Abstract
A collection of Activator (Ac)-containing, near-isogenic W22 inbred lines has been generated for use in regional mutagenesis experiments. Each line is homozygous for a single, precisely positioned Ac element and the Ds reporter, r1-sc:m3. Through classical and molecular genetic techniques, 158 transposed Ac elements (tr-Acs) were distributed throughout the maize genome and 41 were precisely placed on the linkage map utilizing multiple recombinant inbred populations. Several PCR techniques were utilized to amplify DNA fragments flanking tr-Ac insertions up to 8 kb in length. Sequencing and database searches of flanking DNA revealed that the majority of insertions are in hypomethylated, low- or single-copy sequences, indicating an insertion site preference for genic sequences in the genome. However, a number of Ac transposition events were to highly repetitive sequences in the genome. We present evidence that suggests Ac expression is regulated by genomic context resulting in subtle variations in Ac-mediated excision patterns. These tr-Ac lines can be utilized to isolate genes with unknown function, to conduct fine-scale genetic mapping experiments, and to generate novel allelic diversity in applied breeding programs.
TRANSPOSON tagging is a powerful tool for gene isolation and characterization (Fedoroff et al. 1984). It has been used extensively in a number of plant species including Arabidopsis and rice, where large collections of modified Ds elements have been generated and precisely positioned throughout their genomes (Muskett et al. 2003; Zhu et al. 2003; Kim et al. 2004; Kolesnik et al. 2004; Kuromori et al. 2004). In Arabidopsis and rice, Ac and Ds have been used to isolate genes (Bancroft et al. 1993; James et al. 1995; Bhatt et al. 1996; Meissner et al. 1999), define promoter and enhancer elements (Sundaresan et al. 1995; Chin et al. 1999; Greco et al. 2003; Jin et al. 2004; Wu et al. 2003), and create genetic mosaics for clonal analysis (Peng and Harberd 1997; Jenik and Irish 2001).
Although Ac/Ds transposon tagging has proven to be a valuable tool in many plant species, two features of Ac/Ds have restricted their utility in large-scale mutagenesis programs in maize (Walbot 2000; Brutnell 2002). For one, Ac/Ds elements transpose at rates 50- to 100-fold lower than those of the prolific Mutator family of transposable elements (Walbot 2000). This has precluded the use of Ac in reverse genetic programs that exploit a high forward mutation rate to generate a large collection of transposon insertions in many genes with relatively few plants. A second feature of Ac/Ds elements that has restricted their use in gene-tagging experiments is their propensity for transposition to linked sites in the genome (Van Schaik and Brink 1959; Greenblatt 1984; Dooner and Belachew 1989). In studies of Ac transposition at the p1 and bz1 loci, ∼60% of transpositions were to genetically linked sites and the majority of these transpositions were to regions within 10 cM of the donor element (Greenblatt 1984; Dooner and Belachew 1989). Thus, the utility of Ac/Ds as insertional mutagens is limited by the proximity of the transposon to a target locus.
Despite the current limitations of Ac for genome-wide mutagenesis, several molecular and genetic characteristics make the Ac/Ds system an extremely attractive resource. Ac elements are maintained at a relatively low copy number in the genome, facilitating molecular characterization (Fedoroff et al. 1983). In maize, increasing copies of Ac result in a developmental delay of Ac and Ds transposition (McClintock 1951). Therefore, the copy number of Ac elements can be monitored using many well-characterized Ds elements as reporters of Ac dosage, including several in genes for anthocyanin and starch biosynthesis (McClintock 1955; Dooner and Kermicle 1971). The ability to monitor increases in Ac copy number also provides a powerful tool in gene-mapping experiments (Greenblatt 1984; Van Schaik and Brink 1959; Dooner and Belachew 1989). Another feature of Ac/Ds that is extremely useful for gene tagging is a high frequency of somatic excision. Revertant alleles can be used to confirm the identity of an Ac-tagged allele, thereby alleviating the need for transgenic complementation or the recovery of multiple insertion alleles (Schultes et al. 1996; Schauser et al. 1999). Furthermore, somatic transposition events can be exploited to obtain additional gene sequence (Singh et al. 2003). Imprecise Ac or Ds excision events also have the potential to generate “footprints” resulting in stable alleles that create novel proteins with altered activities (Wessler et al. 1986; Giroux et al. 1996).
Although a few Ac elements have been precisely positioned in the maize genome, the utility of Ac/Ds would be greatly improved if elements were distributed at 10- to 20-cM intervals and positioned on physical and genetic maps. Ac insertions at the p1 (Emerson 1917; Athma et al. 1992), bz1 (McClintock 1955; Ralston et al. 1988), and wx1 (McClintock 1964; Klosgen et al. 1986) loci have been utilized as platforms for random mutagenesis (Dellaporta and Moreno 1994; Cowperthwaite et al. 2002). However, only a few genes have been cloned using closely linked Ac or Ds elements as donor loci in regional mutagenesis experiments (Dellaporta et al. 1988; Hake et al. 1989; DeLong et al. 1993; Colasanti et al. 1998; Shen et al. 2000; Singh et al. 2003). Through the precise placement of Ac elements throughout the maize genome, it should be possible to greatly expand the scope of Ac mutagenesis programs in maize.
In this study, we have utilized genetic procedures to distribute ∼158 Ac elements throughout the maize genome. DNA sequence flanking 69 of these elements has been cloned and used to position 41 of the Ac elements on one of three publicly available recombinant inbred populations (Burr et al. 1988; Lee et al. 2002). Ac elements have been mapped to all 10 of the maize chromosomes and are primarily located in single- or low-copy genomic sequence. However, insertions into retrotransposons and other repetitive elements were also identified. Characterization of the Ac-mediated variegation patterns revealed that genomic context may influence Ac activity. These near-isogenic lines along with the cloning methodologies presented here offer powerful resources for efficient Ac-based mutagenesis programs in maize.
MATERIALS AND METHODS
Description of maize stocks:
All stocks were maintained in a color-converted W22 inbred (Dooner and Kermicle 1971; Kermicle 1984). Approximately 400 primary transpositions were generated from either P1-vv or bti97156::Ac. The maize p1 locus controls flavonoid production in floral tissues including pericarp and glumes (Styles and Ceska 1977). P1-vv confers a variegated pericarp and cob due to an unstable Ac insertion in a P1-RR allele of the p1 locus (Lechelt et al. 1989). bti97156::Ac is located on chromosome 5 bin 5.04 (Singh et al. 2003). The Ds tester line used was r1-sc:m3, which contains a Ds insertion in the r1 locus (Alleman and Kermicle 1993) and renders the kernel colorless. Ac-mediated excisions of Ds restore R function, resulting in sectors of purple aleurone. The size and number of sectors reflect the copy number of Ac. In general, increasing copies of Ac result in fewer and smaller sectors (McClintock 1951). When four copies of Ac are present in the triploid endosperm, Ds excisions are rare or absent, resulting in a few small colored sectors or a colorless aleurone, respectively. When one copy of the r-sc:m3 reporter is present in the endosperm (see Figure 1), the frequency of colored sectors is reduced, but the average size of colored sectors is similar to what is observed when two or three copies of the Ds reporter allele r-sc:m3 are present. Thus, our selection for finely spotted and colorless kernels in F1 testcross progeny ensures that we detect all transposition events that contribute to dosage. An average transposition frequency of 2–4% was calculated from a total of ∼12,400 kernels generated from 10 different Ac lines.
Figure 1.—
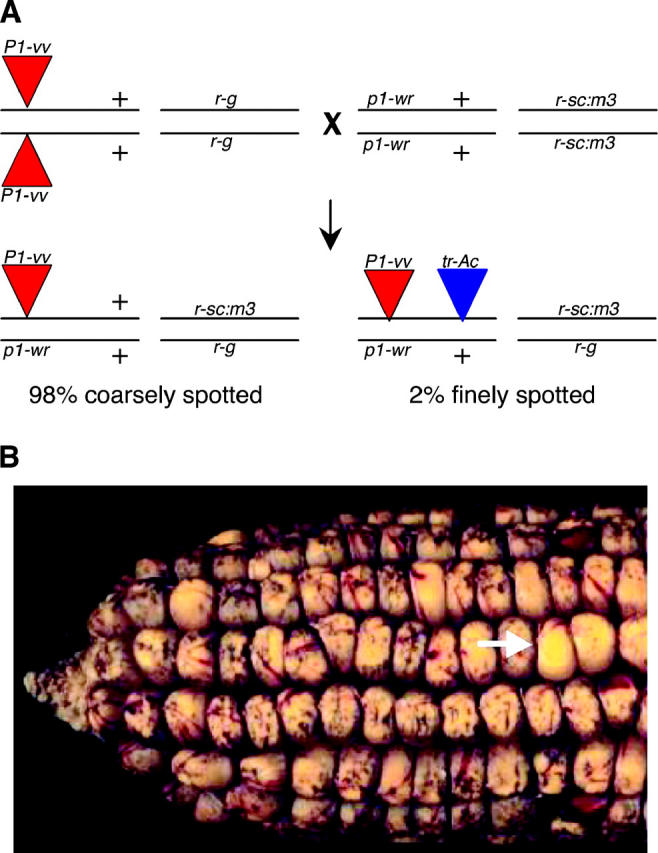
Generation and selection of tr-Acs. (A) Lines homozygous for an Ac insertion (P1-vv illustrated) were crossed to a Ds tester reporter line containing the r1-sc:m3 allele. Approximately 98% of the progeny kernels inherit the original “donor” Ac, resulting in coarsely spotted kernels. Transpositions inherited with the donor Ac result in an increase in Ac copy number in ∼2% of the progeny and finely spotted or colorless kernels. (B) Typical F1 ear showing p1 locus variegation (red stripes) and aleurone variegation (purple sectors) mediated by Ac or Ds excision events, respectively. Transmission of two Ac elements through the female gametophyte results in four copies of Ac in the triploid endosperm and a finely spotted or colorless aleurone (white arrow in B).
Identification of newly transposed Ac elements:
To identify transposed Ac elements (tr-Acs), genomic DNA from ∼1 g of pooled leaf tissue was isolated from 5–10 individuals homozygous for the tr-Ac. DNA pools were made from two families that carried the same tr-Ac to ensure that the Ac-containing band was heritable. DNA extraction was performed as described by Chen and Dellaporta (1994). Restriction enzyme digests were performed according to the manufacturer's recommendation using 3–5 μg of genomic DNA in a 20-μl total reaction volume (Promega, Madison, WI). Digests were fractionated overnight on 0.8% standard electrophoresis-grade Low EEO agarose (Fisher Scientific, Fair Lawn, NJ) gels containing ethidium bromide in Tris-acetate EDTA (TAE) buffer along with digoxygenin (DIG)-labeled Roche Ladder VII (Roche Applied Science, Indianapolis, IN). The digests were then transferred to Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Piscataway, NJ). Filters were hybridized sequentially with the internal 700 bp (Ac700) and 900 bp (Ac900) EcoRI-HindIII fragments of Ac. DIG-labeled DNA fragments were synthesized according to the manufacturer's recommendations, using the DIG Probe Synthesis kit (Roche Applied Science) with Ac-specific primers TBp38 and TBp39 for Ac700 and TBp40 and TBp41 for Ac900 fragments (Table 1). Hybridizations were performed as described (Sawers et al. 2002) and the filters were imaged using the Kodak Image Station 440 CR chemiluminescence detection system (Eastman Kodak, Rochester, NY). All of the lines were examined using the restriction enzyme EcoRI, and in some cases additional digestions were performed with the methylation-sensitive enzymes PstI, SalI, or BamHI.
TABLE 1.
PCR primer sequences
| First-round PCR primers | Protocola | Second-round PCR primers | Protocola |
|---|---|---|---|
| Inverse PCR | |||
| Ac10: TGAACTTGGTTGCAAAGGATG GCTTG |
iPCR-2a, iPCR-3a |
Ac11: GGGTTCGAAATCGATCGGG ATAAAACT |
iPCR-2a |
| Ac14: TCCACTCCTCGGCTTTAGGA CAAATTG |
iPCR-2b, iPCR-3b |
Ac12: GCAGGAACAATTGAGAAA ATCAAAGCG |
iPCR-2a |
| Ac18: ACGAAACGGGATCATCCCGAT TAAAAAC |
iPCR-2a, iPCR-3a, iPCR-3c |
Ac15: AGGTATTTTACCGACCGTT ACCGACCG |
iPCR-2b |
| JGp2: CCGGTTCCCGTCCGATTTCG | iPCR-1b, iPCR-1c |
Ac16: AATTGAGACAAACATACCTG CGAGGA |
iPCR-2b |
| JGp3: ACCCGACCGGATCGTATCGG | iPCR-2b, iPCR-3b, iPCR-3c |
JGp3: ACCCGACCGGATCGTA TCGG |
iPCR-1b, iPCR-1c |
| TBp35: GTCGGGAAACTAGCTCT ACCG |
iPCR-1a, iPCR-1c |
TBp32: CAAACATACCTGCGAGGA TCAC |
iPCR-1b |
| TBp42: GGCTGTAATTGCAGGAAC AATTG |
iPCR-1a | TBp34: ACCTCGGGTTCGAAATC GATCGG |
iPCR-1a, iPCR-1c |
| TBp43: GAATTTATAATGATGACATG TACAAC |
iPCR-1b | TBp37: TAATGAAGTGTGCTAGTG AATGTG |
iPCR-1a |
| Ac700 fragment | Protocolb | Ac900 fragment | Protocolb |
| Probe synthesis and PCR verification | |||
| Ac700 (5′): TTTCCCATCCTACTTTCA TCCCTG |
PCR verification | Ac900 (3′): CGTTACCGACCGTTTT CATCCCTA |
PCR verification, probe synthesis |
| TBp38: AAGCTTCATTTGTCAATAAT CATG |
Ac700 probe | TBp40: GAATTCAACCTATTTGAT GTTGAG |
Ac900 probe |
| TBp39: CATCTAGTTGAGACATCATA TGAG |
Ac700 probe | TBp41: CAACAATCTCCGAACCAA GACG |
Ac900 probe |
Ac-specific primers for iPCR-1, iPCR-2, and iPCR-3 are listed; primers used to amplify sequences flanking (a) 5′ (BamHI) end of Ac of EcoRI-digested fragments, (b) 3′ end of Ac of EcoRI-digested fragments, or (c) both ends of Ac of PstI-digested fragments are shown.
Ac-specific primers for use in PCR verification protocol and for probe synthesis for use in DNA blot analysis.
Amplification of Ac flanking sequences:
Several methods were employed to obtain sequences flanking tr-Acs, including a modified amplification of insertion mutagenized sites protocol (AIMS; Frey et al. 1998) and three inverse PCR (IPCR) methods (detailed protocols are available at http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html). For all protocols, a 200-μl restriction digest was performed according to the manufacturer's recommendations (Promega) using 15 μg of DNA. The digest was fractionated overnight on an agarose gel as described above. DNA from the appropriately sized region was gel extracted and purified using the GeneCleanIII kit (Qbiogene, Vista, CA), following the protocol provided by the manufacturer. For all three IPCR techniques, ∼20 ng of the purified product was self-ligated using T4 DNA ligase in a 50-μl reaction (Promega). The ligation reactions were used as template DNA for one of the following methods. All PCR reactions were performed using Mastercycle Gradient PCR machines (Eppendorf, Westbury, NY).
iPCR-1:
A standard IPCR protocol (Ochman et al. 1988) using two rounds of PCR with nested primers (Table 1) was executed to amplify flanking sequences up to ∼2 kb in size. Approximately 20 ng of ligation products was added to a 50-μl reaction mix containing 2 μl DMSO (Fisher Scientific), 2.5 units Taq (Promega), 0.2 mm dNTPs (Promega), 0.5 μm primers, and 1× Promega Buffer with MgCl2. The DNA was denatured at 94° for 2 min, followed by 30 cycles of 94° for 30 sec, 57° for 30 sec, and 72° for 1 min (+ 1 min for every kilobase of flanking sequence) and one cycle of 72° for 10 min. Products from the first round of iPCR-1 were diluted in water (1:200), and 1 μl of the dilution was used for the second round of PCR with nested primers (Table 1), using the same cycling program as round one.
iPCR-2:
This method is a modified touchdown PCR protocol (McPherson and Møller 2000), using two rounds of PCR with nested primers (Table 1). Fifty microliters of 1× TE pH 8.0 was added to the ligation reaction and placed at 70° for 10 min. The samples were purified using the QIAGEN (Valencia, CA) nucleotide removal kit and resuspended in 50 μl of 1× TE pH 8. Approximately 0.5 ng of purified ligation product was added to a 50-μl reaction mix containing 2 μl DMSO (Fisher Scientific), 2.5 units Taq (Promega), 0.2 mm dNTPs (Promega), 0.2 μm primers, and 1× Promega Buffer containing 15 mm MgCl2. The PCR reaction was denatured at 96° for 3 min; followed by 15 cycles of 94° for 45 sec, 72° for 1 min less 1° per cycle, and 72° for up to 5 min (1 min per kilobase); followed by 16 cycles of 94° for 45 sec, 58° for 1 min, and 72° for up to 5 min with a final cycle of 72° for 10 min. Products from the first round of IPCR were diluted (1:50), and 10 μl of dilution was used for the second round of PCR with nested primers (Table 1), using the same cycling protocol as round one.
iPCR-3:
A long-range IPCR protocol (Barnes 1994) was used to amplify fragments with flanking sequences that were up to 8.0 kb. Approximately 3 ng of the ligated DNA template was added to a 20-μl PCR mix containing 1× buffer, 2 mm MgSO4, 0.3 mm dNTPs, 0.8 μl DMSO, 0.5 mm beteine, 0.3 μm primers, 0.5 units of Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA), and dH2O to a final volume of 20 μl. Reaction conditions included 1 cycle of 94° for 4 min; followed by 10 cycles of 94° for 10 sec, 58° for 1 min, and 68° for up to 12 min (1 min per kilobase); 25 cycles of 94° for 10 sec, 58° for 1 min, and 68° for up to 12 min + 10 sec per cycle; and 1 cycle of 72° for 20 min. Only one round of inverse long-range PCR was performed with the Ac primers listed in Table 1.
AIMS:
The AIMS protocol used in this study was based on the method developed by Frey et al. (1998) with several modifications. Following a 200- to 300-ng genomic DNA digestion, adaptors with one of four selective bases were ligated to the ends of the fragment. After 12 cycles of PCR with a biotinylated primer complementary to one of two Ac end fragments, the products were bound to Streptavidin-coated magnetic beads to enrich for PCR products containing the Ac end. Sequences flanking the Ac insertion were amplified using a nested Ac end primer and an adaptor primer.
Cloning Ac flanking sequences:
Following PCR amplification, products were separated on a 0.8% agarose gel and purified using the QIAquick gel extraction kit (QIAGEN). DNA was cloned into either pGEM-T Easy (Promega) or the TOPO TA cloning kit for sequencing (Invitrogen) and sequenced as previously described (Singh et al. 2003).
Verification of cloned flanking sequences:
For each flanking sequence, a primer pair was designed to the corresponding Ac end and the flanking sequence using PrimerSelect (DNASTAR, Madison, WI) or Primer3 (www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi/) (see Table 1 for Ac end primers and http://bti.cornell.edu/Brutnell_lab2/BMGG_home.html for sequences of flanking primers). Primers were selected to yield a 150- to 300-bp product. PCR conditions were as described above for iPCR-1 and amplification products were fractionated on a 1% agarose gel. DNA blot analysis was performed as described above using DIG-labeled DNA fragments generated using primers designed for PCR verification.
Recombinant inbred mapping:
Panels of filters were created that contained parental DNA samples from the IBM94 (B73 × Mo17; Lee et al. 2002) and BNL96 (CM37 × T232 and Co159 × Tx303; Burr et al. 1988) populations. Each parental DNA sample was digested with EcoRI, HindIII, EcoRV, SacI, PstI, BglII, BamHI, and XhoI and hybridized with DIG-labeled Ac flanking sequences described above to identify RFLPs. Progeny blots containing DNA from 94 recombinant inbred (RI) lines of the IBM94 population, 47 lines of the CM37 × T232 population, or 41 lines of the Co159 × Tx303 population were used to score segregation. When polymorphisms existed between B73 and Mo17, the IBM94 population was scored because this population is now serving as a framework for multiple maize genomics projects (Sanchez-Villeda et al. 2003). However, we also utilized the BNL96 populations to place nine Ac elements. All map position data can be obtained at our project website (http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html), as well as at the Maize Genetics and Genomics Database website (MaizeGDB; http://www.maizegdb.org). To access the map position data, the complete Ac name (e.g., mon00150::Ac) should be used as a search term on the MaizeGDB website. The coordinate position for each Ac will be listed relative to markers on the IBM RI populations.
Genomic analysis of sequences flanking tr-Acs:
The sequences flanking tr-Acs were submitted to the NCBI for TBLASTX analysis to the NR, EST, GSS, HTGS, and dbSTS databases. Sequence scores were compared for most significant hit (e < 10−15) and putative function (Tables 2 and 3). A chi-square goodness-of-fit was used to examine distribution of Ac elements throughout the genome. Each of the Ac flanking sequences was also submitted to MaizeGDB for assembly into genome annotations of GSS assemblies (http://www.plantgdb.org/prj/AcDsTagging/).
TABLE 2.
Description of clonedtr-Acs, including the most significant TBLASTX homology to GenBank
| No. | tr-Ac | GenBank accession |
Bin location | Restriction enzyme |
Cloning method |
Fragment size (kb) |
Zea mays Accessiona |
E-value | Library sourceb |
|---|---|---|---|---|---|---|---|---|---|
| Mapped tr-Ac | |||||||||
| 1 | bti00191 | AY559204 | 2.02 | PstI | iPCR-1 | 6.1 | CC630662 | e−113 | MF |
| 2 | bti00207 | AY618476 | 4.07/4.08 | EcoRI (Ac900) | iPCR-1 | 4.6 | BZ788264 | 0.0 | High CoT |
| 3 | bti00209 | AY559195 | 4.06 | EcoRI (Ac900) | iPCR-1 | 2.7 | CG379427 | 3e−88 | MF |
| 4 | bti00226 | AY559177 | 6.00 | EcoRI (Ac900) | iPCR-1 | 4.1 | CC384514 | 3e−45 | High CoT |
| 5 | bti00245 | AY559181 | 4.08 | EcoRI (Ac700) | iPCR-1 | 3.1 | CC368252 | e−104 | High CoT |
| 6 | bti00252 | AY559208 | 1.04/1.05 | PstI | iPCR-1 | 5.4 | CG111052 | 2e−80 | High CoT |
| 7 | bti00257 | AY559233 | 1.05 | EcoRI (Ac900) | iPCR-1 | 3.1 | CG319773 | e−148 | MF |
| 8 | bti95004 | AY559216 | 1.02/1.03 | EcoRI (Ac900) | iPCR-1 | 3.8 | CG178263 | 6e−75 | High CoT |
| 9 | bti95006 | AY559192 | 1.03 | EcoRI (Ac900) | iPCR-1 | 3.8 | BZ819698 | e−116 | High CoT |
| 10 | bti95076 | AY559207 | 6.05 | EcoRI (Ac900) | iPCR-1 | 3.0 | CG007394 | e−118 | UF |
| 11 | bti99224 | AY559201 | 10.01/10.02 | EcoRI/KpnI | iPCR-1 | 2.0 | CC637600 | 6e−76 | MF |
| 12 | mon02901 | AY559179 | 6.04 | EcoRI (Ac900) | iPCR-1 | 3.2 | BZ532959 | e−118 | MF |
| 13 | mon03068 | AY618479 | 7.02 | EcoRI (Ac900) | iPCR-1 | 3.2 | CC617664 | e−175 | MF |
| 14 | mon03073 | AY559213 | 4.06 | EcoRI (Ac900) | iPCR-1 | 3.4 | CC415995 | 4e−80 | High CoT |
| 15 | mon03077 | AY618478 | 1.01 | EcoRI (Ac900) | iPCR-1 | 3.4 | CF633631 | e−122 | cDNA |
| 16 | mon03080 | AY618477 | 1.02 | EcoRI (Ac700) | iPCR-2 | 3.1 | CG159118 | 3e−75 | High CoT |
| 17 | mon03082 | AY559210 | 5.08/5.09 | EcoRI (Ac700) | iPCR-1 | 4.0 | CC649598 | e−180 | MF |
| 18 | mon00004 | AY559178 | 9.06/9.07 | EcoRI (Ac900) | iPCR-1 | 3.0 | CC613709 | e−134 | MF |
| 19 | mon00012 | AY559231 | 6.01 | EcoRI (Ac700) | iPCR-1 | 3.6 | BH895218 | 2e−99 | Mu |
| 20 | mon00030 | AY559197 | 5.03/5.04 | EcoRI (Ac900) | iPCR-2 | 3.8 | CG129443 | 4e−55 | High CoT |
| 21 | mon00038 | AY559229 | 6.01/6.02 | EcoRI (Ac900) | iPCR-2 | 6.2 | CG164865 | e−161 | High CoT |
| 22 | mon00044 | AY559219 | 5.06 | EcoRI (Ac900) | iPCR-1 | 3.0 | CC626710 | e−170 | MF |
| 23 | mon00060 | AY559220 | 7.04 | PstI | AIMS | 6.3 | CC624521 | 5e−18 | MF |
| 24 | mon00068 | AY559191 | 1.05 | PstI | AIMS | 5.1 | CC698616 | e−125 | MF |
| 25 | mon00072 | AY618471 | 7.03 | EcoRI (Ac700) | iPCR-3 | 6.1 | CG436891 | 3e−77 | MF |
| 26 | mon00084 | AY618474 | 2.08 | EcoRI (Ac700) | iPCR-3 | 6.1 | — | — | — |
| 27 | mon00092 | AY559212 | 9.07/9.08 | EcoRI (Ac700) | iPCR-2 | 4.6 | CG130353 | e−132 | High CoT |
| 28 | mon00098 | AY559226 | 7.04 | EcoRI (Ac700) | iPCR-1 | 2.9 | CG187250 | 6e−24 | High CoT |
| 29 | mon00106 | AY559176 | 1.03 | EcoRI (Ac900) | iPCR-3 | 8.0 | CC607430 | e−174 | MF |
| 30 | mon00108 | AY559206 | 8.07 | PstI | AIMS | 6.3 | CC709951 | 3e−38 | MF |
| 31 | mon00110 | AY559183 | 8.02 | EcoRI (Ac900) | AIMS | 2.8 | CC711298 | 5e−88 | MF |
| 32 | mon00126 | AY559211 | 4.02/4.03 | PstI | AIMS | 5.0 | CG375880 | 1e−61 | MF |
| 33 | mon00152 | AY559217 | 5.04/5.05 | EcoRI (Ac900) | iPCR-3 | 10.0 | BZ637224 | e−117 | MF |
| 34 | mon00178 | AY559203 | 3.05 | EcoRI (Ac900) | iPCR-2 | 3.4 | CG045942 | 6e−42 | High CoT |
| 35 | mon00186 | AY559221 | 1.06/1.07 | EcoRI (Ac700) | iPCR-3 | 5.7 | CC804870 | 1e−37 | UF |
| 36 | mon00192 | AY559223 | 1.03 | EcoRI (Ac700) | iPCR-3 | 7.4 | CC435538 | 1e−29 | High CoT |
| 37 | mon00200 | AY559202 | 1.10/1.11 | EcoRI (Ac900) | iPCR-3 | 9.5 | CC011003 | 6e−48 | High CoT |
| 38 | mon00212 | AY559232 | 5.01 | PstI | AIMS | 5.4 | CC375028 | e−141 | High CoT |
| 39 | mon00218 | AY559185 | 10.07 | EcoRI (Ac700) | iPCR-2 | 5.5 | CC718212 | 8e−93 | MF |
| 40 | mon00236 | AY559188 | 9.03 | EcoRI (Ac700) | iPCR-3 | 7.4 | CC441960 | 2e−71 | High CoT |
| 41 | mon00238 | AY559194 | 5.06 | EcoRI (Ac900) | AIMS | 5.2 | CC614740 | 7e−66 | MF |
| Repetitive tr-Ac | |||||||||
| 42 | bti00190 | AY559199 | Unknown | EcoRI (Ac900) | iPCR-1 | 3.1 | CC328152 | e−153 | MF |
| 43 | bti00194 | AY559209 | Unknown | EcoRI (Ac900) | iPCR-1 | 2.8 | CG291158 | e−151 | MF |
| 44 | bti00220 | AY559218 | Unknown | EcoRI (Ac700) | iPCR-1 | 4.7 | CC646925 | e−169 | MF |
| 45 | bti00225 | AY559205 | Unknown | EcoRI (Ac700) | iPCR-1 | 3.7 | — | — | — |
| 46 | bti00228 | AY618475 | Unknown | PstI | iPCR-1 | 6.8 | CG018677 | e−156 | MF |
| 47 | bti00238 | AY559172 | Unknown | EcoRI (Ac900) | iPCR-1 | 3.5 | CG311429 | 0.0 | MF |
| 48 | bti00242 | AY559198 | Unknown | EcoRI (Ac700) | iPCR-1 | 3.0 | CC808672 | 2e−36 | BAC |
| 49 | bti00256 | AY559227 | Unknown | EcoRI (Ac900) | iPCR-1 | 3.7 | CC735442 | 1e−44 | MF |
| 50 | bti99221 | AY618472 | Unknown | EcoRI (Ac900) | iPCR-1 | 2.7 | CC890275 | 2e−72 | BAC |
| 51 | mon00002 | AY559182 | Unknown | EcoRI (Ac700) | iPCR-2 | 5.2 | CG308013 | 1e−76 | MF |
| 52 | mon00020 | AY559174 | Unknown | EcoRI (Ac900) | AIMS | 3.0 | CC004576 | 2e−48 | Genomic |
| 53 | mon00028 | AY559200 | Unknown | EcoRI (Ac900) | iPCR-2 | 4.4 | CG222531 | 5e−71 | MF |
| 54 | mon00042 | AY559180 | Unknown | EcoRI (Ac900) | iPCR-2 | 2.4 | — | — | — |
| Repetitive tr-Ac | |||||||||
| 55 | mon00054 | AY559184 | Unknown | BamHI | AIMS | 5.2 | CG031998 | 4e−47 | High CoT |
| 56 | mon00058 | AY559230 | Unknown | PstI | AIMS | 5.0 | CC445930 | 2e−19 | High CoT |
| 57 | mon00066 | AY559186 | Unknown | EcoRI (Ac900) | iPCR-2 | 4.2 | CG240822 | 1e−34 | MF |
| 58 | mon00070 | AY559193 | Unknown | EcoRI (Ac900) | iPCR-1 | 2.7 | CC756012 | 4e−25 | cDNA |
| 59 | mon00080 | AY559196 | Unknown | EcoRI (Ac900) | iPCR-2 | 4.0 | BZ974435 | 3e−57 | High CoT |
| 60 | mon00102 | AY559173 | Unknown | EcoRI (Ac900) | iPCR-1 | 2.8 | CG371472 | e−111 | MF |
| 61 | mon00128 | AY618473 | Unknown | EcoRI (Ac900) | iPCR-3 | 3.3 | BZ809673 | e−139 | High CoT |
| 62 | mon00132 | AY559189 | Unknown | EcoRI (Ac900) | iPCR-3 | 10.0 | CC428556 | e−103 | High CoT |
| 63 | mon00160 | AY559190 | Unknown | EcoRI (Ac700) | iPCR-3 | 7.0 | BH408973 | 3e−85 | Mu |
| 64 | mon00166 | AY559225 | Unknown | PstI | iPCR-3 | 8.4 | BZ801148 | e−100 | High CoT |
| 65 | mon00168 | AY559214 | Unknown | EcoRI (Ac900) | iPCR-2 | 3.0 | CC696403 | 2e−71 | MF |
| 66 | mon00194 | AY559234 | Unknown | EcoRI (Ac700) | iPCR-3 | 6.2 | CG178282 | 2e−79 | High CoT |
| 67 | mon00204 | AY559228 | Unknown | EcoRI (Ac900) | AIMS | 3.6 | CC357240 | e−111 | High CoT |
| 68 | mon00210 | AY559187 | Unknown | EcoRI (Ac900) | iPCR-2 | 3.3 | BZ828425 | 2e−70 | High CoT |
| 69 | mon00240 | AY559224 | Unknown | PstI | iPCR-3 | 8.6 | BZ817976 | 3e−51 | High CoT |
Most significant homologies identified in TBLASTX searches of public databases.
Methylation filtration (MF), unfiltered (UF), and high-CoT libraries; rescue Mu flanking sequence libraries (Mu); or GenBank database.
TABLE 3.
Putative function ofAc flanking sequences and insertion site location (in parentheses), if known
| tr-Ac | Database | E-value | Accession | Putative function |
|---|---|---|---|---|
| Mapped tr-Acs | ||||
| bti00207 | nr | 5e−25 | AF466202 | Maize putative pol protein gene |
| bti00209 | est | 3e−22 | AU067887 | Rice unknown gene |
| bti00226 | nr | 2e−43 | AF413200 | Maize d8 gene (promotor and 5′-UTR region) |
| bti00245 | est | 6e−46 | BQ163759 | Maize unknown gene |
| mon03077 | est | e−122 | CF633631 | Maize unknown gene |
| mon03082 | nr | 2e−76 | AY105664 | Maize unknown gene |
| mon00012 | nr | 5e−69 | AY105294 | Maize unknown gene |
| mon00044 | nr | 1e−59 | NM_122227 | Arabidopsis serine carboxypeptidase-related gene (coding sequence) |
| mon00060 | nr | 9e−18 | CA619151 | Wheat unknown gene |
| mon00110 | nr | 8e−45 | AF100769 | Maize receptor-like kinase gene (5′-UTR region) |
| mon00200 | est | 2e−23 | BF728928 | Maize unknown gene |
| mon00212 | est | 5e−37 | AW231322 | Maize unknown gene |
| mon00218 | est | 4e−40 | CF244390 | Maize unknown gene |
| Repetitive tr-Acs | ||||
| bti00190 | nr | e−151 | AF466203 | Maize gypsy-type retrotransposon RIRE2 |
| bti00194 | nr | 3e−15 | AY105224 | Maize unknown gene |
| bti00220 | nr | e−110 | NM_105440 | Arabidopsis leucine-rich repeat family protein (coding sequence) |
| bti00228 | nr | e−106 | AP003022 | Pseudogene, transposable element Txlc protein 2 (putative coding sequence) |
| bti00242 | nr | 1e−25 | AF448416 | Maize retrotransposon Xilon2 LTR in bz genomic region |
| bti00256 | nr | 7e−32 | D63956 | Maize gmlip15 gene (upstream of TATA box) |
| mon00002 | nr | 1e−54 | AP002869 | Rice putative polygalacturonase PG2 gene (coding sequence) |
| mon00020 | nr | 2e−48 | AF546189 | Maize repetitive region in 19-kD zein gene family |
| mon00028 | nr | 1e−29 | AP004693 | Rice putative dioscorin class A precursor (coding sequence) |
| mon00054 | nr | 2e−31 | AY109416 | Maize unknown gene |
| mon00066 | nr | 2e−30 | AF464738 | Maize copia-type pol polyprotein |
| mon00070 | nr | 1e−18 | AF050455 | Maize gypsy/Ty3-type retrotransposon Tekay LTR |
| mon00102 | nr | 2e−63 | AC027037 | Rice putative cellulose synthase (coding sequence) |
| mon00128 | nr | 1e−60 | U57899 | Maize repressor-like protein (in1) gene |
| mon00132 | est | 5e−55 | AI939783 | Maize unknown gene |
| mon00160 | nr | 3e−50 | AP005245 | Rice putative DNA-damage-repair toleration protein (coding sequence) |
| mon00168 | nr | 7e−17 | AY144442 | Sorghum: located between Kaema-2 retrotransposon and MITE |
| mon00194 | nr | 1e−15 | AF010283 | Sorghum: next to solo LTR of retroelement Leviathan |
| mon00204 | est | 6e−46 | BG901272 | Maize unknown gene |
| mon00210 | nr | 4e−40 | AC116033?? | Maize repetitive region in 19-kD zein gene family |
| mon00240 | nr | 7e−32 | L22344 | Maize globulin-1 gene (promoter region) |
Accession numbers:
The sequences flanking the tr-Ac (Table 2) have been submitted to GenBank under the following accession numbers: bti00191, AY559204; bti00207, AY618476; bti00209, AY559195; bti00226, AY559177; bti00245, AY559181; bti00252, AY559208; bti00257, AY559233; bti95004, AY559216; bti95006, AY559192; bti95076, AY559207; bti99224, AY559201; mon02901, AY559179; mon03068, AY618479; mon03073, AY559213; mon03077, AY618478; mon03080, AY618477; mon03082, AY559210; mon00004, AY559178; mon00012, AY559231; mon00030, AY559197; mon00038, AY559229; mon00044, AY559219; mon00060, AY559220; mon00068, AY559191; mon00072, AY618471; mon00084, AY618474; mon00092, AY559212; mon00098, AY559226; mon00106, AY559176; mon00108, AY559206; mon00110, AY559183; mon00126, AY559211; mon00152, AY559217; mon00178, AY559203; mon00186, AY559221; mon00192, AY559223; mon00200, AY559202; mon00212, AY559232; mon00218, AY559185; mon00236, AY559188; mon00238, AY559194; bti00190, AY559199; bti00194, AY559209; bti00220, AY559218; bti00225, AY559205; bti00228, AY618475; bti00238, AY559172; bti00242, AY559198; bti00256, AY559227; bti99221, AY618472; mon00002, AY559182; mon00020, AY559174; mon00028, AY559200; mon00042, AY559180; mon00054, AY559184; mon00058, AY559230; mon00066, AY559186; mon00070, AY559193; mon00080, AY559196; mon00102, AY559173; mon00128, AY618473; mon00132, AY559189; mon00160, AY559190; mon00166, AY559225; mon00168, AY559214; mon00194, AY559234; mon00204, AY559228; mon00210, AY559187; and mon00240, AY559224.
RESULTS
Distribution of Ac elements:
Ac elements were distributed throughout the maize genome utilizing the genetic scheme illustrated in Figure 1. An estimated 400 Ac transpositions (tr-Acs) were selected from two loci, P1-vv (Emerson 1917) located on chromosome 1 (1.03) and bti97156::Ac located on chromosome 5 (5.04; Singh et al. 2003). Transpositions were selected from testcross progeny obtained by crossing plants homozygous for the donor Ac as females with pollen from a reporter line containing a Ds insertion at the r1 locus, r1-sc:m3 (see materials and methods). As illustrated in Figure 1A, the majority of testcross progeny inherit a single active Ac element through the female gametophyte. This results in two copies of Ac in cells of the triploid endosperm corresponding to large and frequent sectors of colored aleurone (coarsely spotted). However, tr-Acs that are inherited with the donor element result in an increased copy number of Ac elements in the endosperm tissue and a dramatic delay in the timing of Ds excision (McClintock 1951). In fact, the presence of four Ac copies in the endosperm often conditions a completely colorless or very finely spotted aleurone (Figure 1B, white arrow). These “near-colorless” kernels were observed at a frequency of ∼2–4% on testcross ears homozygous for Ac insertions in W22 (see materials and methods). As the endosperm and embryo tissues share a common gametophytic lineage, selection of near-colorless aleurone was expected to enrich for Ac insertions that were transmitted to both endosperm and embryo tissues.
To genetically map tr-Ac elements relative to the donor Ac, near-colorless selections were testcrossed to the Ds reporter (Figure 2A). If a tr-Ac shows complete linkage to the donor Ac, then kernels inheriting the parental chromosome with two copies of Ac are expected to condition a colorless or near-colorless aleurone, whereas kernels that inherited the nonrecombinant chromosome are completely colorless due to the absence of Ac activity. However, recombination between the donor Ac and the tr-Ac results in a decrease in Ac copy number and a coarsely spotted aleurone. Thus, by simply counting the number of kernels with coarsely spotted aleurone and dividing by the total number of spotted and near-colorless kernels, this two-point linkage test can be used to approximate the genetic distance between Ac elements. Examples of ears segregating linked and unlinked Ac elements are shown in Figure 2B. The presence of kernels with fully colored aleurone is indicative of premeiotic or gametophytic excision of the Ds reporter from r1-sc:m3 and thus they were excluded from the estimates of linkage. It is important to note that this two-point testcross is a rough estimate of genetic distance. Ac excision or transposition to unlinked sites may also result in coarsely spotted kernels and the overestimation of genetic distance. Nevertheless, it is likely that the linkage estimates are accurate to within 2–4 cM, corresponding to the transposition frequency of Ac (Singh et al. 2003).
Figure 2.—
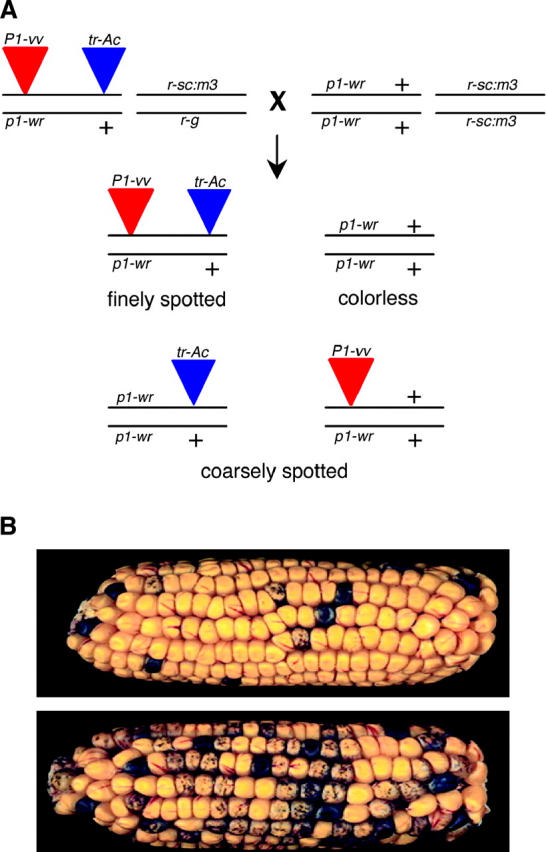
Genetic mapping and segregation of novel tr-Ac. (A) Finely spotted kernels from F1 ears (Figure 1) are testcrossed to the Ds tester (r1-sc:m3) to map the tr-Ac relative to the donor Ac element. The resulting ear will contain kernels that are doubly hemizygous for both Ac elements, hemizygous for one Ac element, or that do not contain any Ac insertions (r1 alleles not shown in schematic). (B) Crosses in which the tr-Ac is linked to the donor Ac will result in few coarsely spotted kernels (top ear). If the tr-Ac is unlinked to the donor Ac, approximately one-half of the kernels will be coarsely spotted (bottom ear).
To select tr-Acs unlinked to the donor Ac, coarsely spotted kernels were chosen from 158 testcross ears in which the donor and tr-Ac were determined to be genetically unlinked. Ten coarsely spotted kernels were selected from each of the testcross ears and these plants were self-pollinated to generate ears that would contain either the segregating donor Ac or the tr-Ac. When P1-vv was used as the donor Ac, ∼50% of the ears inherited the parental Ac and were easily discernible as P1-vv conditions variegated cob and pericarp tissues. Variegated ears were discarded and a single p1-wr ear was selected to represent the unlinked tr-Ac. However, the parental line bti97156::Ac does not condition any obvious mutant phenotype and could be distinguished from the tr-Ac only by using a molecular assay. Leaf tissue was collected and DNA was digested with the methylation-insensitive restriction enzyme EcoRI. DNA blot analysis was performed as described in materials and methods to identify plants that carried either bti97156::Ac or the unlinked tr-Ac. Plants that inherited bti97156::Ac carried a 2.7-kb EcoRI fragment that was detected using the internal 700-bp EcoRI-HindIII fragment of Ac and were discarded. Families that did not contain this band were assumed to carry the unlinked transposition events. All self-pollinated ears were examined for the presence of the p1-wr allele and an aleurone variegation pattern that was consistent with the segregation of a single active Ac element. One representative ear from each of the 158 families that met the above criteria was chosen and kernels with few sectors were selected as representing kernels homozygous for a single Ac insertion (Figure 3). These plants were self-pollinated and seed from 3–5 ears from each line was collected.
Figure 3.—
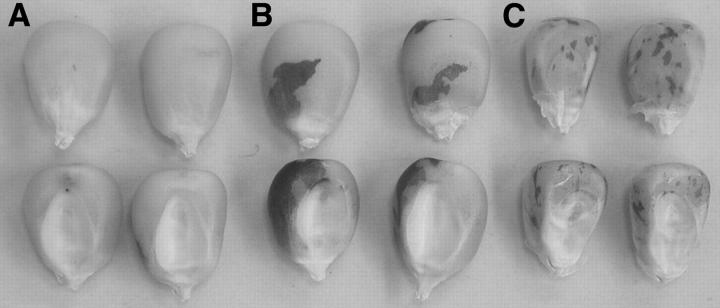
Variation in Ds-mediated variegation pattern. Kernels homozygous for three independent Ac insertions, (A) mon00044::Ac, (B) mon00060::Ac, (C) mon03073::Ac, display different patterns of aleurone variegation. All kernels are homozygous for the Ds reporter at r1 (r1-sc:m3) and are maintained in the W22 inbred.
As mentioned above, the selection for transposition was based on the negative dosage effect, whereby increasing copies of Ac results in a delay in the timing of Ds or Ac excision events in maize. This dominant inhibition of transposition is likely to be mediated in part by post-translational control of Ac activity (Heinlein 1996), but the mechanism remains poorly understood. Close observation of aleurone tissues revealed a continuum of variegation patterns conditioned by independent tr-Ac insertions. At one extreme, the aleurone variegation pattern consisted of a few large colored sectors and no fine spots due to Ds excision events that occurred early but not late in endosperm development. In other lines, a finely spotted aleurone variegation pattern was observed, suggesting a suppression of Ac activity early but not late in endosperm development. Figure 3 shows representative kernels homozygous for each of three different Ac insertions. Although the Ac copy number is the same in each kernel, there are clear differences in the Ds-mediated aleurone spotting patterns. In each case, triploid endosperm cells carry three copies of a single active Ac, yet the timing of Ds excision varies among lines. As these lines have been maintained in a uniform genetic background and are homozygous for the Ds reporter gene, r1-sc:m3, the variation is unlikely to be attributable to segregating modifier loci. This variation may be attributable to spontaneous alterations of transposon activity associated with changes in DNA methylation (Brutnell and Dellaporta 1994; Brutnell et al. 1997). Alternatively, it is possible that cis effects corresponding to the proximity of tr-Ac to enhancer elements or heterochromatic regions of the genome mediate these differences in variegation pattern. Similar position-dependent Ac variegation patterns have been reported in maize (Chomet 1988), suggesting that the weak promoter activity of Ac is modulated to some extent by the chromosomal context of the element. It is important to note that if a tr-Ac did not contribute to the negative dosage effect, it would have escaped our visual screen that relied on reduced kernel variegation. That is, “silenced” tr-Acs inherited with the donor Ac would be phenotypically indistinguishable from kernels that inherited the donor Ac alone or would have resulted in ears that lacked any Ac activity if the silenced tr-Ac cosegregated with the empty donor site. In either case, these potential Ac inactivation events would not have been recovered.
Isolation and characterization of sequences flanking active Ac elements:
The two-point testcross mapping described above enabled selection for 158 Ac transpositions that were genetically unlinked to donor elements. To precisely determine the location of each tr-Ac, we developed a two-step procedure to first identify the tr-Ac through DNA blot analysis and then amplify the flanking DNA for cloning, using several PCR protocols. DNA was first extracted from pooled seedling tissues and DNA blot analysis was performed as described in materials and methods. Both methylation-sensitive and -insensitive restriction enzymes were used to increase the likelihood of detecting an Ac-containing band. Ac-containing bands not detected in either progenitor line were candidates for representing the tr-Ac (Figure 4). Use of the methylation-insensitive enzyme EcoRI resulted in the fractionation of both heavily methylated cryptic Ac elements (Chomet et al. 1987) and hypomethylated active Ac elements (Figure 4). However, because the lines are highly inbred and the cryptic Ac elements are by definition incapable of transposition, they can be easily distinguished from the active Ac bands as those that are common among all Ac lines. DNA blot analysis was performed on progeny from two ears for each of the 158 tr-Acs. A putative Ac insertion that was present in only one of the two families was considered likely to represent either a novel transposition event or a somatic transposition event and the corresponding ears were discarded (Figure 4, lane 1). Lines that carried an Ac-containing restriction fragment present in two families but not in either progenitor were advanced for cloning, sequencing, and mapping (e.g., Figure 4, lanes 9 and 10). Of the 158 tr-Acs, a novel Ac fragment was identified in 114 tr-Ac lines. We then attempted to clone and sequence the DNA flanking each of these unique Ac insertions through one of several PCR-based methods.
Figure 4.—
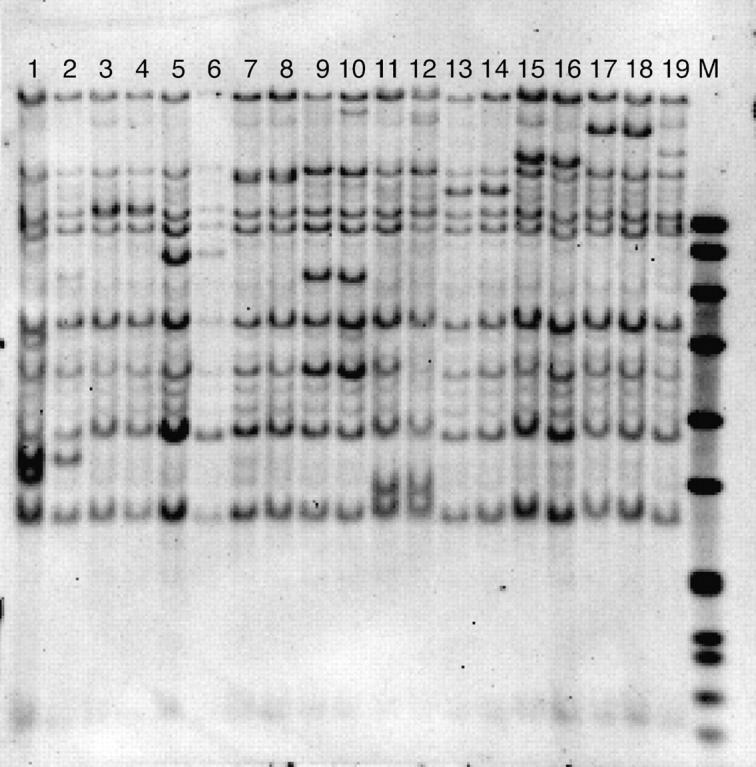
Identification of novel tr-Ac elements in DNA blot analysis. DNA blot analysis was performed with genomic DNA from pooled seedlings derived from two independent ears that each carry the same tr-Ac (lanes 1–18; e.g., lanes 1 and 2 are expected to carry the same tr-Ac, lanes 3 and 4 are expected to carry another tr-Ac), P1-vv (lane 19), and DIG-labeled DNA VII ladder (lane 20; Roche). DNA was digested with EcoRI and hybridized to the internal 700-bp EcoRI/HindIII fragment of Ac. The active, germinally inherited tr-Acs are detected as novel bands present only in the two lines harboring the Ac insertion.
To clone Ac flanking sequences, several PCR-based methods were utilized over the course of this project (detailed protocols available at http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html). All strategies relied on first detecting an Ac-containing restriction fragment in DNA blot analysis. Ultimately, two methods were deemed highly robust and used to clone the majority of Ac-flanking sequences that varied in size from 200 bp to 8 kb (see materials and methods). A modified AIMS protocol was used to amplify the flanking sequence from 12 tr-Acs (Frey et al. 1998), but was expensive and labor intensive, whereas iPCR-1 was an unreliable method. We now routinely use iPCR-2 to amplify sequences from 200 bp to 4 kb of sequence flanking Ac. iPCR-3 has been used to amplify sequences up to ∼8.0 kb in size. Following PCR amplification, DNA fragments were cloned and sequenced (see materials and methods).
As mentioned above, cryptic Ac and Ds elements with high sequence similarity to active elements are distributed throughout the maize genome (Fedoroff et al. 1983; Leu et al. 1992). In addition, somatic transposition of Ac will create a population of sequences that flank active Ac elements, but that are not inherited. Thus, a PCR verification method was developed to discriminate between sequences flanking active and either Ds or cryptic Ac elements and to distinguish between somatic and germinally transmitted events. Examples of the verification PCR assay are shown in Figure 5B. Regions flanking each tr-Ac were sequenced and used to design a tr-Ac flanking sequence-specific primer (see materials and methods). PCR reactions were performed with this tr-Ac flanking sequence primer and an Ac end primer using DNA derived from progenitor lines in addition to DNA pools derived from two families carrying the same tr-Ac. Amplification products from the progenitor lines would indicate that sequences flanking either a Ds or cryptic Ac element were cloned. Forty-five of 114 cloned fragments were determined to be flanking cryptic elements on the basis of this assay and were not characterized further. Amplification of a band of the predicted size in one of the two progeny samples and the absence of bands in the progenitor lines indicated that a somatic transposition event had been recovered. Two flanking sequences were scored as somatic transposition events on the basis of this assay. Only when PCR products of the appropriate size were amplified from both progeny samples and a band was absent from the progenitor lanes was a tr-Ac considered to be an active, germinally heritable element (as shown in Figure 5B). Sixty-nine tr-Acs from the 114 tr-Acs identified by DNA blot fulfilled these criteria and were considered likely to represent novel and heritable tr-Acs.
Figure 5.—
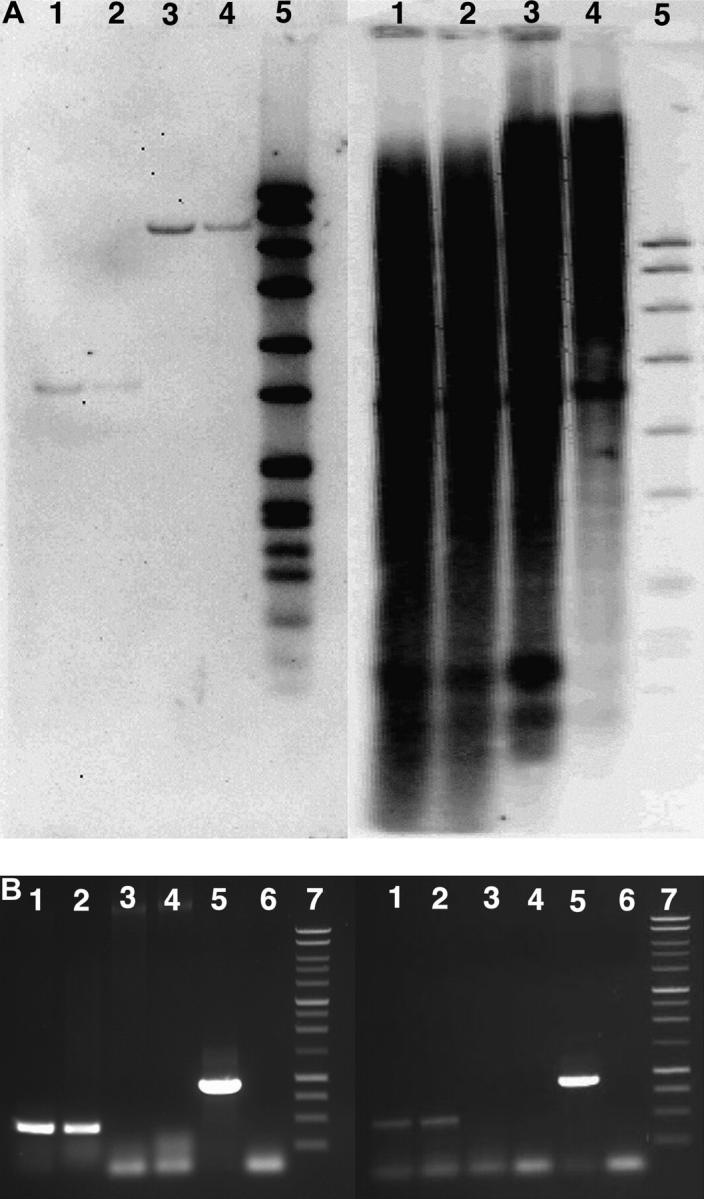
Verification analysis. (A) DNA blot verification. Genomic DNA was isolated from plants carrying bti00245::Ac (left, lanes 1 and 2) or bti00194::Ac (right, lanes 1 and 2) and from progenitor Ac-containing P1-vv (lanes 3) and the Ds reporter plants (lanes 4). Lane 5 contains DIG-labeled DNA VII ladder (Roche). DNA was digested with EcoRI and fractionated on 0.8% agarose gels. DNA blot analysis was performed with a DIG-labeled flanking sequence probe of 443 bp (bti00245::Ac) or 432 bp (bti00194::Ac) generated using the Ac-end primer and flanking sequence-specific primer pairs [DIG-labeled VII ladder (lane 5)]. (B) PCR verification for the presence of bti00245::Ac (left) and bti00194::Ac (right). Verification PCR was performed using DNA from plants that carried the tr-Ac element (lanes 1 and 2), the progenitor P1-vv allele (lane 3), and the r1-sc:m3 allele (lane 4). An Ac-end primer was used with a gene-specific primer to amplify fragments of 443 and 432 bp from bti00245::Ac and bti00194::Ac, respectively. Lanes 5 and 6 represent positive (plasmid template) and negative (no DNA) controls; lane 7 contains a 1-kb DNA ladder (Roche).
To confirm the PCR results, DNA blot analysis was also performed using the DNA flanking sequences as molecular probes. DNA blot analysis was performed using a DIG-labeled flanking sequence probe (see materials and methods). As shown in Figure 5A (left), a shift in RFLP size between DNA from progenitor plants (Figure 5A, lanes 3 and 4) and from plants carrying the bti00245::Ac (Figure 5A, lanes 1 and 2) is observed that corresponds to the introduction of an EcoRI restriction site from the Ac element. This result is consistent with the PCR verification result and indicates that the cloned DNA flanks an active, germinally transmitted Ac element. The presence of a single band in DNA blot analysis also indicates that bti00245::Ac had inserted into a single-copy region of the maize genome.
Surprisingly, sequences flanking several tr-Acs that were verified by the PCR assay were highly repetitive in the maize genome, even though the corresponding PCR amplicon was used as the DNA blot probe. An example of one such hybridization result is shown in Figure 5A (right). Although the PCR verification (Figure 5B, right) indicates that bti00194::Ac is a tr-Ac, the intervening DNA between the Ac end and the flanking sequence primer site is present in multiple copies in the maize genome, as indicated by the smear in DNA blot analysis. Despite our ability to amplify sequences flanking 69 active elements, ∼41% (28) contained DNA sequences that were too repetitive to map using conventional RFLP analysis. Because it was not possible to obtain an independent confirmation through DNA blot analysis, these flanking sequences were not placed on genetic maps. Thus, all Ac flanking sequences that were mapped, as described below, were confirmed as flanking an active Ac element through two independent assays, a PCR verification test and DNA blot analysis.
Mapping and distribution of Ac elements:
As precisely defined map positions are essential to effectively utilizing Ac in gene-tagging experiments, one of three RI populations was utilized to map the tr-Ac insertion sites (see materials and methods). The vast majority of tr-Ac flanking sequences have been placed on an intermated B73 × Mo17 population that provides ∼0.5-cM resolution (Lee et al. 2002). However, 9 of the 41 tr-Ac lines were mapped relative to markers developed for the BNL96 population (Burr et al. 1988). All tr-Ac elements were positioned using the same DIG-labeled flanking sequence probes that were used to show the element was active and heritable in DNA blot analysis. Map bin locations for all 41 tr-Ac lines are listed in Table 2. The precise location of the tr-Ac lines with reference to specific markers can be found on the project website (http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html) or at MaizeGDB (http://www.maizegdb.org).
In summary, ∼400 tr-Acs were selected from two donor loci, of which 158 tr-Acs were determined to be unlinked to their donor Ac. Through DNA blot analysis, we identified RFLPs for 114 of the 158 tr-Acs that were of an appropriate size for cloning, using one of three PCR techniques. Sixty-nine fragments were cloned, sequenced, and used to develop PCR primer pairs to confirm their identity as germinal transposition events. Twenty-eight sequences contained elements that were too repetitive to position using RFLP analysis and 41 were represented in the genome as single- or low-copy DNA (Table 2). These 41 Ac flanking sequences were positioned on one of three recombinant inbred mapping populations. Homozygous stocks of each of these lines have been propagated and have been made available through the Maize Genetics Stock Center (http://www.maizegdb.org/stock.php).
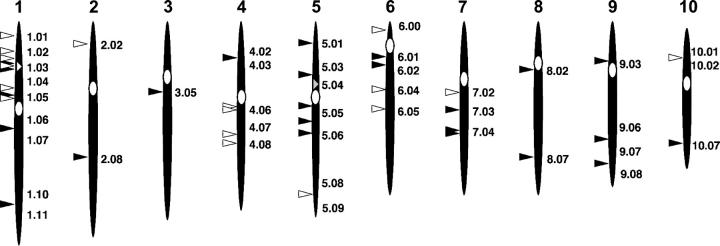
A composite map showing the position of each of the mapped tr-Acs is shown in Figure 6. The tr-Acs are shaded to reflect the donor Ac from which each was derived. One-third of the tr-Acs were derived from P1-vv (Figure 6, open arrowheads), whereas two-thirds of the tr-Acs were derived from bti97156::Ac (Figure 6, solid arrowheads). Insertions were placed on all 10 chromosomes.
Figure 6.—
Schematic of maize chromosomes showing the approximate bin locations of the tr-Ac elements. The locations of the two original donor Acs used to develop new tr-Acs are highlighted by the small open arrowhead and bin location (P1-vv) at 1.03 or the small shaded arrowhead and bin location (bti97156::Ac) at 5.04. The tr-Acs are distributed on each of the 10 chromosomes and are indicated as either open (P1-vv, donor Ac) or solid arrowheads (bti97156::Ac, donor Ac).
A previous study had shown that Ac transpositions unlinked to the bz1 locus often transposed to physically linked sites (i.e., the opposite arm of the same chromosome; Dooner et al. 1994). To examine potential Ac insertion site biases in this study, we examined the distribution of 39 of the 41 tr-Acs among the 100 defined bins of the maize genome. Two Ac elements were genetically mapped as linked to the donor Ac and were not included in this analysis. A chi-square test revealed that the distribution of Ac elements was consistent with a random (Poisson) distribution (χ2 = 0.17; P = 0.919) and suggests that there is no strong regional insertion site bias for Ac transposition from the two donor Acs. However, due to our small sample size only an extremely strong bias would be detectable. Nevertheless, these results suggest that a high-density distribution of Ac throughout the maize genome is feasible using relatively few donor Acs.
Ac inserts preferentially into hypomethylated sequences in the maize genome:
The flanking sequences of all 69 heritable active tr-Ac elements were analyzed for sequence homology through searches of GenBank. All flanking sequences, with the exception of three, bti00225::Ac, mon00042::Ac, and mon00084::Ac, identified a highly significant (e < 10−15) homology to database sequences (Table 2). The majority of sequences with the highest similarity to DNA flanking the 41 mapped tr-Acs were evenly distributed between those found in methylation-filtered (19 sequences) and high-CoT (17 sequences) libraries (Rabinowicz et al. 1999; Yuan et al. 2003). Four of the mapped tr-Acs inserted in DNA sequenced from an unfiltered library, a Mu library, or a cDNA library, and a single mapped Ac flanking sequence did not match any published sequence (e < 10−15; Table 2). Surprisingly, of the 28 flanking sequences that were too repetitive to map by RFLP analysis, 11 were highly similar to sequences present in methylation-filtered libraries and 10 were most similar to sequences present in high-CoT libraries. Five sequences were distributed among several libraries as shown in Table 2 and 2 sequences had no strong sequence similarity to GenBank entries. These results are consistent with previous findings indicating Ac has a strong insertion site bias for hypomethylated, low-copy sequences in the genome (Cowperthwaite et al. 2002). However, our findings suggest that highly repetitive stretches of DNA can be imbedded in these low-copy or hypomethylated islands and that repetitive regions serve as suitable targets for Ac insertion in this context.
BLAST searches were also performed of the GenBank nr and est databases to identify potential Ac insertions into genes of known or unknown function. Of the 41 mapped flanking sequences surveyed, 4 inserted into sequences with strong similarity to genes of known function and 9 were to genes of unknown function (Table 3). The high number of hits to genes with unknown function is reflective of the relatively uncharacterized gene space in maize. GenBank searches of the 28 tr-Acs flanking sequences that were too repetitive to map resulted in 22 significant hits to sequences in the nr database. These included 7 sequences that show high similarity to maize retroelement sequences. Interestingly, 5 of these sequences were also present in the methylation-filtered and high-CoT libraries (see Tables 2 and 3). These results suggests that hypomethylated and low-copy sequences are preferred targets for Ac insertion, but that short stretches of highly repetitive sequence within these hypomethylated clusters are also suitable targets for Ac insertion.
DISCUSSION
Development of Ac insertional mutagenesis in maize:
Through combined classical and molecular genetic approaches we have distributed 158 Ac elements throughout the maize genome. DNA flanking 69 of these elements has been cloned and sequenced and 41 tr-Acs have been placed on one of three recombinant inbred populations. These lines are publicly available (http://w3.aces.uiuc.edu/maize-coop/) and have been developed to facilitate the implementation of Ac-based regional mutagenesis programs in maize. Each Ac line is homozygous for a unique tr-Ac insertion that can be molecularly tracked using both PCR and DNA blot assays and monitored phenotypically with a Ds reporter gene (r1-sc:m3). Detailed molecular and genetic characteristics for each tr-Ac insertion can be found at http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html
Several gene-tagging strategies have been described in maize to disrupt and clone target genes using Ac (Brutnell and Conrad 2003; Dellaporta and Moreno 1994). Because of the low germinal transposition frequency of Ac, these strategies exploit reporter genes to either monitor Ac or Ds excision events from a donor locus or detect an increase in Ac copy number. These strategies also exploit the tendency of Ac/Ds elements to insert at closely linked sites in the genome. This tendency of Ac to insert at physically and genetically closely linked sites has been demonstrated at several loci throughout the maize genome (McClintock 1948; Greenblatt 1984; Dooner and Belachew 1989). Due to this general property of Ac, the 41 tr-Ac lines described here are ideally suited to clone and characterize genes that are closely linked to these donor Acs. To demonstrate the utility of these lines, we have recently cloned and characterized multiple Ac-induced alleles of the Vp7 gene using an Ac donor that mapped to within 4 cM of the Vp7 gene (Singh et al. 2003).
Ac insertions into a multigene family member or the disruption of a single gene controlling a multigenic trait will likely lead to subtle mutant phenotypes that may be detected only in a highly uniform population. Therefore, an extremely important feature of these materials is that they have been maintained in the inbred W22. The uniformity of the genetic materials also facilitates the molecular cloning of Ac flanking sequences as both methylation-sensitive and methylation-insensitive restriction enzymes can be readily utilized to identify novel Ac insertions in the genome (Figure 4). Unlike many lines that harbor highly prolific Mutator elements (May et al. 2003), the mutational load of these Ac lines is relatively low due to the infrequent transposition of Ac and Ds elements. Another important feature of these lines is that they exploit endogenous transposable elements, utilizing classical genetic techniques to mobilize and map insertions. Thus, the growth and propagation of these nontransgenic lines does not necessitate the need for an APHIS permit and compliance with the associated regulations for ensuring genetic isolation of transgenes (http://www.aphis.usda.gov/brs/perstds.html). Thus, these lines can be easily distributed and utilized by researchers in the United States and abroad, facilitating their use in the public research domain.
In addition to developing the genetic resources for regional mutagenesis in maize, we have described two PCR techniques to facilitate the cloning and characterization of Ac-induced mutations. Both techniques exploit the use of an agarose gel purification step to fractionate active Ac insertions from cryptic elements. Nevertheless, despite this purification step and one or two rounds of PCR amplification using two or four Ac-specific primers, several sequences flanking cryptic elements were amplified. Sequences flanking cryptic elements were isolated when both methylation-sensitive and -insensitive restriction enzymes were used, suggesting that some percentage of the inactive Ac derivatives are found in hypomethylated regions of the genome. Although methylation-sensitive enzymes were more effective for cloning flanking sequences linked to active Ac elements, only ∼15% of the fragments cloned could be fractionated with these enzymes, while 85% could be visualized using methylation-insensitive enzymes. Thus, we prefer the use of methylation-insensitive enzymes to identify and clone sequences flanking Ac elements in maize.
The differences between the two favored PCR techniques utilized lie in the use of high-fidelity Taq polymerase (iPCR-3) or a low-cost Taq polymerase (iPCR-2) and one (iPCR-3) vs. two (iPCR-2) rounds of PCR amplification. We found that the incorporation of a high-fidelity Taq and long-range protocol enabled the cloning of sequences up to 8.0 kb from the donor locus. The use of a relatively nonprocessive Taq (Promega) proved satisfactory to amplify flanking sequences of up to 4.0 kb. Importantly, both techniques can be used to efficiently clone sequences flanking most Ac insertions in the maize genome and thus are effective for recovering Ac-flanking sequences in gene-tagging experiments.
Ac insertion site preferences:
Previous studies of Ac transposition in maize have shown that Ac transposes during the replication of mitotic chromosomes (Brink and Nilan 1952; Greenblatt and Brink 1962) into hypomethylated regions of the maize genome (Chen et al. 1987). In a recent survey of 46 Ac insertion sites, Dooner and colleagues found that all Ac transposition events occurred to low- or single-copy regions of the maize genome (Cowperthwaite et al. 2002). However, as reported here, a number of Ac insertions were located in repetitive sequences. A number of possibilities may explain this apparent discrepancy. For one, most of the Ac-flanking sequences cloned in this study were identified using EcoRI, a methylation-insensitive restriction enzyme that rarely discriminates between the hypomethylated low-copy sequences in the genome and the hypermethylated repetitive sequences in the genome. The majority of Ac-flanking sequences characterized in previous studies were identified using methylation-sensitive restriction enzymes that would enrich for Ac insertions in single- or low-copy sequences in the genome. In addition, we utilized a two-step approach to first verify that sequences flanking an Ac element were indeed flanking a heritable, active element and then determined if the sequences were repetitive. In previous studies, sequences that were highly repetitive in DNA blot verification may have been discarded as representing sequences flanking cryptic Ac elements, thus biasing the population for low- or single-copy insertions.
Our finding that Ac occasionally inserts into repetitive elements in the genome that are present in high-CoT and MF libraries suggests that at least a portion of the highly repetitive sequences in the maize genome is hypomethylated and thus a target for Ac insertion. Indeed, our recent characterization of the phytochrome gene family in maize has shown that a Ty3-like insertion is present in the second intron of PhyB2 (Sheehan et al. 2004) and miniature inverted repeat transposable elements (MITES) are often located throughout genic regions (Tikhonov et al. 1999; Zhang et al. 2000; Fu et al. 2001). Ac insertions into retroposons or MITES located within genes could explain why some of the Ac flanking sequences (Table 3) contained repetitive DNA despite a preference for hypomethylated regions of the genome. Despite our inability to position these Ac elements on a genetic map using RFLP analysis, placement to a physical map may be possible as additional maize genomic sequences are assembled.
Variation in Ac dosage effects:
One of the surprising findings was the degree of variation in Ac-mediated Ds variegation patterns. Kernel variegation patterns ranged from nearly colorless to showing relatively large and frequent excision events (Figure 3). As all Ac elements in this study were monitored using the same Ds reporter in near-isogenic lines, it is unlikely that the variation observed is due to variation at the r1-sc:m3 locus or segregating modifier loci. Previous studies of Ac-mediated variegation patterns have indicated that methylation plays an important role in determining the timing of Ac or Ds excision in metastable Ac alleles, corresponding to altered transcriptional activities of Ac (Brutnell and Dellaporta 1994). Furthermore, Ac derivatives such as Ac-st1 (McClintock 1948) and Ac-st2 (Brutnell et al. 1997) that are structurally identical to Ac but display dosage-independent (Ac-st1) or positive dosage effects (Ac-st2) are also altered in their transcriptional profiles relative to active Ac elements (Brutnell 1995). Thus, it is likely that the variation in Ac-mediated excision patterns is due to small changes in the transcriptional activity of Ac across the genome. As the Ac element itself contains weak promoter sequences (Kunze et al. 1987; Fusswinkel et al. 1991), and both Ac and Ds excision is extremely sensitive to small (less than twofold) changes in Ac transcript levels (Brutnell and Dellaporta 1994), these lines may serve as an ideal resource for probing cis-acting elements that control gene expression throughout the maize genome.
Ac-based gene-tagging programs in maize:
The distribution of tr-Ac elements on all 10 chromosomes of maize represents an important first step toward the development of an Ac-based insertional mutagenesis program in maize. To further distribute Ac elements throughout the genome, transposition events have been generated from each linkage group and/or arm from a subset of the 41 “donor” Ac lines and are being precisely positioned on the IBM94 population (http://bti.cornell.edu/Brutnell_lab2/Projects/Tagging/BMGG_pro_currentmap.html). The ultimate goal of this program is to distribute Ac elements at 10–20 cM throughout the maize genome. As genome sequencing efforts proceed (Palmer et al. 2003; Whitelaw et al. 2003), it is likely that tr-Acs will be rapidly placed on physical maps using Ac flanking sequences (e.g., http://www.plantgdb.org/prj/AcDsTagging/). A complete genome sequence of maize will also greatly aid in the development of efficient Ac-based gene-tagging strategies (Brutnell and Conrad 2003) to identify candidate genes that are tightly linked to Ac insertions and thus serve as ideal targets for regional mutagenesis (Fu et al. 2001). Ultimately, the development of an Ac-based forward genetics program to identify genes on the basis of phenotype will greatly complement ongoing reverse genetic programs in maize that exploit gene sequence data to identify Mutator insertions in candidate genes (Brutnell 2002).
Acknowledgments
We thank undergraduate students Kelly Dusinberre, Lauren Putnam, Marika Olson, and Beth Pennell for help in performing several thousand crosses and with genotyping Ac lines. This work was supported by a National Science Foundation Plant Genome Award (DBI-0076892) to T.P.B. and by support from Monsanto Company.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY559172, AY559173, AY559174, AY559175, AY559176, AY559177, AY559178, AY559179, AY559180, AY559181, AY559182, AY559183, AY559184, AY559185, AY559186, AY559187, AY559188, AY559189, AY559190, AY559191, AY559192, AY559193, AY559194, AY559195, AY559196, AY559197, AY559198, AY559199, AY559200, AY559201, AY559202, AY559203, AY559204, AY559205, AY559206, AY559207, AY559208, AY559209, AY559210, AY559211, AY559212, AY559213, AY559214, AY559215, AY559216, AY559217, AY559218, AY559219, AY559220, AY559221, AY559223, AY559224, AY559225, AY559226, AY559227, AY559228, AY559229, AY559230, AY559231, AY559232, AY559233, AY559234, and AY618471, AY618472, AY618473, AY618474, AY618475, AY618476, AY618477, AY618478, AY618479.
References
- Alleman, M., and J. L. Kermicle, 1993. Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics 135: 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma, P., E. Grotewold and T. Peterson, 1992. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I., J. D. Jones and C. Dean, 1993. Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5: 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, W. M., 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. USA 91: 2216–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A. M., T. Page, E. J. Lawson, C. Lister and C. Dean, 1996. Use of Ac as an insertional mutagen in Arabidopsis. Plant J. 9: 935–945. [DOI] [PubMed] [Google Scholar]
- Brink, R. A., and R. A. Nilan, 1952. The relation between light variegated and medium variegated pericarp in maize. Genetics 37: 519–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., 1995 Epigenetic regulation of Activator (Ac) in maize. Ph.D. Thesis, Yale University, New Haven, CT.
- Brutnell, T. P., 2002. Transposon tagging in maize. Funct. Integr. Genomics 2: 4–12. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and L. J. Conrad, 2003. Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236: 157–176. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and S. L. Dellaporta, 1994. Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., B. P. May and S. L. Dellaporta, 1997. The Ac-st2 element of maize exhibits a positive dosage effect and epigenetic regulation. Genetics 147: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, B., F. A. Burr, K. H. Thompson, M. C. Albertson and C. W. Stuber, 1988. Gene mapping with recombinant inbreds in maize. Genetics 118: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., and S. L. Dellaporta, 1994 Urea-based plant DNA miniprep, pp. 526–527 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Chen, J., I. M. Greenblatt and S. L. Dellaporta, 1987. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, H. G., M. S. Choe, S. H. Lee, S. H. Park, J. C. Koo et al., 1999. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 19: 615–623. [DOI] [PubMed] [Google Scholar]
- Chomet, P. S., 1988 Characterization of stable and metastable changes of the maize transposable element, Activator. Ph.D. Thesis, State University of New York, Stony Brook, NY.
- Chomet, P. S., S. Wessler and S. L. Dellaporta, 1987. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., Z. Yuan and V. Sundaresan, 1998. The Indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- Cowperthwaite, M., W. Park, Z. Xu, X. Yan, S. C. Maurais et al., 2002. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S. L., and M. A. Moreno, 1994 Gene tagging with Ac/Ds elements in maize, pp. 219–233 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Dellaporta, S. L., I. M. Greenblatt, J. L. Kermicle, J. B. Hicks and S. R. Wessler, 1988 Molecular cloning of the maize R-nj allele by transposon tagging with Ac, pp. 263–282 in Chromosome Structure and Function: Impact of New Concepts, edited by J. P. Gustafson and R. Appels. Plenum Press, New York.
- DeLong, A., A. Calderon-Urrea and S. L. Dellaporta, 1993. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74: 757–768. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., and A. Belachew, 1989. Transposition pattern of the maize element Ac from the Bz-m2(Ac) allele. Genetics 122: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and J. L. Kermicle, 1971. Structure of the R r tandem duplication in maize. Genetics 67: 437–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., A. Belachew, D. Burgess, S. Harding, M. Ralston et al., 1994. Distribution of unlinked receptor sites for transposed Ac elements from the bz-m2(Ac) allele in maize. Genetics 136: 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, R., 1917. Genetical studies of variegated pericarp in maize. Genetics 2: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N., S. Wessler and M. Shure, 1983. Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235–242. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N. V., D. B. Furtek and O. E. Nelson, 1984. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc. Natl. Acad. Sci. USA 81: 3825–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., C. Stettner and A. Gierl, 1998. A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J. 13: 717–721. [Google Scholar]
- Fu, H., W. Park, X. Yan, Z. Zheng, B. Shen et al., 2001. The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98: 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusswinkel, H., S. Schein, U. Courage, P. Starlinger and R. Kunze, 1991. Detection and abundance of mRNA and protein encoded by transposable element Activator (Ac) in maize. Mol. Gen. Genet. 225: 186–192. [DOI] [PubMed] [Google Scholar]
- Giroux, M. J., J. Shaw, G. Barry, B. G. Cobb, T. Greene et al., 1996. A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93: 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco, R., P. B. Ouwerkerk, R. J. De Kam, C. Sallaud, C. Favalli et al., 2003. Transpositional behaviour of an Ac/Ds system for reverse genetics in rice. Theor. Appl. Genet. 108: 10–24. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, Modulator, in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, I. M., and R. A. Brink, 1962. Twin mutations in medium variegated pericarp maize. Genetics 47: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake, S., E. Vollbrecht and M. Freeling, 1989. Cloning Knotted, the dominant morphological mutant in maize using Ds2 as a transposon tag. EMBO J. 8: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein, M., 1996. Excision patterns of Activator (Ac) and Dissociation (Ds) elements in Zea mays L.: implications for the regulation of transposition. Genetics 144: 1851–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, Jr., D. W., E. Lim, J. Keller, I. Plooy, E. Ralston et al., 1995. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P. D., and V. F. Irish, 2001. The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128: 13–23. [DOI] [PubMed] [Google Scholar]
- Jin, W. Z., S. M. Wang, M. Xu, R. J. Duan and P. Wu, 2004. Characterization of enhancer trap and gene trap harboring Ac/Ds transposon in transgenic rice. J. Zhejiang Univ. Sci. 5: 390–399. [DOI] [PubMed] [Google Scholar]
- Kermicle, J., 1984. Recombination between components of a mutable gene system in maize. Genetics 107: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. M., H. L. Piao, S. J. Park, N. S. Chon, B. I. Je et al., 2004. Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J. 39: 252–263. [DOI] [PubMed] [Google Scholar]
- Klosgen, R. B., A. Gierl, S. Schwarz-Sommer and H. Saedler, 1986. Molecular analysis of the waxy locus of Zea mays. Mol. Gen. Genet. 203: 237–244. [Google Scholar]
- Kolesnik, T., I. Szeverenyi, D. Bachmann, C. S. Kumar, S. Jiang et al., 2004. Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J. 37: 301–314. [DOI] [PubMed] [Google Scholar]
- Kunze, R., U. Stochaj, J. Laufs and P. Starlinger, 1987. Transcription of the transposable element Activator (Ac) of Zea mays L. EMBO J. 6: 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori, T., T. Hirayama, Y. Kiyosue, H. Takabe, S. Mizukado et al., 2004. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37: 897–905. [DOI] [PubMed] [Google Scholar]
- Lechelt, C., T. Peterson, A. Laird, J. Chen, S. L. Dellaporta et al., 1989. Isolation and molecular analysis of the maize P locus. Mol. Gen. Genet. 219: 225–234. [DOI] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Leu, J. Y., Y. H. Sun, Y. K. Lai and J. Chen, 1992. A maize cryptic Ac-homologous sequence derived from an Activator transposable element does not transpose. Mol. Gen. Genet. 233: 411–418. [DOI] [PubMed] [Google Scholar]
- May, B. P., H. Liu, E. Vollbrecht, L. Senior, P. D. Rabinowicz et al., 2003. Maize-targeted mutagenesis: a knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1948. Mutable loci in maize. Carnegie Inst. Wash. Year Book 47: 155–169. [PubMed] [Google Scholar]
- McClintock, B., 1951. Chromosome organization and gene expression. Cold Spring Harbor Symp. Quant. Biol. 16: 13–47. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1955. Controlled mutation in maize. Carnegie Inst. Wash. Year Book 54: 245–255. [PubMed] [Google Scholar]
- McClintock, B., 1964. Aspects of gene regulation in maize. Carnegie Inst. Wash. Year Book 63: 592–602. [Google Scholar]
- McPherson, M. J., and S. G. Møller, 2000 PCR. Springer-Verlag, New York.
- Meissner, R. C., H. Jin, E. Cominelli, M. Denekamp, A. Fuertes et al., 1999. Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11: 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett, P. R., L. Clissold, A. Marocco, P. S. Springer, R. Martienssen et al., 2003. A resource of mapped dissociation launch pads for targeted insertional mutagenesis in the Arabidopsis genome. Plant Physiol. 132: 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., A. S. Gerber and D. L. Hartl, 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, L. E., P. D. Rabinowicz, A. L. O'Shaughnessy, V. S. Balija, L. U. Nascimento et al., 2003. Maize genome sequencing by methylation filtration. Science 302: 2115–2117. [DOI] [PubMed] [Google Scholar]
- Peng, J. R., and N. P. Harberd, 1997. Transposon-associated somatic gai-loss sectors in Arabidopsis. Plant Sci. 130: 181–188. [Google Scholar]
- Rabinowicz, P. D., K. Schutz, N. Dedhia, C. Yordan, L. D. Parnell et al., 1999. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23: 305–308. [DOI] [PubMed] [Google Scholar]
- Ralston, E. J., J. J. English and H. K. Dooner, 1988. Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Villeda, H., S. Schroeder, M. Polacco, M. McMullen, S. Havermann et al., 2003. Development of an integrated laboratory information management system for the maize mapping project. Bioinformatics 19: 2022–2030. [DOI] [PubMed] [Google Scholar]
- Sawers, R. J. H., P. J. Linley, P. R. Farmer, N. P. Hanley, D. E. Costich et al., 2002. elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol. 130: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser, L., A. Roussis, J. Stiller and J. Stougaard, 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schultes, N. P., T. P. Brutnell, A. Allen, S. L. Dellaporta, T. Nelson et al., 1996. Leaf permease1 gene of maize is required for chloroplast development. Plant Cell 8: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan, M. J., P. R. Farmer and T. P. Brutnell, 2004. Structure and expression of maize phytochrome family homeologs. Genetics 167: 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B., Z. Zheng and H. K. Dooner, 2000. A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 97: 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M., P. E. Lewis, K. Hardeman, L. Bai, J. K. Rose et al., 2003. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., and O. Ceska, 1977. The genetic control of flavonoid synthesis in maize. Can. J. Genet. Cytol. 19: 289–302. [Google Scholar]
- Sundaresan, V., P. Springer, T. Volpe, S. Haward, J. D. Jones et al., 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tikhonov, A. P., P. J. SanMiguel, Y. Nakajima, N. M. Gorenstein, J. L. Bennetzen et al., 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96: 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaik, N. W., and R. A. Brink, 1959. Transposition of Modulator, a component of the variegated pericarp allele in maize. Genetics 44: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V., 2000. Saturation mutagenesis using maize transposons. Curr. Opin. Plant Biol. 3: 103–107. [DOI] [PubMed] [Google Scholar]
- Wessler, S. R., G. Baran, M. Varagona and S. L. Dellaporta, 1986. Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J. 5: 2427–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, C. A., W. B. Barbazuk, G. Pertea, A. P. Chan, F. Cheung et al., 2003. Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120. [DOI] [PubMed] [Google Scholar]
- Wu, C., X. Li, W. Yuan, G. Chen, A. Kilian et al., 2003. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35: 418–427. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., P. J. SanMiguel and J. L. Bennetzen, 2003. High-Cot sequence analysis of the maize genome. Plant J. 34: 249–255. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., J. Arbuckle and S. R. Wessler, 2000. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc. Natl. Acad. Sci. USA 97: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. G., Y. P. Fu, H. Xiao, G. C. Hu, H. M. Si et al., 2003. Ac/Ds transposition activity in transgenic rice population and DNA flanking sequences of Ds insertion site. Acta Bot. Sin. 45: 102–107. [Google Scholar]