/TM6 animals, taken as 100%, that survive in each individual cross. (B) Animals of the listed genotypes were analyzed for first instar larval lethality. Homozygous mutant animals were identified by the absence of the balancer chromosome, while males/females were distinguished anatomically.
To better characterize the lethal phase of each sex in different bon mutant backgrounds, we examined the percentage of homozygous males and females that die during the first instar larval stage for bon434 and bon241; partial loss-of-function alleles; and bon21B, a null allele (Figure 1B; Beckstead et al. 2001). We observed that all bon434 and bon241 homozygous males died as first instar larvae, while all mutant females survived at least to the second instar larval stage. In contrast, a strong loss-of-function allele, bon21B, causes lethality in both male and female first instar larvae. Thus, partial loss-of-function alleles of bon have a more deleterious effect on males than on females, while severe loss-of-function alleles affect males and females more equally.
Male lethality is not due to defects in the sex determination and dosage compensation pathways:
In Drosophila development, Sex lethal (Sxl) is upregulated in females and remains silent in males (Cline 1993). Sxl directs sexual development in females by controlling the female-specific splicing of the transformer (tra) gene (Lucchesi 1978; Boggs et al. 1987). Expression of Sxl in males is lethal, as it will lead to a lack of dosage compensation through its regulation of splicing and translation of male-specific-lethal 2 (msl2; Bashaw and Baker 1997; Kelley et al. 1997). The sex-specific requirement of Sxl is seen in the loss-of-function (SxlF1) and the gain-of-function (SxlM1) Sxl mutations. Loss of Sxl associated with SxlF1 results in female-specific lethality, while ectopic expression of Sxl in the gain-of-function mutation SxlM1 results in male-specific lethality (Muller and Zimmering 1960; Skripsky and Lucchesi 1982).
To determine whether the bon male-specific lethality is due to aberrant expression of the Sxl gene in bon males, we created homozygous bon434 males that lacked Sxl gene function by crossing SxlF1/FM7; bon434/TM6 females to SxlF1/Y; bon434/TM6B males. As seen in Figure 2A, almost no SxlF1/Y; bon434/bon434 escapers were observed whereas 764 SxlF1/Y; bon434/TM6 flies survived to adulthood. Similar crosses performed with bon241 showed that no SxlF1/Y; bon241/bon241 male animals survived beyond the first instar larval stage (data not shown). These observations demonstrate that ectopic expression of Sxl in males is not the primary cause of death.
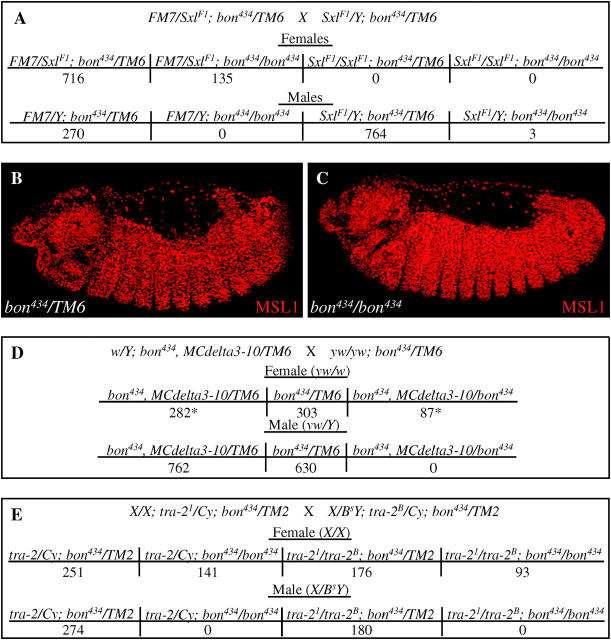
Figure 2.—
bon434 male-specific lethality is not due to defects in sex determination and dosage compensation. (A) Numbers of offspring from the cross FM7/SxlF1; bon434/TM6 × SxlF1/Y; bon434/TM6. Note that the SxlF1 mutation is lethal to females, but not to males except in the homozygous bon434 background where only three males were detected. (B and C) Immunohistochemistry using and antibody against MSL1 in bon434/TM6 (control) and bon434/bon434 late-stage embryos. (D) Numbers of offspring from w/Y; bon434, MCdelta3-10/TM6 × y w/y w; bon434/TM6. MCdelta encodes for a constitutively active MSL2 protein. An asterisk denotes female animals with a 3X2A-like pattern. (E) Number of offspring from X/X; tra-21/Cy; bon434/TM2 × X/BsY; Tra-2B/Cy; bon434/TM2. Note that tra-21/tra-2B females are still viable in the bon mutant background.
We next tested whether defects in X chromosome dosage compensation could be the basis of the bon434 male-specific lethality. In Drosophila males, dosage compensation increases the transcriptional rate of genes on the X chromosome to compensate for the presence of only one X chromosome. This process is mediated by five msl genes: maleless (mle), the male-specific-lethal genes (msl1, msl2, msl3), and males absent on the first (mof; Cline and Meyer 1996). Protein products of the msl genes form a complex with the noncoding RNA products of the roX1 and roX2 genes to mediate dosage compensation by regulating the chromatin structure on the male X chromosome (Franke et al. 1996). To determine whether msl genes were properly expressed in the bon male embryos, we stained embryos with anti-MSL1 antibodies. As seen in Figure 2, B and C, similar nuclear MSL localization of MSL1 was detected in both bon434/TM6 and bon434/bon434 embryos. Similar results were obtained for the protein products of other msl genes (msl1, msl2, msl3, and mof; data not shown), suggesting that the dosage compensation complex is present in bon male embryos.
To test whether reduced levels of Bon impede the mechanism of dosage compensation, we expressed a MSL2 (MCdelta3′-10) in bon females (Bashaw and Baker 1997). Expression of MSL2 in females results in the activation of the dosage compensation pathway. Some cells that express MSL2 upregulate dosage compensation, giving rise to females with a 3X2A phenotype. As shown in Figure 2D, constitutively active MSL2 in females with reduced levels of Bon still leads to the production of numerous 3X2A females, suggesting that dosage compensation is not affected. We conclude that there is no evidence of a role for bon in dosage compensation.
To determine whether the somatic sex of the animals plays a role in bon-induced male lethality, we tested whether 2X female animals that are somatically male are lethal when homozygous for bon434. We generated homozygous bon434 females that are mutant for the tra-2 gene and hence develop somatically as males (Belote and Baker 1987). As seen in Figure 2E, tra-21/tra-2B; bon434/bon434 animals that are genetically female, but phenotypically male, are viable, showing that somatic sex of the animal is not the cause of the lethality associated with a partial loss of Bon. In summary, the experiments shown in Figure 2 indicate that bon434 male lethality is not due to defects in dosage compensation or somatic sex differentiation.
Male lethality is due to the presence of the Y chromosome:
Because the bon male lethality is not due to defects in sex determination pathways, we tested whether the presence or absence of the Y chromosome influences the phase of lethality. In Drosophila, the Y chromosome, which accounts for ∼12% of the male genome, is made up almost entirely of heterochromatin and functions as a Su(var) (Pimpinelli et al. 1978; Dimitri and Pisano 1989). The Y chromosome itself is not necessary for male viability, but contains genes that are required for male fertility, as well as the bobbed (bb) locus, which encodes rRNA genes (Stern 1927; Brosseau 1960; Kennison and Ripoll 1981; Carvalho et al. 2001).
To test whether male lethality of bon434 is caused by the Y chromosome, we crossed bon434/TM6 males that contained a compound X-Y chromosome (C(1:Y) y1 BS) to bon434/TM6 females to produce males that lacked the Y chromosome (XO) and females that contain the Y chromosome (XXY; Figure 3A). The presence of a Y chromosome in bon434/bon434 females results in 100% lethality, while the absence of the Y chromosome in bon434/bon434 males rescues the lethality. Similar data were obtained for other bon alleles (data not shown). Thus, the presence of the Y chromosome in a partial loss-of-function bon background causes lethality, irrespective of the sex of the fly.
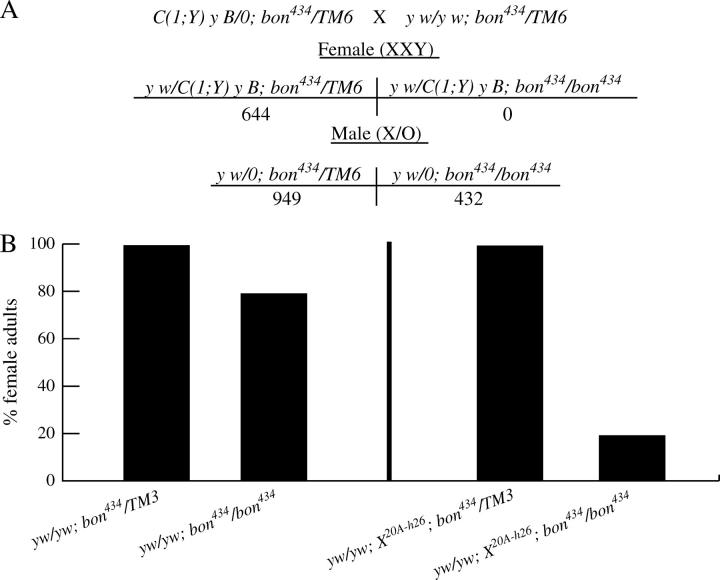
Figure 3.—
bon434 male-specific lethality is due to the presence of the Y chromosome. (A) Number of offspring from the cross C(1:Y) y1 BS/0; bon434/TM6 × y w/y w; bon434/TM6. (B) The percentage of female animals that survived to the adult stage in different genetic backgrounds. Percentages for y w/y w; bon434/bon434 was determined by comparison to the number of y w/y w; bon434/TM3, taken as 100%, and y w/y w; X20A-h26; bon434/bon434 was determined by comparison to the number of y w/y w; X20A-h26; bon434/TM3, taken as 100%. n > 50 for each genotype.
The bon Y-conditional lethality suggested that the viability of bon434/bon434 animals may depend on the levels of heterochromatin present in the genome. To test this hypothesis, we generated female flies that contained an additional copy of heterochromatic DNA that is associated with the X chromosome (20A-h26) (Dobzhansky 1932). The presence of this additional heterochromatin resulted in a 60% decrease in viability of bon434/bon434 female adults as compared to those that lack the additional X heterochromatic DNA (Figure 3B). In summary, the data indicate that the presence of the Y chromosome or X-heterochromatin decreases viability of bon mutants.
bonus acts as both an Enhancer and a Suppressor of position-effect variegation:
Because of the interaction of bon with heterochromatin, we wished to determine if bon affects heterochromatin formation or spreading. We therefore tested the ability of different loss-of-function bon mutations to modify the expression pattern of euchromatic genes that were inserted on the Y chromosome. Two SUPor-P-element lines, which map to the Y chromosome, were obtained (Roseman et al. 1995). Data for y1 P{y+mDint2 wBR.E.BR=SUPor-P}25-4-3 and y1 P{y+mDint2 wBR.E.BR=SUPor-P}222-1 were similar, and only data for y1 P{y+mDint2 wBR.E.BR=SUPor-P}25-4-3 are shown. SUPor-P elements (Figure 4A) are transposons that contain both the yellow (y+) gene with body (B) and wing enhancers (W), and the white gene (w+) with an eye enhancer (E). The w+ gene and its enhancer are flanked on either side by Su(Hw)-binding sites. These sites insulate the w+ gene from the effects of other enhancers and local heterochromatin domains (Roseman et al. 1993). As seen in Figure 4, B and D, both the w+ and y+ genes are expressed in a variegated pattern. The loss of expression of both the w+ and y+ genes is believed to be due to the effects of domains of heterochromatin that silence gene expression. The w+ gene (Figure 4B) is less affected by heterochromatin than the y+ gene (Figure 4D), probably because of the insulation of the Su(Hw)-binding sites (Roseman et al. 1993). Removal of one copy of bon+ (bon21B) resulted in a weak but significant dominant suppression of w+ expression as compared to the control (Figure 4, B and C). Thus bon is an enhancer of variegation. The other bon alleles could not be tested in this assay as they contain a copy of the w+ gene in the bon locus. These data suggest that bon+ plays an inhibitory role in heterochromatin formation and/or spreading. As shown in Figure 4, D–G, decreasing the levels of bon protein with bon241, bon487, and bon21B resulted in an increased expression of y+ as compared to the control. The level of suppression of y+ variegation also correlates with the strength of the bon allele (Beckstead et al. 2001). Hence, for the y+ gene, present in the same P element as w+, bon mutations act as suppressors of variegation. These data suggest that bon+ plays a positive role in heterochromatin formation and/or spreading.
Figure 4.—
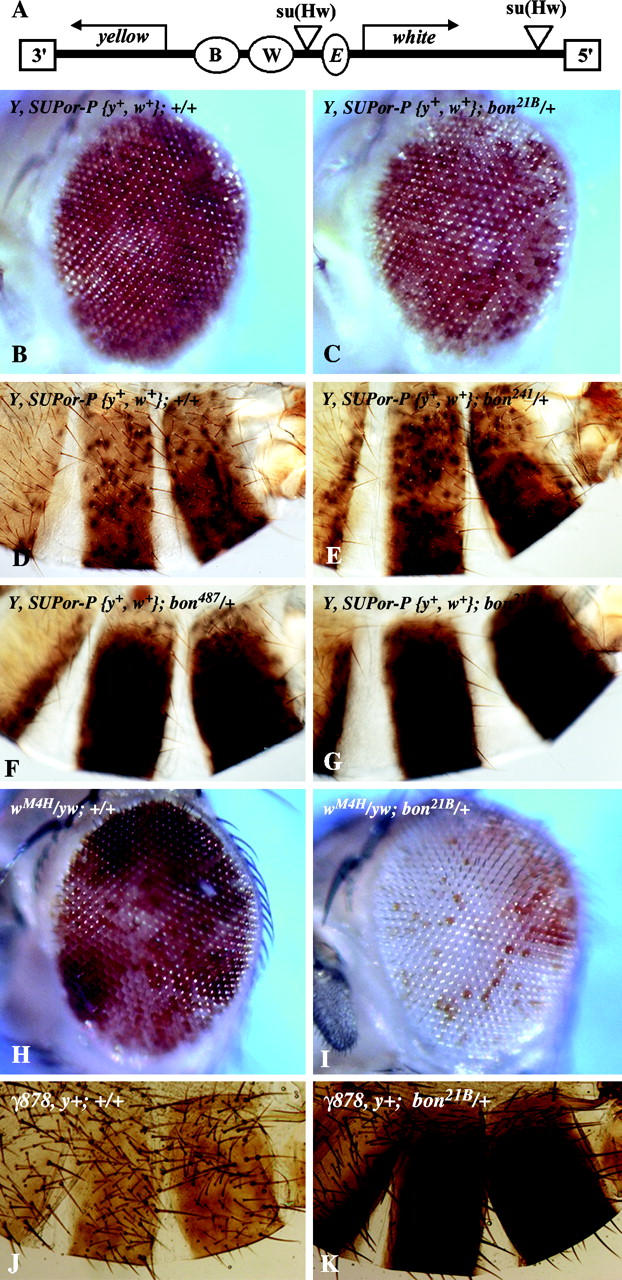
Bon acts as a Su(var) for the yellow gene and an E(var) for the white gene. (A) The SUPor-P element. Boxes represent 3′- and 5′-inverted repeats, ovals represent the yellow enhancers for body [B] and wing [W] and the eye enhancer [E] for the white gene, and triangles represent Su(Hw)-binding sites (Roseman et al. 1995). (B and C) Eyes from control flies (+/+) and bon21B/+ flies with a SUPor-P {y+, w+} inserted into the Y chromosome. (D–G) Abdominal cuticles from control (+/+), bon241/+, bon487/+, and bon21B/+ in the SUPor-P {y+, w+} backgrounds. (H and I) Eyes from control (+/+) and bon21B/+ in the ln(1)wM4H background. (J and K) Abdominal cuticles from y1; ry506; γ878, y+ ;+/+ and y1; ry506; γ878, y+ ; bon21B/+ animals.
The observation that the wild-type allele of bon is simultaneously an E(var) and Su(var) may be due to the properties of the SUPor-P element. The Su(Hw)-binding sites flanking the w+ gene may buffer or alter the effect of the loss of Bon. We therefore determined if removal of a copy of bon could affect the variegation of a w+ gene that is in close proximity heterochromatin at the base of the X chromosome, using the white-mottled (l(1)wM4H) chromosome (Reuter and Wolff 1981). As shown in Figure 4H, l(1)wM4H results in a phenotype that is characterized by red dots in a brown background (Reuter and Wolff 1981). In the presence of one copy of bon21B, variegation associated with l(1)wM4H is dramatically enhanced (Figure 4I) and the eyes appear mostly white with a few orange and red spots. To determine if loss of bon could affect the variegation of the y+ gene, we assayed the effect that loss of bon could modify the y+ variegation associated with the minichromosome γ878 (Le et al. 1995). As seen in Figure 4J, y1; ry506; γ878, y+ results in an abdominal cuticle phenotype characterized by a severe reduction in y+ expression with a few y+ patches observed. In the presence of one copy of bon21B, variegation associated with y1; ry506; γ878, y+ is dramatically suppressed (Figure 4K) and the abdominal cuticle appears mostly y+ due to the increase in size of the y+ patches. As with the SUPor-P experiments, decreasing the levels of bon protein with bon241, bon487, and bon21B resulted in an increased expression of y+ as compared to the control (data not shown). Hence, bon mutations can act as either enhancers or suppressors of PEV, depending upon the gene contexts.
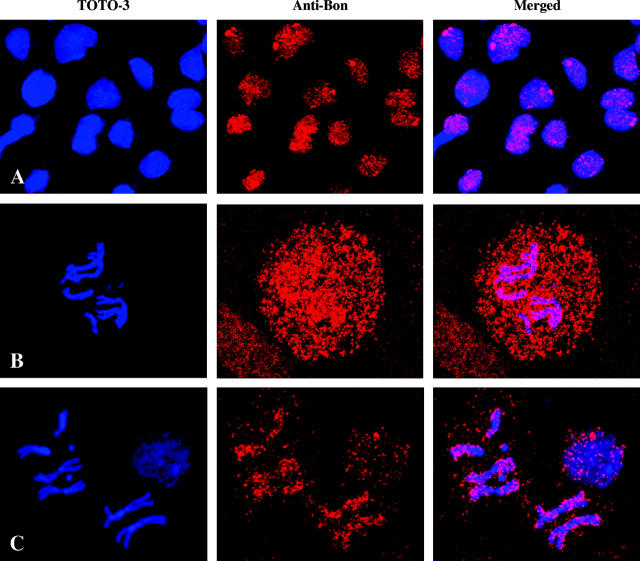
Bonus is associated with both interphase and prometaphase chromosomes:
Because of the genetic interaction between Bon and heterochromatin and the observations that Bon mutants can act as both a Su(var) and an E(var), we were interested in determining whether Bon is localized to regions of heterochromatin or euchromatin in interphase and prometaphase cells. We therefore stained 0-hr prepupal brains with a Bon antibody (Beckstead et al. 2001). The specificity of the Bon antibody was previously demonstrated by the almost complete lack of staining seen in the bon21B/bon21B mutant embryos and the significant decrease in protein levels as determined by Western analysis of bon487/bon21B prepupae (4 hr old) as compared to the bon21B/+ control. This time point and tissue were chosen due to the high levels of Bon that are detected during this stage and the ease of viewing brain cells that undergo numerous rounds of cell division. As shown in Figure 5A, Bon is a nuclear protein that is mostly localized to the nucleus of interphase cells in a punctate staining pattern. We observed one to five regions of the nucleus that show high levels of Bon staining. In prometaphase cells, Bon is localized throughout the cytoplasm of the cell (Figure 5B), making it very difficult to detect regions of the chromosome to which Bon may bind. To circumvent this problem, squashed brain tissue was incubated with detergent prior to fixation to remove most soluble Bon. As shown in Figure 5C, incubation with detergent resulted in the removal of the majority of Bon and allowed detection of the Bon protein that is associated with the mitotic chromosomes. In addition, there was a dramatic decrease in the amount of Bon detected in the interphase nucleus. The merged image with TOTO-3 shows that Bon is associated with all prometaphase chromosomes, including the Y, along the entire length of the chromosomes with no obvious preference for centromeric or telomeric regions of heterochromatin. These data suggest that Bon does not appear to be limiting in the cell, as there is much soluble Bon that can be removed with pretreatment with detergent. In addition, the Bon localization suggests that it may be playing a role in the organization of chromatin at numerous sites.
Figure 5.—
Immunostaining of brain cells from 0-hr prepupae with a Bon antibody. Interphase (A) and prometaphase (B) cells from brains fixed with formaldehyde prior to staining. (C) Interphase and prometaphase cells from brains washed in PBS + 1% Triton X-100 to remove soluble Bon and then fixed in formaldehyde prior to staining.
Bonus is associated with many loci, including the histone cluster and the Stellate locus:
Previous immunostaining of salivary gland polytene chromosomes and staining presented here of larval mitotic chromosomes and interphase nuclei with the Bon antibody reveals that the Bon protein is present at numerous sites (Beckstead et al. 2001). Further examination of the polytene staining suggested that the histone cluster (HIS-C) located at the 39D-E region of chromosome 2 is a site of Bon localization (data not shown). To confirm this observation and to gain an insight into the Bon distribution at the histone cluster, we performed a ChIP analysis using the Bon antibody on chromatin prepared from 8- to 16-hr embryos. HIS-C is a large multicopy histone gene complex and comprises ∼0.5 Mb of DNA containing 110 copies of the five histone genes (his unit; Samal et al. 1981). It is also the site of localization of Su(var)3-9 (Ner et al. 2002) and HP1 (van Steensel et al. 2001), two proteins involved in organizing chromatin structure.
Chromatin immunoprecipitations were performed using the Bon antibody, the anti-diMe-Lys9 H3 antibody (Belote and Baker 1987), and a control anti-T7 antibody. Any enrichment of DNA fragments corresponding to those of the HIS-C was detected by PCR. Seven pairs of oligonucleotide primers that hybridize within the 5-kb his units were used to detect DNA fragments pulled down by the antibodies. Two primer pairs amplified coding regions of HIS2B and HIS4 (Figure 6A, P4 and P13, respectively), and five primer pairs amplified intergenic sequence upstream and downstream of the HIS coding regions (Figure 6A, P6, P8, P14, P2, and P5). The ChIP analysis shows sequences corresponding to the intergenic regions and the coding regions of the histone genes are enriched with the Bon antibody. The enrichment is specific to this antibody since the control α-T7 antibody and the protein A sepharose alone (data not shown) failed to immunoprecipitate HIS-C sequences above background levels. In addition, the Bon antibody failed to immunoprecipitate sequences corresponding to the 5S rRNA cluster or to a ubiquitin protein gene (Figure 6D). We next repeated the immunoprecipitation analysis but used chromatin prepared from a bon mutant line (bon434/TM3). The bon434/TM3 embryos should contain less wild-type protein. We show only the detection of the P2 product as representative data for this analysis (Figure 6B). As predicted, using chromatin prepared from the bon434/TM3 line, the P2 region is still enriched by the Bon antibody; however, the level of enrichment is reduced compared with our positive control antibody that detects methylated lysine 9 of histone H3. From this analysis we conclude that Bon is associated with HIS-C and that it is distributed over the 5-kb his sequences.
Next, we examined the Stellate locus on the X chromosome (12E1-2). We examined this region to ask if other reiterated euchromatic loci are also sites for Bon. The Stellate complex is significantly smaller than HIS-C (∼30 kb in size) and comprises ∼20 copies of the Stellate gene. We examined for enrichment of Stellate sequences that are unique only to the X chromosome cluster. Immunoprecipitation using the Bon antibody was performed on chromatin prepared from wild-type flies and bon434/TM3 flies. Figure 6C shows that there is significant enrichment of Stellate sequences, suggesting that Bon protein associates with this reiterated cluster as well. Similar to the results with the HIS-C sequences, Bon localization to the Stellate locus is reduced in the bon434/TM3 mutant line. Taken together, our ChIP data show that two sites of Bon localization are the large reiterated loci, HIS-C and Stellate.
DISCUSSION
These studies define a new role for Bon in the organization of chromatin and provide new insights into its possible regulation of transcription. We show that the enhanced male lethality in bon partial loss-of-function alleles is due to the presence of the Y chromosome. We further show an interaction between bon and X-heterochromatin, demonstrating a broader interaction with heterochromatic sequences, and thus uncover a new function for this transcription cofactor. Interestingly, depending on the gene, bon can function as either a Su(var) or an E(var). Localization of Bon along the entire pro-metaphase chromosome suggests that Bon may play a more general role in chromatin organization near many genes. Using ChIP data, we show that Bon is associated with chromatin. Specifically, we demonstrate that Bon is at the HIS-C and Stellate loci. Clearly, the absence of Bon at heterochromatin, which is highly enriched in repetitive DNA sequences, along with its association with two reiterated euchromatic loci that are highly expressed, suggests that the function of Bon is not simply in the packaging of repeat sequences. One possibility is that Bon has a role in chromatin organization of transcriptionally competent loci such as HIS-C and Stellate, as well as the numerous euchromatic loci where Bon is present. Additionally, our data provide the first in vivo example of the requirement of a TIF1 family member in the regulation of chromatin.
The Y chromosome may act as a sink for Bonus:
Two other Su(var) genes, Su(var)2-1 and Su(var)3-3, show a similar lethal interaction with the Y chromosome as bon (Reuter et al. 1982; Dorn et al. 1986). Interestingly, through Y chromosome deletions, it was demonstrated that the strength of the genetic interaction between the Y chromosome and Su(var)2-1 was related to the amount of Y heterochromatin and not to a discrete Y region (Dimitri and Pisano 1989). Our data, demonstrating a genetic interaction between bon and X-heterochromatin, suggest that the bon Y-conditional lethality, like Su(var)2-1, is most likely due to the heterochromatic nature of the Y chromosome and not to a specific Y region.
On the basis of our knowledge of Bon function, we propose the following model to account for the dramatic effects that the presence/absence of the Y chromosome (and other heterochromatin) has on the viability of flies carrying weak alleles of bon. In wild-type animals, Bon is not limiting and occupies both heterochromatic and euchromatic sites, playing an essential role in gene regulation. As the amount of functional Bon activity becomes limiting in bon mutant animals, some of the Bon protein is sequestered to heterochromatic sites, thus making a fraction of Bon unavailable to euchromatic sites where it is essential for viability. The removal of the Y chromosome or other chromatin allows more Bon to be available for those sites where it plays essential roles, hence suppressing the lethality associated with the partial loss of function of bon. In other words, the presence of the Y chromosome makes a weak bon loss-of-function allele in the male act as a strong loss-of-function or null allele. Thus, the stage of lethality for bon partial loss-of-function males is similar to both Bon null females and male animals. This model is supported also by the localization of Bon to the Y chromosome, as well as by the genetic interaction observed between Bon and X-heterochromatin.
Bonus participates in the regulation of chromatin:
To determine if Bon plays a role in the regulation of chromatin packaging, we assayed the effect of bon mutations on variegation of the white and yellow genes found in several SUPor-P elements located on the Y chromosome. Interestingly, loss of bon enhanced variegation of the white gene and suppressed variegation of the yellow gene in the same animal. We also observed that bon mutations act as an E(var) in the white-mottled (l(1)wM4H) background and a Su(var) in a γ878 background. These results indicate that Bon plays a role in the regulation of heterochromatin, but also suggest that the role of Bon is gene specific.
Finally, we note that HP1 and TIF1(Bonus) share several features. First, both Bon and HP1 can function as Su(var)'s and have been shown to play both positive and negative roles in the regulation of transcription (Eissenberg et al. 1990; Beckstead et al. 2001; Piacentini et al. 2003). Second, both Bon and HP1 participate in the transcriptional response to ecdysone (Beckstead et al. 2001; Piacentini et al. 2003). Third, TIF1 members have been shown to interact with and phosphorylate HP1 (Le Douarin et al. 1998; Nielsen et al. 1999; Ryan et al. 1999). Fourth, ChIP assays indicate that Bonus and HP1 are associated with the same DNA fragments in the histone cluster (van Steensel et al. 2001) and Stellate loci. The in vivo relationship between Bonus and HP1 should therefore be explored.
Acknowledgments
R.B.B. thanks Carl Thummel for allowing him to finish this work in his laboratory. We thank the Bloomington Stock Center for numerous fly strains and Karen Schulze for comments on the manuscript. R.B.B. was supported by a National Aeronautics and Space Administration grant and is currently a Howard Hughes Medical Institute (HHMI) associate, B.S.B. is supported by the National Institute of General Medical Sciences, T.A.G. and S.S.R. are supported by National Science and Engineering Council of Canada operating grant A3005 and National Cancer Institute of Canada research grant no. 013378, and H.J.B. is an Investigator of the HHMI.
References
- Bashaw, G. J., and B. S. Baker, 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798. [DOI] [PubMed] [Google Scholar]
- Beckstead, R., J. A. Ortiz, C. Sanchez, S. N. Prokopenko, P. Chambon et al., 2001. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol. Cell 7: 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote, J. M., and B. S. Baker, 1987. Sexual behavior: its genetic control during development and adulthood in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84: 8026–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs, R. T., P. Gregor, S. Idriss, J. M. Belote and M. McKeown, 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50: 739–747. [DOI] [PubMed] [Google Scholar]
- Brosseau, G. E., 1960. Genetic analysis of the male fertility factors on the Y chromosome of Drosophila melanogaster. Genetics 45: 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas, F., M. Mark, P. Dolle, A. Dierich, P. Chambon et al., 2000. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127: 2955–2963. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B., B. A. Dobo, M. D. Vibranovski and A. G. Clark, 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98: 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T. W., 1993. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 9: 385–390. [DOI] [PubMed] [Google Scholar]
- Cline, T. W., and B. J. Meyer, 1996. Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet. 30: 637–702. [DOI] [PubMed] [Google Scholar]
- Deak, P., M. M. Omar, R. D. Saunders, M. Pal, O. Komonyi et al., 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E–87F. Genetics 147: 1697–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec, M., and H. Slizynska, 1937. Mottled white 258–18 of Drosophila melanogaster. Genetics 22: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., and C. Pisano, 1989. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics 122: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1932. Cytological map of the X-chromosome of Drosophila melanogaster. Biol. Zent. Bl. 52: 493–509. [Google Scholar]
- Dorn, R., S. Heymann, R. Lindigkeit and G. Reuter, 1986. Suppressor mutation of position-effect variegation in Drosophila melanogaster affecting chromatin properties. Chromosoma 93: 398–403. [Google Scholar]
- Eissenberg, J. C., T. C. James, D. M. Foster-Hartnett, T. Hartnett, V. Ngan et al., 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., G. D. Morris, G. Reuter and T. Hartnett, 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, A., A. Dernburg, G. J. Bashaw and B. S. Baker, 1996. Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development 122: 2751–2760. [DOI] [PubMed] [Google Scholar]
- Fraser, R. A., D. J. Heard, S. Adam, A. C. Lavigne, B. Le Douarin et al., 1998. The putative cofactor TIF1alpha is a protein kinase that is hyperphosphorylated upon interaction with liganded nuclear receptors. J. Biol. Chem. 273: 16199–16204. [DOI] [PubMed] [Google Scholar]
- Friedman, J. R., W. J. Fredericks, D. E. Jensen, D. W. Speicher, X. P. Huang et al., 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10: 2067–2078. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and S. C. Elgin, 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12: 178–187. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and D. Moazed, 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- Grigliatti, T. A., 1992 Position-effect variegation: an assay for non-histone chromosomal proteins and chromatin assembly and modifying factors, pp. 587–627 in Functional Organization of the Nucleus, edited by B. Hamkalo and S. C. Elgin. Academic Press, San Diego.
- Henikoff, S., 2000. Heterochromatin function in complex genomes. Biochim. Biophys. Acta 1470: 1–8. [DOI] [PubMed] [Google Scholar]
- Kelley, R. L., J. Wang, L. Bell and M. I. Kuroda, 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199. [DOI] [PubMed] [Google Scholar]
- Kellum, R., 2003. HP1 complexes and heterochromatin assembly. Curr. Top. Microbiol. Immunol. 274: 53–77. [DOI] [PubMed] [Google Scholar]
- Kennison, J. A., and P. Ripoll, 1981. Spontaneous mitotic recombination and evidence for an X-ray-inducible system for the repair of DNA damage in Drosophila melanogaster. Genetics 98: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. S., Y. M. Chen, E. O'Leary, R. Witzgall, M. Vidal et al., 1996. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl. Acad. Sci. USA 93: 15299–15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, M. H., D. Duricka and G. H. Karpen, 1995. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141: 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin, B., C. Zechel, J. M. Garnier, Y. Lutz, L. Tora et al., 1995. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 14: 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin, B., J. You, A. L. Nielsen, P. Chambon and R. Losson, 1998. TIF1alpha: a possible link between KRAB zinc finger proteins and nuclear receptors. J. Steroid Biochem. Mol. Biol. 65: 43–50. [DOI] [PubMed] [Google Scholar]
- Lifton, R. P., M. L. Goldberg, R. W. Karp and D. S. Hogness, 1978. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harbor Symp. Quant. Biol. 42: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., L. Sandler, B. S. Baker, A. T. Carpenter, R. E. Denell et al., 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J., 1990. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 124: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J., M. A. Kotarski and K. D. Tartof, 1988. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi, J. C., 1978. Gene dosage compensation and the evolution of sex chromosomes. Science 202: 711–716. [DOI] [PubMed] [Google Scholar]
- Matsuo, Y., and T. Yamazaki, 1989. tRNA derived insertion element in histone gene repeating unit of Drosophila melanogaster. Nucleic Acids Res. 17: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., and S. Zimmering, 1960. A sex-linked lethal without evident effect in Drosophila males but partially dominant in females. Genetics 45: 1001–1002. [Google Scholar]
- Ner, S. S., M. J. Harrington and T. A. Grigliatti, 2002. A role for the Drosophila SU(VAR)3-9 protein in chromatin organization at the histone gene cluster and in suppression of position-effect variegation. Genetics 162: 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, A. L., J. A. Ortiz, J. You, M. Oulad-Abdelghani, R. Khechumian et al., 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18: 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V., H. Strutt and R. Paro, 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11: 205–214. [DOI] [PubMed] [Google Scholar]
- Piacentini, L., L. Fanti, M. Berloco, B. Perrini and S. Pimpinelli, 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli, S., G. Santini and M. Gatti, 1978. 3h-actinomycin-D binding to mitotic chromosomes of Drosophila melanogaster. Chromosoma 66: 389–395. [DOI] [PubMed] [Google Scholar]
- Pimpinelli, S., S. Bonaccorsi, L. Fanti and M. Gatti, 2000 Preparation and analysis of Drosophila mitotic chromosomes, pp. 3–24 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Reuter, G., and I. Wolff, 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182: 516–519. [DOI] [PubMed] [Google Scholar]
- Reuter, G., R. Dorn and H. J. Hoffmann, 1982. Butyrate sensitive suppressor of position-effect variegation mutations in Drosophila melanogaster. Mol. Gen. Genet. 188: 480–485. [DOI] [PubMed] [Google Scholar]
- Roseman, R. R., V. Pirrotta and P. K. Geyer, 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R. F., D. C. Schultz, K. Ayyanathan, P. B. Singh, J. R. Friedman et al., 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19: 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal, B., A. Worcel, C. Louis and P. Schedl, 1981. Chromatin structure of the histone genes of D. melanogaster. Cell 23: 401–409. [DOI] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, R. Dorn and G. Reuter, 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14: 67–75. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., V. K. Lloyd and T. A. Grigliatti, 1989. Characterization of mutations that enhance position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 216: 328–333. [DOI] [PubMed] [Google Scholar]
- Skripsky, T., and J. C. Lucchesi, 1982. Intersexuality resulting from the interaction of sex-specific lethal mutations in Drosophila melanogaster. Dev. Biol. 94: 153–162. [DOI] [PubMed] [Google Scholar]
- Stern, C., 1927. Ein genetischer und zytologischer Beweis fur Vererbung im Y-Chromosom von Drosophila melanogaster. Z. indukt. Abstamm.-u. VererbLehre 44: 187–231. [Google Scholar]
- van Steensel, B., J. Delrow and S. Henikoff, 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27: 304–308. [DOI] [PubMed] [Google Scholar]
- Venturini, L., J. You, M. Stadler, R. Galien, V. Lallemand et al., 1999. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene 18: 1209–1217. [DOI] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- Wustmann, G., J. Szidonya, H. Taubert and G. Reuter, 1989. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 217: 520–527. [DOI] [PubMed] [Google Scholar]
- Zhao, T., T. Heyduk and J. C. Eissenberg, 2001. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem. 276: 9512–9518. [DOI] [PubMed] [Google Scholar]