Abstract
In the pairing-site model, specialized regions on each chromosome function to establish meiotic homolog pairing. Analysis of these sites could provide insights into the mechanism used by Drosophila females to form a synaptonemal complex (SC) in the absence of meiotic recombination. These specialized sites were first established on the X chromosome by noting that there were barriers to crossover suppression caused by translocation heterozygotes. These sites were genetically mapped and proposed to be pairing sites. By comparing the cytological breakpoints of third chromosome translocations to their patterns of crossover suppression, we have mapped two sites on chromosome 3R. We have performed experiments to determine if these sites have a role in meiotic homolog pairing and the initiation of recombination. Translocation heterozygotes exhibit reduced gene conversion within the crossover-suppressed region, consistent with an effect on the initiation of meiotic recombination. To determine if homolog pairing is disrupted in translocation heterozygotes, we used fluorescent in situ hybridization to measure the extent of homolog pairing. In wild-type oocytes, homologs are paired along their entire lengths prior to accumulation of the SC protein C(3)G. Surprisingly, translocation heterozygotes exhibited homolog pairing similar to wild type within the crossover-suppressed regions. This result contrasted with our observations of c(3)G mutant females, which were found to be defective in pairing. We propose that each Drosophila chromosome is divided into several domains by specialized sites. These sites are not required for homolog pairing. Instead, the initiation of meiotic recombination requires continuity of the meiotic chromosome structure within each of these domains.
MEIOTIC recombination usually occurs between similar or identical sequences on homologous chromosomes. In most organisms, at least part of the meiotic recombination pathway occurs within the context of the synaptonemal complex (SC), which holds aligned homologous chromosomes together along their entire lengths (von Wettstein et al. 1984). Surprisingly, organisms can be classified into at least two types on the basis of the relationship of double-strand-break (DSB) formation to the SC. In Saccharomyces cerevisiae, DSB formation occurs prior to, and is required for, SC formation (Padmore et al. 1991). A similar course of events occurs in the mouse (Baudat et al. 2000; Romanienko and Camerini-Otero 2000) and in Arabidopsis (Grelon et al. 2001). In contrast, Drosophila and Caenorhabditis elegans form SC in the absence of recombination (Dernburg et al. 1998; McKim et al. 1998), suggesting that the temporal order of events in these organisms might be different. Cytological studies in Drosophila have supported the view that SC formation occurs prior to DSB formation (Jang et al. 2003).
The presence of single-strand tails at DSB sites provides a mechanism for homology searching and chromosome alignment (Roeder 1997). In organisms like Drosophila and C. elegans, however, another mechanism aside from recombination must exist to precisely align homolgous chromosomes during meiosis. Indeed, DSB-independent mechanisms for aligning meiotic chromosomes appear to be widespread. Similar to Drosophila and C. elegans, homolog pairing in fission yeast involves a DSB-independent component (Ding et al. 2004). While homolog pairing is observed in Drosophila somatic chromosomes (Fung et al. 1998) and male meiotic chromosomes that do not recombine (Vazquez et al. 2002), studies of meiotic chromosome pairing prior to or in the absence of SC formation in Drosophila females have not been reported.
Classical studies on chromosome pairing in Drosophila have involved the analysis of crossover suppression or segregation patterns in chromosome rearrangement heterozygotes. Translocation heterozygosity suppresses crossing over in a variety of organisms and has been extensively studied in Drosophila (e.g., Roberts 1976). Although the mechanism of crossover suppression is not known, a pairing defect has often been implicated. Dobzhansky (1931) proposed that crossover reductions were the result of competitive pairing between the partial homologs in translocation heterozygotes. Roberts (1970)(1972) concluded that crossover reductions in translocation heterozygotes were due to disturbed pairing and not due to elimination of the crossover strands. He also proposed that synapsis initiates in the distal regions of each chromosome arm. Consistent with these genetic observations, cytological studies have found a reduction in recombination nodules in some translocation heterozygotes (reviewed in Herickhoff et al. 1993). Effects on synapsis, however, are usually limited to the regions surrounding the breakpoints.
Pairing-site models have been proposed in C. elegans and Drosophila. Although the details of these models differ, both suggest that one or more sites on each chromosome are required for normal levels of crossing over and may have a role in the pairing or synapsis of homologs. Experiments using chromosome rearrangements in C. elegans have mapped a single site at one end of each chromosome that is required for crossing over (McKim et al. 1988, 1993; Villeneuve 1994; Zetka and Rose 1995). In Drosophila, Hawley (1980) investigated the relationship between X chromosome translocation breakpoints and the crossover-suppressed region. Heterozygosity for a translocation suppressed crossing over within an interval defined by specific sites or boundaries and had little effect on crossing over in adjacent intervals. It was proposed that these sites mediated the initial interactions between homologous chromosomes leading to synapsis and recombination. Currently, however, there is no direct evidence that these sites play a role in homolog pairing. In this study, we carried out an analysis of crossing-over suppression on chromosome 3R in translocation heterozygotes to test two predictions of the pairing-site hypothesis. First, does the function of these sites play a role in the initiation of meiotic recombination? And second, are these sites required for the pairing of homologous chromosomes?
MATERIALS AND METHODS
Genetic crosses and translocations on the third chromosome:
All cultures were raised at 25°. Translocation breakpoints are listed in Table 1 and were obtained from three sources. First, a variety of translocations isolated in previous studies and described in Lindsley and Zimm (1992) were obtained from the Bloomington Stock Center. Second, T(2;3)ltx13 and T(2;3)ltx16 were obtained from B. Wakimoto (Wakimoto and Hearn 1990). Third, we isolated a group of translocations in a screen for dominant crossover suppressors of chromosome 3R. These translocations were derived from a uniform genetic background and were induced on a ry531 chromosome to facilitate the gene conversion analysis (see below). ry531 homozygous males were irradiated with 4000 R gamma radiation and crossed to ru h th st cu sr e ca/TM6B, Tb Hu females. Females that were ry531/ru h th st cu sr e ca were selected and crossed to ru h th st cu sr e Pr ca/TM6B, Tb Hu males. The resulting progeny were screened for crossover suppression on chromosome 3R between the ca-e, the e-cu, or the cu-st intervals. Genetic (pseudolinkage) and cytological tests determined whether the crossover suppressor was a translocation, and if so, which other chromosome was involved [either a T(1;3) or a T(2;3)]. For cytological determination of the breakpoints, T(1;3)/+ or T(2;3)/+ larvae were dissected in 45% acetic acid and the salivary glands were separated and stained in lactic acid/acetic acid/orcein for 30 sec. The glands were transferred to a clean slide, a coverslip was placed on top, and the glands were gently squashed.
TABLE 1.
Breakpoints of translocations
| Breakpoint
|
||
|---|---|---|
| Translocation | Chromosome 3R | Chromosome X, 2, or 4 |
| T(2;3)P607 | 82F | 57C-D |
| T(2;3)82Fi2 | 82F10-83A1 | 57A10-B1 |
| T(2;3)MAP3 | 84A4-5 | 26D-F |
| T(2;3)ScrWrv1 | 84B1-2 | 58F1-2 |
| T(1;3)JA29 | 85A | 1E |
| T(3;4)p42 | 85A6 | 101h |
| T(2;3)DP77 | 85C | 26E/27A |
| T(1;3)OR60 | 88A | 4B |
| T(2;3)C202 | 89D | 56D |
| T(2;3)C287 | 89F | 56D |
| T(2;3)gl63d | 91A | 47B |
| T(2;3)DP19 | 93D | 56D |
| T(3;4)A2 | 94A3-4 | 101F |
| T(2;3)ltx16 | 95A3 | 40 |
| T(2;3)dpD | 95B-D | 25A |
| T(3;4)A30 | 96E5-12 | 102BC |
| T(2;3)ltx13 | 97C | 40 |
| T(2;3)DP92a | 87-88; 97A | 58A; ? |
| T(2;3)DP49 | 98F-99A | 60A |
This chromosome has two translocation breakpoints on chromosome 3R.
Each translocation was tested for crossover suppression across various intervals along the right arm of the third chromosome. The crosses to measure crossing over between standard visible markers (Lindsley and Zimm 1992) are described in the footnotes to Tables 3–5. In some cases an interval could not be measured because the translocation chromosome also carried one of the genetic markers. In addition, two P-element insertions were also used as genetic markers: P{ry+t7.2=PZ}abs00620 is a P-element insertion at 82A1-2 and carries a wild-type rosy gene, and P{w+mC=lacW}ksrj5E2 is inserted at 83A4-6 and carries a white mini-gene.
TABLE 3.
Crossing over in thecu-e region on chromosome 3R (cM)
| Genotype | th-kar | kar–cv-c | cv-c–Sb | Sb-gl | gl-e | Total cv-c–e | Total progeny |
|---|---|---|---|---|---|---|---|
| +/+a | 3.9 | 4.3 | 923 | ||||
| +/+b | 2.7 | 5.4 | 7.8 | 16.0 | 807 | ||
| T(2;3)ScrWrv1c | 1.2 | 5.1 | 928 | ||||
| T(3;4)p42c | 1.8 | 4.2 | 674 | ||||
| T(2;3)DP77a | 7.6 | 0.4 | 1236 | ||||
| T(2;3)DP77b | 0.1 | 0.0 | 1.5 | 1.6 | 1050 | ||
| T(2;3)C202d | 0.6 | 1194 | |||||
| T(2;3)C287d | 0.1 | 773 | |||||
| T(2;3)DP19e | 1.3 | 3.7e | 5.0 | 757 | |||
| T(2;3)DP19d | 2.9 | 931 | |||||
| T(2;3)dpDb | 1.6 | 0.7 | 0.3 | 2.6 | 582 | ||
| T(3;4)A2b | 1.0 | 0.9 | 0.3 | 2.2 | 787 | ||
| T(3;4)A30b | 2.1 | 2.1 | 2.9 | 7.1 | 481 | ||
| T(2;3)DP92a | 8.6 | 0.2 | 1015 |
T(2;3) or +/th kar cv-c females were crossed to th kar cv-c males.
T(2;3) or +/cv-c Sb gl e females were crossed back to cv-c Sb gl e males.
T(2;3)/cv-c Sb gl females were crossed back to cv-c Sb gl males.
T(2;3)/gl e females were crossed back to gl e males.
T(2;3)/cv-c Sb e females were crossed to cv-c Sb e males. The Sb to e distance is reported.
TABLE 5.
Crossing over in proximal chromosome 3R
| Genotype | Translocation | cM | Recombinants | Total |
|---|---|---|---|---|
| P{ry+t7.2=PZ}abs00620 ry506a | + | 3.1 | 21 | 1374 |
| T(2;3)82Fi2 | 3.3 | 15 | 909 | |
| T(2;3)ScrWrv1 | 3.2 | 18 | 1134 | |
| P{w+mC=lacW}ksrj5E2 cub | + | 1.3 | 20 | 1562 |
| T(2;3)82Fi2 | 1.3 | 17 | 1330 | |
| T(2;3)ScrWrv1 | 0.8 | 9 | 1070 |
This P element carries a wild-type copy of the ry gene. The distance between the insertion site (82A1-2) and the rosy gene was measured when P{ry+t7.2=PZ}abs00620 ry506/T(2;3) females were crossed to ry/ry males. Approximately two-thirds of this interval is distal to 85A-C and not expected to be affected by proximal translocations.
This P element carries a white mini-gene. This was used as a marker to measure crossing over between the insertion site (83A4-6) and cu when w/w; P{w+mC=lacW}ksrj5E2 cu/T(2;3) females were crossed to w/Y; cu/cu males.
Intragenic recombination experiments:
Intragenic recombination was measured using the ry606 and ry531 chromosomes (Hilliker et al. 1991). T(2;3), ry531/TM6B, ry males were crossed to th kar ry606 cv-c females and the th kar ry606 cv-c/T(2;3) ry531 male and female progeny were then crossed together in bottles containing 25 ml of food and transferred every 3 days for five broods. Immediately after brooding, 750 μl of a 0.2% purine solution was added to each bottle, which was the lowest dose that selected against rosy mutants. A subset of the bottles was not treated with purine and the flies were counted to estimate the total number of progeny. Any non-rosy progeny were selected and crossed to thr kar ry606 cvc flies to categorize the recombination event as either a gene conversion or a crossover. A parental arrangement of flanking markers indicated a gene conversion. This is a reliable method to classify the recombination event because the flanking markers are close to the ry locus and therefore it was unlikely that a gene conversion and a separate crossover between ry and a flanking marker would occur in the same meiosis (Chovnick et al. 1970).
Preparation of DNA probes:
Cy3-labeled probes were made from BAC genomic DNA clones (Hoskins et al. 2000). As described by Marshall et al. (1996), ∼10 μg of DNA was fragmented by digestion with six four-cutter restriction enzymes (AluI, HaeIII, MseI, MspI, RsaI, Sau3AI) and then labeled with Cy3-dCTP (Amersham, Buckinghamshire, UK) using terminal deoxynucleotidyl transferase (Invitrogen, San Diego). The identifiers and cytological locations of the clones are shown in Figure 1.
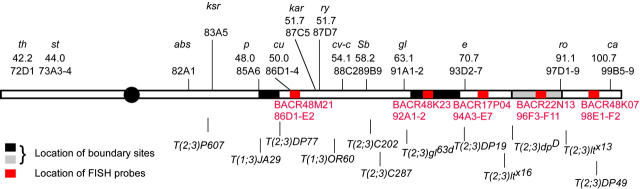
Figure 1.—
Map of chromosome 3R. The genetic markers used in the study are shown above the line. Superimposed on the chromosomes are rectangles showing the positions of boundary sites (black) and BAC clones (Hoskins et al. 2000) used as FISH probes (red). The size of the black rectangles indicates the uncertainty in the mapping of the boundary sites. The boundary site between e and ca is gray because the data supporting its existence are not conclusive. Below the line are the breakpoints of representative translocations.
Hybridization to whole-mount ovarioles:
For fluorescent in situ hybridization (FISH), a protocol modified from Dernburg et al. (1996) was used. Females of the genotype +/+ or ry531/+ (wild-type controls), T(?;3)/+, or c(3)G68ca/c(3)G68 ca were aged 24–48 hr and then the ovaries were dissected in modified Robb's saline (MRS) containing 0.2% Tween-20. Following the dissection of the ovaries, the MRS was removed and cacodylate fixative buffer was added for 4 min. During the fixation, the ovaries were teased apart toward the germarium tip. After the fixative was removed, the ovaries were washed four times in 2× SSCT (0.3 m NaCl, 0.03 m NaCitrate, 0.1% Tween-20). After washing, the ovaries were completely separated into individual ovarioles before being transferred from the dissecting dish to a 0.5-ml Eppendorf tube. Ovarioles were then washed in 2× SSCT three times and then gradually exchanged into 2× SSCT/50% formamide with 10-min washes in 2× SSCT/20% formamide, then in 2× SSCT/40% formamide, and then two washes in 2× SSCT/50% formamide. The ovarioles were then prehybridized in 2× SSCT/50% formamide for 4 hr at 37°. Ovarioles were then allowed to settle and the 2× SSCT/50% formamide was removed prior to the addition of 36 μl of hybridization solution [3× SSC/50% formamide/10% (w/v) dextran sulfate] and up to 4 μl of probe. The tissue and solution was gently mixed by flicking the tube and then heated to 91° in a thermal cycler for 3 min followed by incubation overnight at 37° in the dark. Following the overnight incubation, 2× SSCT/50% formamide was added to the sample and inverted several times to mix thoroughly. Ovarioles were allowed to settle and then samples were washed in fresh 2× SSCT/50% formamide. Two more 30-min washes in 2× SSCT/50% formamide were done at 37° followed by one wash in 2× SSCT/25% formamide at room temperature and four washes in 2× SSCT. Ovarioles were then blocked in 6 mg/ml normal goat serum in 2× SSCT for 4 hr at room temperature and then washed three times quickly in 2× SSCT. Anti-oo18 RNA-binding protein monoclonal antibodies (6H4 and 4H8; Lantz et al. 1994) at 1:30 or anti-C(3)G antibody (guinea pig or mouse) at 1:500 (Page and Hawley 2001) were added in 2× SSCT and incubated overnight at room temperature. The following day, ovarioles were washed three times in 2× SSCT for 10 min, 1 hr, and 1.5 hr. Fluorescein-conjugated secondary antibody (Vector, Burlingame, CA, or Jackson Labs, West Grove, PA) was added and incubated for 4 hr. Ovarioles were then washed two times quickly in 2× SSCT, once for 3 hr, and then overnight at room temperature. After settling, excess 2× SSCT was removed and the ovarioles were mounted in Vectashield (Vector).
Image analysis:
Images were collected and analyzed on two systems. The first was a DeltaVision restoration microscopy system (Applied Precision) equipped with a Nikon ×60 N.A.1.4 oil immersion objective. The restoration and modeling was performed with softWoRx software (Applied Precision) on an Octane Workstation (Silicon Graphics). The second was a Zeiss Axioplan II imaging microscope equipped with a ×60 or ×100 N.A.1.4 oil immersion objective and a Sensicam CCD camera (Cooke). These images were analyzed using deconvolution and 3D analysis software from Vaytek (Fairfield, IA). Similar results were obtained using either system. In FISH experiments using ORB staining, oocytes were scored only where ORB was clearly localized to one cell, usually in regions 2b and 3 and thus in midpachytene. In experiments with C(3)G staining, all oocytes could be observed from region 2a (early pachytene) to region 3. No differences in homolog pairing were observed between early pachytene (region 2a) and late pachytene (region 3). Foci of hybridization were typically brightest in the surrounding nurse cell nuclei, and in some oocytes (<10%) no hybridization signal could be found. The failure to detect a signal had only a minor impact on the conclusions because of its low frequency and the experiments with c(3)G mutant females demonstrated the frequency at which pairing defects were detected. The distance between a homolog pair was measured between the brightest pixels of two foci. In a minority of nuclei, three foci were observed; two of these foci were usually close together and probably sister chromatids. The distance reported in these cases was the largest of the three possible measurements.
RESULTS
Crossover suppression by translocations on chromosome 3R:
Using genetic markers spanning all of chromosome 3R (Figure 1) to measure crossing over, the translocations were separated into three groups on the basis of patterns of crossover suppression (Table 2). The first group suppressed crossing over most severely in the cu-e region, the second group suppressed crossing over most severely in the e-ca region (Figure 2), and the third group with breaks closest to the centromere did not suppress crossing over in any region, even between st and cu, which included the breakpoints of this group. In addition, complex rearrangements were identified with more extensive effects on crossing over. For example, T(2;3)DP92 involves two translocation breakpoints on chromosome 3R and almost no crossovers were recovered.
TABLE 2.
Crossing over on chromosome 3R in translocation heterozygotes (cM)
| Break region | Genotype | st-cu | cu-e | e-ca | st-ca | Total progeny | 3R breakpoint |
|---|---|---|---|---|---|---|---|
| ry531/+ | 3.2 | 17.3 | 30.4 | 51.3 | 618 | ||
| e-ca | T(2;3)DP92 | 10.4 | 0.0 | 0.4 | 12.1 | 770 | 87-88; 97A |
| T(3;4)A2 | 4.1 | 4.5 | 5.0 | 9.1 | 242 | 94A 3-4 | |
| T(2;3)ltx16 | 3.6 | 8.6 | 1.8 | 14.0 | 511 | 95A3 | |
| T(2;3)dpD | 4.3 | 7.0 | —a | 715 | 95B-D | ||
| T(3;4)A30 | 2.2 | 9.7 | 4.8 | 16.8 | 536 | 96E 5-12 | |
| T(2;3)ltx13 | 4.9 | 16.0 | 5.9 | 26.8 | 977 | 97C | |
| cu-e | T(2;3)DP77 | 2.3 | 5.0 | 34.6 | 42.4 | 563 | 85C |
| T(1;3)OR60 | 3.1 | 2.1 | 32.1 | 38.4 | 617 | 88A | |
| T(2;3)C202 | 3.1 | 0.8 | 27.5 | 33.0 | 382 | 89D | |
| T(2;3)C287 | 4.0 | 0.3 | 25.4 | 30.7 | 374 | 89F | |
| T(2;3)gl63d | 6.2 | 0.7 | 14.4 | 22.4 | 548 | 91A | |
| cu-cent | T(2;3)P607 | 4.4 | 18.2 | 36.4 | 59.1 | 1181 | 82F |
| T(2;3)82Fi2 | 4.5 | 43.9b | 48.8 | 551 | 82F10-83A1 | ||
| T(2;3)MAP3 | 1.6 | 13.0 | 34.7 | 49.3 | 377 | 84A4-5 | |
| T(2;3)ScrWrv1 | 5.0 | 38.5b | 43.8 | 714 | 84B 1-2 | ||
| T(1;3)JA29 | 2.8 | 18.0 | 29.2 | 50.0 | 178 | 85A | |
| T(3;4)p42 | 4.8 | 46.0b | 50.8 | 559 | 85A6 |
Value for the e-ca region not determined (ca was homozygous).
Value for the cu-ca region (e not scored because it was homozygous).
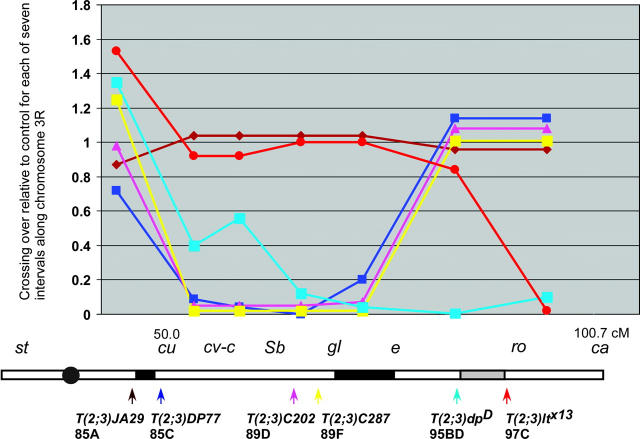
Figure 2.—
Summary of crossover suppression in selected translocations. The effect of each translocation on crossing over is shown relative to the normal sequence chromosome control. The x-axis is in centimorgans and shows the genetic intervals measured. Below the graph is a schematic of chromosome 3R and the locations of each breakpoint and boundary sites. The lines are color coded for each translocation.
As described in detail below and summarized in Figure 2, our results can be explained by the existence of specialized sites at 85A-C and 91A-93D, and possibly another at 95B-97C, which have a role in meiotic recombination. These conclusions are based on the idea that a discontinuity due to heterozygosity for a translocation breakpoint causes crossover suppression within the interval between two sites but not in other regions (Hawley 1980). Throughout this article, we refer to these sites as boundary sites to reflect the observation that crossover suppression in translocation heterozygotes is strongest in the interval containing the breakpoint and weaker or absent in other intervals. These sites can be mapped by observing the patterns of crossover suppression in a series of translocations with breakpoints spanning a chromosome arm. For example, the 91A-93D site near e was mapped using the following criteria. Translocation breakpoints to the left of this site suppressed crossing over in the cu-e but not e-ca intervals whereas translocation breakpoints to the right of this site suppressed crossing over in the e-ca interval.
Crossover suppression in one genetic interval did not result in compensatory increases in other intervals. For example, when the cu-e region was suppressed, there was no increase in crossing over in the e-ca region (Table 2, Figure 2). Thus, crossover suppression by translocations did not activate a system that regulates the total number of exchanges per chromosome arm. Crossing over increased in the centric st-cu region with some translocations, but these increases did not compensate for the observed decreases in other regions and may be related to the interchromosomal effect (Williamson 1966; K. McKim, unpublished results).
Evidence for a boundary in 85A-C:
Translocations with breakpoints between divisions 85 and 91, such as T(2;3)DP77 (85C) and T(2;3)C287 (89F), suppressed crossing over between cu and e but not e and ca (Tables 2 and 3). However, translocations with breakpoints proximal to 85A6 had no effect on crossing over in the cu-e interval. For example, T(2;3)P607 (82F), T(2;3)MAP3 (84A4-5), T(2;3)ScrWrv1 (84B1-2), T(1;3)JA29 (85A), and T(3;4)p42 (85A6) did not suppress crossing over in the cu-e region (Tables 2 and 3). Crossing over within the cu-e region was investigated using kar, cv-c, Sb, gl, and e (Figure 1). Crossover suppression was not restricted to the region around the breakpoint; the reductions in crossing over were uniform throughout the cu-e interval (Figure 2). For example, T(2;3)DP77 breaks close to cu but has similar crossover suppression effects within the kar–cv-c, cv-c–Sb, Sb–gl, and gl–e intervals (Table 3). Similarly, T(2;3)C202 and T(2;3)C287 suppressed crossing over to a frequency lower than that of any of the subintervals, indicating that they suppress crossing over throughout the cu-e region.
Evidence for a boundary in 91A-93D:
The translocations that suppressed crossing over in the cu-e region, such as T(2;3)C202 and T(2;3)C287, had a normal frequency of crossing over in the e-ca region (Table 2). The location of the boundary is distal to 91A because T(2;3)gl63d (91A1-2) strongly suppressed the cu-e region. T(2;3)gl63d heterozygotes also caused a relatively mild reduction of crossing over in the e-ca region (47% of wild type), but this is not severe enough to indicate that it has a breakpoint to the right of the boundary site. More distal breakpoints suppressed crossing over when measured in the e-ca region. The distal limit for the boundary site is 93D because T(2;3)DP19 (93D) strongly suppressed crossing over in the e-ca region but mildly suppressed it in the cu-e region [Table 4 and compare gl-e in Table 3 for T(2;3)DP19 (37.0% of control) and T(2;3)C287 (1.6% of control)]. The mild effects of T(2;3)DP19 in the cu-e region are consistent with the effects of other distal translocations discussed below. That T(2;3)C202 and T(2;3)C287 suppressed crossing over in the gl-e region has two implications (Table 3). First, the location of the boundary site is closer to e (93D). Second, crossover suppression occurs on both sides of a translocation breakpoint (e.g., cu-breakpoint and breakpoint-e).
TABLE 4.
Crossing over betweenst, ro, andca on chromosome 3R (cM)
| Genotype | st-Sb | Sb-e | e-ro | ro-ca | Total st-ca | Total progeny |
|---|---|---|---|---|---|---|
| +/+ | 7.2 | 10.5 | 25.5 | 12.1 | 55.3 | 1123 |
| T(2;3)DP19 | 12.9 | 5.6 | 1.0 | 2.3 | 21.8 | 737 |
| T(2;3)dpD | 9.4 | 1.3 | 0.1 | 0.1 | 11.0 | 818 |
| T(2;3)ltx16 | 14.1 | 5.3 | 0.9 | 0.6 | 20.9 | 923 |
| T(2;3)ltx13 | 18.8 | 10.6 | 6.6 | 0.3 | 36.2 | 775 |
| T(2;3)DP49 | 14.5 | 7.1 | 6.8 | 0.3 | 28.6 | 780 |
T(2;3)/st Sb e ro ca females were crossed back to st Sb e ro ca males.
To investigate crossover suppression in the e-ca region, we subdivided the interval using ro (Table 4). Translocations with the most distal breakpoints, such as T(2;3)ltx13 and T(2;3)DP49, caused mild crossover suppression in the e-ro interval but more severe crossover suppression in the ro-ca interval, suggesting that there may be a boundary site between e and ca at 95B-97C (Figure 1, Figure 2). In contrast, T(2;3)ltx16 and T(2;3)dpD suppressed crossing over in both e-ro and ro-ca regions, suggesting that there are no additional sites or boundaries. This contradiction could be explained, however, by the observation described below that translocations with breaks in the 94-96 region, such as T(2;3)ltx16 and T(2;3)dpD, suppress crossing over along the whole arm.
Translocations that suppress crossing over along the whole arm:
While most of our data are consistent with a boundary at 91A-93D, some translocations in the e-ca region showed mild crossover suppression in the cu-e region. Crossing over was mildly reduced throughout the e-cu region in T(2;3)dpD and T(2;3)ltx16 (Tables 2 and 3). Similar results were also observed in two translocations involving chromosome 4 [T(3;4)A2 and T(3;4)A30] (Tables 2 and 3), and Roberts (1972) identified several translocations with breaks between e and ca, which, on the basis of measuring crossing over between st and ca, probably reduced crossing over between cu and e. This effect was most severe with translocation breakpoints in the middle of the e-ca region. For example, T(2;3)ltx16 and T(3;4)A2 suppressed crossing over in the cu-e region to a larger degree than T(2;3)ltx13 and T(3;4)A30. These results suggest that distal regions, especially the 94-96 region, are important for crossing over throughout chromosome 3R. We have excluded a centromere effect as an explanation for our results since the translocations that were characterized did not bring a centromere closer to the proximal regions.
The effects of distal translocation breaks on proximal regions such as the cu-e interval does not contradict the evidence for a boundary in 91A-93D. Distal translocations caused mild crossover suppression in the cu-e region (∼50% of wild type; Figure 2), whereas translocation breaks between the sites at 85A-C and 91A-93D such as T(2;3)C202 caused much more severe reductions on crossing over (∼5% of wild type). Therefore, the reductions in crossing over by breaks in the 94-96 region were tempered by the boundary in 91A-93D.
Translocations in centromere proximal regions do not suppress crossing over:
Most translocations did not suppress crossing over in the st-cu region, which includes the most proximal region on chromosome 3R. Even the T(2;3) chromosomes with breaks between the centromere and the 85A-C boundary site did not suppress crossing over anywhere on the right arm of the third chromosome (Table 2). One translocation, T(2;3)MAP, reduced crossing over in the st-cu interval, but this may be an effect of genetic background since the reduction was mild and crossing over was also low in the adjacent cu-e interval. One interpretation of these results is that the region between the centromere and the 85A-C boundary site is not sensitive to translocation heterozygosity. This conclusion assumes, however, that not all the crossing over between st and cu occurs to the left of the centromere. To directly measure the frequency of crossing over in proximal 3R, we used two P-element insertions (abs at 82A1-2 and ksr at 83A4-6; Table 5). No significant crossover suppression was observed with the translocations that break proximal to the 85A-C boundary. Similarly, as part of a study on crossover interference, Denell and Keppy (1979) measured crossing over within proximal chromosome 3R of T(Y;3)B155 (82C) heterozygotes and it was not suppressed. These results support the conclusion that crossing over in the proximal euchromatin is not sensitive to translocation heterozygosity.
Gene conversion is reduced in translocation heterozygotes:
Crossover suppression in translocation heterozygotes could occur because of either an early defect in the recombination pathway such as a failure to make DSBs or a defect in the system that controls whether a DSB is repaired as a crossover or as a noncrossover. If initiation of recombination is defective, then both recombination products, noncrossover (gene conversion) and crossover, should be affected. We tested this hypothesis by measuring the frequency of gene conversion at the rosy locus (Hilliker et al. 1988) in two translocation heterozygotes (Table 6). T(2;3)DP92, which is a two-break rearrangement that suppresses crossing over throughout the entire right arm of the chromosome, was chosen because of its strong crossover suppression. T(2;3)DP77, which suppresses crossing over only within the cu-e region, was chosen because it had a more restricted crossover suppression pattern. Both suppressed crossing over in the region around the rosy gene (Table 3).
TABLE 6.
Intragenic recombination at therosy locus in translocation heterozygotes
| Translocation | Gene conversion |
Crossovers | Total progeny |
Gene conversion frequency (10−6) |
|---|---|---|---|---|
| +/+ | 7 | 6 | 574,600 | 12.2 |
| T(2;3)DP77 | 1 | 0 | 416,300 | 2.4 |
| T(2;3)DP92 | 0 | 0 | 403,600 | 0 |
Parental females were T(2;3) ry531/th kar ry606 cvc.
A reduction in crossovers was expected since the translocations suppress crossing over in the rosy region. In addition, among ∼820,000 progeny from T(2;3)DP92, ry531/ry606 and T(2;3)DP77, ry531/ry606 females, only one gene conversion event recovered compared to seven gene conversion events from ∼575,000 progeny in the normal chromosome control. These data demonstrate that the frequency of both products of DSB repair is reduced, suggesting that translocation heterozygotes have a defect in the initiation of meiotic recombination.
Analysis of meiotic homolog pairing using FISH:
If crossover suppression in translocation heterozygotes is due to defects in homolog pairing, as predicted by the pairing-site model, then the homologs should be unpaired in the crossover-suppressed regions. We tested this prediction in three sets of experiments by using FISH to observe the locations of homologous loci within the oocyte nucleus. In the first experiment, we examined homolog pairing in wild-type oocytes. In the second, we examined homolog pairing in translocation heterozygotes to determine if crossover-suppressed regions failed to pair. Finally, we examined c(3)G68 mutant females, where we hypothesized that homolog pairing defects would be observed.
Whole-mount ovaries were probed with one of five fluorescently labeled BAC clones on chromosome 3R (Figure 1). Within the ovary, prophase oocytes develop among 15 nurse cells and can be identified using antibodies to either the ORB protein, which accumulates during pachytene in the oocyte cytoplasm (Lantz et al. 1994), or the C(3)G protein, which is a component of the synaptonemal complex that forms between synapsed meiotic chromosomes (Page and Hawley 2001). On the basis of the staining patterns of these two antibodies and relative position within the ovary, oocytes can be identified and staged (for details see Figure 3).
Figure 3.—
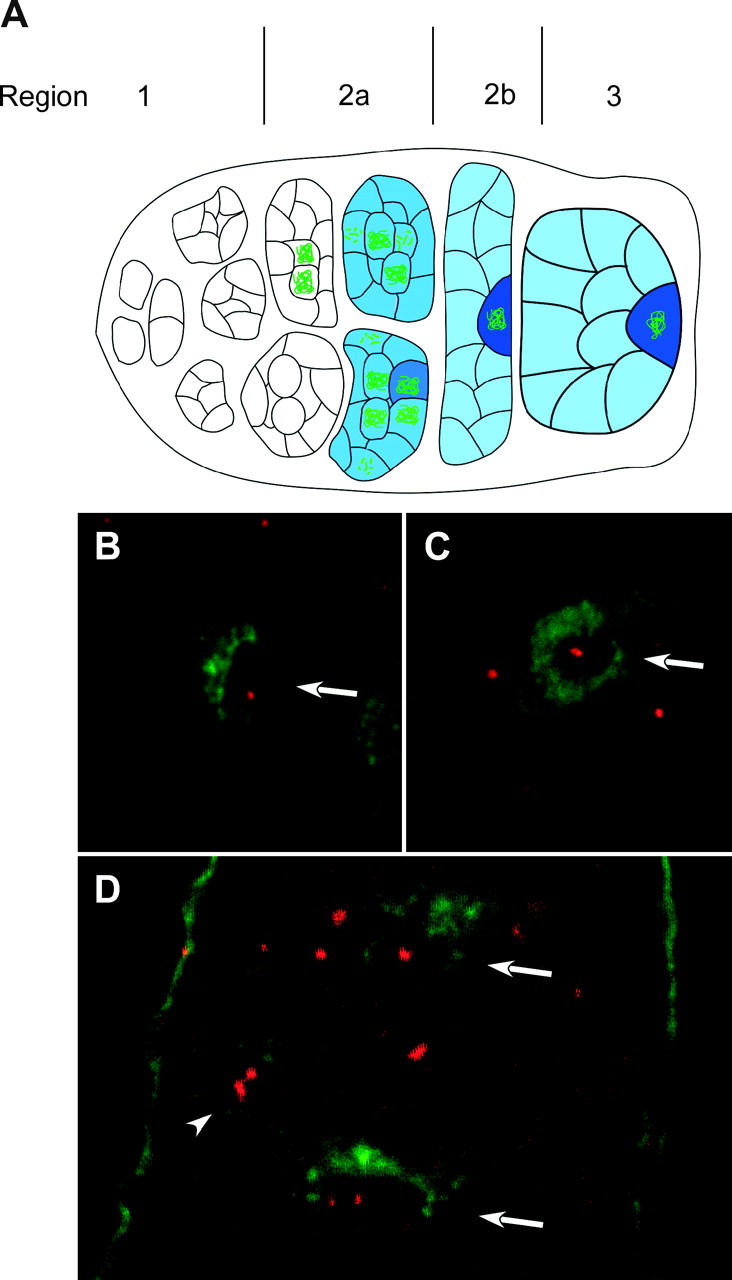
FISH analysis in wild-type and c(3)G mutant oocytes. (A) Schematic of the germarium in the female ovary. Oocytes develop within a 16-cell cyst, which forms from four incomplete mitotic cell divisions (region 1). Two of the 16 cells have four interconnections, or ring canals, and become the pro-oocytes. SC [detected with antibody to C(3)G (green)] and cytoplasmic protein ORB (blue) are not detected in premeiotic cells or the very earliest meiotic (16-cell cyst) cells in region 2a. Since ORB stains all germline cells, this could be used to distinguish these cells from the surrounding somatic follicle cells. Even in cases where ORB staining was not used, when examining premeiotic cells or nurse cells in experiments with C(3)G staining, the germline cells could be identified on the basis of DNA staining. The somatic follicle cells form a single layer around the outside of the developing germline cysts. Changes in cyst morphology differentiate regions 2a, 2b, and 3. In addition, the 16-cell cysts move anterior to posterior within the ovary and are usually arranged in developmental order. In region 2a, both pro-oocytes enter meiosis, first in zygotene and then early pachytene, where the SC assembles between homologs and meiotic recombination initiates. Region 2a cysts are round and while cytoplasmic ORB is usually present, it usually does not concentrate in the oocyte. Region 2b cysts flatten out and are surrounded by somatic follicle cells, and region 3 cysts are round. In some region 2b and all region 3 cysts, one cell is identifiable as the oocyte by localization of the cytoplasmic ORB protein. In addition, by this time usually one of the two pro-oocytes has reverted to a nurse cell fate, leaving one cell in pachytene with C(3)G staining. (B and C) Hybridization of a FISH probe (red) and ORB staining (green) in wild-type oocytes. The oocytes are identified by the strongest ORB staining (arrows). The FISH staining in C is an example where two foci appear to be touching. (D) In c(3)G mutant oocytes, unpaired homologs can be observed in a region 3 oocyte (bottom arrow) and in a nurse cell (arrowhead). A region 2b oocyte (top arrow) has a single focus of hybridization. The anterior end of this germarium is toward the top.
In wild-type oocytes with normal sequence chromosomes the homologous loci were usually paired (Table 7). A single focus of staining was observed in the majority of nuclei and when there were two foci, they were close together. FISH in combination with C(3)G antibody staining to detect the SC in wild type revealed that when a probe detected two foci, they were on either side of a thread of C(3)G staining (see below). These foci were usually closer than 1.0 μm and two foci could be distinguished at a distance of as little as ∼0.4 μm (Figure 3). Since most wild-type oocyte nuclei contained only a single focus of hybridization, the homologous loci were usually ≤0.4 μm apart. And when there were two foci, they were probably within the context of homologs joined by SC. Similar results in wild-type females with an X chromosome probe have been reported by Webber et al. (2004).
TABLE 7.
Third chromosome pairing in wild-type andc(3)G68 mutants
| No. of foci
|
||||||
|---|---|---|---|---|---|---|
| Probe | 1 | 2 | 3 or 4 | Multiple foci (%) |
Average distance between foci (μm)a |
|
| +/+ | M21 | 13 | 2 | 13.3 | 0.51 (0.01)b | |
| c(3)G68 | M21 | 6 | 5 | 3 | 57.1 | 1.47 (0.55) |
| +/+ | K23 | 10 | 5 | 33.3 | 1.0 (0.81) | |
| c(3)G68 | K23 | 1 | 2 | 1 | 75.0 | 1.37 (0.53) |
| +/+ | N13 | 29 | 10 | 25.6 | 0.66 (0.13) | |
| +/+c | N13 | 75 | 2 | 2.6 | ND | |
| +/+d | N13 | 21 | 2 | 8.7 | ND | |
| c(3)G68 | N13 | 12 | 15 | 2 | 58.6 | 1.92 (2.00) |
| c(3)G68c | N13 | 59 | 27 | 7 | 36.6 | ND |
| c(3)G68d | N13 | 16 | 7 | 4 | 40.7 | ND |
| +/+ | K7 | 18 | 1 | 5.3 | 0.62 (0.12) | |
| c(3)G68 | K7 | 7 | 3 | 30.0 | 2.13 (0.42) | |
Probes: BACR48M21 (86D1-E2), M21; BACR48K23 (89A1-5), K23; BACR22N13 (96F3-11), N13; and BACR48K07 (98E1-F2), K7. ND, not determined.
See materials and methods for how these distances were measured.
Standard deviation in parentheses.
Nurse cells.
Prepachytene cells were defined as those in the germarium that lacked ORB or C(3)G staining. These could be cells within a 16-cell cyst very early in region 2a or in region 1 and thus truly premeiotic.
Homolog pairing was also examined in the nuclei without complete SC formation by characterizing the FISH signals in germline cells that lacked C(3)G staining. We looked at prepachytene cells (either early meiotic prophase or premeiotic cells), which were defined as those that failed to stain with ORB or SC in region 1 and early 2a of the germarium and thus that had not yet developed SC. Although there may be differences between the prepachytene cells due to asymmetric mitotic cell divisions, we did not use markers to differentiate these cells. In addition, the classification of “premeiotic” included cells that were still mitotic and those had completed the last mitotic division to become a 16-cell cyst and possibly entered meiotic prophase. The 16-cell cysts in S-phase or early prophase (e.g., leptotene) were not detected with C(3)G and ORB staining. Despite the heterogeneity of these prepachytene cells, the majority of cells had only a single focus of hybridization using BACR22N13 as a probe (Table 7). Similar results were observed with the four other probes (data not shown), indicating that the homologs enter meiosis tightly paired along their lengths.
We also looked at nurse cells, which are the sister cells of the proocytes that form little or no SC, and found that the majority of these cells had a single focus of staining (Table 7). Since most nurse cells lack or have substantially reduced C(3)G staining (Page and Hawley 2001; data not shown), these results are consistent with the results using premeiotic cells in showing that complete SC formation is not required for homolog pairing. However, since short stretches of SC are often detected by EM (Carpenter 1975) and may not be detected by C(3)G staining, these experiments did not determine if the homolog pairing in nurse cells was SC independent. As described below, however, efficient pairing in nurse cells is partially dependent on c(3)G.
Homolog pairing in translocations that suppress crossing over:
The probe BACR48M21 detects a locus at 86D1-E2, which is within the crossover suppressed region (cu-e) of translocations like T(2;3)C287 and T(2;3)DP77 with breaks between the 85A-C and 91A-93D sites. BACR48M21 detected a single focus of hybridization in the majority of T(2;3)C287/+ (89F) oocytes (27/35), which was similar to the frequency of nuclei with a single focus (66/78) in wild type (Table 8). In addition, the average distance between separated foci in T(2;3)C287/+ oocytes (0.96 μm) was similar to wild type. Similar results were obtained with T(2;3)DP77; 70/77 oocytes contained a single focus of hybridization and the distances between separated foci were similar to those seen in wild type. Experiments were also performed with T(2;3)dpD, which suppresses crossing over between e and ca. We used probes BACR22N13 and BACR17P04, which are within the crossover-suppressed region, and BACR48K23, which is close to the boundary site at 91A-93D and may be within the crossover-suppressed region. Again, the frequency of nuclei with a single focus of hybridization and the distance between separated foci was similar to the wild-type controls. These results suggest that crossover suppression in translocation heterozygotes may not be due to pairing defects. As described below, these results contrast with c(3)G mutant females, in which homolog pairing defects were observed.
TABLE 8.
Third chromosome pairing in translocation heterozygotes
| No. of foci
|
||||||
|---|---|---|---|---|---|---|
| Probe | 1 | 2 | 3 | Multiple foci (%) |
Average distance between foci (μm)a |
|
| +/+ | M21 | 66 | 12 | 15.4 | 0.84 (0.43)b | |
| T(2;3)C287 | M21 | 27 | 8 | 22.8 | 0.96 (0.69) | |
| T(2;3)DP77 | M21 | 70 | 7 | 9.1 | 0.92 (0.36) | |
| +/+ | K23 | 9 | 1 | 1 | 18.2 | 2.1 (1.1) |
| T(2;3)dpD | K23 | 48 | 6 | 11.1 | 0.85 (0.37) | |
| +/+ | N13 | 42 | 9 | 17.6 | 0.74 (0.30) | |
| T(2;3)dpD | N13 | 42 | 8 | 16.0 | 0.78 (0.40) | |
| +/+ | P04 | 34 | 8 | 19.0 | 0.57 (0.20) | |
| T(2;3)dpD | P04 | 25 | 6 | 19.3 | 0.67 (0.16) | |
Probes: BACR48M21 (86D1-E2), M21; BACR48K23 (89A1-5), K23; BACR22N13 (96F3-11), N13; BACR17P04 (94A3-E7), P04.
See materials and methods for how these distances were measured.
Standard deviation in parentheses.
Although homologs are usually paired in translocation heterozygotes, it remained possible that there were defects in SC formation that were not detectable using a pairing assay such as FISH. To determine if the paired loci in translocation heterozygotes were associated with SC formation, we performed FISH experiments in T(2;3)dpD/+ oocytes also stained for C(3)G as a marker for SC formation (Figure 4). Probes for BACR22N13 or BACR17P04 were used because they are on different sides of the T(2;3)dpD breakpoint (Figure 1) and could reveal defects specific to one side of a breakpoint. Page and Hawley (2001) measured the length of SC in the Drosophila oocyte using the C(3)G antibody and the average length of a chromosome arm is ∼22.3 μm. Given that there are 20 cytological divisions per chromosome, there may be ∼2 μm from the T(2;3)dpD breakpoint to the BACR22N13 site. If SC formation was disrupted by a translocation breakpoint, we expected an absence of C(3)G staining in the vicinity of the homologous loci.
Figure 4.—
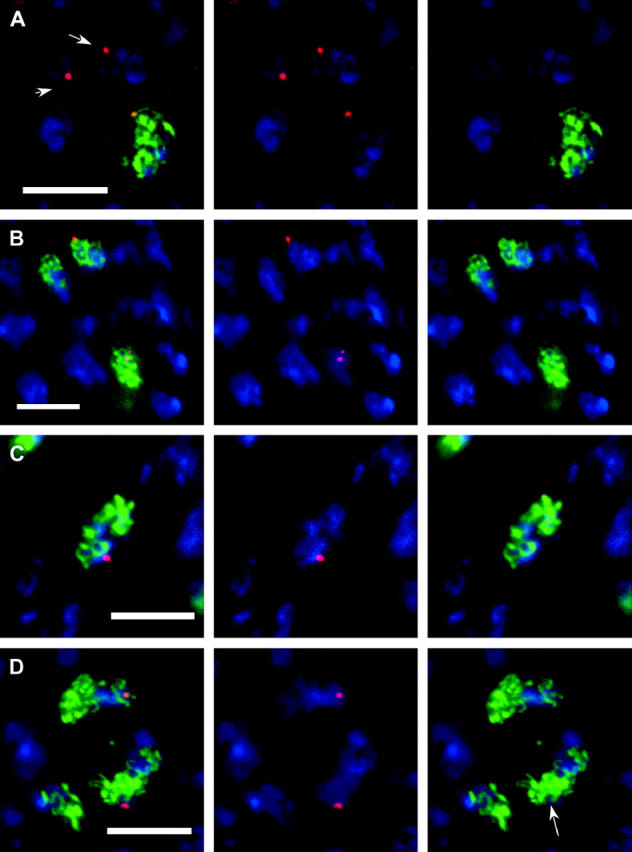
Homolog pairing in translocation heterozygotes. FISH using probe BACR22N13 (red) in conjunction with C(3)G (green) and DNA (blue) staining. (A) Wild-type early pachytene. The arrowed nuclei are in a cyst prior to SC formation. (B) Wild-type oocytes in pachytene. The oocyte at the bottom of the image has two foci separated by a thread of C(3)G staining. (C) Oocyte nucleus from T(2;3)dpD/+ female. The single focus of staining is adjacent to a thread of SC. (D) A less common example from a T(2;3)dpD/+ oocytes where the SC appears disorganized in the region of the probe signal. In the image showing only C(3)G, the arrow marks the region where the probe signal should be. Each image is a single section or projection of a small number of sections. Bar, 5 μm; B is at a slightly lower magnification.
In wild-type oocytes, the foci detected by the probe for BACR22N13 were always associated with C(3)G (n = 51, Table 8). In T(2;3)dpD/+ oocytes, there were no gross defects in SC formation, the majority of oocytes had a single focus of BACR22N13 hybridization (94%), and the foci were associated with C(3)G staining in 30/33 oocyte nuclei. Similarly, with BACR17P04, the hybridization foci were associated with C(3)G staining in 26/31 T(2;3)dpD/+ oocyte nuclei compared to 41/42 in wild type. Furthermore, in favorable nuclei where the FISH signal was on the outside of the nucleus, the foci were clearly adjacent to a single strand of C(3)G staining (Figure 4). Therefore, a region of chromosome 3R associated with crossover suppression in a translocation heterozygote was usually associated with C(3)G staining, suggesting that the SC could form between homologs.
However, these results did not rule out an effect of translocation heterozygotes on SC formation, especially if there are multiple initiation sites. Indeed, there were some indications that SC formation may be affected by the breakpoints. The C(3)G staining associated with the FISH foci often stained lightly and in a minority of nuclei was entirely absent near the hybridization signals for BACR22N13 (3/33; Figure 4) or BACR17P04 (5/31). Thus, while SC may form between homologous sequences in the crossover-suppressed regions of translocation heterozygotes, its structure or the organization of the chromosomes may be disturbed.
c(3)G mutant females have defects in the maintenance of homolog pairing:
Since homolog pairing defects were not detected in translocation heterozygotes, FISH experiments were performed in a mutant where we predicted that the homologs would be farther apart than in wild type. We tested if pairing defects could be detected in c(3)G mutant females that lack synapsis between homologs (Smith and King 1968; Rasmussen 1975). These experiments were performed using the c(3)G68 allele, which is a nonsense mutation that does not produce detectable protein (Page and Hawley 2001). On the basis of two criteria, significant pairing defects were observed in c(3)G mutant oocytes (Table 7). First, there were an increased number of nuclei with greater than one focus of hybridization. Second, when there were multiple foci in c(3)G mutant oocytes, the distance between them was greater than what was observed in wild type (Figure 3). Using the third chromosome probe BACR48M21, 8/14 c(3)G nuclei had multiple foci compared to 2/15 in wild type, and the distance between them (an average of 1.3 μm; Table 7) was greater than that seen in wild type (0.51 μm). Similar results were obtained with additional third chromosome probes (Table 7). Using BACR22N13, there were twice as many c(3)G nuclei with multiple foci (17/29, 58.6%) compared to wild type (10/39, 25.6%). When there were multiple foci in a c(3)G mutant, they were usually farther apart than in wild type.
In some c(3)G mutant nuclei, there were three or four foci, indicating separation of the sister chromatids (see materials and methods). The genetic consequences of this observation are probably minimal since c(3)G mutants do not exhibit meiosis II nondisjunction and crossing over is eliminated. Therefore, cohesion is probably not affected at the sister centromeres and, without crossing over, defects in arm cohesion would not be expected to increase the frequency of meiosis I nondisjunction (Hall 1972).
The absence of C(3)G did not eliminate homolog pairing during meiotic prophase. Roughly half (12) of the c(3)G nuclei exhibited only a single focus of BACR22N13 hybridization, indicating that homolog pairing was not completely abolished. With probe BACR48K07, 7/10 c(3)G nuclei had a single focus of staining. Nonetheless, the separated foci in c(3)G nuclei were farther apart (2.1 μm) when compared with wild type (0.62 μm). The frequent occurrence of a single focus of hybridization in c(3)G mutant oocytes suggests that there is a SC-independent mechanism for meiotic pairing of the homologs.
Similar to the experiments described above in wild type, homolog pairing was examined in prepachytene cells (early meiotic prophase or premeiotic cells lacking ORB staining) and nurse cells of c(3)G mutant females (Table 7). In these cells as well, homolog pairing was partially c(3)G dependent. In both the prepachytene and nurse cells, an increase in separated foci was observed in c(3)G mutant females. Thus, even though antibody staining does not reveal C(3)G on the chromosomes of these cells, these results suggest that small amounts of C(3)G can function to promote or maintain homolog pairing. Due to the rapid transition from premeiotic to early meiotic prophase, we could not determine if the prepachytene cells requiring C(3)G for homolog pairing were premeiotic or in early meiotic prophase. Since C(3)G protein is first detected in early meiotic prophase, it is reasonable to suggest that the homologs enter meiosis paired in c(3)G mutant females, but then can rapidly become unpaired. Interestingly, these results suggest that somatic pairing mechanisms are not sufficient to explain the tight alignment along the entire length of meiotic chromosomes. From the earliest stages of meiotic prophase, homolog pairing is partially dependent on C(3)G.
DISCUSSION
Several studies have described crossover suppression in translocation heterozygotes of Drosophila (Roberts 1976), but the mechanism for this phenomenon is not known. The assumption in these prior studies of translocation heterozygotes has been that the reductions in crossing over are caused by a failure to pair or synapse the homologs. For example, the pairing-site model is based on results from studies of the X chromosome and proposes that a small number of sites on each chromosome promote alignment and pairing of the homologs (Hawley 1980). However, there has been no demonstration that translocations cause pairing defects during meiotic prophase in Drosophila.
Boundary sites on chromosome 3R:
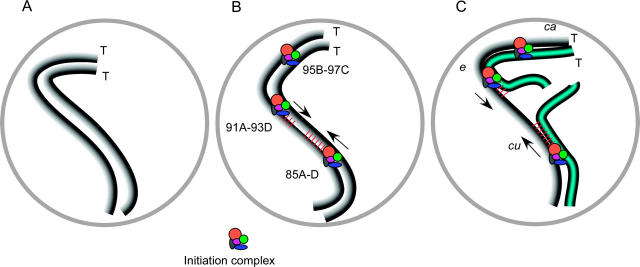
Translocations in Drosophila are region-specific crossover suppressors, where crossover suppression is most severe between discrete sites or boundaries. On the other side of these sites, crossing over is either unaffected or only mildly reduced. On the basis of this principle, sites important for meiotic recombination can be mapped by determining which translocations suppress crossing over in a given interval (Figure 5). Hawley (1980) examined crossing over within small regions in X chromosome translocations and mapped four sites. We have definitive evidence for two sites on chromosome 3R at 85A-C and 91A-93D and have tentatively defined a third site at 95B-97C. Also, a fourth site may be near the telomere if we assume that all recombination must occur between two boundary sites (Hawley 1980).
Figure 5.—
Model for the function of boundary sites and the effects of translocations on synapsis and DSB formation. (A) Prior to SC formation, homologs are aligned along their lengths at <0.5 μm. (B) In wild type, a change in chromosome structure initiates from the boundary sites. This could be SC formation although this has not been shown. The mapped boundary sites are located at cytological locations 85A-D, 91A-93D, and 95B-97C and the telomeres (T) are indicated. (C) The boundary sites are mapped by determining where crossing over is suppressed in a series of translocations. In a T(2;3)C287 heterozygote (blue; chromosome 2 portion of translocation not shown), crossing over is reduced in the cu-e but not in the e-ca regions. We propose that efficient DSB formation requires the continuity of a structure between the boundary sites. It is not known, however, if this structure is an SC component(s). Due to the translocation break, this structure cannot be continuous and SC formation cannot be completed.
Translocation heterozygotes have an early defect in meiotic recombination:
Previous studies on translocation heterozygotes did not differentiate between two possible defects in the meiotic recombination pathway that would lead to the reductions in crossing over. Translocations could affect the initiation of recombination (DSB formation). Alternatively, since DSB repair can result in either a crossover or a simple gene conversion, translocation heterozygosity could influence how the DSBs are repaired. Gene conversion was significantly reduced in the two translocation heterozygotes that were tested, suggesting an early defect in the DSB repair pathway. The reduction of both recombination products is most simply explained by a severe reduction in double-strand-break formation in translocation heterozygotes. However, we cannot rule out that DSBs are formed in translocation heterozygotes and are either not repaired or repaired using a sister chromatid. It is unlikely that the breaks are not repaired since Robert's (1970) study concluded that crossover suppression was not due to loss of chromatids.
DSB-independent synapsis of homologs during meiotic prophase and the role of boundary sites:
On the basis of the strong effect of distal breakpoints, Roberts (1972) concluded that the pairing of homologs initiates in the distal regions of each chromosome arm. In the pairing-site model, homologous chromosomes are brought together through the interactions of specialized regions (Hawley 1980). These models are attractive because the proposed activity of these sites provides a mechanism for a homology search and SC formation in the absence of DSBs (McKim et al. 1998). To directly test the pairing-site model, we employed FISH analysis to investigate homolog pairing in the meiotic nucleus of translocation heterozygotes. If the pairing-site model is correct, we would expect pairing defects in the crossover-suppressed region of translocation heterozygotes.
In contrast to these predictions, no significant homolog pairing defects specifically associated with recombination suppression were observed in translocation heterozygotes. The frequency of FISH signals appearing as two distinct foci and the distance between these foci were usually similar in wild-type and translocation heterozygotes. Furthermore, the FISH foci in crossover-suppressed regions were usually associated with C(3)G staining, indicating that SC can form in the crossover-suppressed regions of translocation heterozygotes. The behavior that we have observed with Drosophila translocations may be similar to other species, with the caveat that light microscopy may not detect small defects in synapsis. Electron microscopy of translocation heterozygotes in tomato, for example, showed that asynapsis was limited to the region around the breakpoints but involved 10–16% of the chromosome arm length (Herickhoff et al. 1993). These data suggest that a defect unrelated to homolog pairing should be considered as the cause of crossover suppression. Indeed, since Drosophila homologs enter meiotic prophase already paired, there may be no need to propose the existence of additional generalized pairing mechanisms during meiosis.
Because homolog pairing appeared normal in translocation heterozygotes, we propose that the reduction in meiotic recombination is due to defects in chromosome structure or organization (Figure 5). Since a single translocation breakpoint suppresses crossing over throughout the region between two boundary sites, structural continuity between these sites appears to be crucial for normal levels of meiotic recombination. We propose that these sites establish chromosomal domains that regulate DSB formation. The nature of these domains is not known, but the boundary sites could help to establish a chromatin structure that facilitates recombination, or there could be a signal for recombination that must travel between two sites, analogous to the phenomenon of interference. Since SC formation defects were observed in some nuclei of translocation heterozygotes, establishing these domains could involve establishing continuous SC between two boundary sites. In addition, a translocation break could cause defects in chromosome structure that affect DSB formation, such as disruption of the transverse or lateral elements. A role for the assembly of SC components in the initiation of recombination is consistent with the observations that mutants in some Drosophila SC components have reduced DSB formation (Jang et al. 2003; S. Mehrotra and K. McKim, unpublished results).
There were two exceptions to the idea that crossing over depends on two flanking boundary sites. First, translocations with breaks in the 94-96 region reduced crossing over throughout chromosome 3R whereas breakpoints proximal to the 91A-93D site suppressed crossing over only within their interval. Roberts (1972) also noted that chromosome 3R translocations with the most severe crossover suppression had breakpoints between divisions 91 and 96. The results with these translocations imply that, while recombination is affected primarily by factors operating between two boundary sites, there are also factors that regulate recombination on a chromosome-wide basis. This effect may represent an important difference between the X chromosome and the autosomes. Hawley (1980) found that distal translocation breakpoints generally did not have an effect on proximal regions.
Second, crossing over within the interval between the centromere and the 85A-D site was not suppressed by translocation breakpoints and in some cases was increased. The proximal regions of most chromosome arms have several other exceptional properties. Crossing over in this region is very low relative to the genome average (McKim et al. 2002) and yet crossing over is often increased in this region in mutants that reduce crossing over in most other regions (Baker et al. 1976; Carpenter 1988; Bhagat et al. 2004). Crossovers in proximal regions may also exhibit positive interference (Green 1975; Sinclair 1975; Denell and Keppy 1979).
Mechanism of SC formation in Drosophila:
Previous studies have suggested that chromosomes are homologously paired prior to meiosis in the female germline (Grell and Day 1970), but these studies were based on the analysis of spread metaphase chromosomes. Our FISH analysis of prophase chromosomes demonstrates the accuracy attained by this alignment in relation to SC formation. We found that prior to SC formation most homologous loci are aligned at a distance of ≤0.4 μm, which is reminiscent of the presynaptic alignment observed in other organisms prior to SC formation (Zickler and Kleckner 1998). Similarly, only a single focus of hybridization was observed in many c(3)G mutant oocytes, indicating that SC-independent pairing forces can bring the homologs together. The SC-independent pairing mechanism could be related to the forces that act in somatic cells (Hiraoka et al. 1993; Fung et al. 1998).
c(3)G mutant oocytes were unable to achieve the accuracy of meiotic homolog pairing observed in wild type. Therefore, somatic pairing mechanisms are not sufficient for homolog pairing during meiotic prophase. We cannot rule out a role for C(3)G in somatic pairing, but this is unlikely since C(3)G staining is not observed until meiotic prophase (Page and Hawley 2001). Since homologs were observed to be paired prior to SC formation in wild type, it is possible that the pairing defects in c(3)G mutant oocytes reflect dissociation of pairing rather than the initial failure to pair. In C. elegans syp-1 mutants, a c(3)G homolog, chromosomes initially pair but then dissociate prematurely (MacQueen et al. 2002). These results suggest that, while mechanisms similar to those that operate in somatic cells might be involved in the initial establishment of pairing, they are not sufficient to maintain meiotic chromosome pairing.
A somatic pairing mechanism does not provide insights into the mechanism of how Drosophila forms SC. In budding yeast, DSB formation has been proposed to be directly involved in SC formation (Zickler and Kleckner 1998; Henderson and Keeney 2004). Another role for DSBs is suggested by the analysis of spo11 mutants in Sordaria, in which both presynaptic alignment of homologs and SC formation are absent (Storlazzi et al. 2003). These results suggest that an important function for DSBs is to bring homologs together prior to SC formation. The exceptional ability of Drosophila females to align chromosomes prior to prophase may be the basis for the difference between organisms that can form SC in the absence of DSBs and those that cannot. SC formation may initiate in the absence of DSB formation once the homologous chromosomes come within ∼0.4 μm of each other. In this model, SC formation could initiate either at many random sites or at specialized sites (see below). SC formation in budding yeast has been proposed to initiate at prospective crossover sites (Fung et al. 2004).
Above we proposed that the boundary sites are involved in established chromosomal domains that regulate DSB formation and that SC components may play a role in this process. An intriguing possibility is that the boundary sites are where SC formation initiates, filling the role ascribed to crossover sites in budding yeast (Figure 5). While only a minority of the translocation heterozygote nuclei had defects in SC formation, our experiments probably underestimated these effects due to the limited resolution of light microscopy. For example, the failure of a FISH signal to be associated with SC may not be detected in all nuclei due to the close proximity of the SCs to other chromosomes. In addition, more pervasive defects in assembling the SC and associated chromatin structure around a translocation breakpoint could go undetected in our studies since they may not be observed at the resolution of light microscopy. Interestingly, Drososphila ord mutants lack classical SC structure by electron microscopy although there is relatively normal C(3)G staining by immunofluorescence (Webber et al. 2004). Further high-resolution experiments using electron microscopy will allow the structure of the SC in the crossover-suppressed regions of translocation heterozygotes to be determined. Finally, understanding the role of these sites will require molecular characterization of their sequences.
Acknowledgments
We are grateful to Li Nguyen for technical assistance and Abby Dernburg for advice early in this project. We also thank Scott Page and Scott Hawley for providing C(3)G antibodies. Some stocks used in this study were obtained from the Bloomington Stock Center and Barbara Wakimoto. The ORB antibodies were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. A grant from the National Science Foundation (MCB-0077705) to K. McKim supported this work.
References
- Baker, B. S., A. T. C. Carpenter, M. S. Esposito, R. E. Esposito and L. Sandler, 1976. The genetic control of meiosis. Annu. Rev. Genet. 10: 53–134. [DOI] [PubMed] [Google Scholar]
- Baudat, F., K. Manova, J. P. Yuen, M. Jasin and S. Keeney, 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Bhagat, R., E. A. Manheim, D. E. Sherizen and K. S. McKim, 2004. Studies on crossover specific mutants and the distribution of crossing over in Drosophila females. Cytogenet. Genome Res. 107: 160–171. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1975. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51: 157–182. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1988 Thoughts on recombination nodules, meiotic recombination, and chiasmata, pp. 529–548 in Genetic Recombination, edited by R. Kucherlapati and G. Smith. American Society for Microbiology, Washington, DC.
- Chovnick, A., G. H. Ballantyne, D. L. Baillie and D. G. Holm, 1970. Gene conversion in higher organisms: half-tetrad analysis of recombination within the rosy cistron of Drosophila melanogaster. Genetics 66: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell, R. E., and D. O. Keppy, 1979. The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics 93: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A. F., J. W. Sedat and R. S. Hawley, 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 85: 135–146. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Ding, D. Q., A. Yamamoto, T. Haraguchi and Y. Hiraoka, 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6: 329–341. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1931. The decrease of crossing-over observed in translocations, and its probable explanation. Am. Nat. 65: 214–232. [Google Scholar]
- Fung, J. C., W. F. Marshall, A. Dernburg, D. A. Agard and J. W. Sedat, 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J. C., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Green, M. M., 1975. Conversion as a possible mechanism of high coincidence values in the centromere region of Drosophila. Mol. Gen. Genet. 139: 57–66. [DOI] [PubMed] [Google Scholar]
- Grell, R. F., and J. W. Day, 1970. Chromosome pairing in the oogonial cells of Drosophila melanogaster. Chromosoma 31: 424–445. [DOI] [PubMed] [Google Scholar]
- Grelon, M., D. Vezon, G. Gendrot and G. Pelletier, 2001. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J. C., 1972. Chromosome segregation influenced by two alleles of the meiotic mutant c(3)G in Drosphila melanogaster. Genetics 71: 367–400. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, K. A., and S. Keeney, 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herickhoff, L., S. Stack and J. Sherman, 1993. The relationship between synapsis, recombination nodules and chiasmata in tomato translocation heterozygotes. Heredity 71: 373–385. [Google Scholar]
- Hilliker, A. J., S. H. Clark and A. Chovnick, 1988 Genetic analysis of intragenic recombination in Drosophila, pp. 73–90 in The Recombination of Genetic Material, edited by K. B. Low. Academic Press, New York.
- Hilliker, A. J., S. Clark and A. Chovnick, 1991. The effect of DNA sequence polymorphisms on intragenic recombination in the rosy locus of Drosophila melanogaster. Genetics 129: 779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka, Y., A. F. Dernburg, S. J. Parmelee, M. C. Rykowski, D. A. Agard et al., 1993. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 120: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. R. Nelson, B. P. Berman, T. R. Laverty, R. A. George et al., 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287: 2271–2274. [DOI] [PubMed] [Google Scholar]
- Jang, J. K., D. E. Sherizen, R. Bhagat, E. A. Manheim and K. S. McKim, 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Lantz, V., J. S. Chang, J. I. Horabin, D. Bopp and P. Schedl, 1994. The Drosophila ORB RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8: 598–613. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, San Diego.
- MacQueen, A. J., M. P. Colaiacovo, K. McDonald and A. M. Villeneuve, 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F., A. F. Dernburg, B. Harmon, D. A. Agard and J. W. Sedat, 1996. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell 7: 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., A. M. Howell and A. M. Rose, 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., K. Peters and A. M. Rose, 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., B. L. Green-Marroquin, J. J. Sekelsky, G. Chin, C. Steinberg et al., 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., J. K. Jang and E. A. Manheim, 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Padmore, R., L. Cao and N. Kleckner, 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S. W., 1975. Ultrastructural studies of meiosis in males and females of the c(3)G mutant in Drosophila melanogaster meigen. C.R. Trav. Lab. Carlsberg 40: 163–173. [Google Scholar]
- Roberts, P. A., 1970. Screening for X-ray-induced crossover suppressors in Drosophila melanogaster: prevalence and effectiveness of translocations. Genetics 65: 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P. A., 1972. Differences in the synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics 71: 401–415. [DOI] [PubMed] [Google Scholar]
- Roberts, P. A., 1976 The genetics of chromosome aberration, pp. 68–184 in The Genetics and Biology of Drosophila, edited by M. Ashburner and E. Novistski. Academic Press, New York.
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Romanienko, P. J., and R. D. Camerini-Otero, 2000. The mouse spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., 1975. Crossing over between closely linked markers spanning the centromere of chromosome 3 in Drosophila melanogaster. Genetics Res. 11: 173–185. [DOI] [PubMed] [Google Scholar]
- Smith, P. A., and R. C. King, 1968. Genetic control of synaptonemal complexes in Drosophila melanogaster. Genetics 60: 335–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner et al., 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, J., A. S. Belmont and J. W. Sedat, 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Villeneuve, A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein, D., S. W. Rasmussen and P. B. Holm, 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18: 331–413. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B. T., and M. G. Hearn, 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, H. A., L. Howard and S. E. Bickel, 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, J. H., 1966. Interchromosomal effects of autosomal translocations on recombination in Drosophila melanogaster. Genetics 54: 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka, M., and A. Rose, 1995. The genetics of meiosis in Caenorhabditis elegans. Trends Genet. 11: 27–31. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]