Abstract
An early genetic study showed that most radiation-induced mutations are not transmitted to progeny. In recent molecular studies in plants, mainly M2 plants or their progeny, which contain only transmissible mutations, have been analyzed, but the early results imply that these studies are insufficient as comprehensive descriptions of radiation-induced mutations. To study radiation-induced mutations caused by low-LET γ-rays and high-LET carbon ions at the molecular level, we used the pollen-irradiation method and the plant Arabidopsis thaliana to study various mutations, including nontransmissible mutations. This analysis revealed that most mutants induced with irradiation with γ-rays (150–600 Gy) or carbon ions (40–150 Gy) carried extremely large deletions of up to >6 Mbp, the majority of which were not transmitted to progeny. Mutations containing 1- or 4-bp deletions, which were transmitted normally, were also found. Comparison of the deleted regions in the mutants showing various manners of transmission suggests that the nontransmissibility of the large deletions may be due to the deletion of a particular region that contains a gene or genes required for gamete development or viability.
SINCE the discovery by Muller (1927) in Drosophila and by Stadler (1928) in barley that X rays can induce mutations, radiation-induced mutants have been extensively studied and utilized in various ways, such as in the analysis of gene function and for mutation breeding. Later, it was found that ionizing radiation causes DNA damage, including base damage and single- and double-strand breaks (SSBs and DSBs, respectively), and that misrepair of DSBs is a major contributing factor to mutations induced by ionizing radiation (Britt 1999; Gorbunova and Levy 1999; Sachs et al. 2000; Lieber et al. 2003).
Ionizing radiation is typically classified into low linear energy transfer (LET) radiation, such as γ-rays and X rays, and high-LET radiation, such as α-particles and heavy ion particles. Low-LET radiation gives relatively low energy per unit length of the particle's path and transfers energy evenly to the irradiated area. High-LET radiation gives relatively high energy per unit length of the particle's path and transfers energy to very limited regions. This difference is thought to cause different types of damage to chromosomal DNA. For example, fluorescence in situ hybridization has shown that high-LET radiation generates complicated rearrangements of chromosomal structures, whereas low-LET radiation generates complicated structural alterations only at high doses (Sachs et al. 2000). However, the difference in the effects of low-LET and high-LET radiation has not been characterized well at the DNA sequence level.
In plants, sequence analyses of radiation-induced mutations have been widely carried out. These mutations include deletions, translocations, and sometimes insertions (Shirley et al. 1992; Bruggenmann et al. 1996; Buschges et al. 1997; Shikazono et al. 2000; Yano et al. 2000; Nakazaki et al. 2003; Shikazono et al. 2003). In these studies, however, mainly M2 plants or their progeny were analyzed, which could lead to biased results, because mutations with defective transmissibility to progeny are excluded from this type of analysis.
In the infancy of genetic analysis using induced mutation, Stadler and Roman (1948) studied X-ray-induced mutations by an interesting method. They irradiated maize pollen with X rays and used the pollen to fertilize a recessive mutant line carrying the a mutation, which has a defect in anthocyanin synthesis, then screened the M2 plants for the a phenotype (here we refer to the irradiated pollen as the M1 generation). This method enables the detection of even nontransmissible mutations, because a complete set of normal genes from the female plant is retained in the M2 plants, and mutagenesis is performed after gamete formation is complete. Large-scale screening revealed that only a few of several hundred mutations were transmitted to the progeny, implying that the molecular analyses of radiation-induced mutations mentioned above are insufficient as descriptions of mutations induced by ionizing radiation.
In this study, we employed Stadler and Roman's sophisticated method to study mutations caused by low-LET γ-rays and high-LET carbon ions. We call this “the pollen-irradiation method.” We chose Arabidopsis thaliana as the plant material, because the availability of a complete genome sequence and a large collection of mutants for this species make it feasible to analyze mutations at the DNA level. In addition, the applicability of the pollen-irradiation method to Arabidopsis has been demonstrated (Clark et al. 1993; Vizir et al. 1994). Our results showed that most mutants, whether induced by γ-ray (150–600 Gy) or carbon-ion (40–150 Gy) irradiation, carried extremely large deletions, that is, up to >6 Mbp. Furthermore, we found that most of these deletions were not transmitted to progeny and that the nontransmissibility can be attributed to the deletion of a particular region. These analyses provide a molecular basis for Stadler and Roman's observations.
MATERIALS AND METHODS
Plant materials and growth conditions:
A gl1-1 ms1-1 double-mutant line of A. thaliana was generated by crossing ms1-1 [Landsberg erecta (Ler) background] with gl1-1 (Ler background). Both lines and the Columbia (Col-0) line were obtained from the Arabidopsis Information Management System. The ttg1-1 ms1-1 double-mutant line (Ler background) was provided by N. Goto (Miyagi Education College).
Seeds suspended in 0.1% agar were sown one by one on moistened Golden Peat Plates (Sakatanotane, Yokohama, Japan). After cold treatment at 4° for 4 days, plants were grown at 20° with a 16-hr light/8-hr dark photoperiod.
Mutagenesis by γ-rays and carbon ions:
Just before flowering, buds were harvested and placed in a plastic bag to avoid desiccation during irradiation. Right after harvesting, the buds in the plastic bag were placed under an acrylic pane (4 mm thick) and were irradiated at room temperature with various doses of γ-rays from a 60Co source.
Carbon ions (220 MeV) penetrate only 1.1 mm in water (Tanaka et al. 1997; Morishita et al. 2003). To ensure that the carbon ions would pass through pollen, buds were taken just before flowering; their petals, sepals, and pistils were removed with tweezers; and they were sandwiched between Kapton films (7.5 μm thickness) and then irradiated. Other procedures for irradiation were as described by Tanaka et al. (1997). The irradiated pollen was transferred to open flowers of gl1-1 ms1-1 or ttg1-1 ms1-1 mutants within a day after irradiation.
Molecular markers and estimation of deletion size:
All cleaved amplified polymorphism (CAP) and derived cleaved amplified polymorphism (dCAP) markers around GL1 and TTG1 were newly constructed. Polymorphisms between Col and Ler were identified by comparing sequences of PCR products from Col and Ler or by using polymorphism information from the A. thaliana database (http://mips.gsf.de/proj/thal/db/) at the Munich Information Center for Protein Sequences and The Arabidopsis Information Resource (http://www.arabidopsis.org/). The primers and the restriction enzymes for each marker are listed in Tables 1 and 2. SSLP marker NGA139, which is 50 kbp downstream of the TTG1 locus, was designed by Bell and Ecker (1994). Because there is no DNA polymorphism in the TTG1 gene between Col and Ler, the marker TTG1 was designed to distinguish mutations in ttg1-1 from the wild-type TTG1 (Walker et al. 1999; Table 2). Deletion sizes were estimated by the distance of the farthest Col markers that were lost in a mutant.
TABLE 1.
CAPs/dCAPs markers around theGL1 locus
| Marker | 5′-forward primer-3′ | 5′-reverse primer-3′ | Enzyme | Distancea |
|---|---|---|---|---|
| moe17 | agaatcataccccctacttagtgcaga | tagagtttccatggtgaaatgtgg | MboI | 2.9 Mbp |
| k14b15 | cgatacgtgtgaacgcgttggtc | attacaaatttgcaactactgtttgagc | AluI | 1.9 Mbp |
| muj8-1 | tctagtggcagtaaaactacagagg | acaacatctactgacacctgcacaaagt | AfaI | 1.6 Mbp |
| t5m7-1 | cttctgaggctaactctgaaagcg | attcttcgtgtctatatagtcgacc | Hsp92III | 1.0 Mbp |
| mdj14-2 | taaagtgtcttttaaccggaatcc | cacgagaacaagcagagattacc | Hsp92III | 440 kbp |
| k1g2-1 | tgttggaaacattaggatcaatgaaactag | atgcaaggaagcacaacatgatctg | SpeI | 180 kbp |
| mgf10-1 | ggagtctgcgatccgaacc | gaatcagtacagagcttgcc | AfaI | 80 kbp |
| k16n12-9 | tgcctatttctcctccatgc | gctctatatacatataaaacaaattaa | PshBI | 54 kbp |
| k16n12-8 | ttttaacaacgtattatcctctcta | aaatagcacaaattcctttggc | XspI | 47 kbp |
| k16n12-5 | gaaagacgatgcaacggcg | aacaagatcttgcttgtacgg | MboI | 23 kbp |
| k16n12-2 | catgaagaagaccctatttgg | atctctccttgttgtcctcg | AfaI | 5 kbp |
| GL1 | tactggaacactcatctcagc | cagtatccgcggtaactaac | — | 0 kbp |
| k16n12-3 | gtctttgccattggcaatcc | acgttttgttgactgaagtcc | AfaI | 5 kbp |
| k24a2-1 | ggagtagggtcacaaagcc | cttgatacattggtctcttcc | TaqI | 7 kbp |
| k24a2-2 | aatgaatgcaaacccatgcg | ctaggaatcttcaaccacac | AluI | 13 kbp |
| k24a2-3 | ggagaatcggatgcacgag | ttgtcccaacaacatactcc | AluI | 18 kbp |
| k24a2-4 | cctatatgaatcgaacgcaaggatat | ttggagaccgtgatgacgtg | EcoRV | 27 kbp |
| mmg15-3 | agttttgttggatggggatgacctagttaa | ccgccaacctatattacacc | HincII | 55 kbp |
| mmg15-2 | agaatccttttaaatccatcgc | atccttgtagatgtccttagg | AluI | 90 kbp |
| mzf16-2 | ggatctatgggagattatacttgc | ttaaacactcctcaccgtgctagg | XspI | 280 kbp |
| t19n8-1 | cggagctggagattctactgccag | gtagcgcctccagctgtggcacc | MspI | 520 kbp |
| mmf24-1 | tttagccgttttctcgtggttacg | cggatgtgttctgcacgagtgttattgtac | Bst1407I | 950 kbp |
| mil15-1 | tacatcacctagggaagtcaatcg | tggaactcttgtgaggtgaactcc | HincII | 1.5 Mbp |
| f21a17-1 | tgaagtccgtagaaatgaattcc | gagcttacactcttacatagatgtcagc | Hsp92III | 2.0 Mbp |
| f1d9-1 | atctctccttctaccagaacgtgc | gaagaatataccttcaacacaagc | AccII | 3.1 Mbp |
Distance from the GL1 locus.
TABLE 2.
CAPs/dCAPs and SSLP markers around theTTG1 locus
| Marker | 5′-forward primer-3′ | 5′-reverse primer-3′ | Enzyme | Distancea |
|---|---|---|---|---|
| f5e19-2 | atacaccatatatatacgtcatac | gttctctatcaatattctaaactctggccc | HinfI | 2.9 Mbp |
| t1a4-1 | taaacatgttgggccgattatcct | tgagtataatgttatttcttattttggtac | KpnI | 2.0 Mbp |
| f5o24-1 | tttcgggttatgaccgcattcgtt | caaagtgttaaatgtggatatggatgaaat | SspI | 1.5 Mbp |
| mye28-1 | attacatttccatcagaatgatta | tatacaagggaggagatggc | StyI | 1.1 Mbp |
| t32g24-1 | ggcaggtcatggatgcattag | cactcccaacgtgagggaggagtac | DraI | 500 kbp |
| mzf18-1 | ggccactctctgccacgtgg | aagcgcaccaataacctacctgtgggctct | XhoI | 190 kbp |
| NGA139 | accgaaccgattgatttcagaagg | ggcggtacgacctcctatgg | — | 80 kbp |
| TTG1 | tactggaacactcatctcagc | cagtatccgcggtaactaac | MboI | 0 kbp |
| k18p6-3 | ggtttcgtttcactatccagg | agagctaccagatccgatgg | — | 50 kbp |
| t11h3-3 | actagtaagacaacaatgtttacc | tattctattcaaaatgttacgagg | — | 220 kbp |
| che24 1 | tcaaaccggtttgaaggatgtatacgg | ctcaagattctccatctgacccaacga | Eco52I | 560 kbp |
| wue99-1 | atgttattgagaagcacatctgct | ttaggatgatgcggtaatagaagt | SspI | 1.0 Mbp |
| tig16-3 | gaatttaagaggaaacttgatagg | cacactgagtttgtctctaaaaac | HhaI | 1.5 Mbp |
| t8m17-1 | tttattttaatgttttaaaacttatggatc | attacttttagattgcaagtataaaa | BamHI | 2.1 Mbp |
| t3p01-1 | atccaaaggaaccttatctccccgcttgcg | tgaagcagagtgcagtagtaggtc | HhaI | 3.0 Mbp |
Distance from the TTG1 locus.
Polymerase chain reaction and DNA sequence determination:
The conditions for PCR for the CAPS/dCAPS analyses were 94° for 3 min, 30 cycles of 95° for 30 sec, 52° for 30 sec, and 72° for 1 min, followed by 72° for 5 min. The primers used for PCR amplification and sequence determination of GL1 were GL1F1 (5′-TAGAATGTAAGTGTATACAGTGC-3′), GL1F2 (5′-CACTGGCCAATGGAACCGCATCG-3′), GL1R1 (5′-TTTATGATTCTTTTTATTATTTCG-3′), F3 (5′-TACTGGAACACTCATCTCAGC-3′), and R2 (5′-CAGTATCCGCGGTAACTAAC-3′). Inverse PCR (Ochman et al. 1988) was performed to identify the breakpoints of the translocation in a gl1 mutant (G300f2). Genomic DNA was triple digested with NheI, SpeI, and XbaI and was subjected to self-ligation. The resultant DNA was used for PCR amplification using the primer set GL1F2 and GL1IR1 (5′-ACTATTTAACTAAATAATTGGAGG-3′). The conditions for PCR were 95° for 1 min, 30 cycles of 98° for 10 sec, 55° for 30 sec, and 72° for 3 min, followed by 72° for 5 min. To confirm the break/rejoining points in G300f2, genomic PCR was performed with the primers GL1F2 (GL1) and mwf20F1 (BAC clone MWF20) and GL1R3 (GL1) and mwf20R1 (BAC clone MWF20). The sequences of the PCR primers were mwf20F1 (5′-ATGGATTAGAGACGAAACCAAACG-3′), mwf20R1 (5′-GTACCGACTAATTTGGTAAAGC-3′), and GL1R3 (5′-AGATTTGTTTTTGGAGGAAACACC-3′).
PCR products were purified with a QIAquick PCR purification kit (QIAGEN, Tokyo) and then directly sequenced with a Prism 377 genetic analyzer (Applied Biosystems, Tokyo).
Principle of the experiment:
To analyze various mutations induced by ionizing radiation, including nontransmissible mutations, we employed the pollen-irradiation method (Stadler and Roman 1948). The experimental procedure was as follows (Figure 1): (1) The pollen of wild-type plants was irradiated; (2) the irradiated pollen (M1 generation) was used to pollinate plants homozygous for a recessive marker mutation; and (3) M2 plants were screened for the recessive marker phenotype. Because the whole body of an M2 plant is derived from a single fertilized egg, this method can avoid chimeras, which make the molecular analysis of mutations difficult (Figure 1).
Figure 1.—
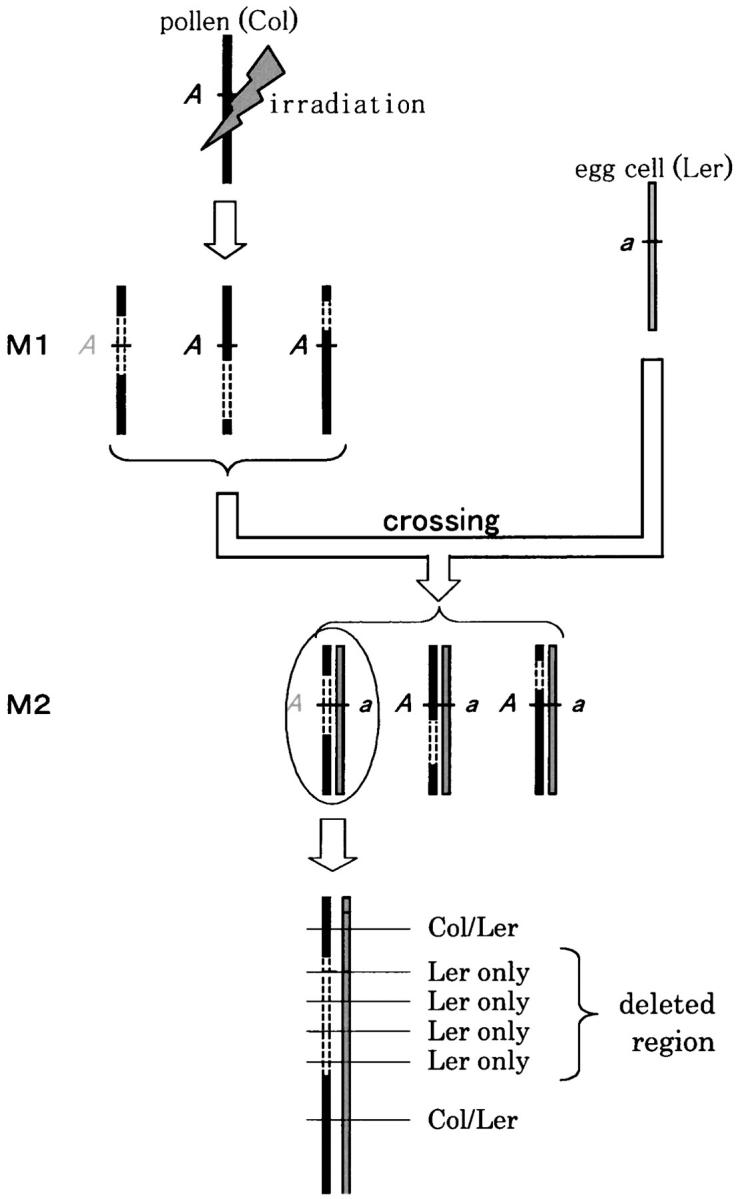
Principle of the experiment. Pollen from A. thaliana Col plants (M1) was irradiated and used to fertilize Ler plants carrying a recessive marker mutant gene (a). The resultant M2 plants were screened for the marker phenotype. These mutants were analyzed by using molecular markers around the marker locus that distinguish the Col and Ler genomes. This enables the deleted regions of the Col genome caused by pollen irradiation to be determined in the M2 plants. Col chromosomes are solid, Ler chromosomes shaded, and deleted regions open.
In this experiment, A. thaliana ecotype Col was used as the pollen donor for mutagenesis, and ecotype Ler as the pollen recipient. This approach enables the identification of radiation-induced mutations in pollen by using molecular markers that distinguish the Col and Ler genomes. As recessive visible marker mutations, the trichomeless mutations gl1-1 (chromosome 3) and ttg1-1 (chromosome 5) on the Ler background were used. gl1-1 carries an ∼10-kbp deletion spanning the whole coding region (Oppenheimer et al. 1991). ttg1-1 harbors a single nucleotide replacement that generates a premature stop codon (Walker et al. 1999). The male-sterile mutation ms1-1 on the Ler background (Wilson et al. 2001) was also introduced into the pollen recipient lines to eliminate the need for emasculation. SSLP markers F21B7-1334 on chromosome 1 and AtBIO2b on chromosome 2 were used to confirm that the mutants were hybrids of Col and Ler. Only M2 plants that grew true leaves were used for calculation of mutation rates.
RESULTS
Mutation rate:
Ler gl1-1 ms1-1 parent lines were pollinated with Col pollen irradiated with γ-rays of various dosages. The mutation rate rose as the radiation dose increased: 0% at 75 Gy irradiation, 0.20% at 150 Gy irradiation, 0.57% at 300 Gy irradiation, and 1.2% at 600 Gy irradiation (Table 3A). Vizir et al. (1994) observed a similar mutation rate (0.89%) using the pollen-irradiation method with 600 Gy γ-ray irradiation at the GL1 locus. Irradiation at 600 Gy resulted in a high rate of seed abortion. In total, 46 plants among 11,448 M2 plants showed the trichomeless phenotype. Col pollen irradiated with 300 Gy γ-rays was also used to pollinate Ler ttg1-1 ms1-1 lines. Five trichomeless plants among 1040 were obtained (0.48%), which was similar to the mutation rate at the GL1 locus resulting from 300 Gy irradiation (Table 3B).
TABLE 3.
Mutation rates in plants grown from pollen irradiated with γ-rays or carbon ions
| A. γ-ray/GL1 | ||||
|---|---|---|---|---|
| Irradiation dose (Gy) | 75 | 150 | 300 | 600 |
| No. of mutants | 0 | 10 | 30 | 6 |
| No. of M2 plants | 2542 | 4953 | 5272 | 485 |
| Mutation rate (%) | 0 | 0.20 | 0.57 | 1.24 |
| B. γ-ray/TTG1 | ||||
| Irradiation dose (Gy) | 300 | |||
| No. of mutants | 5 | |||
| No. of M2 plants | 1040 | |||
| Mutation rate (%) | 0.48 | |||
| C. Carbon ions/GL1 | ||||
| Irradiation dose (Gy) | 40 | 150 | ||
| No. of mutants | 4 | 9 | ||
| No. of M2 plants | 770 | 349 | ||
| Mutation rate (%) | 0.52 | 2.58 | ||
Carbon-ion (220 MeV) irradiation at 40 and 150 Gy was also performed using Col pollen. Again, a higher mutation rate was observed at the higher dosage of irradiation (Table 3C): 0.52% at 40 Gy irradiation and 2.6% at 150 Gy irradiation. At 150 Gy irradiation, a higher mutation rate with carbon-ion irradiation (2.6%) than with γ-ray irradiation (0.20%) was observed. This is consistent with the high relative biological effectiveness and mutagenic effect of carbon ions (Shikazono et al. 2003).
Shikazono et al. (2003) estimated that the mutation rate caused by irradiation of Arabidopsis dry seeds with 150-Gy carbon ions was 0.029% in a large-scale screening of an M2 population, but a mutation rate of 2.6% (9 of 349 M2 plants) was observed under the same irradiation conditions in this study. The different results could be due to various factors, such as differing sensitivity to irradiation of different tissues (dry seeds and pollen; Shikazono et al. 2002). However, the main reason for the increased mutation rate in our study may be that we captured mutations that are undetectable in ordinary M2 screening and that constitute the majority (>90%) of mutations induced by ionizing radiation (see below).
Mutations induced by γ-rays:
In the gl1 mutants induced with γ-rays, only 3 of 46 mutants retained the GL1 sequence from the pollen donor (Col). CAPs/dCAPs analysis revealed that all the remaining mutants carried large deletions, ranging from 80 kbp (G600b1) to >6 Mbp (G300d1 and G300e3); the latter corresponds to >5% of the whole genome. The average sizes of the deletions induced with 150, 300, and 600 Gy irradiation were 2.2, 2.7, and 1.8 Mbp, respectively (Table 4A). In this article, we define “large” deletions as any deletions with a size on the order of megabase pairs or kilobase pairs rather than base pairs.
TABLE 4.
Sizes of large deletions observed in the mutants induced by γ-ray or carbon-ion irradiations
| A. γ-ray/GL1 | |||
|---|---|---|---|
| Irradiation dose (Gy) | 150 | 300 | 600 |
| No. of mutantsa | 9 | 28 | 6 |
| Max (Mbp)b | >4.5 | >6.0 | >4.5 |
| Min (Mbp)c | 1 | 0.7 | 0.08 |
| Ave (Mbp)d | 2.2 | 2.7 | 1.8 |
| B. γ-ray/TTG1 | |||
| Irradiation dose (Gy) | 300 | ||
| No. of mutantsa | 5 | ||
| Max (Mbp)b | >3.5 | ||
| Min (Mbp)c | 0.5 | ||
| Ave (Mbp)d | 2.1 | ||
| C. Carbon ions/GL1 | |||
| Irradiation dose (Gy) | 40 | 150 | |
| No. of mutantsa | 3 | 9 | |
| Max (Mbp)b | 1.1 | >4.5 | |
| Min (Mbp)c | 0.5 | 0.5 | |
| Ave (Mbp)d | 0.7 | 2.5 | |
Number of mutants with a large deletion.
Size of the largest deletion.
Size of the smallest deletion among the large deletions.
Average size of the large deletions.
In the ttg1 mutants induced with 300 Gy γ-ray irradiation, none of the mutants retained the TTG1 sequence from Col. The size of the deletions ranged from 550 kbp to >3.5 Mbp, and the average was ∼2.1 Mbp (Table 4B).
Three mutants (G150c3, G300e2, and G300f2) induced by γ-ray irradiation retained all Col markers around the GL1 locus, including GL1 itself. To obtain more detailed information about these mutations, the mutated GL1 gene in each mutant plant was amplified by PCR, and its DNA sequence was determined. G150c3 and G300e2 had 4- and 1-bp deletions, respectively, in the third exon, which caused frameshifts and premature stop codons (Figure 2).
Figure 2.—
Positions of mutations in the gl1 alleles having small deletions. Small letters indicate the second intron and the 3′-untranslated region of GL1. Capital letters indicate the third exon. Nucleotides deleted in the mutants are shaded.
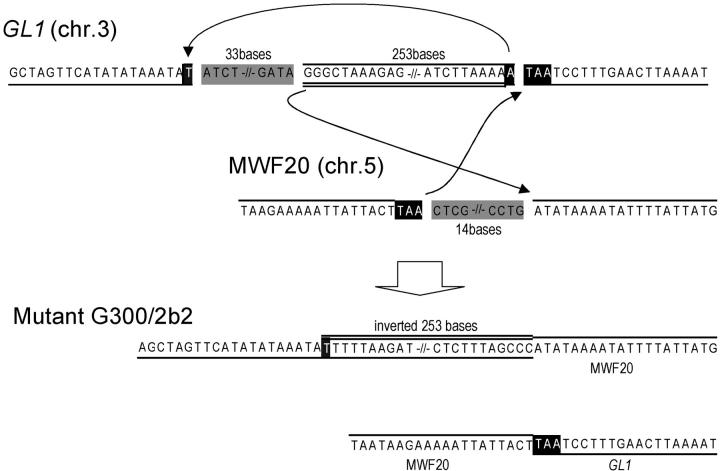
In G300f2, DNA fragments corresponding to the first and third exons of GL1 could be amplified by PCR, but no DNA fragment was amplified with the primer sets designed to amplify the DNA sequence between the first exon and the third exon (data not shown), perhaps due to a sequence rearrangement around this region. An inverse PCR experiment revealed that 33 bp of the first intron (from the 215th to the 247th base pair) were deleted and that the next 253 bp, including the whole second exon (from the 247th to the 499th base pair), were inverted (Figure 3). The downstream DNA sequence did not correspond to any region of the GL1 locus. A database search of the whole Arabidopsis genome revealed that part of BAC clone MWF20 (chromosome 5) is identical to this sequence. Further analysis by genomic PCR using a primer set designed in GL1 and MWF20 revealed that the other broken end of the GL1 gene was connected to another part of MWF20, suggesting that cotranslocation between chromosomes 3 and 5 had occurred at this point. Fourteen base pairs of MWF20 were deleted at the breakpoint, which was located in an intergenic region. In this mutation, microhomologies existed between two of the three rejoining sites (Figure 3). Similarly, in G150c3 and G300e2, a 1-bp homology might be used for DSB repair (Figure 2).
Figure 3.—
Structural changes in the DNA of mutant G300b2. Deleted nucleotides are shaded. Gaps indicate broken ends within chromosomes, and arrows indicate the rejoinings of broken ends. Letters on black backgrounds indicate microhomologies at the rejoining sites.
Mutations induced by carbon-ion irradiation:
Large deletions were frequently found in carbon-ion-induced mutants: only 1 of 13 retained the GL1 sequence. This mutant (C40a2) retained all Col markers around the GL1 locus. Sequence analysis of its GL1 locus revealed a 1-bp deletion in the third exon, which caused a frameshift and a premature stop codon (Figure 2). In this mutant, no microhomology was found at the rejoining site. The average sizes of the remaining large deletions were 650 kbp at 40 Gy irradiation and 2.5 Mbp at 150 Gy irradiation. The minimum size of the deletion was 495 kbp, and the maximum was >4.5 Mbp (Table 4C).
Fertility of the mutants:
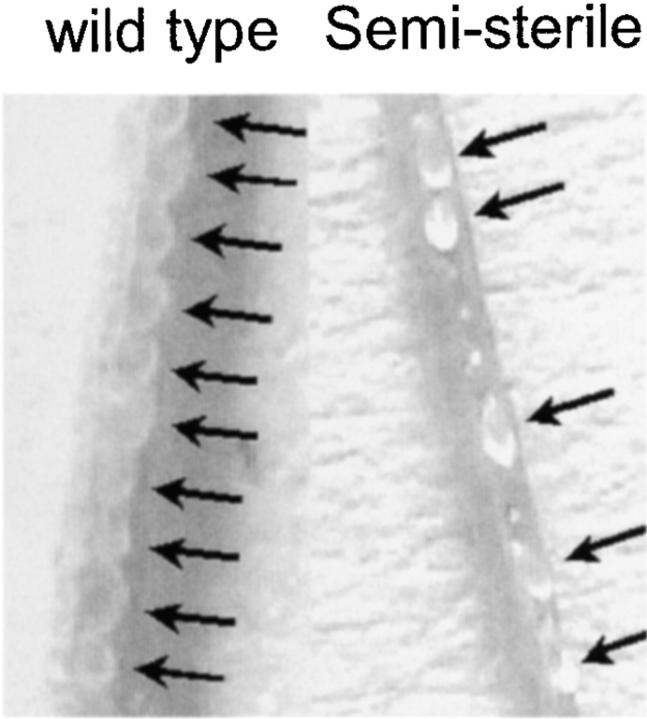
Irregularly shaped siliques were observed in most of the γ-ray- and carbon-ion-induced gl1 mutants. In these siliques, only about half the seeds were developing normally (Figure 4). Interestingly, all of these semisterile mutants carried large deletions. Similarly, all the ttg1 mutants were semisterile and had large deletions. In contrast, the fertility of all mutants having small deletions (G150c3, G300e2, and C40a2) was normal. In addition, both C40a1 and C40a3, carrying 495-kbp deletions around the GL1 locus, and C40b2, having a 1050-kbp deletion around the GL1 locus, showed normal fertility. Complete sterility was also observed in some mutants induced by 600-Gy γ-rays (G600b1 and G600b2) and by 150-Gy carbon ions (C150b2). G600b2 carried an 80-kbp deletion around the GL1 locus, the smallest among the large deletions. The high extent of chromosomal aberration caused by high-dose irradiation might have prevented normal meiosis in these mutants, thereby preventing normal gamete development. Unexpectedly, the reciprocal translocation mutant (G300f2) was fully fertile, even though reciprocal translocation generally causes reduced fertility in heterozygotes (data not shown; van Harten 1998). It is possible that another reciprocal translocation between the two chromosomes occurred close to the translocation point in this mutant and compensated for its effects on semisterility.
Figure 4.—
Developing seeds in semisterile mutants produced by the pollen-irradiation method. Opened siliques from a wild-type plant and from a typical semisterile mutant are shown. Arrowheads indicate normally developing seeds.
Transmissibility of the radiation-induced mutations:
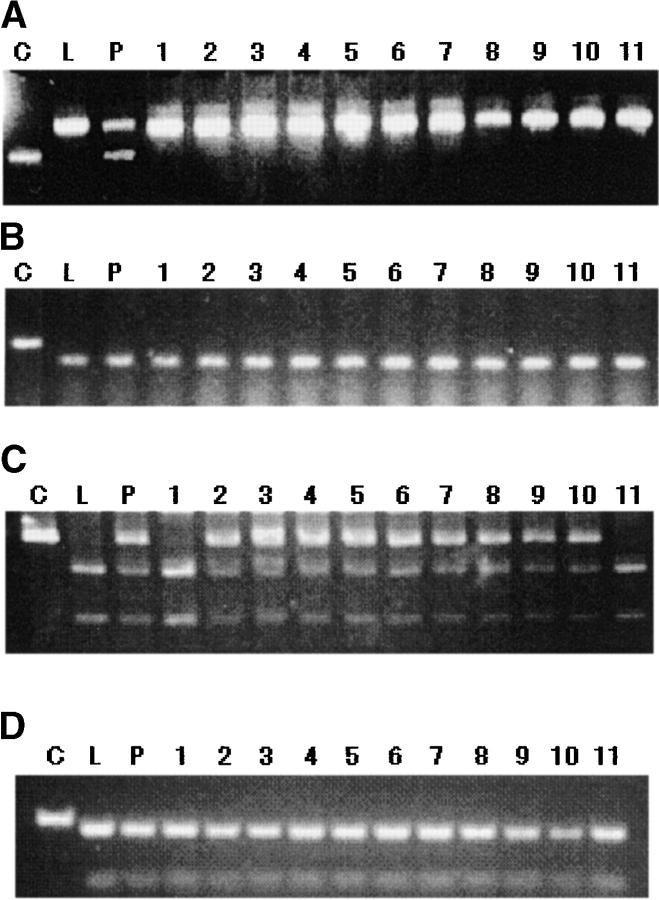
Because all the selfed M3 progeny from a mutant M2 plant showed the trichomeless phenotype, the transmissibility of the mutation was examined by genotyping the M3 plants with the molecular markers. In the gl1 alleles having 1- and 4-bp deletions (G150c3, G300e2, and C40a2), no distortion in transmissibility was observed among 40 M3 progeny of each line: about three-fourths of the M3 progeny had the Col GL1 sequence (data not shown). The reciprocal translocation mutation (G300f2) was also transmitted normally among 40 M3 progeny (data not shown). However, when selfed M3 progeny of the semisterile mutants were analyzed, no Col genotypes for markers closely linked to the GL1 locus were detected. For example, in 40 M3 progeny of the semisterile mutant G300a2, only Ler marker bands were detected in CAPS/dCAPS analysis using t5m71-l, mdj14-2, and k1g2-1 (Figure 5A; data not shown), suggesting that no or very few gl1 alleles associated with semisterility were transmitted to the M3 plants. In addition, all the 40 M3 progeny examined carried the Ler genotype for k24a2-4, whose Col allele was deleted in G300a2 (Figure 5B). In the fully fertile mutants having large deletions (C40a1, C40a3, and C40b2), no Col-genotype homozygotes for mmg15-2, which is closely linked to the deletions, were found in 40 M3 progeny of each mutant, although heterozygotes were found (Figure 5C; data not shown). These observations suggest that these mutations could be transmitted to progeny only heterozygously, in spite of their full fertility. Among 40 M3 progeny of each mutant, all progeny had the Ler genotype for k24a2-4, whose Col allele was deleted in these mutants, supporting this hypothesis (Figure 5D; data not shown). One explanation for these observations is that these deletions affect the viability of only male gametes. However, in germination tests, only 73 of 100 M3 seeds of C40a1 germinated, although 98 of 100 F2 seeds of a hybrid between Col and Ler germinated. Although we were not able to determine the genotype of seeds that did not germinate, it appears that homozygotes for these deletions cannot germinate. We should note that these transmissible large deletions cannot be detected in ordinary M2 screening.
Figure 5.—
Transmissibility of the large deletions. (A and B) dCAPs analysis of the M3 plants of the semisterile mutant G300a2 (see Figure 6) using t5m7-1 (A) and k24a2-4 (B). (C and D) dCAPs and CAPs analysis of the M3 progeny of the fully fertile mutant C40a3 (see Figure 6) using k24a2-4 (C) and mmg15-2 (D). Lanes C, L, and P indicate Col, Ler, and the mutant M2 plants, respectively. Lanes indicated by numbers are the M3 progeny.
The semisterile ttg1 mutants also did not transmit the Col ttg1 mutation to the M3 progeny. For example, among 40 M3 progeny of tG300a1, which carried a 550-kbp deletion from t32g24-1 to k18p6-3, no Col genotype was found for the marker t11h3-3, which is closely linked to the deletion (Table 2; data not shown).
DISCUSSION
Mutations induced by ionizing radiation:
Although DSBs are not the major DNA damage induced by irradiation, they have greater importance in mutations than do other types of DNA damage (Britt 1999; Gorbunova and Levy 1999; Tuteja et al. 2001). DSBs are thought to be repaired in three ways. The first is homologous recombination, which copies the homologous sequence from the undamaged sister chromatid. The second is single-strand annealing, which is mainly involved in the repair of repeated sequences. The third is nonhomologous end joining (NHEJ), which rejoins one DNA broken end with another without specificity (Britt 1999; Gorbunova and Levy 1999; Lieber et al. 2003). NHEJ can be precise but is often accompanied by these small deletions. The deletion of one or a few base pairs observed in G150b3, G300d2, and C40a2 was probably due to DSB repair by NHEJ of two broken ends that were originally linked. Alternatively, replication slippage at damaged bases can be involved in the generation of small deletions (Kunkel 1990).
Most of the mutations identified in this study were very large deletions (>80 kbp) in both GL1 and TTG1 loci at various irradiation doses (40–600 Gy) and with both high- and low-LET irradiation (carbon ions and γ-rays, respectively). The largest deletion was >6 Mbp around the GL1 locus, which should involve the deletion of >1300 genes, on the basis of the average gene density in Arabidopsis chromosome 3 (a gene every 4.5 kbp; Arabidopsis Genome Initiative 2000). Vizir et al. (1994) also characterized the size of deletions in A. thaliana induced by γ-rays using the pollen-irradiation method. In this classic genetic study, they estimated that the average size of deletions was 160 kbp, which are large deletions, but the average size of deletions in our study was much larger (>2 Mbp, even if point mutations are included).
Very large deletions are thought to be generated by NHEJ (Sachs et al. 2000). When both terminal DNA fragments of the three DNA fragments generated by two DSBs in one chromosome are joined together, the consequent omission of the middle fragment constitutes a deletion. When the omitted fragment is large, a large deletion occurs. In this context, the observation that most of the mutations found in our study were large deletions suggests that two DSBs were not frequently generated in nearby positions, even with high-LET carbon-ion irradiation, despite its high density of excited/ionized molecules. This could be a consequence of the higher-order structure of chromatin. Importantly, this study shows that both low-LET γ-rays and high-LET carbon ions generate large deletions, but it does not necessarily suggest that there is no difference between them regarding the mechanism responsible for the generation of mutations. Furthermore, we should note that the generation of large deletions vs. point-like mutations by the DSB repair process may be less frequent than expected from the frequency of large deletions found in our mutants, because large deletions have a higher mutagenic effect and are thus detected more often; that is, it is more likely for a gene to be affected by one large deletion than by one point mutation.
Transmissibility of mutations:
All the semisterile gl1 mutants carried large deletions around the GL1 locus, suggesting a correlation between large deletions and semisterility. These large deletions did not appear to be transmitted even heterozygously. The most likely explanation for these observations is that the large deletions contained a gene or genes required for the formation or viability of pollen and egg cells: only egg cells and pollen that do not carry the large deletions and are able to survive can undergo successful fertilization, resulting in semisterility and nontransmissibility of the deletions. Consistent with this, no seed development was observed visually in the semisterile mutants in about half the placentas, implying either the absence of fertilization or very early abortion of seeds (Figure 4). It is unlikely that a dominant lethal mutation affecting embryogenesis is involved in the semisterility, because this would result in no M2 seed formation. It is also possible that DNA rearrangement in another region contributed to the semisterility, but the distortion in transmissibility of the gl1 alleles strongly suggests that the semisterility can be attributed to the large deletion around the GL1 locus. Similar observations were made for the TTG1 locus, suggesting that genes involved in gamete formation or viability are not rare and are dispersed throughout the genome.
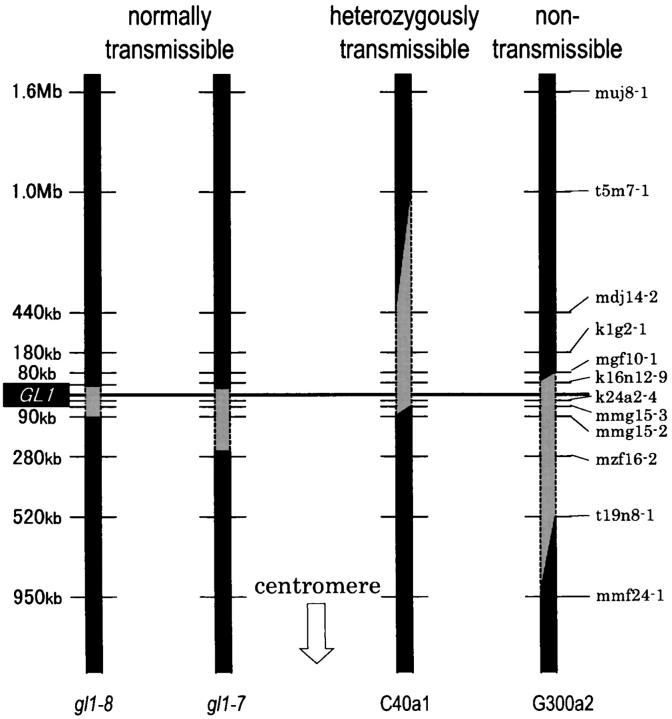
Interestingly, three mutants showed full fertility even though they carried large deletions around the GL1 locus (Figure 6). These deletions were transmitted only heterozygously. Taking into consideration the observation that one-fourth of the M3 seeds of C40a1 did not germinate, we hypothesize that fertilized eggs homozygous for the deletions can develop into seeds but lack the ability to germinate. Shikazono et al. (2003) isolated gl1 alleles carrying a large deletion, which were transmitted normally (Figure 6; N. Shikazono, C. Suzuki, S. Kitamura, H. Watanabe, S. Tano and A. Tanaka, unpublished results). Comparison of the deleted region in C40a1 with those in mutants gl1-7 and gl1-8 suggests that the gene is located between 37 kbp upstream of GL1 and t5m7-1 (1 Mb upstream of GL1). On the other hand, comparison of the deleted regions in the semisterile and nontransmissible mutant, G300a2, with those in C40a1 and gl1-7 suggests that the region between 194 kb downstream of GL1 and mmf24-1 (950 kbp downstream of GL1) contains at least one gene essential for gamete development or viability.
Figure 6.—
Deleted regions in the gl1 mutants showing various manners of transmission. Deleted regions are shaded. Positions of molecular markers are on the left and marker names are on the right.
By the pollen-irradiation method using X rays and applied to maize, Stadler and Roman (1948) obtained a very small number of transmissible mutations (2 of 415), and both mutations showed low transmissibility. Nuffer (1957) and Mottinger (1970) made similar observations for another locus using the same method and concluded that intragenic mutation, which usually shows complete transmissibility, cannot be induced by X rays. Our results are similar to those observations: most mutations induced by ionizing radiation had defective transmissibility. However, some apparent “intragenic” mutations were isolated (3 of 64) in our study. The difference may be attributed to the use of different species, because an intragenic mutation induced by X rays using the pollen-irradiation method has been reported in Arabidopsis (Clark et al. 1993, 1997). Consistent with this, the DNA repair system may differ between plant species having different genome sizes, e.g., a very small genome size such as Arabidopsis and a large genome size such as tobacco (Kirik et al. 2000). Simple repair of a single DSB by NHEJ, which often results in deletion of a small number of nucleotides, might be rare in maize in comparison with Arabidopsis.
In this study, we showed that most radiation-induced mutants carry extremely large deletions, regardless of the irradiation dose, radiation type, or locus. On the basis of our results, we hypothesize that most such deletions are not transmitted to progeny because the deletions remove a gene or genes involved in gamete development or viability. This means that most mutations induced by ionizing radiation have not been used in the mutation breeding of seed-propagated crops. However, “nontransmissible” mutations can survive over generations in vegetatively propagated crops and have probably contributed to the genetic improvement of such crops.
Acknowledgments
We thank N. Goto for providing the ttg1-1 ms1-1 mutant line. This work was supported by a grant for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
This article is dedicated to Toshiya Takano, who passed away in December 2003.
References
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bell, C. J., and J. R. Ecker, 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144. [DOI] [PubMed] [Google Scholar]
- Britt, A. B., 1999. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4: 20–25. [DOI] [PubMed] [Google Scholar]
- Bruggenmann, E., K. Handwerger, C. Essex and G. Storz, 1996. Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J. 10: 755–760. [DOI] [PubMed] [Google Scholar]
- Buschges, R., K. Hollricher, R. Panstruga, G. Simons, M. Wolter et al., 1997. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., M. P. Running and E. M. Meyerowitz, 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., R. W. Williams and E. M. Meyerowitz, 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Gorbunova, V., and A. A. Levy, 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4: 263–269. [DOI] [PubMed] [Google Scholar]
- Kirik, A., S. Salomon and H. Puchta, 2000. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 19: 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T. A., 1990. Misalignment-mediated DNA synthesis errors. Biochemistry 29: 8003–8011. [DOI] [PubMed] [Google Scholar]
- Lieber, M. R., Y. Ma, U. Pannicke and K. Schwarz, 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4: 712–720. [DOI] [PubMed] [Google Scholar]
- Morishita, T., H. Yamaguichi, K. Degi, N. Shikazono, Y. Hase et al., 2003. Dose response and mutation induction by ion beam irradiation in buckwheat. Nucl. Inst. Meth. 206: 506–569. [Google Scholar]
- Mottinger, J. P., 1970. The effects of X rays on the bronze and shrunken loci in maize. Genetics 64: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1927. Artificial transmutation of the gene. Science 66: 84–87. [DOI] [PubMed] [Google Scholar]
- Nakazaki, T., Y. Okumoto, A. Horibata, S. Yamahira, M. Teraishi et al., 2003. Mobilization of a transposon in the rice genome. Nature 421: 170–172. [DOI] [PubMed] [Google Scholar]
- Nuffer, M. G., 1957. Additional evidence on the effect of X-ray and ultraviolet radiation on mutation in maize. Genetics 42: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., A. S. Gerber and D. L. Hartl, 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D. G., P. L. Herman, S. Sivakumaran, J. Esch and M. D. Marks, 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493. [DOI] [PubMed] [Google Scholar]
- Sachs, R. K., L. R. Hlatky and B. J. Trask, 2000. Radiation-produced chromosome aberrations. Trends Genet. 16: 143–146. [DOI] [PubMed] [Google Scholar]
- Shikazono, N., A. Tanaka, H. Watanabe and S. Tano, 2000. Rearrangements of the DNA in carbon ion-induced mutants of Arabidopsis thaliana. Genetics 157: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikazono, N., A. Tanaka, S. Kitayama, H. Watanabe and S. Tano, 2002. LET dependence of lethality in Arabidopsis thaliana irradiated by heavy ions. Radiat. Environ. Biophys. 41: 159–162. [DOI] [PubMed] [Google Scholar]
- Shikazono, N., Y. Yokota, S. Satoshi, G. Suzuki, H. Watanabe et al., 2003. Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 163: 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B. W., S. Hanley and H. M. Goodman, 1992. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, L. J., 1928. Mutation in barley induced by X-rays and radium. Science 67: 186–187. [DOI] [PubMed] [Google Scholar]
- Stadler, S. J., and H. Roman, 1948. The effect of X-rays upon mutation of the gene A in maize. Genetics 33: 273–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., N. Shikazono, Y. Yokota, H. Watanabe and S. Tano, 1997. Effects of heavy ions on the germination and survival of Arabidopsis thaliana. Int. J. Radiat. Biol. 72: 121–127. [DOI] [PubMed] [Google Scholar]
- Tuteja, N., M. B. Singh, M. K. Misra, P. L. Bhalla and R. Tuteja, 2001. Molecular mechanisms of DNA damage and repair: progress in plants. Crit. Rev. Biochem. Mol. Biol. 36: 337–397. [DOI] [PubMed] [Google Scholar]
- van Harten, A. M., 1998 Mutation Breeding. Cambridge University Press, Cambridge, UK.
- Vizir, I. Y., M. L. Anderson, Z. A. Wilson and B. J. Mulligan, 1994. Isolation of deficiencies in the Arabidopsis genome by γ-irradiation of pollen. Genetics 137: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A. R., P. A. Davison, A. C. Bolognesi-Winfield, C. M. James, N. Srinivasan et al., 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, Z. A., S. M. Morroll, J. Dawson, R. Swarup and P. Tighe, 2001. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39. [DOI] [PubMed] [Google Scholar]
- Yano, M., Y. Katayose, M. Ashikari, U. Yamanouchi, L. Monna et al., 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]