Abstract
To integrate genetic, physical, and cytological perspectives of the Sorghum bicolor genome, we selected 40 landed bacterial artificial chromosome (BAC) clones that contain different linkage map markers, 21 from linkage group 2 (LG-02) and 19 from linkage group 8 (LG-08). Multi-BAC probe cocktails were constructed for each chromosome from the landed BACs, which were also preevaluated for FISH signal quality, relative position, and collective chromosome coverage. Comparison to the corresponding linkage map revealed full concordance of locus order between cytological and prior segregation analyses. The pericentromeric heterochromatin constituted a large quasi-uniform block in each bivalent and was especially large in the bivalent corresponding to LG-08. Centromere positions in LG-02 and LG-08 were progressively delimited using FISH to identify landed BACs for which the FISH signals visibly flanked the centromere. Alignment of linkage and cytological maps revealed that pericentromeric heterochromatin of these sorghum chromosomes is largely devoid of recombination, which is mostly relegated to the more distal regions, which are largely euchromatic. This suggests that the sorghum genome is thus even more amenable to physical mapping of genes and positional cloning than the C-value alone might suggest. As a prelude to positional cloning of the fertility restorer, Rf1, FISH of BAC clones flanking the Rf1 locus was used to delimit the chromosomal position of the gene. FISH of BACs that contain the most proximal linkage markers enabled localization of Rf1 to a ∼0.4-Mbp euchromatic region of LG-08. Cytogenetic analyses of Rf1 and other trait loci will aid in assessing the feasibility of positional cloning and help formulate strategies required for cloning this and other agriculturally critical genes.
SORGHUM (2n = 20) is the fifth most important cereal (Doggett 1988). Although used worldwide as a grain and forage, it is especially important in the semiarid tropics because of its unusual tolerance of hot, dry environments. The relatively small size of its genome (750–818 Mbp; Arumuganathan and Earle 1991; Price et al. 2005) empirically suggests that sorghum will be highly amenable to structural genomics. Since rice (2n = 24) is related to sorghum and its genome also relatively small (∼62% that of sorghum; Johnston et al. 1999), there are opportunities for rapid advancement and insight concerning comparative genomics between these two grasses. However, a holistic comparison of their genomes will require the assembly of well-integrated resources that include not only sequence data and genetic/physical maps, but also cytological maps.
Integrated genome maps enable map-based isolation of genes, targeted genome sequencing, detailed investigation of genome architecture, comparisons among genomes of related species, and association studies that link DNA markers and genes to important phenotypes. However, efficacy of these applications can be undermined by discrepancies between linkage and physical maps. Although linkage maps are usually good indicators of order among recombinationally resolved genes or markers, a linkage map per se is a poor indicator of molecular sizes and physical distances. Variation of recombination density among grass chromosome regions can be extreme (Künzel et al. 2000; Islam-Faridi et al. 2002). Plant molecular genetic manipulations that are dependent on physical distances, e.g., positional or map-based cloning, have a greater probability of success if they are undertaken with a priori knowledge of the physical size of the trait locus.
Rapid progress has been made in the construction of an integrated Sorghum bicolor genome map. Contributing factors have been the use of a combination of high-throughput amplified fragment length polymorphism (AFLP) DNA marker technology (Klein et al. 2000; Menz et al. 2002), six-dimensional pooling of BAC clones (Klein et al. 2000), cDNA capture technology (Childs et al. 2001), sequence-based alignment of the genomes of sorghum and rice (Klein et al. 2003), and BAC-based fluorescence in situ hybridization (FISH; Islam-Faridi et al. 2002; Kim et al. 2002). The latter two studies demonstrated the feasibility of using FISH of landed BACs to associate all linkage groups with specific sorghum chromosomes and to visibly integrate the genetic recombination frequencies with physical distances along each chromosome at high resolution. A recently constructed 24-point cytogenomic map of mitotic chromosomes has established an integrated nomenclature for chromosomes, arms, and linkage groups of S. bicolor, where all linkage groups are assigned to specific chromosomes and oriented with respect to arms, on the basis of estimated molecular size (Kim et al. 2005).
Physical maps assembled from FISH to pachytene bivalents provide directly visible physical evidence of the order and physical positions on a chromosome of the associated molecular markers and/or genes of interest. Meiotic pachytene bivalents are much longer than somatic metaphase chromosomes and offer greater cytological resolution of the chromosomes (McClintock 1930), especially in plants. Thus, in situ hybridization to pachytene bivalents is being applied increasingly for high-resolution physical and integrative mapping of plant genomes (Shen et al. 1987; Wu 1992; Xu and Earle 1996; Fransz et al. 1998, 2000; Peterson et al. 1999; Zhong et al. 1999; Chen et al. 2000; Song et al. 2000; Islam-Faridi et al. 2002). Detailed pachytene BAC-FISH analysis of sorghum chromosome 1 by Islam-Faridi et al. (2002) demonstrated that multiprobe FISH methods are readily capable of revealing the relationships between physical distances and linkage distances.
In this study, we have improved the high-resolution mapping strategy of Islam-Faridi et al. (2002) and extended application to two additional sorghum linkage groups. Using multiprobe BAC cocktails, the molecular architectures of sorghum linkage groups 2 (LG-02) and 8 (LG-08) were analyzed. In addition, the position and physical size of the fertility restoration locus, Rf1, located on sorghum LG-08 were delimited using FISH analysis with BAC clones containing the most proximal linkage markers. This information is being used to aid in the positional cloning and identification of Rf1.
MATERIALS AND METHODS
Selection of marker-anchored BACs and BAC DNA purification:
The BACs used in this study were from two large-insert genomic libraries derived from sorghum (S. bicolor [L.] Moench.) inbred BTx623 (Woo et al. 1994; Tao and Zhang 1998). Molecular markers spaced ∼5 cM apart across LG-02 and LG-08 (Menz et al. 2002; Kim et al. 2005) were used to select BACs for FISH analysis. In some regions of the genome, clones were selected on the basis of BAC gene content as revealed by sequence scanning (Klein et al. 2003). BAC DNA was isolated by alkaline lysis, digested with EcoRI, and further purified by Plant DNeasy spin columns (QIAGEN, Valencia, CA) as detailed elsewhere (Childs et al. 2001).
Southern hybridization:
EcoRI-digested BAC DNA was resolved on 1% agarose slab gels and blotted to membrane filters (Hybond N+; Amersham Biosciences, Piscataway, NJ). Filters were hybridized to sorghum genomic DNA, processed, and images were detected by autoradiography (Sambrook et al. 1989).
Pachytene chromosome preparation and in situ hybridization:
Pachytene chromosomes were prepared from immature anthers of BTx623 according to the protocol of Zhong et al. (1996) with the exception that chromosomes were spread with a drop of ethanol-acetic acid (3:1) fixative rather than with 60% acetic acid. In situ hybridization techniques were a modification of the protocol by Jewell and Islam-Faridi (1994), as described by Hanson et al. (1996).
Microscopy:
Images were captured from an Olympus AX-70 epifluorescence microscope using a 1.3 megapixel Sensys camera (Roper Scientific, Trenton, NJ) with the MacProbe v.4.2.3 digital image system (Applied Imaging Corp., Santa Clara, CA). To assess relative strengths of FISH signals and their distributions, blue (4′, 6-diamidino-2-phenylindole, DAPI, signal from chromosomal DNA), green (FITC from probe), and red (Cy3 from probe) signals were measured from digital images using Optimas v. 6.0 (Media Cybernetics, Carlsbad, CA). Luminance values were sampled along lines spanning the lengths of the somatic chromosomes or meiotic bivalents. Data were extracted for the Optimas “linear morphology default data collection set” and exported by DDE to a spreadsheet (Microsoft Excel).
RESULTS
BAC probe evaluation by Southern blot hybridization:
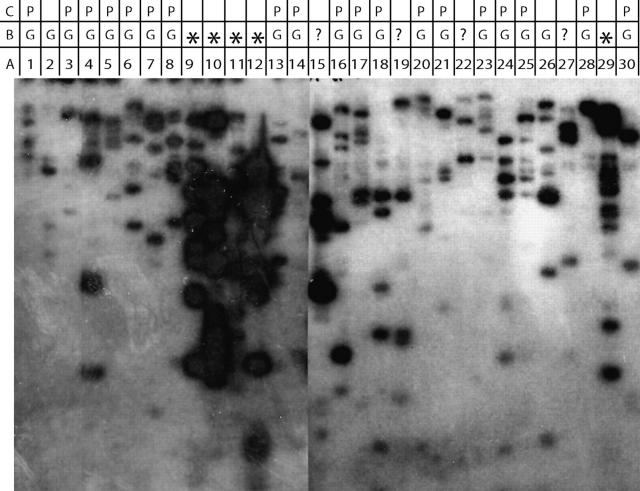
Southern blot hybridization and individual BAC FISH were used to determine if a more rapid analysis such as Southern blots could be used to predict the quality of a FISH signal that would be obtained for a selected BAC. To determine if the amount of sequence repetitiveness in sorghum BACs influenced their ability to yield locus-specific FISH signals without suppression by “blocking” with C0t 1 DNA, Southern blots of BAC DNA were probed with genomic DNA (Hanson et al. 1995) and compared to FISH results. More than 30 BAC clones were selected with LG-08 markers for this comparison (Table 1). Twenty-one of the 30 BACs produced a clean pair of FISH signals while 5 BACS resulted in a “chromosome painting” pattern (not shown) not unlike that expected from FISH of one or more interspersed repetitive elements. The remaining BACs (sbb23799, sbb26136, sbb24657, and sbb16555) yielded no FISH signal. The five BACs (sbb1433, sbb9324 sbb11593, sbb20161, and sbb12305) that yielded strong background signals in FISH experiments also yielded Southern blot signals that were relatively more intense and complex (Figure 1) and strongly indicated that these BACs contain abundant repetitive sequence(s). BACs that yielded a less intense Southern blot signal (sbb10760, sbb18861, sbb9171, sbb7724, sbb24521, sbb16523, sbb10990, sbb4303, sbb18981, sbb23303, sbb18578, sbb12329, sbb19883, sbb18071, sbb14482, sbb23575, sbb16700, 66E20, sbb14774, sbb10453, and sbb2887) yielded good, clean FISH signals and strongly indicated that these BACs contain few or no abundant repetitive sequence(s). These results suggest that initial screening of BAC clones by Southern blot analysis may provide a rapid, cost-effective means of determining the utility of a BAC clone as a FISH probe.
TABLE 1.
List of BACs used for FISH and Southern hybridization analysis, their associated markers, and location on linkage group 8
| Marker | Distance (cM) | BAC clonea |
BAC name | FISH results |
BACs used in probe cocktail |
|---|---|---|---|---|---|
| Xtxp273 | 0 | 1 | sbb10760 | G | P |
| Xtxa3525 | 6.1–13.1 | 2 | sbb18861 | G | Other chr. |
| Xtxa3686 | 20.1–22.9 | 3 | sbb9171 | G | P |
| Xtxp47 | 38.7 | 4 | sbb7724 | G | P |
| Xtxa4117 | 45.5 | 5 | sbb24521 | G | P |
| Xtxa3638 | 51.0–55.3 | 6 | sbb16523 | G | P |
| Xtxa3682 | 63.8–68.1 | 7 | sbb10990 | G | P |
| Xtxa6081 | 72.7–76.4 | 8 | sbb4303 | G | P |
| Xtxa2864 | 77.5–80.2 | 9 | sbb1433 | * | |
| Xtxa3667 | 77.5–80.2 | 10 | sbb9324 | * | |
| Xtxa2711 | 72.7–80.2 | 11 | sbb11593 | * | |
| Xtxa174 | 72.7–80.2 | 12 | sbb20161 | * | |
| Xtxa3968 | 82.5–86.0 | 13 | sbb18981 | G | P |
| Xtxa3856 | 86 | 14 | sbb23303 | G | P |
| Xtxa73 | 88.2–90.7 | 15 | sbb23799 | ? | |
| cdo459 | 99.2 | 16 | sbb18578 | G | P |
| Xtxa388 | 99.2–104.2 | 17 | sbb12329 | G | P |
| Xtxs2065 | 99.2–104.2 | 18 | sbb19883 | G | P |
| Xtxa2582 | 104.2–109.5 | 19 | sbb26136 | ? | |
| Xtxp18 | 109.5–111.2 | 20 | sbb18071 | G | P |
| Xtxa606 | 111.6–113.4 | 21 | sbb14482 | G | P |
| Xtxa3827 | 116.6–122.7 | 22 | sbb24657 | ? | |
| Xtxp105 | 126.1 | 23 | sbb23575 | G | P |
| Xtxa6107 | 129.4–132.9 | 24 | sbb16700 | G | P |
| Xtxa6346 | 132.9–136.8 | 25 | 66E20 | G | P |
| Xtxa588 | 136.8 | 26 | sbb14774 | G | Other chr. |
| Xtxa3876 | 140.4 | 27 | sbb16555 | ? | |
| Xtxa4024 | 143.3–146.3 | 28 | sbb10453 | G | P |
| Xtxa2332 | 147.1 | 29 | sbb12305 | * | |
| Xgap34 | 147.1–149.4 | 30 | sbb2887 | G | P |
G, good locus specificity; *, high background signal; ?, not determined; P, BACs used in probe cocktail; Other chr., BACs FISHed to a chromosome not corresponding to LG-08.
BAC clone numbers correspond to the lane numbers in Figure 1.
Figure 1.—
Southern hybridization of EcoRI-digested DNA of BACs mapped to LG-08 probed with sorghum genomic DNA. (A) Lane numbers correspond to the BAC clone number in Table 1. (B) FISH results: G, good locus specificity; *, high background signal; ?, not determined. (C) BACs used to construct a chromosome-specific multi-BAC probe cocktail for LG-08.
Cytogenetic maps of chromosomes corresponding to linkage groups 2 and 8:
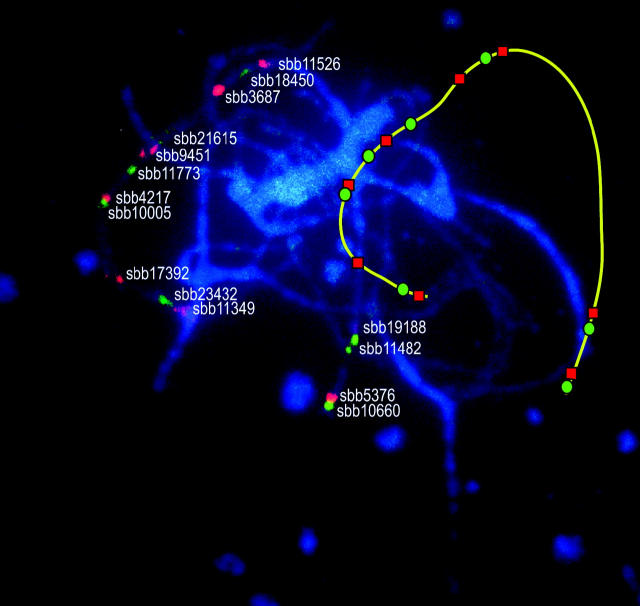
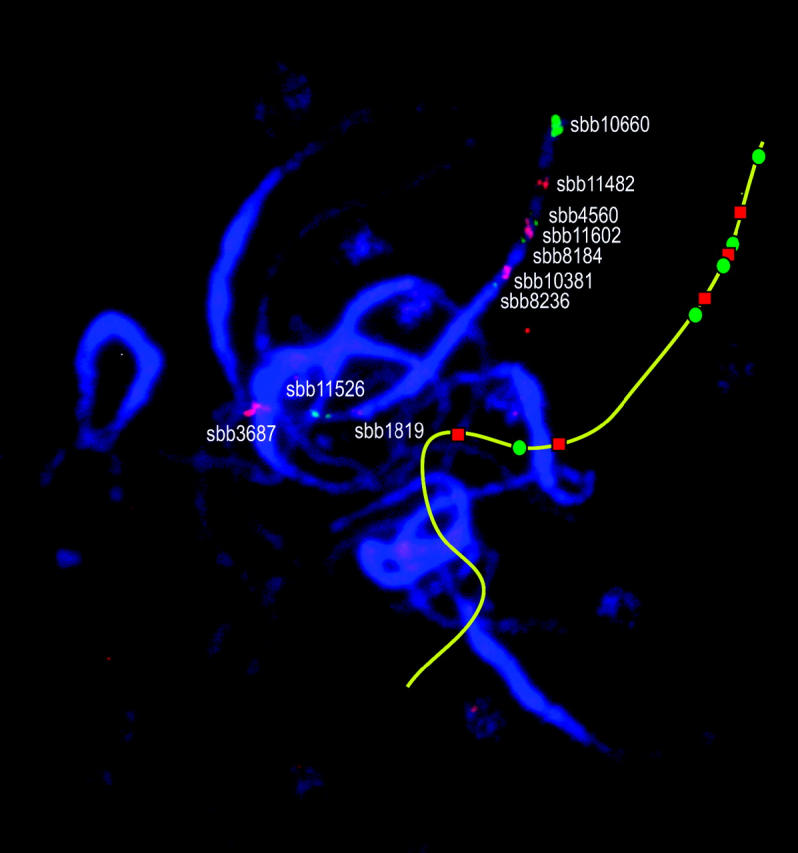
To obtain multiple BAC-FISH signals for LG-02 and LG-08, an initial set of BACs was chosen that included markers at or near the ends of each LG, as well as BACs dispersed at ∼5-cM intervals along the entire LG. For genomic regions where an initial FISH signal was poor or coverage was considered insufficient, additional BACs were chosen for FISH on the basis of BAC gene content (P. E. Klein, personal communication). To create multi-BAC probe cocktails for simultaneous evaluation of multiple BAC-FISH signals, only BACs that resulted in highly locus-specific signals were selected. Of the 40 BACs analyzed from LG-02 by single-color FISH, 21 were selected for FISH-based mapping, 9 from the short arm and 12 from the long arm. Given the cytological proximity of certain BACs, two multi-BAC FISH cocktails with overlapping subsets of these BACs were constructed instead of one complex cocktail (Figures 2 and 3). Four BACs were common to both probe cocktails, namely sbb10660 and sbb11482 from the short arm and sbb3687 and sbb11526 from the long arm (Figures 2 and 3).
Figure 2.—
FISH signals on sorghum pachytene bivalents using a 15-probe cocktail. The identity of each individual BAC clone used in the probe cocktail is given next to its representative FISH signal. An additional line drawing to the side depicts the spatial arrangement of the bivalent corresponding to LG-02 and the FISH signals associated therewith.
Figure 3.—
FISH signals on sorghum pachytene bivalents using a 10-probe cocktail. The identity of each individual BAC clone used in the probe cocktail is given next to its representative FISH signal. An additional line drawing to the side depicts the spatial arrangement of the bivalent corresponding to LG-02 and the FISH signals associated therewith.
FISH signals on pachytene bivalents revealed that all of the 21 BACs originated from euchromatic regions as indicated by the fainter DAPI signal intensity. Linear positions of the FISH signals from the four BACs common to both BAC FISH cocktails were concordant and their alignment enabled a combined analysis of the different BAC-FISH signals (Figure 4, C and D). Peak luminance values were associated with the respective BAC-FISH probes by analysis of luminance levels along a segmented line that collectively spanned the FISH-adorned bivalent. The peaks were used to assign linear positions along the pachytene bivalent and create a cytogenetic map of the pachytene bivalent corresponding to LG-02 (Figure 4B). The order of individual BAC-FISH loci along the chromosome was fully concordant with that of marker loci along the linkage map (Figure 4, A and B). Furthermore, the FISH results resolved the relative order of several DNA markers previously unresolved by marker segregation analysis, specifically Xtxp84 vs. Xtxp50 (25–29 cM) and Xtxa6252 vs. Xtxa3424 (141–145 cM). FISH analysis of BACs linked to these linkage markers clearly showed that SSR marker Xtxp50 is distal to Xtxp84 and AFLP marker Xtxa3424 is distal to Xtxa6252 on LG-02.
Figure 4.—
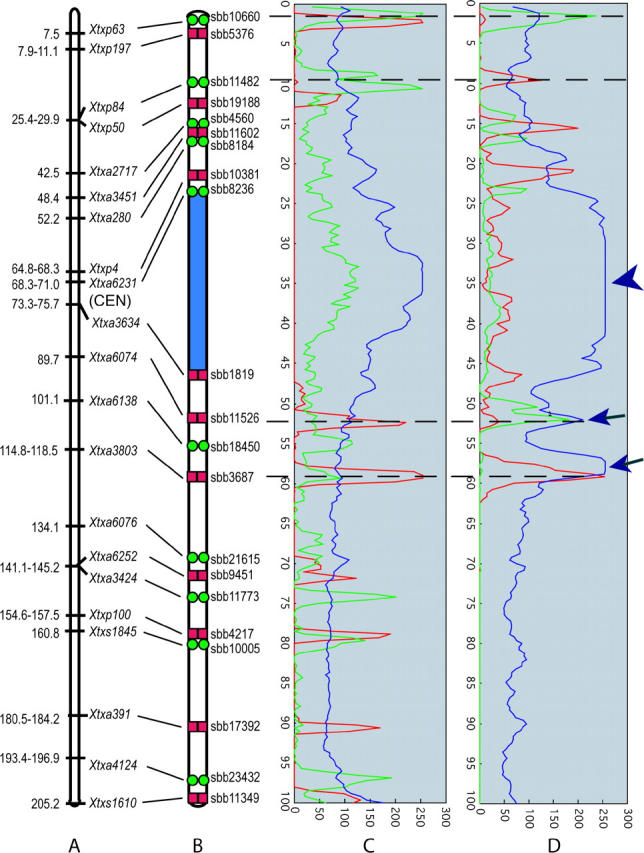
A diagrammatic representation of the cytogenetic locations of 21 sorghum BACs on sorghum LG-02 and the corresponding marker positions (Menz et al. 2002). (A) Linkage group 2 markers used to select BACs for FISH (Menz et al. 2002). (B) Diagram of LG-02 pachytene bivalent indicating the positions of signals from BACs that contain LG-02 marker loci. Green circles, FITC detection; red boxes, Cy3 detection. Signals were located on the basis of the positions of signal peaks from Figure 3. (C and D) Estimation of the strength and location of BAC probes (C for Figure 2 and D for Figure 3). Graph peaks represent relative strengths of fluorescence signals and their distributions; blue represents 4′,6-diamidino-2-phenylindole (DAPI) signal from chromosomal DNA, green represents FITC signal, and red represents Cy3 signal. Dashed lines spanning C and D indicate peaks for probes common to both BAC-FISH cocktails. Arrowhead indicates saturated signal from DAPI and arrows indicate signals of DAPI from overlapping strands of the pachytene bivalents.
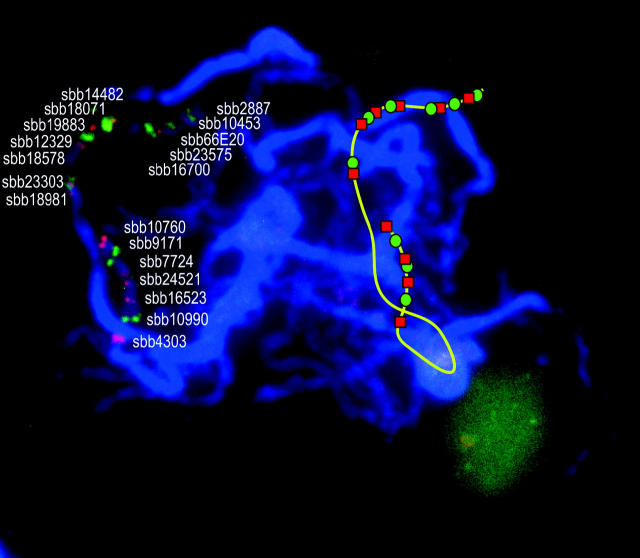
The 21 BACs mapped to LG-08 that yielded single-locus specificity (low background) after single-probe FISH were ordered along the chromosome and the resulting sequence was compared to that of marker loci along the linkage map. Signals from two BACs, sbb14774 and sbb18861, occurred on chromosome(s) other than that corresponding to LG-08. The relative order and synteny of the remaining 19 BACs on the cytomolecular map were determined by pairwise analysis of adjacent BACs using dual-color FISH to pachytene chromosomes. On the basis of these results, a multi-BAC FISH probe cocktail containing the 19 BAC clones was developed. On spreads of pachytene bivalents, FISH signals were readily resolved for each of the 19 BAC components in the multi-BAC probe cocktail (Figure 5). All signals were located in the DAPI-dim euchromatic regions of LG-08 with 12 BACs hybridizing to the long arm and 7 BACs hybridizing to the short arm of LG-08 (Figure 5). Cytogenetic locations were measured as described for LG-02 and used to create a cytogenetic map of LG-08 (Figure 6B). The order of individual BAC-FISH loci along the chromosome was fully concordant with that of molecular marker loci along the linkage map (Figure 6, A and B). Moreover, FISH analysis resolved the order of two LG-08 markers that previously had been unresolved by segregation analysis, namely Xtxs2065 and Xtxa388.
Figure 5.—
FISH signals on sorghum pachytene bivalents using a 19-probe cocktail. The identity of each individual BAC clone used in the probe cocktail is given next to its representative FISH signal. An additional line drawing to the side depicts the spatial of the bivalent corresponding to LG-08 and the FISH signals associated therewith.
Figure 6.—
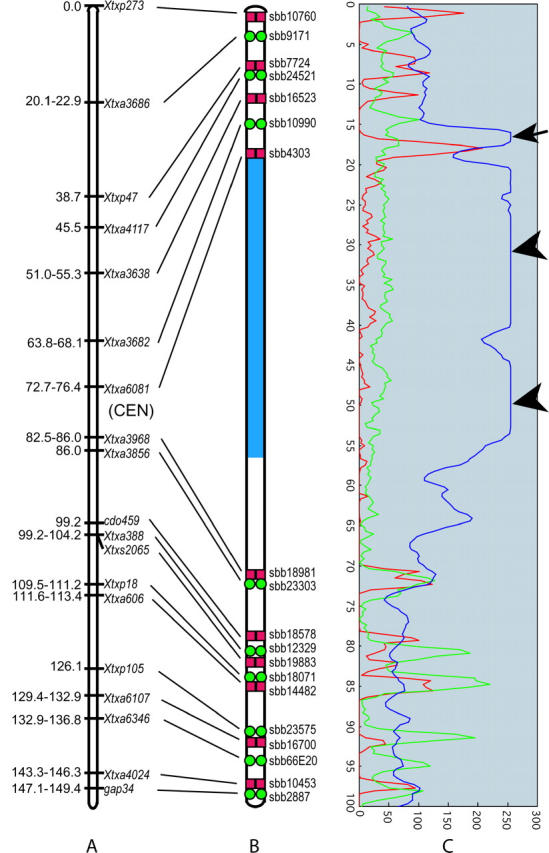
A diagrammatic representation of the cytogenetic locations of 19 sorghum BACs from sorghum LG-08 and the corresponding marker positions (Menz et al. 2002). (A) Linkage group 8 markers used to select BACs for FISH (Menz et al. 2002). (B) Diagram of the pachytene bivalent indicating the positions that contain LG-08 marker loci. Green circles, FITC detection; red boxes, Cy3 detection. (C) Estimation of the strength and location of BAC probes for Figure 5. Graph peaks represent relative strengths of fluorescence signals and their distributions; blue represents DAPI signal from chromosomal DNA, green represents FITC signal, and red represents Cy3 signal. Arrowheads indicate saturated signal from DAPI and arrow indicates signals of DAPI from overlapping strands of the pachytene bivalents.
Physical coverage of chromosomes corresponding to linkage groups 2 and 8 by molecular marker loci:
To assess whether distal ends of the chromosome arms were covered by molecular marker loci, BAC probes corresponding to the end of each LG arm were hybridized to pachytene bivalents and the physical location of the FISH signals was examined. BAC clone sbb10660 containing the LG-02 marker Xtxp63 (mapped to position 7.5 cM; Menz et al. 2002) was revealed by FISH to have originated from very near the end of the short arm of this chromosome, where the physical segment distal to Xtxp63 was ∼1% of the total bivalent length. BAC clone sbb11349 containing LG-02 marker Xtxs1610 (mapped to position 205.0 cM; Menz et al. 2002) was revealed by FISH signal to be syntenic with that from sbb10660 FISH. Signal from sbb11349 was located very near the end of the long arm, where the distal segment was only ∼1% of the total bivalent length. The segment delimited by BAC clones sbb10760 (LG-08 marker Xtxp273) and sbb2887 (LG-08 marker Xgap34) spans map positions 0 to 147.1–149.4 cM, which is nearly all of LG-08 (152.3 cM; Menz et al. 2002). The corresponding FISH signals for these two BACs were very near opposing ends of the chromosome, with distal segments being just 1.2 and 1.6% of the total length of the chromosome bivalents. These results indicate that the molecular markers composing the linkage maps for LG-02 and LG-08 provide excellent coverage of the corresponding chromosomes.
The average ratio between linkage map and physical distances was also estimated for the two chromosomes. The amounts of DNA in the chromosomes that correspond to LG-02 and LG-08 were estimated as 82.5–90 Mbp and 66–72 Mbp, respectively, using relative length measurements of mitotic chromosomes [11% (LG-02) and 8.8% (LG-08); Kim 2003; Kim et al. 2005] and the estimated total genomic DNA content (750–818 Mbp). The estimated DNA content of each respective chromosome (megabase pairs) or segment was then divided by the length of the respective linkage map [205.2 cM (LG-02) and 152.3 cM (LG-08)] to determine the average amount of DNA per unit recombination. The mean ratio observed for LG-02 was 0.40–0.44 Mbp/cM (2.3–2.5 cM/Mbp) and that for LG-08 was 0.43–0.47 Mbp/cM (2.1–2.3 cM/Mbp), while the overall genome average was 0.46–0.51 Mbp/cM (2.0–2.2 cM/Mbp; Kim 2003).
Delimitation of the physical size of centromeric regions:
Linkage group 2:
The location of the centromere on LG-02 was determined by observing the locations of FISH signals of BACs that contain LG-02 marker loci relative to pachytene bivalent pericentromeric heterochromatin. The signal from FISH of BAC sbb8236 revealed that LG-02 marker Xtxa6231 (mapped to 68.3–71.0 cM; Menz et al. 2002) is in the short arm near the heterochromatin-euchromatin junction (Figures 3 and 4). Signal from FISH of BAC sbb1819 indicated that Xtxa3634 (mapped to 73.3–75.7 cM; Menz et al. 2002) is in the long arm, also near the heterochromatin-euchromatin junction (Figures 3 and 4). The interval defined by markers Xtxa6231 and Xtxa3634 spans ∼2.4% of LG-02 (4.85 cM/205.0 cM), whereas the corresponding heterochromatic segment defined by BACs sbb8236 and sbb1819 accounts for ∼22% of the physical length of the pachytene bivalent (Figure 3).
Linkage group 8:
The BAC-FISH signal corresponding to Xtxa6081 (sbb4303) occurred in the short arm near the heterochromatin-euchromatin junction, whereas BAC-FISH signal for Xtxa3968 (sbb18981) occurred in the long arm a short distance from the heterochromatin-euchromatin junction (Figures 5 and 6). Whereas the interval defined by markers Xtxa6081 and Xtxa3968 spans only 6.3% of the total length of LG-08 (9.7 of 152.3 map units; Menz et al. 2002), FISH of BACs sbb4303 and sbb18981 revealed that the corresponding physical segment accounts for >50% of the length of the corresponding pachytene bivalent (Figure 6).
Estimation of the physical size of the Rf1 trait locus on pachytene bivalents:
The Rf1 locus was previously mapped to LG-08 of the high-density linkage map of sorghum (Klein et al. 2001; Menz et al. 2002). BAC clones that contained one of two molecular markers closely flanking the Rf1 locus were used for FISH to assess the physical size of the Rf1 region at its present level of linkage map resolution. FISH of BACs sbb18071 and sbb14482, respectively, revealed that Xtxp18 (2.3 cM from Rf1) and Xtxa606 (∼1.7 cM from Rf1) are physically very close to each other in the middle of the euchromatic region of the long arm of the chromosome corresponding to LG-08 (Figures 5 and 6). The 4-cM segment between sbb18071 and sbb14482 occupied just 0.58% of the total length of the chromosome at pachytene. Under the simplifying assumption of a constant DNA density along chromosomes, the calculated molecular size of the marker-delimited segment that spans Rf1 is 66–72 Mbp × 0.58% = 0.383–0.418 Mbp. This estimate of ∼0.4 Mbp is much shorter than the estimate of 1.8 Mbp that is obtained by simple conversion of the linkage map distance (4 cM) to base pairs on the basis of the estimated average DNA density for the chromosome (∼0.45 Mbp/cM).
DISCUSSION
Several molecular marker-based linkage maps of sorghum have been constructed (e.g., Peng et al. 1999; Bhattramakki et al. 2000; Kong et al. 2000; Menz et al. 2002). Recently, two key steps were taken toward integration of sorghum linkage and physical maps. First, Kim et al. (2002) established a skeletal relationship between linkage group markers and each of the 10 S. bicolor chromosomes. Second, Islam-Faridi et al. (2002) established the first high-resolution integrated map for a single S. bicolor chromosome and demonstrated that detailed information for each sorghum chromosome was feasible and useful.
In this study, sorghum BACs were hybridized to pachytene bivalents to assign their cytological location on the chromosomes corresponding to linkage groups 2 and 8 (Menz et al. 2002). FISH of the BACs selected with markers in LG-02 and LG-08 yielded signals on the corresponding chromosomes, except for two BACs that were selected with linkage group 8 markers but yielded signals on other chromosome(s). These BACs were likely assigned to the incorrect contig due a false-positive AFLP signal (∼5%) during marker analyses of BAC DNA pools (see Klein et al. 2000). The physical order of signals from FISH of the other 40 BACs was fully concordant with the two linkage maps. Therefore, FISH analysis confirmed the relative order of the DNA markers that had been established through segregation analysis (Menz et al. 2002). Results from BAC FISH indicated that the two chromosomes are physically, and thus genetically, well covered by molecular marker loci from the high-density genetic map (Menz et al. 2002), except for the central region occupied by pericentromeric heterochromatin.
The positions of centromeres on the linkage maps were delimited by FISH of marker-containing BACs that yielded locus-specific FISH signals. In sorghum, most BACs giving good single-locus FISH signal originated from euchromatin. Collective results from successive sampling of landed BACs enabled the centromeres to be delimited on the linkage maps. The centromere related to LG-02 occurs in a large heterochromatic region that spans ∼25% of the chromosome, but is confined to a segment of the linkage map that is very short and accounts for only ∼2.4% of its meiotic recombination. The centromere related to LG-08 is situated in a large heterochromatic region that spans ∼50% of the chromosome, but integration with the linkage map revealed that it accounts for merely 6.1% of the meiotic recombination within this chromosome. Disproportionately low rates of recombination also occur in the pericentromeric region of sorghum chromosome 1 (Islam-Faridi et al. 2002). Thus, this similarity across these three chromosomes could suggest that very low recombination is characteristic of the pericentromeric regions of all sorghum chromosomes. A similar phenomenon occurs in chromosomes of wheat, barley, and tomato (Tanksley et al. 1992; Delaney et al. 1995a,b; Sherman and Stack 1995; Künzel et al. 2000). Variation in recombination density and structural rearrangements significantly constrains the utility of linkage maps for molecular endeavors. Upon integration with each other, physical and linkage maps become synergistically useful. Linkage maps become more effective for molecular genetic manipulation, and physical maps become more effective for recombination-based genetic manipulation.
FISH resolved the relative order of several DNA markers that had previously not been resolved by segregation analysis of linkage group 2 (Xtxp84 and Xtxp50; Xtxa6252 and Xtxa3424). FISH also resolved the relative order of two DNA markers that were colocalized in LG-08 (Xtxs2065 and Xtxa388). The physical resolution of linkage maps is subject to regional variation in the level of recombination. In physical regions that are low in recombination, the exact order of DNA markers and hence, associated BAC clones, cannot be easily resolved without complementary information. This study indicates that cytological analysis of somatic and/or pachytene sorghum chromosomes can eliminate ambiguity in at least certain regions of sorghum linkage maps. The value of such analyses will vary by map and population and will tend to be highest for maps derived from wide crosses. Interspecific crosses, for example, are commonly used to facilitate map development due to elevated rates of marker polymorphism (Bowers et al. 2003). However, the high incidence of translocations, inversions, and other abnormalities between genetically distant parents has typically been ignored in terms of the impact on resulting maps. In general, physical mapping can help reveal such cryptic problems in linkage maps and resolve some of the recognized ones. The wide applicability of FISH across plant taxa suggests that FISH of marker-landed probes could enhance development and usefulness of plant structural genomics resources.
Most BAC libraries are composed of genomic clones that are 100–200 kbp in size, so the targets of BAC FISH are relatively large and easy to detect. Such large genomic clones, however, are more likely than small clones to contain dispersed repetitive sequences that cause high background signal after FISH (Hanson et al. 1995). With the aim of simultaneously localizing many BACs on individual sorghum chromosome spreads, we deemed it especially important to select BACs with relatively low repetitive sequence content and relatively high gene content or at least high unique sequence content. Detailed integration of maps across the entire genome will require the identification of many BACs that are amenable to multiprobe FISH. Evaluation of each BAC by single-probe FISH would be very time-consuming, so overall efficacy can be enhanced by development of facile non-FISH methods that enable selection of BACs likely to yield locus-specific FISH signals and/or enhance their evaluation prior to FISH-based testing. BACs were selected on the basis of two different methods, Southern hybridization (Hanson et al. 1995) and BAC DNA sequence scan analyses. Five of the six sorghum BACs that yielded exceptionally strong (highest intensity and complexity) signal after Southern hybridization were also characterized by FISH; all five also yielded the strongest background signals after FISH, reflective of a higher content of repetitive sequences. BACs that yielded only moderate, less intense signals after Southern hybridization with genomic DNA yielded a good, clean signal after FISH. Since numerous BACs can be tested per membrane, Southern hybridization enabled a facile means to screen large numbers of sorghum BAC clones likely to be amenable to FISH. However, all six BACs that were selected on the basis of sequence scan data also yielded good FISH signals. Thus, in species like sorghum where sequence data is already available (Klein et al. 2003), in silico sequence scanning methods will also be a powerful means to select BACs.
A key goal of integrative mapping is alignment of linkage group markers relative to major chromatin features. Four (nos. 9–12) of the five BACs that yielded high background FISH signal were shown by integrative mapping to have originated from the pericentromeric heterochromatin. Linkage mapping indicated that four of the five respective marker loci are recombinationally clustered and indicated which loci flank them, but not where they physically reside in the chromosome. FISH signal from the BACs that contained any of these four markers was so widely dispersed that we did not attempt to map them directly by FISH. Instead, we delimited their locations to pericentromeric heterochromatin regions by FISH of BACs that contain flanking linkage markers and yield single-locus FISH signals. This indirect approach might be used systematically to target BACs and contigs from specific heterochromatin regions and to identify boundaries between hetero- and euchromatin. Given the relatively high density of repetitive elements among BACs from pericentromeric heterochromatin, it seems likely that smaller genomic clones and selection for relatively low repetitive sequence content and/or high gene content will be required to find clones that yield single-locus FISH signal and enable physical mapping within pericentromeric heterochromatin blocks.
There are several possible approaches to physical map development through FISH of multiple probes. Among these are single-, dual-, and multiprobe FISH. In some instances, a single slide has been used for multiple rounds of FISH, each with a different probe or small set of probes (Cheng et al. 2001). The purposeful selection of BACs for multiprobe FISH (Islam-Faridi et al. 2002; Kim et al. 2002) offers several advantages. This approach enables greater efficacy, as a single FISH run requires less time to conduct and is simpler to interpret. Additionally, repeated FISH of a slide progressively reduces the quality of preparations in each subsequent round. On the other hand, simultaneous FISH of multi-BAC cocktails can lead to confluence of independent FISH signals. The problem is greatest at mitotic metaphase, when chromosomes are shortest and neighboring signals are thus closest. Such confluence of signals among closely spaced BACs also indicates that BAC-based cocktails offer a highly flexible approach for creating “paints” for specific chromosomes, arms, and segments, i.e., by coordinated use of linkage maps and ordered large-insert libraries (Schubert et al. 2001). Although pachytene bivalents are much longer than mitotic chromosomes, the problem of signal confluence arises with greater numbers of BACs in a chromosome-specific cocktail. Strategies to address this problem will be needed as the number and thus physical density of FISH signals rises. We successfully used a simple strategy that can be employed to help address such situations; i.e., we used “overlapping” probe cocktails that included some common BACs, which enabled facile multipoint alignment across chromosome (bivalent) spreads. By extension, any number of BACs could be used to create a common map.
Two methods were used to estimate the size of the segment defined by the two molecular markers most closely flanking the Rf1 locus. One measure was based on the linkage map distances and the average megabase pair/centimorgan ratio for this chromosome; it yielded an estimate of 1.8 Mbp. The other measure was based on cytological distance and the average DNA density per chromosome length; it yielded an estimate of 0.4 Mbp. FISH revealed that the Rf1 locus is located in euchromatin, so the DNA density in its vicinity is likely much lower than the chromosomal average. Indeed, the disparity between the estimates (0.4 Mbp vs. 1.8 Mbp) supports this view and the ratio of 0.4 Mbp/4 cM suggests that the density between Xtxp18 and Xtxa606 is ∼0.100 Mbp/cM. Considering that the distance between the two BACs spanning the Rf1 locus was near the limit of resolution along sorghum pachytene bivalents, our findings are quite similar to reports from other plants. In tomato, pachytene FISH can resolve probes separated by 1.2 Mbp in heterochromatic regions and 120 kbp in euchromatic regions (de Jong et al. 1999) while the resolution in euchromatic and heterochromatic regions in Arabidopsis thaliana is reportedly 60 and 140 kbp, respectively (de Jong et al. 1999). In rice, BAC clones separated by ∼100 kbp were spatially resolvable on rice pachytene chromosomes (Cheng et al. 2001).
The data indicate that the Rf1 gene is in a highly recombinant segment of euchromatin, which suggests that it is a realistic target for cloning using a map-based strategy. We are currently generating further recombinants and anchoring BACs in this region for this purpose. Once isolated, the Rf1 gene will serve as an important tool for elucidating the developmental regulation of this fertility-restorer gene. In addition, this cytogenetic analysis can be applied to other genes targeted for positional cloning to assess the feasibility of this arduous task. Conventional methods for positional cloning are not well suited for heterochromatic regions and hence an early assessment of the chromosomal position and physical size of the targeted locus is warranted. Clearly, FISH mapping of specific markers to meiotic pachytene chromosomes adds an extra dimension to contemporary sorghum genomics. FISH can provide crucial physical information to positional cloning projects that might otherwise be fruitlessly aimed at a target gene on the basis of markers that are very tightly linked but physically distant. Moreover, the integrated cytogenetic molecular map permits the estimation of distances up to megabase pairs between markers along the chromosome.
Acknowledgments
We thank William L. Rooney, Department of Soil and Crop Sciences, for providing inflorescences for meiotic preparations and seed for mitotic preparations. We gratefully acknowledge support by the Texas Agricultural Experimental Station, Texas A&M University, and the Perry Adkisson Chair (J.E.M.) and grants from the Texas Higher Education Coordinating Board Advanced Technology Program (D.M.S. and H.J.P.), the National Science Foundation Plant Genome Grants DBI-0077713 (J.E.M. and P.E.K.) and DBI-0321578 (P.E.K., J.E.M. and R.R.K.), and the United States Department of Agriculture Agricultural Research Service (R.R.K.).
References
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Bhattramakki, D., J. Dong, K. Chhabra and G. E. Hart, 2000. An integrated SSR and RFLP linkage map of Sorghum bicolor (L.) Moench. Genome 43: 988–1002. [PubMed] [Google Scholar]
- Bowers, J. E., C. Abbey, S. Anderson, C. Chang, X. Draye et al., 2003. A high-density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165: 367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. C., C. M. Chen, F. C. Hsu, C. J. Wang, J. T. Yang et al., 2000. The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101: 30–36. [Google Scholar]
- Cheng, Z., G. G. Presting, C. R. Buell, R. A. Wing and J. Jiang, 2001. High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157: 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs, K. L., R. R. Klein, P. E. Klein, D. T. Morishige and J. E. Mullet, 2001. Mapping genes on an integrated sorghum genetic and physical map using cDNA selection technology. Plant J. 27: 243–255. [DOI] [PubMed] [Google Scholar]
- de Jong, J. H., P. Fransz and P. Zable, 1999. High resolution FISH in plants—techniques and applications. Trends Plant Sci. 4: 258–263. [DOI] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. a Cytogenetically based physical maps of the group-2 chromosomes of wheat. Theor. Appl. Genet. 91: 568–573. [DOI] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. b Cytogenetically based physical maps of the group-3 chromosomes of wheat. Theor. Appl. Genet. 91: 780–782. [DOI] [PubMed] [Google Scholar]
- Doggett, H., 1988 Sorghum, Ed. 2. John Wiley & Sons, New York.
- Fransz, P. F., S. Armstrong, C. Alonso-Blanco, T. C. Fischer, R. A. Torres-Ruiz et al., 1998. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13: 867–876. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., S. Armstrong, J. H. de Jong, L. D. Parnell, C. van Drunen et al., 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376. [DOI] [PubMed] [Google Scholar]
- Hanson, R. E., M. S. Zwick, S. D. Choi, M. N. Islam-Faridi, T. D. McKnight et al., 1995. Fluorescent in situ hybridization of a bacterial artificial chromosome. Genome 38: 646–651. [DOI] [PubMed] [Google Scholar]
- Hanson, R. E., M. N. Islam-Faridi, E. A. Percival, C. F. Crane, T. D. McKnight et al., 1996. Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105: 55–61. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi, M. N., K. L. Childs, P. E. Klein, G. Hodnett, M. A. Menz et al., 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell, D. C., and M. N. Islam-Faridi, 1994 Details of a technique for somatic chromosome preparation and C-banding of maize, pp. 484–493 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Johnston, J. S., M. D. Bennett, A. L. Rayburn, D. W. Galbraith and H. J. Price, 1999. Reference standards for determination of DNA content of plant nuclei. Am. J. Bot. 86: 609–613. [PubMed] [Google Scholar]
- Kim, J.-S., 2003 Genomic analysis of sorghum by fluorescence in situ hybridization. Ph.D. Dissertation, Texas A&M University, College Station, TX.
- Kim, J.-S., K. L. Childs, M. N. Islam-Faridi, M. A. Menz, R. R. Klein et al., 2002. Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45: 402–412. [DOI] [PubMed] [Google Scholar]
- Kim, J.-S., P. E. Klein, R. R. Klein, H. J Price, J. E. Mullet et al., 2005. Chromosome identification and nomenclature of Sorghum bicolor. Genetics 169: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, S. W. Cartinhour, P. E. Ulanch, J. Dong et al., 2000. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 10: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, J. Vrebalov and J. E. Mullet, 2003. Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34: 605–621. [DOI] [PubMed] [Google Scholar]
- Klein, R. R., P. E. Klein, A. K. Chhabra, J. Dong, S. Pammi et al., 2001. Molecular mapping of the rf1 gene for pollen fertility restoration in sorghum (Sorghum bicolor L.). Theor. Appl. Genet. 102: 1206–1212. [DOI] [PubMed] [Google Scholar]
- Kong, L., J. Dong and G. E. Hart, 2000. Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs). Theor. Appl. Genet. 101: 438–448. [Google Scholar]
- Künzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1930. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of Zea mays. Proc. Nat. Acad. Sci. USA 16: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz, M. A., R. R. Klein, J. E. Mullet, J. A. Obert, N. C. Unruh et al., 2002. A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol. Biol. 48: 483–499. [DOI] [PubMed] [Google Scholar]
- Peng, Y., K. F. Schertz, S. Cartinhour and G. E. Hart, 1999. Comparative genome mapping of Sorghum bicolor (L.) Moench using an RFLP map constructed in a population of recombinant inbred lines. Plant Breed. 118: 225–235. [Google Scholar]
- Peterson, D. G., N. L. V. Lapitan and S. M. Stack, 1999. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, H. J., S. L. Dillon, G. Hodnett, W. Rooney, L. Ross et al., 2005. Genome evolution in the genus Sorghum (Poaceae). Ann. Bot. 95: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schubert, I., P. F. Fransz, J. Fuchs and J. H. de Jong, 2001. Chromosome painting in plants. Methods Cell Sci. 23: 57–69. [PubMed] [Google Scholar]
- Shen, D. L., Z. F. Wang and M. Wu, 1987. Gene mapping on maize pachytene chromosomes by in situ hybridization. Chromosoma 95: 311–314. [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141: 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., F. Dong and J. Jiang, 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43: 199–204. [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Q., and H. B. Zhang, 1998. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 26: 4901–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, S. S., J. Jiang, B. S. Gill, A. H. Paterson and R. A. Wing, 1994. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T. P., 1992. B-chromosomes in Sorghum stipoideum. Heredity 68: 457–463. [Google Scholar]
- Xu, J., and E. D. Earle, 1996. High resolution physical mapping of 45S (5.8S, 18S and 25S) rDNA gene loci in the tomato genome using a combination of karyotyping and FISH of pachytene chromosomes. Chromosoma 104: 545–550. [DOI] [PubMed] [Google Scholar]
- Zhong, S. B., J. H. de Jong and P. Zabel, 1996. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 4: 24–28. [DOI] [PubMed] [Google Scholar]
- Zhong, X. B., J. Bodeau, P. F. Fransz, V. M. Williamson, A. van Kammen et al., 1999. FISH to meiotic pachytene chromosomes of tomato locates the root knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor. Appl. Genet. 98: 365–370. [Google Scholar]