Abstract
We report here the first successful use of embryonic nuclear transfer to create viable adult Drosophila melanogaster clones. Given the generation time, cost effectiveness, and relative ease of embryonic nuclear transplant in Drosophila, this method can provide an opportunity to further study the constraints on development imposed by transplanting determined or differentiated nuclei.
CLONING at the organismal level refers to the creation of a genetically identical individual from an existing individual, generally through nuclear transfer. Embryonic and somatic nuclear transplantation has been successful to varying degrees in amphibians (Gurdon and Uehlinger 1966), arthropods (Illmensee 1968), fish (Lee et al. 2002), and mammals (Wilmut et al. 2002). This technology can be exploited to create stem cells for use in therapeutic cloning and is being used to increase the production of transgenic mammals producing pharmacologically important compounds. In many cases, the technology is constrained by a lack of fundamental understanding of the nuclear reprogramming events that occur following transplantation, resulting in a high frequency of developmental defects in the cloned offspring (Shi et al. 2003). We have successfully used embryonic nuclear transfer to create viable adult Drosophila clones. Embryos that hatch but fail to develop to adulthood exhibit characteristic developmental defects; hence we can potentially use this system to identify gene mutations or conditions that encourage complete nuclear reprogramming. The developmental programming of nuclei is a fundamental epigenetic process based, in part, on histone modification and packaging so the events involved in nuclear reprogramming in Drosophila are likely conserved across taxa. The method outlined herein provides a straightforward, cost-effective means of studying the effects of epigenetic interactions on nuclear transplants.
Host embryos laid by white-eyed w1118 females were fertilized by homozygous ms(3)K81 males. These males generate sperm incapable of participating in pronuclear fusion and thus the resulting embryos are unable to complete embryogenesis under control of their own DNA (Yasuda et al. 1995). Embyros donating nuclei possessed green-fluorescent-protein-labeled histone 2AvD (H2AvDGFP; Clarkson and Saint 1999) so donor nuclei were easily distinguishable from those of the host. Less than 2 μl of cytoplasm was aspirated from preblastoderm stage embryos 70–100 min after egg laying. Nuclei were drawn laterally from the ventral face of the embryo and 5–15 nuclei were transplanted to the ventral area of a 10- to 30-min-old recipient embryo. Nuclei drawn from a single donor embryo were injected into one to six recipients, potentially allowing for the generation of more than one clone from a single donor embryo. In the trial reported here, two of the five adult clones, both females, originated from adjacent embryos, suggesting that they may have been derived from one donor. Recipient embryos were incubated at 18° until the completion of embryogenesis at which point larvae were raised on standard Drosophila culture medium.
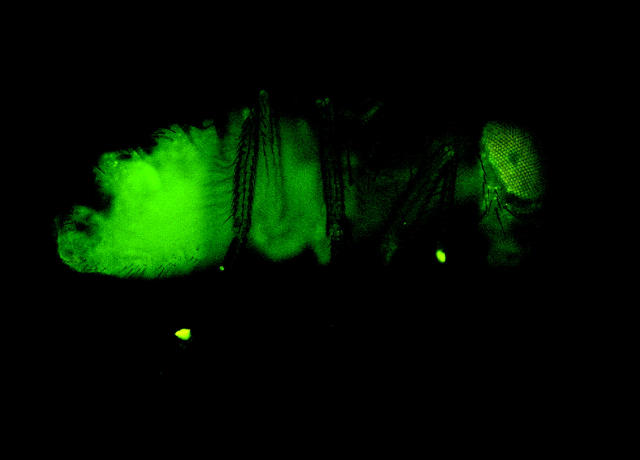
Of the 820 w1118 host embryos into which H2AvDGFP nuclei were injected, 61 (7.4%) expressed H2AvDGFP from the donor nuclei, 14 (1.7%) of those hatched as larvae, and 5 (0.6%) eclosed as fertile adults expressing fluorescence from the H2AvDGFP marker transgene (Figure 1). These individuals represent the first cloned adult Drosophila.
Figure 1.—
Drosophila adult derived from embryonic nuclear transplant expressing H2AvDGFP.
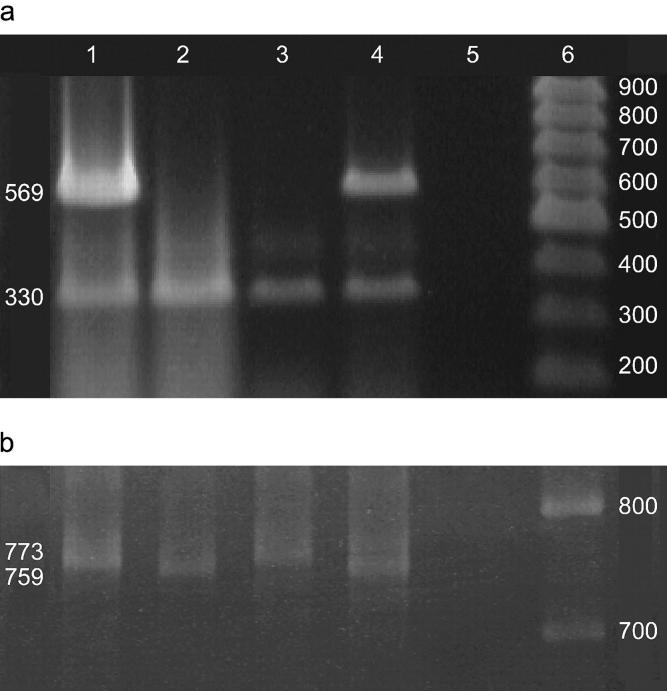
Evidence that these individuals represent animals derived from the injected embryonic nuclei stems from analysis of the mitochondrial and nuclear DNA of the clones. The Drosophila mitochondrial genome is highly variable in size (Lewis et al. 1994), and the length of the A + T-rich region differs between the w1118 and H2AvDGFP strains. This difference was detectable using PCR. Cloned animals possess nuclear DNA derived from the donor embryo but mitochondrial DNA from the host egg. Reciprocal transplantations (w1118 nuclei injected into H2AvDGFP host embryos) exhibited a reciprocal pattern of nuclear and mitochondrial DNA (Figure 2). DNA analysis of adult clones derived from H2AvDGFP donor nuclei yielded identical results.
Figure 2.—
PCR analysis of nuclear and mitochondrial DNA from cloned Drosophila. (a) Analysis of nuclear DNA: primers 5′-ACTGTTTATTGCCCCCTC-3′ and 5′-GTCGTCGAACAAAAGGTG-3′ amplified a 330-bp fragment of white exon 6 present in both w1118 and the CaSpeR-4 P-element transformation vector used to create the H2AvDGFP strain. Primers 5′-ACATCAAATTGTCTGCGG-3′ and 5′-CGCTCGTTGCAGAATAGT-3′ amplified a 569-bp fragment of the CaSpeR-4 P-element transformation vector present in H2AvDGFP but absent in w1118due to a deletion from ∼+2100 to +11,000 relative to the w start codon. PCR allowed for the molecular detection of the CaSpeR-4-based transgene in nuclear transplant recipients. Genomic multiplex PCR at the white locus (1.5% agarose gel) is shown: lane 1, H2AvDGFP genomic control DNA exhibits a 569-bp band from the CaSper-4-derived H2AvDGFP P-element transformation vector and a 330-bp band from white exon 6; lane 2, w1118 genomic control DNA exhibits the 330-bp band but not the 569-bp Casper-4 band; lane 3, w1118 nuclei injected into the H2AvDGFP host embryo. The 569-bp CaSper-4-derived fragment is absent while the 330-bp w1118 fragment is present, indicating only w1118 nuclei in the cloned embryo; lane 4, H2AvDGFP nuclei injected into the w1118 host embryo. The 569-bp CaSpeR-4-derived fragment and the 330-bp w1118 fragment are present, indicating H2AvDGFP nuclei in the cloned embryo; lane 5, DNA negative control; lane 6, 100-bp ladder (MBI Fermentas). DNA was extracted from cloned late-stage embryos and adults using a technique modified from Hatton and O'Hare (http://www.bio.ic.ac.uk/research/ohare/t01816.htm). (b) Analysis of mitochondrial DNA: primers 5′-AATAACAAATTTTTAAGCC-3′ and 5′-GAATAGGGGGAATAAATT-3′ amplified a variable region of the mitochondrial genome ∼759 bp in w1118 and 773 bp in H2AvDGFP, distinguishing host from donor mitochondria. PCR was performed across a variable region of the mitochondrial genome (4% acrylamide gel): lane 1, the H2AvDGFP control amplifies a 773-bp fragment; lane 2, the w1118 control amplifies a 759-bp fragment; lane 3, w1118 nuclei injected into the H2AvDGFP host exhibits the 773-bp fragment from the H2AvDGFP host embryo mitochondria; lane 4, H2AvDGFP nuclei injected into the w1118 host exhibits the 759-bp fragment from the w1118 host embryo mitochondria; lane 5, DNA negative control; lane 6, 100-bp ladder (MBI Fermentas).
The failure of the majority (98.3%) of the cloned embryos to develop normally is likely a consequence of multiple factors. To determine the percentage of embryos rendered inviable from mechanical damage intrinsic to the nuclear transplant procedure, viable w1118 nuclei were injected into diploid w1118 host embryos. Of 202 embryos injected, 21.8% (44) hatched, compared with 1.7% of cloned embryos, suggesting that ∼80% of transplant-recipient embryos die from mechanical damage. Of the remaining 20%, in some cases nonuniform concentrations of GFP expression suggest the failure of donor nuclei to replicate and/or distribute themselves throughout the embryo. Characteristic defects in those expressing GFP, which die shortly after hatching, such as the absence of mouth hooks, defects in the tracheal system, and disorganized or absent spiracles, could potentially be due to epigenetic constraints on reprogramming of donor nuclei.
The first attempts to clone Drosophila by embryonic nuclear transplantation produced ∼1% of embryos able to complete embryogenesis and only one developed as far as the third instar larval stage (Illmensee 1968, 1972). The failure of these cloned Drosophila to survive to adulthood likely resulted from the failure to activate the unfertilized egg. The technique reported here allows for the generation of cloned adult Drosophila.
The rate at which developmental defects arise and the rate at which viable adult Drosophila clones are generated are comparable to that observed in mammals (Wilmut et al. 2002); ∼10% of clones survive through embryogenesis, and ∼1% develop into viable adults. Failure to properly reprogram mammalian embryonic and somatic nuclei in cloning frequently manifests itself as placental abnormalities, fetal overgrowth, and premature death (Shi et al. 2003). Likewise, the abnormalities seen in inviable Drosophila clones could be due to incomplete reprogramming of donor nuclei. As the genetic regulation of early development is well characterized in Drosophila and there is a wealth of mutations affecting early development and maintenance of differentiated cell states, this method can be used to quickly and easily assess constraints on reprogramming of nuclei when cloning.
Acknowledgments
We thank L. A. McEachern, N. Gorguy, M. Hart, and V. Walker for discussion and comments on the manuscript. The H2AvDGFP stock was generously provided by S. Campbell; we thank the Bloomington Drosophila Stock Center for all other stocks. The work was supported by a Natural Sciences and Engineering Research Council discovery grant to V.K.L. and W.A.M. was supported by a fellowship from the Nova Scotia Health Research Foundation.
References
- Clarkson, M., and R. Saint, 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18: 457–462. [DOI] [PubMed] [Google Scholar]
- Gurdon, J. B., and V. Uehlinger, 1966. “Fertile” intestine nuclei. Nature 210: 1240–1241. [DOI] [PubMed] [Google Scholar]
- Illmensee, K., 1968. Transplantation of embryonic nuclei into unfertilized eggs of Drosophila melanogster. Nature 219: 1268–1269. [DOI] [PubMed] [Google Scholar]
- Illmensee, K., 1972. Developmental potencies of nuclei from cleavage, preblastoderm, and syncytial blastoderm transplanted into unfertilized eggs of Drosophila melanogaster. Wilhelm Roux' Arch. 170: 267–298. [DOI] [PubMed] [Google Scholar]
- Lee, K.-Y., H. Huang, J. Bensheng, Z. Yang and S. Lin, 2002. Cloned zebrafish by nuclear transplant from long-term-cultured cells. Nat. Biotechnol. 20: 795–799. [DOI] [PubMed] [Google Scholar]
- Lewis, D. L., C. L. Farr, A. L. Farquar and L. S. Kaguni, 1994. Sequence, organization, and evolution of the A+T region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 11: 523–538. [DOI] [PubMed] [Google Scholar]
- Shi, W., V. Zakhartchenko and E. Wolf, 2003. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation 71: 91–113. [DOI] [PubMed] [Google Scholar]
- Wilmut, I., N. Beaujean, P. A. De Sousa, A. Dinnyes, T. J. King et al., 2002. Somatic cell nuclear transfer. Nature 419: 583–586. [DOI] [PubMed] [Google Scholar]
- Yasuda, G. K., G. Schubiger and B. T. Wakimoto, 1995. Genetic characterization of ms(3)K81, a paternal effect gene of Drosophila melanogaster. Genetics 140: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]