Abstract
In Schizosaccharomyces pombe, meiosis-specific DNA breaks that initiate recombination are observed at prominent but widely separated sites. We investigated the relationship between breakage and recombination at one of these sites, the mbs1 locus on chromosome I. Breaks corresponding to 10% of chromatids were mapped to four clusters spread over a 2.1-kb region. Gene conversion of markers within the clusters occurred in 11% of tetrads (3% of meiotic chromatids), making mbs1 a conversion hotspot when compared to other fission yeast markers. Approximately 80% of these conversions were associated with crossing over of flanking markers, suggesting a strong bias in meiotic break repair toward the generation of crossovers. This bias was observed in conversion events at three other loci, ade6, ade7, and ura1. A total of 50–80% of all crossovers seen in a 90-kb region flanking mbs1 occurred in a 4.8-kb interval containing the break sites. Thus, mbs1 is also a hotspot of crossing over, with breakage at mbs1 generating most of the crossovers in the 90-kb interval. Neither Rec12 (Spo11 ortholog) nor I-SceI-induced breakage at mbs1 was significantly associated with crossing over in an apparently break-free interval >25 kb away. Possible mechanisms for generating crossovers in such break-free intervals are discussed.
IN eukaryotic cells, DNA double-strand breaks (DSBs) are a dangerous form of DNA damage and yet are also crucial for the process of meiosis. During vegetative growth, DSBs are a threat because, left unrepaired, they cause chromosomal fragmentation. In addition, repair of DSBs can promote genomic instability, including DNA rearrangements resulting from homologous recombination. In contrast, the deliberate generation of DSBs appears to be a conserved feature of meiosis precisely because of their ability to promote homologous recombination and generate crossovers. Crossovers, in turn, are critical for correct meiotic chromosome segregation and the promotion of genetic diversity among the products of meiosis. The importance of DSB formation in generating meiotic crossovers has been well established in Saccharomyces cerevisiae and Schizosaccharomyces pombe and inferred in other organisms (Sun et al. 1989; Cao et al. 1990; Cervantes et al. 2000; Keeney 2001). In contrast, the mechanistic relationship between the location and frequency of DSBs and the location and frequency of crossovers is less well understood.
It is commonly held that in S. cerevisiae meiotic breaks and crossovers are coincident, but detailed examination of the location of DSBs and crossovers in S. pombe revealed a different pattern. DSBs are distributed in preferred sites at widely separated physical distances (∼50–100 kb on average). In contrast, crossovers are observed at a relatively constant frequency per unit of physical distance (Young et al. 2002); i.e., crossovers are not consistently more frequent in intervals where strong breaks are observed than in apparently break-free intervals. It was proposed that the DSB and crossover patterns could be reconciled if meiotic DSBs are able to generate crossovers over large distances (tens of kilobases) around their positions (Young et al. 2002).
Two factors control how the frequency of DSBs relates to the frequency of crossovers. First, DSBs can be repaired using either sister chromatids or homologous chromosomes (homologs), but genetically observable crossovers can arise only from interhomolog events. The relative frequency of intersister and interhomolog events will therefore affect the proportion of DSBs that generate observable crossovers. Second, interhomolog recombination does not always generate crossovers. More precisely, gene conversions (which indicate interhomolog recombination events) can occur with or without associated crossovers. For example, at the S. cerevisiae ARG4 locus only ∼30% of gene conversions are accompanied by a crossover (Gilbertson and Stahl 1996), while at the S. pombe ade6 locus ∼60% are accompanied by a crossover (Grimm et al. 1994). In other organisms, values as low as 15% and as high as 75% have been reported (Whitehouse 1982).
It was long thought that gene conversions with crossovers and those without crossovers resulted from random cutting of a single kind of recombination intermediate, the four-strand Holliday junction. Such random cutting should produce equal numbers of crossovers and noncrossovers. Recently, however, models that generate crossover and noncrossover products via two separate pathways have been proposed, only one of which involves resolution of Holliday junctions, with the second proceeding by synthesis-dependent strand annealing (SDSA). Holliday junction resolution may always result in crossovers, with noncrossovers arising solely from the second pathway (Allers and Lichten 2001; Cromie et al. 2001; Hunter and Kleckner 2001; Osman et al. 2003; Smith et al. 2003).
In many organisms the relationship between the frequency of meiotic DNA breakage and crossing over is further complicated by the phenomenon of interference, where the occurrence of one crossover inhibits additional crossovers in a surrounding region of variable size. However, while the budding yeast S. cerevisiae exhibits interference, some other fungi, including the fission yeast S. pombe, do not (Strickland 1958; Munz 1994). It should be simpler to study the relationship between DSB and crossover frequencies in organisms lacking interference.
We set out to examine the location and frequency of recombination events associated with mbs1 (meiotic break site 1), a naturally occurring meiotic break hotspot in S. pombe (Young et al. 2002). We wished to know if mbs1 is a conversion hotspot and, in addition, how important it is for generating nearby and distant crossovers. We also wished to measure how often crossovers are associated with gene conversions at mbs1 and at other loci as a measure of the proportion of interhomolog recombination events that give crossovers. Values consistently higher or lower than 50% would indicate that crossovers and noncrossovers are not generated by random Holliday junction resolution in S. pombe.
MATERIALS AND METHODS
Yeast strains and genetic techniques:
The S. pombe strains used in this study, and their genotypes, are listed in Table 1. Meiotic crosses were carried out by suspending single yeast colonies in 5 ml of supplemented yeast extract liquid medium + adenine (100 μg/ml; Gutz et al. 1974) and growing at 30° until saturated. For each cross, aliquots of 100 μl from two saturated cultures were mixed, washed with water, and spotted on sporulation agar plates (SPA; Gutz et al. 1974). After 2 days incubation at 25°, the cell-ascus mixture from each spot was suspended in 1 ml of water and treated with glusulase and ethanol to kill vegetative cells, essentially as described by DeVeaux et al. (1992). Tetrad dissection is described in Gutz et al. (1974).
TABLE 1.
S. pombe strains used in this study
| Strains | Genotypea |
|---|---|
| GP789 | h+ ade6-M26 ura1-171 |
| GP877 | h− ade6-52 ura1-61 |
| GP1252 | h− ade6-52 ura4-D18 ura4+-aim tps16-23b |
| GP3362 | h+ ade6-M26 his3-D1 ura4-D18 mbs1-1::ura4+ |
| GP3398 | h+ ade6-216 leu2-32 ura4-D18 lys1-37 rec12-171::ura4+ |
| GP3410 | h− ade6-3008 ura1-61 rqh1-h2 pom1::3HA.kan |
| GP3477 | h+ mbs1-2 rqh1-h2 |
| GP3672 | h+ ade6-M26 mbs1-2 rec12-172 |
| GP3673 | h+ ade6-M26 rec12-172 |
| GP3674 | h− ade6-M26 mbs1-2 rec12-172 |
| GP3677 | h− ade6-M26 mbs1-2 rec12-173 |
| GP3696 | h− lys3-37 ura1-61 rqh1-h2 |
| GP3718 | h+ ade6-3049 rad50S pat1-114 end1-458 |
| GP3731 | h+ ade6-M26 mbs1-102 his3-D1 |
| GP3785 | h+ ade6-3034 ura4-D18 arg1-14 leu1-32 |
| GP3801 | h+ ade6-52 lys3-37 ura1-61 mbs1-2 rqh1-h2 rec12-172 |
| GP3802 | h− ade6-52 lys3-37 ura1-61 mbs1-2 rqh1-h2 rec12-172 |
| GP3805 | h− ade6-52 lys3-37 ura1-61 rqh1-h2 rec12-172 |
| GP3806 | h+ ade6-52 lys3-37 ura1-61 mbs1-2 rqh1-h2 rec12-173 |
| GP3839 | h+ ade6-M26 pat1-114 rad50S |
| GP4282 | h+ ade6-210 ura4-D18 mbs1-22::ura4+ |
| GP4514 | h+ ade6-216 ura1-61 mbs1-102 |
| GP4515 | h− ade6-3049 mbs1-103 rqh1-h2 |
| GP5091 | h+ ade7-C8 ura5-294 |
| GP5125 | h− swi5-201::kanMX6 ade7-50 |
| GP5145 | h+ ade6-M26 his3-D1 ura4-D18 mbs1-102 |
Mutations other than commonly used auxotrophies and mating-type alleles are described in the following references: mbs1 alleles (this work), pat1-114 (Iino and Yamamoto 1985), pom1::3HA.kan (Bahler and Pringle 1998), rad50S (Farah et al. 2002), rec12-172 and 173 (this work), rqh1-h2 (Enoch et al. 1992; Stewart et al. 1997), swi5-201::kanMX6 (Ellermeier et al. 2004), tps16-23 (Gygax and Thuriaux 1984), and ura4+-aim (Zahn-Zabal et al. 1995).
tps16 mutations map to the ags1 gene (F. Hochstenbach, personal communication).
Scoring of the restriction site markers 1–4, L, and R (see Figure 4) was done by colony PCR followed by digestion with the appropriate restriction enzyme and gel electrophoresis. Auxotrophic markers were scored by replica plating onto appropriately supplemented nitrogen-base minimal agar (Ponticelli and Smith 1989). Replica plating onto yeast extract agar + phloxin B plates (Moreno et al. 1991) at 37° was used to score tps16. In the case of the cross B tetrad analysis (Figure 2), ura4 was scored by FOAR. The ura1 marker was scored directly among ura4+ colonies. In ura4− spore colonies, the ura1 genotype was inferred from the genotypes of the ura4+ colonies (i.e., if two colonies were ura4+ ura1+, it was assumed that the other two colonies were ura4− ura1−) or scored on the basis of the configuration of flanking markers (i.e., if the flanking markers lys3 and rqh1 were in the parental configuration, then it was assumed that ura1 was also). This left only one ambiguous tetrad.
Figure 4.—
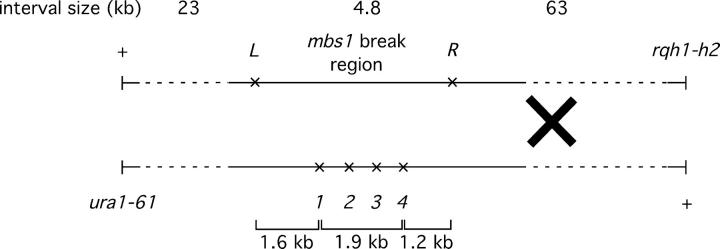
Additional markers at mbs1 used to identify the position of ura1-rqh1 crossovers. PmlI (L) and XbaI (R) recognition sites were generated as shown by single-base-pair mutations (at positions 5213 and 9977 with respect to the SPAC4G8 cosmid). Cross F is shown (GP4514 × GP4515).
Figure 2.—
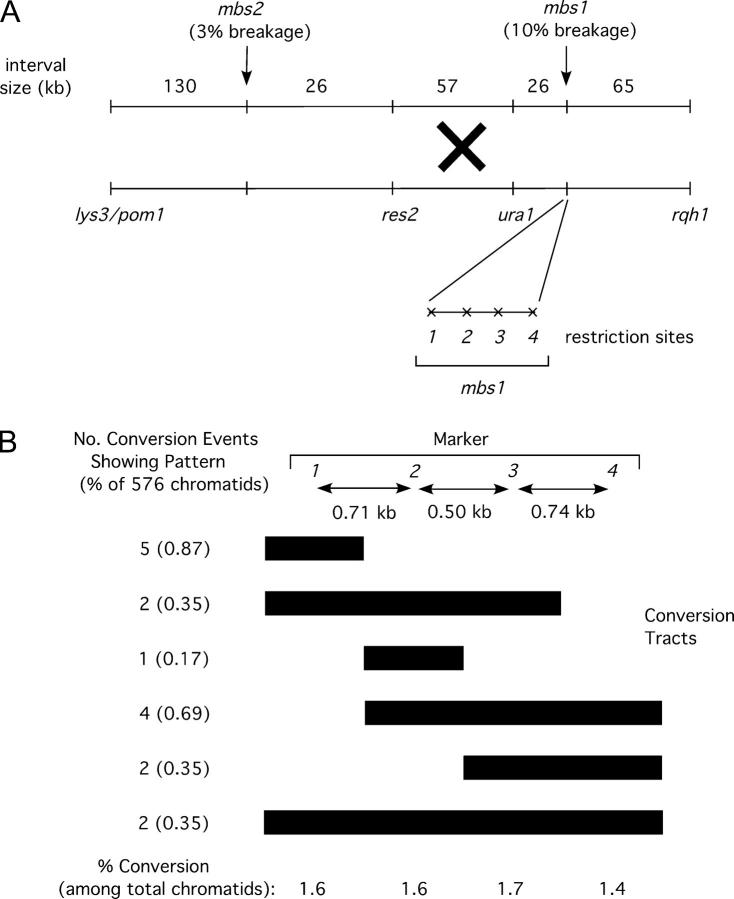
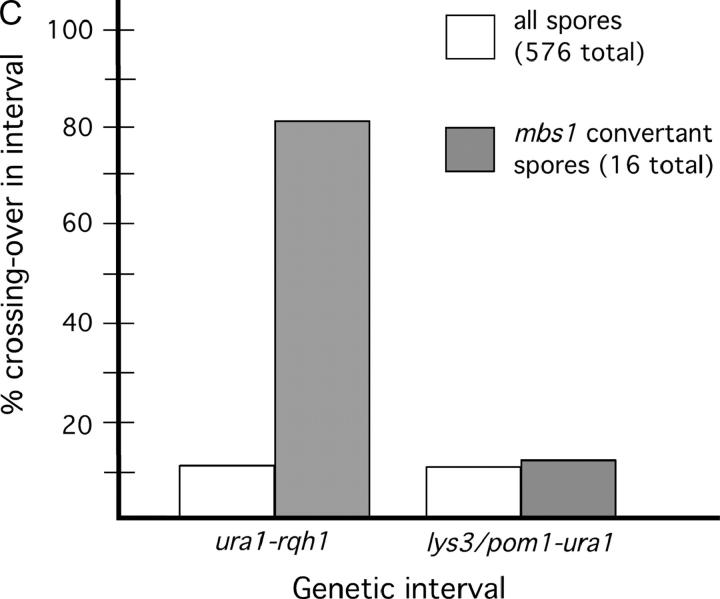
The mbs1 locus is a hotspot for gene conversion, highly associated with ura1-rqh1 crossovers. (A) Map of mbs1 and surrounding region showing markers used to measure crossover frequencies and the four restriction sites used to monitor gene conversion at mbs1. The physical sizes of the genetic intervals are shown along with observed meiotic break frequencies at sites mbs1 and mbs2. The restriction sites (1–4) in Figure 1C were scored by PCR and restriction. (B) Conversion patterns of markers 1–4 observed at mbs1 from 144 tetrads. Tetrads from two crosses (cross A—GP3696 × GP3731, 94 tetrads; cross B—GP3410 × GP5145, 50 tetrads) were scored for restriction sites to identify conversion frequency and distribution (see supplemental Figure 1 at http://www.genetics.org/supplemental/). Ten conversion tetrads were identified in cross A (including one apparent double-conversion tetrad) and five in cross B. Bars extend under the markers converted; endpoints are arbitrarily placed at midpoints between the markers scored. (C) Crossovers in the interval ura1-rqh1, but not in the interval lys3/pom1-ura1, are associated with mbs1 conversions. The frequencies of crossovers in the intervals lys3/pom1-ura1 and ura1-rqh1 are shown among total spores and among mbs1 conversion spores. The marker lys3 was used in cross A and the marker pom1 in cross B; these markers are only ∼4 kb apart but are ∼210 kb from ura1 so the data for the crosses were pooled.
In the I-SceI experiments (Table 2), potential diploids were identified as apparent double crossovers between lys3-ura1 and ura1-rqh1, giving a wild-type phenotype for all of these markers. They were confirmed as diploids by iodine staining on SPA plates (i.e., h+/h−) or by DNA content determined by flow cytometry.
TABLE 2.
Crossover stimulation near and far from an artificialmbs1 break
| % crossing over in interval |
|||
|---|---|---|---|
| Crossa | lys3-ura1 | ura1-rqh1 | No. of colonies analyzed |
| Active (cross C) | 0.33 | 2.4 | 2123c |
| No site (cross D) | <0.25b | 0.25 | 1193d |
| Inverted ORF (cross E) |
0.084 | 0.34 | 1187e |
The frequency of crossing over in the lys3-ura1 and ura1-rqh1 intervals was measured in a meiotic cross with I-SceI cutting at mbs1 (Figure 3) or control versions of that cross. Cross C (GP3674 × GP3801 or GP3672 × GP3802) expressed I-SceI (rec12-172) and possessed the I-SceI target site at mbs1 (mbs1-2) as shown in Figure 3. Cross D (GP3673 × GP3805) expressed I-SceI but lacked the I-SceI target site, and cross E (GP3677 × GP3806) had the I-SceI target site (mbs1-2) but an inverted (inactive) I-SceI ORF (rec12-173).
No colonies were observed; the value is the upper 95% confidence limit based on the Poisson distribution (where three colonies are assumed).
Sum of five experiments (GP3674 × GP3801, three experiments; GP3672 × GP3802, two experiments). Data from the two types of cross were not statistically different.
Sum of three experiments.
Sum of two experiments.
Measurement of crossovers associated with gene conversions:
Crossovers of flanking markers accompanying gene conversion at mbs1 were measured in the same tetrads used to assess mbs1 conversion frequencies. For other loci, gene convertants were selected as intragenic recombinants (prototrophs). The frequency of crossing over of flanking markers was then measured in these spore colonies. In all cases the frequency of crossovers in the total spore population was also measured and was assumed to represent the frequency of incidental crossovers in the convertant spore population. Observed crossovers arise when an associated crossover occurs without an incidental crossover or when an incidental crossover occurs without an associated crossover; i.e., CO = CA(1 − CI) + CI(1 − CA), where CO is the observed frequency of crossovers among convertants, CA is the frequency of crossovers associated with gene conversion, and CI is the frequency of incidental crossovers. The frequency of crossovers associated with gene conversion is then given by CA = (CO − CI)/(1 − 2CI). When CO < 0.5, then CA < CO, and when CO > 0.5, then CA > CO for all values of CI. The flanking markers used with mbs1 conversions were ura1 and rqh1; with ade6, they were ura4+-aim and tps16; with ura1, they were lys3 and rqh1; and with ade7, they were swi5 and ura5.
Engineering restriction site markers at mbs1:
The restriction site markers 1–4 are inactivations of HaeII, BstBI, NciI, and BstBI sites, respectively, by mutations at positions 6850 (G to A), 7564 (C to T), 8061 (G to A), and 8798 (T to C) using the numbering of cosmid SPAC4G8 (GenBank accession no. Z56276.2). These were generated by subcloning the 3.8-kb SpeI fragment containing these sites into a plasmid followed by site-directed mutagenesis using the Stratagene QuikChange kit. The modified SpeI fragment was then excised by restriction and used to transform strain GP3362 to FOAR, resulting in strain GP5145. The markers L and R are creations of PmlI and XbaI sites, respectively. Marker L is a G-to-C mutation at position 5213 of SPAC4G8, and marker R is a T-to-A mutation at position 9977. These changes were made by transforming strain GP4282 with linear DNA carrying the appropriate point mutations (introduced by PCR primer mutagenesis) at the L and R sites to create mbs1-103. The marker mbs1-22 in strain GP4282 is a replacement of the SPAC4G8 sequence 5208–9981 with that of a 1.8-kb HindIII fragment containing ura4+ (Grimm et al. 1988; GenBank accession no. X13976.1). The marker mbs1-1 in strain GP3362 is a replacement of the SPAC4G8 sequence 6486–9213 with the same 1.8-kb HindIII fragment.
Construction of the inducible I-SceI cutting system:
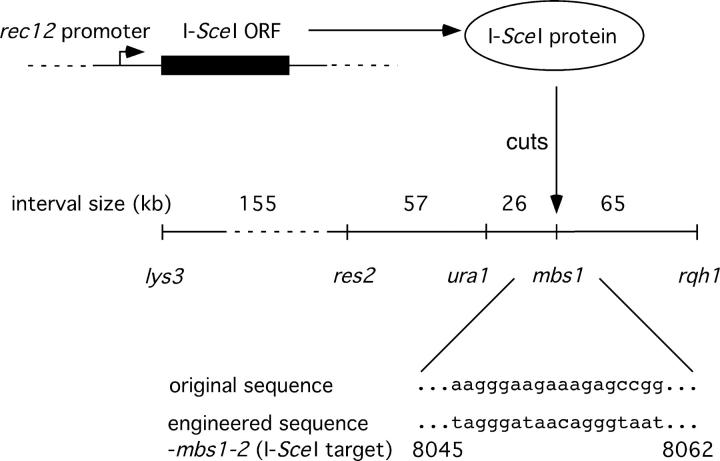
The mbs1-2 allele creates an I-SceI recognition site at the mbs1 locus (SPAC4G8 positions 8045–8062; Figure 3). The mbs1-2 sequence was generated by PCR primer mutagenesis, and a modified 3.8-kb chromosomal SpeI fragment containing this mutation was cloned into a plasmid. The SpeI fragment was then excised and used to transform strain GP3362 to FOAR, resulting in a chromosomal mbs1-2 allele.
Figure 3.—
System for generating a single meiotic DSB, located at the mbs1 region of chromosome I. An exact replacement of the rec12 ORF by that of I-SceI was engineered (see materials and methods). A target site for I-SceI was made by mutating 9 bp in the mbs1 region (numbering refers to the SPAC4G8 cosmid). In controls, the I-SceI ORF was placed in inverted orientation or the target site was absent. The markers used to score crossovers in various intervals are shown, along with the physical sizes of those intervals. Not to scale.
The rec12-172 allele is a replacement of the rec12 ORF with that of I-SceI while the rec12-173 allele has the same replacement, but with the I-SceI ORF in the inverted orientation. A linear fragment containing the I-SceI ORF was produced by PCR from the plasmid pSCM525 (Perrin et al. 1993) and used to replace the rec12 ORF on pJF20 (J. Farah, unpublished results) in both the correct and the inverted orientations. Linear DNA containing the two modified rec12 loci was excised, producing linear DNA with the I-SceI sequence and rec12 flanking DNA. The two kinds of linear DNA were used to transform GP3398 to FOAR, resulting in chromosomal rec12-172 and rec12-173 alleles.
Analysis of meiotic DNA breaks:
Meiotic inductions, flow cytometry, and preparation of DNA in agarose plugs were performed as described (Young et al. 2002). The agarose-embedded DNA was digested with restriction enzymes, separated by gel electrophoresis, Southern blotted, and hybridized with the probes described. Probe c227 is described in Young et al. (2002). Probe Spe1A is identical to the sequence 5972–6330 of cosmid SPAC4G8.
RESULTS
The mbs1 locus is a 2.1-kb cluster of break sites:
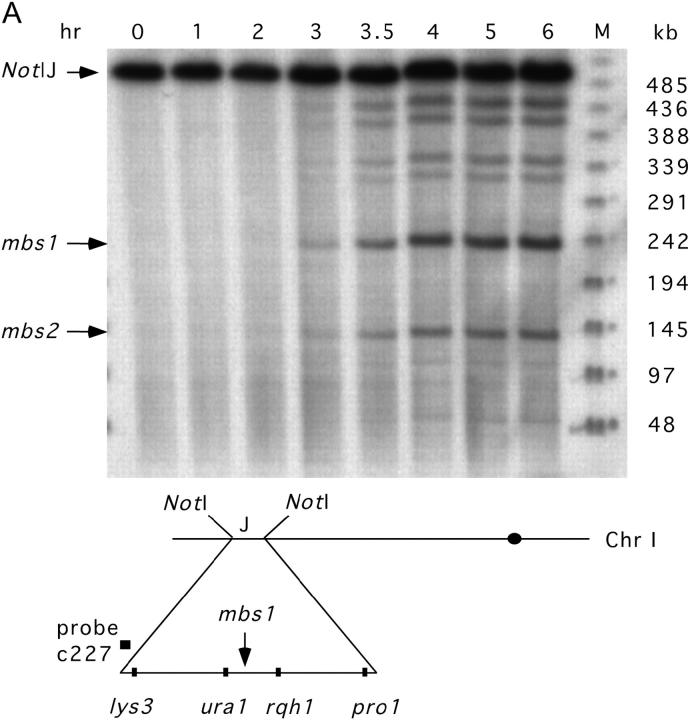
The mbs1 locus is a prominent meiotic break site (∼10% breakage) visible upon Southern blot analysis of the 501-kb NotI “J” fragment of chromosome I (Young et al. 2002; Figure 1A). Analysis of a PmeI digest (Young et al. 2002) localized mbs1 to a 6-kb gene-free region (interval 5305–11,360 of cosmid SPAC4G8; GenBank accession no. Z56276.2). Finer resolution mapping was then undertaken by probing smaller DNA fragments generated by SpeI and HpaI digestion (Figure 1B; data not shown). This analysis revealed the mbs1 locus to consist of four clusters of break sites spread over a 2.1-kb region (Figure 1C; data not shown). The fraction of DNA broken at these four break clusters ranged from 3.3% to 0.9% (Figure 1C), showing a gradient, with the strongest breakage toward the left end as drawn.
Figure 1.—
Mapping DNA breakage at mbs1. (A) The mbs1 locus is visible as an intense meiosis-specific break site (∼10% breakage) in the 501-kb NotI “J” fragment of chromosome I. DNA from the (break-accumulating) rad50S haploid strain GP3718 was harvested at the times indicated after the beginning of pat1-114 (temperature-inducible) meiosis. The DNA was digested with NotI, electrophoresed, and probed as shown. Other meiotic breaks, including mbs2, are visible. M, phage-λ marker ladder with sizes in kilobases at the right. The approximate positions of the NotI “J” fragment and mbs1 on chromosome I are indicated. (B) Finer resolution mapping of a SpeI digest of meiotic GP3839 DNA (probed as shown) revealed that the mbs1 break locus consists of four break clusters. The 0- and 5-hr time points of a meiotic induction are shown. M, 1-kb marker ladder (Invitrogen, San Diego) with sizes in base pairs at the left. (C) The break sites were mapped by probing HpaI and SpeI digests of meiotic GP3839 DNA from each end (additional data not shown). In the first cluster, breaks mapped to positions 6780, 6920, and 7075 with respect to the SPAC4G8 cosmid (numbered telomere-centromere). In the second cluster, they mapped to 7525, 7670, and 7800; in the third to 8020 and 8190, and in the fourth to 8520, 8660, 8740, and 8850. These positions were derived as averages from the four probings, and we estimate their uncertainty to be ∼50 bp. The intensity of breakage at the four clusters was quantified from the SpeI blots. Single-base-pair transitions were introduced to abolish four restriction sites (1, HaeII; 2, BstBI; 3, NciI; 4, BstBI) within each of the four break clusters (see materials and methods). Shown to scale.
The mbs1 locus is a hotspot for gene conversion:
In S. cerevisiae, markers <1 kb from DSB sites show gene conversion at high frequency (Nicolas et al. 1989; Sun et al. 1989; Detloff et al. 1992; Fan et al. 1995). In general, markers in S. pombe convert at low frequency except where mutation or engineering has resulted in high levels of DNA breakage nearby (Gutz 1971; Zahn-Zabal et al. 1995; Steiner et al. 2002; Young et al. 2002, p. 258). To test the natural mbs1 break cluster for gene conversion, we mutated four restriction sites, each within one of the four clusters of break sites, by single-base-pair transitions (Figure 1C and Figure 2A; see materials and methods). Strains carrying this modified mbs1 locus (designated mbs1-102) were crossed with mbs1+ strains. The asci produced from these crosses were dissected, and DNA from the spore colonies was assayed for the four restriction sites by PCR and restriction digestion.
The results of this analysis were as follows. Among the 144 tetrads analyzed, 15 showed conversion of at least one of the four restriction sites. One of these 15 tetrads showed two gene conversion events, giving a frequency of 11% conversion per tetrad (supplemental Figure 1 at http://www.genetics.org/supplemental/). Considering frequencies as events per chromatid, this represents 16 conversion events among 576 chromatids (2.8%). The four markers converted at comparable frequencies from 1.4% to 1.7% of chromatids (Figure 2B and supplemental Figure 1 at http://www.genetics.org/supplemental/). Gene conversion in S. pombe is generally infrequent (0.25% of tetrads per locus on average; Young et al. 2002, p. 258), making the mbs1 cluster of sites the most frequently converting markers reported in this organism, with the exception of previously described hotspot mutations. Co-conversion of adjacent markers was common, accounting for 10 of the 16 conversion events observed (63%; Figure 2B; supplemental Figure 1 at http://www.genetics.org/supplemental/). As three adjacent markers often co-converted, it appears that conversion tracts are frequently >1.2 kb in length (Figure 2B; supplemental Figure 1 at http://www.genetics.org/supplemental/; see discussion). Co-converting markers always converted to the same parental genotype. No discontinuous conversion tracts were observed, consistent with data from S. pombe and other organisms that suggest that such events are rare (Borts and Haber 1989; Grimm et al. 1994).
Crossing over in a 90-kb interval containing mbs1 is highly associated with mbs1 gene conversion:
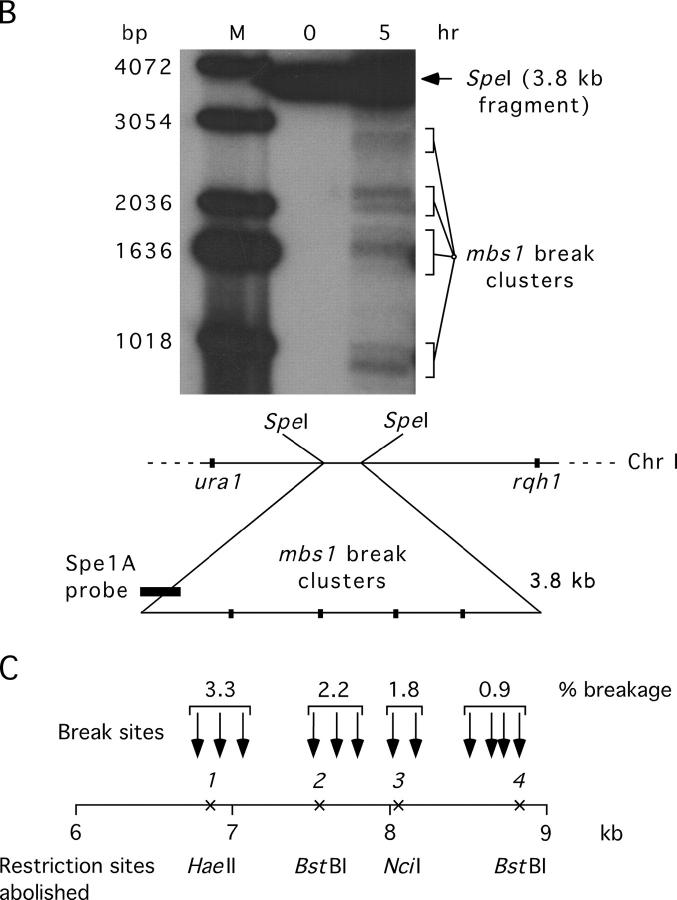
We expected that the breaks observed at mbs1 would generate crossovers in the 90-kb interval ura1-rqh1 (in which mbs1 is located; Figure 2A). To test this, we examined tetrads that could be scored for both mbs1 gene conversions and crossovers between the flanking markers ura1 and rqh1. Gene conversions were used as a signal that recombinational repair of a break at mbs1 had occurred. If ura1-rqh1 crossovers were elevated among mbs1 convertant spores compared to total spores, it would indicate that mbs1 breakage frequently generated crossovers in that interval.
The tetrads we analyzed previously for conversion were scored for crossing over between ura1 and rqh1. No conversions of the ura1 or rqh1 markers were seen among the 144 tetrads, consistent with the typical low frequency of gene conversion in S. pombe. Among the 576 spore colonies analyzed above, 62 showed crossing over in the ura1-rqh1 interval, i.e., 10.8% of chromatids (Figure 2C; supplemental Figure 1 at http://www.genetics.org/supplemental/). This is very similar to the value of 9.7% from random spore analysis (Young et al. 2002). Of the 16 conversion events (see above), 13 (81%) showed an accompanying ura1-rqh1 crossover (Figure 2C). Despite the small number of conversions, the frequency of ura1-rqh1 crossovers among convertant spores (81%) is significantly higher (χ2 = 69.9; d.f. = 1; P < 0.0001) than that seen in the total spore population (10.8%) and demonstrates a high association between conversion (and presumably breakage) at mbs1 and crossing over in the ura1-rqh1 interval (see discussion).
To estimate how important breakage at mbs1 is for crossing over in the ura1-rqh1 interval, we considered the proportion of such crossovers having an associated mbs1 conversion. As a single crossover produces two crossover spore colonies, the 62 crossover colonies observed represent 31 ura1-rqh1 crossovers. Among this total population of 31 crossovers, 42% (13/31) were associated with a conversion event at mbs1 (the crossover involved the converting chromatid). This suggests that at least 40% of all ura1-rqh1 crossovers are initiated by mbs1 breakage. We also noted that only one simple crossover was observed between the four mbs1 restriction markers in the absence of conversion of any of those markers (supplemental Figure 1 at http://www.genetics.org/supplemental/). This has implications for how efficiently conversion of our markers accompanies interhomolog DSB repair at mbs1 (see discussion).
Crossovers >25 kb from mbs1 are not associated with mbs1 gene conversion:
Young et al. (2002) showed that there are genetic intervals in the 501-kb NotI “J” fragment containing mbs1 that lack detectable DSBs but have substantial levels of crossing over. The model of Szostak et al. (1983) predicts that meiotic crossing over is caused by meiotic DSBs, and this is supported by the existence of multiple rec gene mutations that abolish both meiotic crossing over and DSBs in S. pombe. Therefore, Young et al. (2002) hypothesized that breakage at the observed sites, such as mbs1, in the NotI “J” fragment could generate crossovers not only in the intervals containing the breaks but also in the apparently break-free intervals. One mechanism by which this could happen is by the migration of double Holliday junctions from the initiating break to the crossover position, where junction resolution would occur. As an example, in the interval lys3-rqh1 there are only two observed break sites, mbs1 (∼10% breakage) and mbs2 (∼3% breakage; Young et al. 2002), but several subintervals that do not include these break sites still experience crossovers (e.g., ∼3.7% crossing over in res2-ura1; Figure 2A). The hypothesis of Young et al. (2002) predicts that mbs1 (with ∼80% of all lys3-rqh1 breakage) would be important for crossing over in much of the lys3-ura1 interval (including subintervals like res2-ura1) and not just in the immediate subinterval (ura1-rqh1) that contains the mbs1 locus.
To test this hypothesis, the association of gene conversion at mbs1 and crossing over in the interval lys3-ura1 (or the similar interval pom1-ura1) was examined as done previously for the interval ura1-rqh1. Again, gene conversions were used as a signal that recombinational repair of a break at mbs1 had occurred. If crossovers in the lys3/pom1-ura1 interval were elevated among mbs1 convertant spores compared to total spores, it would indicate that mbs1 breakage was responsible for crossovers in that interval.
The same set of tetrads that was studied above was analyzed for lys3/pom1-ura1 crossovers. The ura1 marker lies ∼26 kb from mbs1 (Figure 2A). Among the 576 spore colonies analyzed above, 64 had crossovers in the lys3/pom1-ura1 interval, i.e., 11.1% of chromatids (Figure 2C and supplemental Figure 1 at http://www.genetics.org/supplemental/). This is somewhat lower than the frequency previously reported for random spore data (∼17%; Young et al. 2002). In contrast to the strong linkage between gene conversion at mbs1 and crossing over in the ura1-rqh1 interval, only 2 of 16 conversions at mbs1 were associated with a lys3/pom1-ura1 crossover (12.5%; Figure 2C; supplemental Figure 1 at http://www.genetics.org/supplemental/). This is not significantly different from the level of lys3/pom1-ura1 crossing over in the total population (11.1%) and does not support the hypothesis of Young et al. (2002)(see discussion).
An artificial meiotic DSB at mbs1 does not frequently stimulate crossovers >25 kb away:
As a more direct test of the hypothesis of Young et al. (2002), we reduced meiotic DNA breakage to a single genomic site at the mbs1 locus and observed where this breakage stimulated crossing over. This was done by replacing the coding sequence for Rec12, the S. pombe Spo11 ortholog that generates normal meiotic DSBs, with that of the I-SceI endonuclease (Carroll 2004). As rec12 is meiotically induced (Lin and Smith 1994), this should give meiosis-specific I-SceI expression. The 18-bp I-SceI recognition site (Colleaux et al. 1988), which is absent from the S. pombe genomic sequence (Wood et al. 2002), was introduced into the mbs1 region (Figure 3) so that a single meiotic DSB could be generated there. Control strains were constructed with inactive I-SceI (an inversion of the I-SceI coding sequence at the rec12 locus) or with no I-SceI recognition site at mbs1. Standard meiotic crosses were carried out and the frequency of crossing over among random spores was measured for the lys3-ura1 and ura1-rqh1 intervals (Figure 3). Complementing diploids, which occur in rec12 meioses (Davis and Smith 2003), would be scored as crossovers in both intervals. These were identified and removed from the analysis (see materials and methods).
The results of this study are shown in Table 2. Breakage at mbs1 specifically stimulated crossing over in the ura1-rqh1 interval 10-fold compared to the controls (χ2 = 40.3; d.f. = 1; P < 0.0001; contingency χ2 test with controls pooled). Statistically significant (χ2 = 4.5; d.f. = 1; P < 0.05) stimulation was also seen in the lys3-ura1 interval but the effect was very small. Therefore, it appears that breakage at mbs1 does not frequently generate crossovers in the lys3-ura1 interval, i.e., >25 kb from mbs1 (see discussion).
mbs1 crossovers are concentrated in a 4.8-kb region around mbs1:
The results presented so far indicate that breakage at mbs1 is strongly associated with crossing over in the ura1-rqh1 interval and infrequently, if at all, with crossing over in intervals >25 kb away. This led us to investigate the distribution of crossovers within the ura1-rqh1 interval. If crossovers are located close to the position of their initiating DSBs, then the mbs1 locus should be a hotspot for crossovers and the rest of the ura1-rqh1 interval should be cold. Distinguishing crossover position was not possible using the 1–4 restriction sites at mbs1, because frequent conversion of these markers generally made the location of crossovers ambiguous. Therefore, we introduced two additional restriction sites flanking mbs1, denoted “L” and “R” (Figure 4). Both were single-base-pair changes (see materials and methods). These two markers define a 4.8-kb region containing the mbs1 break sites (Figure 4) and, as they are 1.2 and 1.6 kb from the nearest observed break sites, were expected to convert sufficiently infrequently to reveal unambiguously if crossovers lay between them.
A meiotic cross heterozygous for these two new markers and the original four restriction markers was carried out (Figure 4). Tetrads with ura1-rqh1 crossovers were identified, and analysis of the six markers was restricted to these. Only two-strand double-crossover tetrads, expected to make up ∼5% of all ura1-rqh1 crossovers tetrads, would be missed by study of this subpopulation. In addition, the data already presented showed that the great majority (∼80%) of mbs1 conversion events were associated with ura1-rqh1 crossovers so that examination of crossover tetrads should also detect most mbs1 conversion events.
In the 89 tetrads examined, 20 showed ura1-rqh1 crossovers (one four-strand double, i.e., 11.8% of chromatids, consistent with previous results). Among these 20 crossover tetrads, 5 showed a conversion of either the L or R marker, or both (four R conversions and one tetrad showing independent conversion of L and R; supplemental Figure 2 at http://www.genetics.org/supplemental/). In four of these five, the crossover position could not be located unambiguously. Thus, crossover position could be assigned in 16 tetrads, and among these were three double-crossover tetrads (one four-strand double and two three-strand doubles). This gave 19 crossovers, of which 15 lay between L and R (79%), three lay between ura1 and L (16%), and one lay between R and rqh1 (5%; Table 3; supplemental Figure 2 at http://www.genetics.org/supplemental/). From this analysis the 4.8-kb region containing mbs1 contains about three-quarters of the crossovers found in the whole 90-kb ura1-rqh1 interval. When all crossovers were considered, 83% (19/23) either fell between L and R or were associated with conversion of L or R.
TABLE 3.
Crossovers in the 90-kbura1-rqh1 interval are concentrated in a 48-kb region aroundmbs1
| Crossover position | Tetrad analysis | Random spore analysis |
|---|---|---|
| ura1-L | 3 (16%) | 22 (21%) |
| L-R (mbs1) | 15 (79%) | 57 (54%) |
| R-rqh1 | 1 (5%) | 26 (25%) |
| Total | 19 | 105 |
Crossovers between ura1 and rqh1 were identified either among 89 tetrads (supplemental Figure 2 at http://www.genetics.org/supplemental/) or among 1200 random spores. In the tetrad analysis crossover events were counted, while in the random spore analysis crossover spores were counted. The positions of the crossovers were localized to one of three regions within the ura1–rqh1 interval (ura1-L, L-R, and R-rqh1; Figure 4). In the tetrad study only ura1-rqh1 crossover tetrads without L or R conversions were scored (to avoid ambiguity in crossover position). Crosses using the strains GP4514 and GP4515 (cross F) were used for both analyses. Two independent crosses, giving similar results, were pooled for the random spore analysis.
The frequency with which mbs1 conversions accompanied crossovers in this tetrad analysis was similar to that reported earlier. Among the 23 total ura1-rqh1 crossovers, 11 were associated with conversion of one or more of the central four mbs1 markers (supplemental Figure 2 at http://www.genetics.org/supplemental/), consistent with the earlier tetrad analysis (13 of 28). When all six mbs1 markers were considered (including L and R), 13/23 crossovers were associated with a gene conversion (supplemental Figure 2 at http://www.genetics.org/supplemental/). This further supports the conclusion that 40% or more of ura1-rqh1 crossovers are caused by mbs1 breaks. No significant disparity was observed in the direction of conversion, as would be expected if the single-base-pair mutations did not influence recombination (supplemental Figure 2 at http://www.genetics.org/supplemental/).
The analysis of crossover position was extended by carrying out the same cross as described above, but analyzing random spores for ura1-rqh1 crossovers and then scoring these for crossover position with respect to L and R. The frequency of ura1-rqh1 crossovers was 105/1200 (8.8%), close to that seen in previous random spore analyses (see above and Young et al. 2002). Compared to the tetrad analysis, a somewhat lower proportion (54%) of ura1-rqh1 crossovers lay in the 4.8-kb interval defined by the L and R markers, 21% lay in the ura1-L interval, and 25% lay in the R-rqh1 interval (Table 3). However, the two data sets were not significantly different (χ2 = 4.7; d.f. = 2; 0.1 > P > 0.05). Random spore analysis does not allow crossovers with ambiguous locations to be identified and discounted. Such events can happen when a crossover is accompanied by conversion of L or R. In random spore analysis these events result in two spores, each with an apparent crossover on opposite sides of the converting marker. This would tend to elevate the frequency of apparent R-rqh1 crossovers, as these are otherwise rare. This effect may explain the somewhat higher level of R-rqh1 crossovers in the random spore data compared to the tetrad analysis. Nevertheless, it can be seen that 55–79% of ura1-rqh1 crossovers are located in a 4.8-kb region around mbs1 and only 22–45% in the remaining 85 kb of this interval (Table 3).
Association of gene conversion with crossing over at other loci:
As discussed previously, gene conversion at mbs1 was accompanied at high frequency (∼80%) by crossing over between ura1 and rqh1, markers flanking the mbs1 site (see Figure 2C). As this value is considerably higher than that reported for most tested loci in S. cerevisiae (Whitehouse 1982; Gilbertson and Stahl 1996), we investigated whether crossovers were highly associated with conversion at other loci in S. pombe.
To examine conversions at loci converting less frequently than mbs1, we selected for conversion events as intragenic recombinants. For closely spaced intragenic markers in ade6, with separation similar to those involved in our crosses, all observed recombinants arose from gene conversion (Gutz 1971). Among the intragenic recombinants, the frequency of flanking marker exchange was measured and corrected for incidental events to give the frequency of crossovers associated with the conversion (see materials and methods). The same correction was carried out to calculate the frequency of crossovers associated with mbs1 conversion.
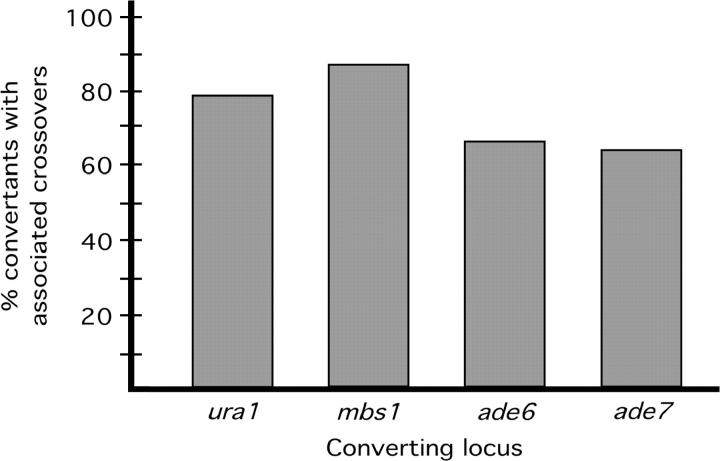
As can be seen from Figure 5, at all of the loci examined the crossovers were associated with gene conversion at high frequency (64–87%). The value derived for ade6 is consistent with published results (Grimm et al. 1994). The high association consistently observed implies bias in S. pombe meiotic recombination toward the production of crossovers (see discussion).
Figure 5.—
Crossovers are highly associated with gene conversion at four loci in S. pombe. Conversion events were identified by tetrad analysis (mbs1) or selection for intragenic recombinants (ura1, ade6, and ade7). Crossovers between flanking markers were counted among convertants and among unselected spores, and the frequency of associated crossovers was determined as described in materials and methods. The ura1 (cross H, GP789 × GP3696), ade6 (cross I, GP1252 × GP3785), and ade7 (cross J, GP5091 × GP5125) crosses were each carried out twice and the results pooled. The mbs1 data come from tetrad analysis of crosses A and B (Figure 2C) and cross G (GP877 × GP3477) and measurement of crossover frequencies among mbs1 convertant spores. Uncorrected (observed) crossover frequencies for the flanking markers at each locus (see materials and methods) were 80% (mbs1), 66% (ura1), 62% (ade6), and 60% (ade7). The total number of convertant spore colonies assayed was 30 (mbs1), 359 (ura1), 1140 (ade6), and 359 (ade7). The frequency of incidental crossovers, in unselected spores, for each interval (see materials and methods) was 10% (mbs1), 22% (ura1), 16% (ade6), and 15% (ade7).
DISCUSSION
We set out to characterize the natural meiotic break hotspot mbs1 and to analyze the frequency and location of recombination events generated by mbs1 breaks. This is the first study of a natural meiotic break hotspot in S. pombe, rather than one generated by point mutations or large insertions (Gutz 1971; Zahn-Zabal et al. 1995). We observed that the mbs1 locus is a hotspot for meiotic gene conversion and crossing over as well as DNA breakage. The determination of conversion frequencies at mbs1, showing it to be a conversion hotspot, allowed us to compare break and conversion rates. This comparison suggested that our converting markers are able to detect a substantial proportion of mbs1 break-repair events and that at least 30% of meiotic breaks are repaired against homologous chromosomes rather than sister chromatids. The use of multiple markers at mbs1 in our conversion study also allowed us to estimate the size of the conversion tracts, which were often >1 kb in length.
As well as a hotspot for gene conversion, we showed that a 4.8-kb region including mbs1 is a hotspot for crossovers, which occur in this interval ∼10 times more frequently (per kilobase) than the S. pombe average. We also observed that crossovers accompany mbs1 conversions well over 50% of the time, a pattern seen at three other loci, suggesting that recombination is biased to produce crossovers. In contrast to the ability of mbs1 to stimulate crossovers in the immediate vicinity of the breaks, no substantial stimulation of crossovers >25 kb away was seen. This observation does not support the hypothesis that meiotic DSBs generate crossovers many kilobases away from the location of the breaks, a possibility raised to explain the difference in the distribution of meiotic breaks and crossovers observed in S. pombe (Young et al. 2002). This leaves the disparity in DSB and crossover distributions unresolved and leads us to suggest that a proportion of meiotic crossovers may result from recombination initiated by single-strand lesions.
More than 30% of mbs1 breaks can be detected as gene conversions:
Our tetrad analysis not only indicated that mbs1 was a hotspot for gene conversion but also suggested that the markers we used were able to detect a significant proportion (>30%) of all mbs1 break-repair events as conversions. Southern blot analysis of meiotic DNA showed that the mbs1 site undergoes breakage at a frequency of ∼10% (Figure 1; Young et al. 2002). We observed that 3% of chromatids showed conversion of at least one of the four central mbs1 restriction markers but there was only one double conversion tetrad (supplemental Figure 1 at http://www.genetics.org/supplemental/); i.e., the conversions arose from separate break events. This indicates that 3% of chromatids would need to be broken to generate the observed conversions, representing 30% of all mbs1 breaks. Some of the remaining 70% of mbs1 breaks may be repaired using a sister chromatid rather than a homologous chromosome. Therefore, it is likely that our markers detect >30% of interhomolog DSB repair events at mbs1.
A single example was seen of an apparent two-strand double-crossover event within the marker 1–4 constellation (supplementary Figure 1 at http://www.genetics.org/supplemental/). This event could have arisen from mismatch repair acting on a region of symmetrical heteroduplex DNA. The existence of this kind of heteroduplex has been inferred in S. pombe (Fleck et al. 1999). Alternatively, the apparent two-strand double crossover could have arisen from two independent conversions. We conclude that events suggestive of symmetric heteroduplex at mbs1 do occur, but they are rare.
Size of conversion tracts at mbs1:
The use of multiple markers at mbs1 allowed us to estimate the length of conversion tracts in our full tetrad analysis (Figure 2B). Of 16 conversions, 10 (63%) involved two or more markers; of these, 10 were >0.75 kb, 8 were >1.2 kb, and 2 were >2 kb. In addition, 5 of the 6 conversions affecting only one marker involved marker 1 (i.e., the extent of the conversion on one side could not be determined). Analysis of markers at the ade6 locus also indicated that conversion tracts >1 kb are common (Grimm et al. 1994). These S. pombe values are also comparable to an average tract length of 1.5 kb reported for S. cerevisiae (Borts and Haber 1989). Additional mbs1 conversions including the L and R markers were seen among the tetrads selected for ura1-rqh1 crossovers (supplemental Figure 2 at http://www.genetics.org/supplemental/). One of these conversion tracts was >3.5 kb (an outside marker converted, allowing no maximum to be set); at ade6 the longest reported conversion tract was >3.6 kb (including conversion of an outside marker). Gene conversions can arise from either asymmetric or symmetric heteroduplex DNA. According to current models of recombination, asymmetric heteroduplexes correspond to regions of single-strand resection at DSBs, whereas symmetric heteroduplexes occur when strand exchange progresses beyond the resected region of DNA. There is evidence for both symmetric and asymmetric heteroduplex formation during S. pombe meiotic recombination (Fleck et al. 1999). Therefore, while longer gene conversion tracts might indicate longer regions of resection, it is difficult to relate the conversion tract and break-resection lengths at mbs1.
mbs1 is a hotspot of crossing over:
Our results indicate that breaks at mbs1 not only cause crossovers in the 90-kb interval ura1-rqh1 but also are responsible for at least 40% of all crossovers in this interval. Crossovers in the interval ura1-rqh1 are greatly elevated among convertants at mbs1, compared to the total population. If it is assumed that conversions at mbs1 indicate repair of mbs1 breaks, then these breaks generate ura1-rqh1 crossovers. In addition, 55–79% of ura1-rqh1 crossovers lie in the 4.8-kb region around mbs1 (Table 3), and ∼40% of ura1-rqh1 crossovers are associated with a conversion event at mbs1 (supplemental Figures 1 and 2 at http://www.genetics.org/supplemental/). This implies that most ura1-rqh1 crossovers are caused by mbs1 breaks and occur close by.
The frequent occurrence of crossovers within a 4.8-kb region around the mbs1 locus means that mbs1 forms a hotspot for crossovers (12–18 cM/10 kb) as well as a hotspot for gene conversion. In contrast, the break-free regions to the left of marker L and especially to the right of marker R are recombinationally much colder (0.7–0.9 cM/10 kb and 0.08–0.4 cM/10 kb). Recombination intensities in these two intervals are also lower than the genome average (1.6 cM/10 kb) or the average of 1.1 cM/10 kb (range 0.6-1.6 cM/kb) reported for the whole lys3-pro1 region (Young et al. 2002). Therefore, in the 90-kb interval ura1-rqh1 the observed double-strand breakage and the observed crossing over are qualitatively consistent with each other. However, as with analysis of the whole lys3-pro1 region (Young et al. 2002), crossovers still occur in break-free subintervals of ura1-rqh1, albeit at lower frequency than the S. pombe average.
Crossovers in apparently break-free intervals:
In contrast to what was seen for the interval ura1-rqh1, our results show no significant enrichment for lys3/pom1-ura1 crossovers among mbs1 convertants. Similarly, an artificial break at mbs1 stimulated frequent ura1-rqh1 crossing over, but little lys3-ura1 crossing over (Table 2). It appears that few mbs1-initiated recombination events lead to crossovers between lys3 and ura1. This is despite mbs1 accounting for ∼80% of all breakage observed in the interval lys3-rqh1 and the existence of apparently break-free subintervals that show normal levels of crossovers and for which mbs1 is the closest observed break site (e.g., res2-ura1; Young et al. 2002).
Reconstruction experiments demonstrate that we can detect DSB levels lower than that required to explain the crossovers in break-free intervals even if they are spread over many sites rather than concentrated in hotspots (data not shown). We are therefore left with the conclusion that, for these intervals, the closest observed DSBs are not responsible for the observed crossovers but neither is there sufficient double-strand breakage seen within the intervals to account for the crossovers. It is possible that in crosses to measure recombination genetically DSBs do occur in these intervals, but that these specific DSBs either are absent or do not accumulate under the conditions used to measure DSBs physically. Alternatively, crossovers in these apparently DSB-free intervals might be generated by something other than DSBs. Such initiating events could be single-strand lesions. Single-strand gaps appear to be recombinogenic in bacteria (Wang and Smith 1984) and recent work suggests that single-strand lesions can be recombinogenic in eukaryotes also. Certain mutant RAG proteins [catalyzing DNA cleavage in V(D)J recombination for mammalian immunoglobulin gene rearrangement] generate nicks rather than DSBs but stimulate homologous recombination even more frequently than do wild-type RAG proteins (Lee et al. 2004). Similarly, certain S. cerevisiae rad52 mutants are proficient for spontaneous mitotic recombination but defective in the repair of DSBs generated by ionizing radiation (Mortensen et al. 2002). Expression of the site-specific single-strand-nicking protein encoded by f1 bacteriophage gene II in S. cerevisiae also stimulates mitotic recombination (Strathern et al. 1991). In S. pombe the lesion at the mat1 locus that initiates recombination for mating-type switching is a single-strand nick (Arcangioli 1998), although this may be converted into a DSB by replication before recombination (Kaykov et al. 2004). Thus, single-strand lesions may stimulate eukaryotic homologous recombination, although direct evidence is lacking.
High frequency of crossing over associated with gene conversion in S. pombe:
Our results demonstrate that gene conversions in S. pombe are frequently accompanied by crossovers. At the four loci that we investigated, 64–87% of gene conversions had an associated crossover. We assume that interhomolog recombination events that cause gene conversion are typical of all interhomolog recombination events. If so, then recombination in S. pombe is biased toward the production of crossovers. This could be explained in two ways. First, there could be a single pathway of recombination that produces crossover and noncrossover outcomes by different resolution of the same Holliday junction intermediates. In S. pombe this resolution would have to be biased toward the production of crossovers. Random resolution would lead to equal frequencies of crossovers and noncrossovers. Second, as has been recently suggested, there could be two pathways of recombination, one progressing via resolution of Holliday junctions or related structures and one via SDSA (Allers and Lichten 2001; Cromie et al. 2001; Hunter and Kleckner 2001; Osman et al. 2003; Smith et al. 2003). The SDSA pathway would produce only noncrossovers and the Holliday junction resolution pathway could give only crossovers. In support of this idea, a mutation in mus81, whose product appears to resolve Holliday junctions or related structures in S. pombe, does not reduce the frequency of gene conversions that lack associated crossovers but does greatly reduce the frequency of gene conversions with associated crossovers (Osman et al. 2003; Smith et al. 2003). The activity of purified Mus81·Eme1 on nicked double Holliday junctions is also consistent with this model (Gaillard et al. 2003; Osman et al. 2003). The observed bias toward crossovers in S. pombe would then suggest that the Holliday junction pathway predominates in that organism.
The observed frequency with which gene conversions have accompanying crossovers in S. pombe is higher than that reported for S. cerevisiae. The higher frequency of noncrossovers in S. cerevisiae may be caused by crossover interference; i.e., any recombination event in the region of interference around a crossover site would be channeled toward a noncrossover. Therefore, in mutants that abolish interference, gene conversions would be more frequently accompanied by crossovers. If so, meiotic recombination in S. cerevisiae and other organisms with interference might proceed as in S. pombe but with the addition of the interference pathway. This would make S. pombe a particularly attractive model organism for studying the fundamental processes of meiotic recombination.
Acknowledgments
We thank Jürg Kohli and Steve Goldman for S. pombe strains; Bernard Dujon for plasmid pSCM525; and Sue Amundsen, Luther Davis, and Andrew Taylor for critical reading of the manuscript. G.A.C. was supported by a long-term European Molecular Biology Organization fellowship (no. ALTF 123-2201). This work was supported by research grant GM31693 from the National Institutes of Health to G.R.S.
References
- Allers, T., and M. Lichten, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Arcangioli, B., 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17: 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., and J. R. Pringle, 1998. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12: 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., and J. E. Haber, 1989. Length and distribution of meiotic gene conversion tracts and crossovers in Saccharomyces cerevisiae. Genetics 123: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., E. Alani and N. Kleckner, 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carroll, D., 2004. Using nucleases to stimulate homologous recombination. Methods Mol. Biol. 262: 195–207. [DOI] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Colleaux, L., L. D'Auriol, F. Galibert and B. Dujon, 1988. Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl. Acad. Sci. USA 85: 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., J. C. Connelly and D. R. Leach, 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8: 1163–1174. [DOI] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163: 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., M. A. White and T. D. Petes, 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier, C., H. Schmidt and G. R. Smith, 2004. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168: 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch, T., A. M. Carr and P. Nurse, 1992. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6: 2035–2046. [DOI] [PubMed] [Google Scholar]
- Fan, Q., F. Xu and T. D. Petes, 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15: 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta and G. R. Smith, 2002. A 160-bp palindrome is a Rad50·Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, O., E. Lehmann, P. Schar and J. Kohli, 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21: 314–317. [DOI] [PubMed] [Google Scholar]
- Gaillard, P. H., E. Noguchi, P. Shanahan and P. Russell, 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12: 747–759. [DOI] [PubMed] [Google Scholar]
- Gilbertson, L. A., and F. W. Stahl, 1996. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., J. Kohli, J. Murray and K. Maundrell, 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215: 81–86. [DOI] [PubMed] [Google Scholar]
- Grimm, C., J. Bahler and J. Kohli, 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 135: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69: 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974 Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Gygax, A., and P. Thuriaux, 1984. A revised chromosome map of the fission yeast Schizosaccharomyces pombe. Curr. Genet. 8: 85–92. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Iino, Y., and M. Yamamoto, 1985. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 198: 416–421. [DOI] [PubMed] [Google Scholar]
- Kaykov, A., A. M. Holmes and B. Arcangioli, 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Lee, G. S., M. B. Neiditch, S. S. Salus and D. B. Roth, 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117: 171–184. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and G. R. Smith, 1994. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Mortensen, U. H., N. Erdeniz, Q. Feng and R. Rothstein, 2002. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz, P., 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, A., D. Treco, N. P. Schultes and J. W. Szostak, 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338: 35–39. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Perrin, A., M. Buckle and B. Dujon, 1993. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO J. 12: 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., M. N. Boddy, P. Shanahan and P. Russell, 2003. Fission yeast Mus81·Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, W. W., R. W. Schreckhise and G. R. Smith, 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9: 847–855. [DOI] [PubMed] [Google Scholar]
- Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr and T. Enoch, 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16: 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern, J. N., K. G. Weinstock, D. R. Higgins and C. B. McGill, 1991. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics 127: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, W. N., 1958. An analysis of interference in Aspergillus nidulans. Proc. R. Soc. Lond. B Biol. Sci. 149: 82–101. [DOI] [PubMed] [Google Scholar]
- Sun, H., D. Treco, N. P. Schultes and J. W. Szostak, 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90. [DOI] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Wang, T. V., and K. C. Smith, 1984. recF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J. Bacteriol. 158: 727–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse, H. L. K., 1982 Genetic Recombination, pp. 319–321. John Wiley & Sons, New York.
- Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne et al., 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Young, J. A., R. W. Schreckhise, W. W Steiner and G. R. Smith, 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Zahn-Zabal, M., E. Lehmann and J. Kohli, 1995. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics 140: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]