Abstract
The meiotic recombination hot spot ura4A (formerly ura4-aim) of Schizosaccharomyces pombe was observed at the insertion of the ura4+ gene 15 kb centromere-proximal to ade6 on chromosome III. Crosses heterozygous for the insertion showed frequent conversion at the heterology with preferential loss of the insertion. This report concerns the characterization of 12 spontaneous ura4A mutants. A gradient of conversion ranging from 18% at the 5′ end to 6% at the 3′ end was detected. A novel phenomenon also was discovered: a mating-type-related bias of conversion. The allele entering with the h+ parent acts preferentially as the acceptor for conversion (ratio of 3:2). Tetrad analysis of two-factor crosses showed that heteroduplex DNA is predominantly asymmetrical, enters from the 5′ end, and more often than not covers the entire gene. Restoration repair of markers at the 5′ end was inferred. Random spore analyses of two-factor crosses and normalization of prototroph-recombinant frequencies to physical distance led to the demonstration of map expansion: Crosses involving distant markers yielded recombinant frequencies higher than the sum of the frequencies measured in the subintervals. Finally, marker effects on recombination were defined for two of the ura4A mutations.
HOMOLOGOUS recombination contributes to the generation of genetic diversity and is required for proper chromosome segregation in meiosis and for the repair of DNA damage. Since homologous recombination occurs most frequently during meiosis, the underlying mechanisms are best studied in ascomycetous fungi in which all four products of single meioses can be individually recovered and analyzed (Paques and Haber 1999). The fate of the four chromatids in a diploid cell undergoing meiosis can be followed by tetrad analysis. Examination of the segregation of genetic markers in tetrads has demonstrated two classes of recombination events: the classical reciprocal exchange of DNA sequences, called crossing over, and the unidirectional transfer of genetic information between chromatids, designated non-Mendelian segregation (NMS). Two types of NMS events are distinguished. One is the nonreciprocal transfer of information of both DNA strands of a donor chromatid to a chromatid of the homologous chromosome. This segregation type is called gene conversion and manifests itself in 6+:2− or 2+:6− segregation of genetic markers. The numbers refer to the eight single strands of the tetrad with + indicating wild type and − indicating mutant information. The other unidirectional transfer changes only one strand of the recipient chromatid. The resulting spore bears both wild-type and mutant information. Since the genetic marker segregates only in the first mitotic division after meiosis, the observed 5+:3− or 3+:5− tetrads are called postmeiotic segregations (PMS).
The frequency of homologous recombination varies widely from interval to interval along the genome. DNA regions with lower or higher than average frequency of aberrant segregation and crossing over are called cold spots or hot spots, respectively. They have been identified and described in many different organisms (Lichten and Goldman 1995; Paques and Haber 1999; Petes 2001). Specific nucleotide sequences have been identified in some cases, which are recognized by proteins promoting, directly or indirectly, a rate-limiting step of the recombination process (Smith 2001).
A phenomenon often seen in intragenic studies is polarity of NMS. The frequency of NMS events falls steadily from one end of the gene to the other, indicating a preferential initiation or termination point for homologous recombination (Nicolas and Petes 1994). In the budding yeast Saccharomyces cerevisiae, hot spots and high NMS ends have been correlated with the occurrence of double-strand breaks (DSB) at nuclease hypersensitive sites in chromatin (Ohta et al. 1994; Wu and Lichten 1994; de Massy et al. 1995; Liu et al. 1995). Double-strand breaks initiating meiotic recombination have also been demonstrated in Schizosaccharomyces pombe (Cervantes et al. 2000; Steiner et al. 2002; Young et al. 2002).
For the characterization of the meiotic hot spot mutation ade6-M26 (Gutz 1971), the flanking marker ura4-aim, in this article renamed ura4A, was constructed between ade6 and the centromere of chromosome III. The insertion 15 kb upstream of ade6 contains the functional S. pombe ura4+ gene on a 1.8-kb DNA fragment (Grimm et al. 1994). The insertion heterology turned out to be a hot spot itself with 3.7% NMS frequency and disparity of conversion loss being 3.5 times more frequent than transfer of the insertion (Zahn-Zabal et al. 1995). When the M26 hot spot is active, NMS frequency at ura4A is reduced, indicating competition for recombination factors by the two hot spots. The hot spot property of ura4A near ade6 is possibly due to a new chromosomal context, since ura4+ at its original locus on the left arm of chromosome III shows no elevated NMS frequency when crossed with a strain carrying the ura4-D18 deletion (Zahn-Zabal et al. 1995).
To see whether ura4A also behaves as a hot spot in homozygous configuration, we isolated and characterized 12 spontaneous mutations within ura4A, tested them with respect to NMS by tetrad analysis, and determined prototroph frequencies in two-factor crosses. The highest frequencies of conversion (18%) so far recorded in fission yeast were observed. In addition, we document a novel phenomenon: a mating-type-related bias of NMS (mat bias) at ura4A and confirm the existence of map expansion, a phenomenon first described in classical fine-structure mapping of genes.
MATERIALS AND METHODS
Strains, media, and general genetic methods:
The construction of the ura4A+ strains GC151a and GC151b was described by Grimm et al. (1994). The spontaneous ura4A− mutants were derived from GC151a (alleles 1–6) and from GC151b (alleles 9–14; Table 1) by selection of uracil-dependent cells on minimal medium containing uracil and 5-fluoroorotic acid as described by Grimm et al. (1988). The isolated ura− clones were classified as ura4A or ura5 using the criss-cross technique described by Gutz et al. (1974). ura5 mutants were not considered further. The ura4A isolates are independent of each other because different starting clones were used. In the two instances in which small clusters of homoalleles were found, only one representative was kept. Standard minimal medium (MMA) consists of 0.67% Difco yeast nitrogen base without amino acids, 1% glucose, and 1.8% agar. The other standard media, yeast extract agar (YEA), malt extract agar (MEA), and the general genetic methods, are described by Gutz et al. (1974). Supplements were added to the media at concentrations of 0.01% (w/v).
TABLE 1.
S. pombe strains used in this study
| Designationsa | Genotypeb |
|---|---|
| GC151a BSC 46-1811 | h−ura4A+ ura4-D18 |
| GC151b BSC 46-1812 | h+ura4A+ ura4-D18 |
| S64 BSC 45-1764 | h−ura4A-1 ura4-D18 |
| S65 BSC 45-1765 | h+ura4A-1 ura4-D18 |
| S66 BSC 45-1766 | h−ura4A-2 ura4-D18 |
| S67 BSC 45-1767 | h+ura4A-2 ura4-D18 |
| S68 BSC 45-1768 | h−ura4A-3 ura4-D18 |
| S69 BSC 45-1769 | h+ura4A-3 ura4-D18 |
| S70 BSC 45-1770 | h−ura4A-4 ura4-D18 |
| S71 BSC 45-1771 | h+ura4A-4 ura4-D18 |
| S72 BSC 45-1772 | h−ura4A-5 ura4-D18 |
| S73 BSC 45-1773 | h+ura4A-5 ura4-D18 |
| S74 BSC 45-1774 | h−ura4A-6 ura4-D18 |
| S75 BSC 45-1775 | h+ura4A-6 ura4-D18 |
| S80 BSC 45-1780 | h−ura4A-9 ura4-D18 |
| S81 BSC 45-1781 | h+ura4A-9 ura4-D18 |
| S82 BSC 45-1782 | h−ura4A-10 ura4-D18 |
| S83 BSC 45-1783 | h+ura4A-10 ura4-D18 |
| S84 BSC 45-1784 | h−ura4A-11 ura4-D18 |
| S85 BSC 45-1785 | h+ura4A-11 ura4-D18 |
| S86 BSC 45-1786 | h−ura4A-12 ura4-D18 |
| S87 BSC 45-1787 | h+ura4A-12 ura4-D18 |
| S88 BSC 45-1788 | h−ura4A-13 ura4-D18 |
| S89 BSC 45-1789 | h+ura4A-13 ura4-D18 |
| S90 BSC 45-1790 | h−ura4A-14 ura4-D18 |
| S91 BSC 45-1791 | h+ura4A-14 ura4-D18 |
| OL497 BSC 84-3346 | h−ura4A+ ura4-D18 his3-D1 swi10::kanMX ade6-M210 |
| MAB039 BSC 58-2295 | h−ura4A+ ura4-D18 pms1::his3+ his3-D1 ade6-M210 |
| MU132 BSC 97-3873 | h+ ura4A-13 ura4-D18 his3-D1 swi10::kanMX |
| MU134 BSC 97-3875 | h+ura4A-10 ura4-D18 his3-D1 swi10::kanMX |
| MU137 BSC 98-3891 | h−ura4A-10 ura4-D18 his3-D1 pms1::his3+ swi10::kanMX |
| MU136 BSC 98-3890 | h+ura4A-13 ura4-D18 his3-D1 pms1::his3+ swi10::kanMX |
The first entry is either the original strain name or a short version of the BSC number. The second entry is the number of the Berne strain collection.
All strains are from this study except GC151a and GC151b (Grimm et al. 1994) and OL497 obtained from O. Fleck. MU132 is a segregant from cross OL497 × S89, MU134 was obtained from OL497 × S83, MU136 from MAB039 × MU132, and MU137 from MAB039 × MU134.
Sequencing the ura4A junctions:
The construction of the ura4A insertion has been published (Grimm et al. 1994). Polymerase chain reaction (PCR) fragments were generated from the genomic DNA of strain GC151b with primers KL1 (5′-gtaatgaagccagccagtcg-3′) and KL4 (5′-catgctcctacaacattaccac-3′) to give the 3′-junction fragment and KL2 (5′-tgggtacccttccaatagtctc-3′) and KL3 (5′-cacaaagtgcaaacattatcatg-3′) to result in the 5′-junction fragment. The DNA was purified using NucleoSpin Extract columns (Macherey-Nagel, Duren, Germany). The 3′-junction fragment was sequenced with KL5 (5′-tggttataaacattggtgttggaac-3′) and the 5′-junction fragment with KL6 (5′-caccatgccaaaaattacacaag-3′), respectively.
Sequencing of ura4A mutations:
Total genomic DNA was isolated from haploid ura4A mutant strains (Table 1) by the small-scale method of Wright et al. (1986). To amplify a 1015-bp ura4A-specific genomic DNA fragment by PCR, two synthetic oligonucleotide primers with artificial 5′ restriction sites were designed (BamHI, 5′-gcgcg∨gATCCCAGTTTAACTATGCTTCGTCGGC-3′; EcoRI, 5′-gccgg∨AATTCTAAATGCCTTCTGACATAAAACGCC-3′). The desired PCR fragment, containing the entire 792-bp ura4 open reading frame (ORF) and including 118-bp upstream and 105-bp downstream regulatory sequences, was amplified in a 100-μl reaction volume containing 750 ng of genomic DNA, 0.25 μm of each primer, 200 μm of each dNTP, 0.5 units Super Taq polymerase (Stehelin, Basel Switzerland), 10 mm Tris·HCl, pH 9.0, 50 mm KCl, 1.5 mm MgCl2, 0.1% Triton X-100, and 0.01% (w/v) gelatin. Reactions were performed in a Perkin-Elmer (Norwalk, CT) Cetus DNA thermal cycler as follows: 4 min at 94°, 30 cycles at 94°, 59°, and 72° for 1 min each, and 10-min extension at 72°. Phenol-extracted PCR fragments were ligated after appropriate restriction digestion into the BamHI and EcoRI sites of pUC18 and analyzed by DNA sequencing. In addition to the two PCR primers, four internal primers had to be synthesized for the entire sequencing of both DNA strands by the dideoxy chain termination method [United States Biochemical (Cleveland) Sequenase kit]. To exclude artifacts, especially to distinguish Taq polymerase errors from the original ura4A mutations, at least three independent PCR products of each mutant were analyzed. In this way, all of the mutations in question could be located within the ura4A ORF.
Tetrad and octad analysis:
Tetrads were dissected on YEA plates using the Singer Instruments MSM system. After 4–5 days at 30°, the grown colonies were treated further. In monofactorial crosses their constitution with respect to mating type and ura4A was established. The latter was done by replica plating onto medium without uracil. In this way conversion, but not PMS, can be detected because a print consisting of approximately half ura+ and half ura− cells will be scored as ura+.
In the heteroallelic cross X1119 = S82 × S89 = h− ura4A-10 ura4-D18 × h+ ura4A-13 ura4-D18, the four colonies of each tetrad were streaked onto a YEA master plate and after growth were replicated on three MEA plates. The first was cross-stamped with the wild-type testers 972 (h−) and 975 (h+), the second with the tester pair S88/S89 (allele 13), and the third with the pair S82/S83 (allele 10). After this second round of mating and sporulation, the first plate was exposed to iodine vapors to determine mating type. The two others were replicated onto medium lacking uracil. Ensuing growth at the crossing between segregant and tester reflects wild-type sequence in the segregant with respect to the mutation carried by the tester. Correspondingly, no growth means that there is a mutant sequence in the segregant. In this way the allelic constitution of spores at the ura4A locus could be assessed: + +, + 10, 13+, and 13 10. Again, single-site PMS cannot be detected in this way. On the other hand, co-PMS spores (13+/+10) are detected. Such streaks do not yield confluent growth on medium without uracil, in contrast to prototrophic recombinants. They show growth with both testers in the limited crossing areas. This interpretation was verified by plating single cells from the original streak and typing the resulting colonies: None were prototrophic; approximately half were of one parental genotype (13+) and half of the other type (+10).
An octad analysis of cross MX1124 was done. MX1124 = MU137 × MU136 = h− ura4A-10 ura4-D18 × h+ ura4A-13 ura4-D18, as in X1119, but here, in contrast to X1119, both parents had inactivated repair genes, pms1::his3+ and swi10::kanMX. The cross was set up in the usual way on MEA. Tetrads were dissected on YEA in the afternoon and kept 3 hr at 30°. Then plates were transferred to 18°. The next day many spores appeared pear shaped, indicating germination. Plates were placed at 30° and observed at regular intervals. If a spore produced two daughters, they were separated by micromanipulation. In the best case, upon incubation eight colonies, organized in four pairs, resulted. Since in MX1124 the same ura4A alleles were involved (13 and 10) as were involved in X1119, the same procedures and tester strains as were used to analyze spore clones in X1119 could be used to type the postmeiotic segregants.
Random spore analysis:
The frequency of prototrophic recombinants was determined in heteroallelic crosses between mutants listed in Table 1. Cell material of both parental strains was suspended, mixed, and plated in a dense lawn on MEA. Incubation was 3 days at 25°. To kill vegetative cells, the sporulated material was treated over night at 30° with an aqueous snail enzyme solution [1:1000 (v/v) Helix pomatia juice, Biosepra]. For titer estimation of prototrophic recombinants and total spores, samples of appropriately diluted spore suspensions were spread on MMA and MMA + uracil, respectively. Incubation was 5 days at 30°.
RESULTS
Insertion junctions and ura4A mutants:
To verify correct insertion of the ura4A gene according to the published procedure (Grimm et al. 1994), PCR fragments of the ura4A junctions were generated and sequenced as described in materials and methods. It was found that the ura4A gene is transcribed in the opposite direction to the ade6 gene (Figure 1). While the 5′-junction (AAGCT-ATC) was exactly as predicted from the construction, the 3′-junction was found to contain an additional 15 bp of unknown origin: GAT-cctggtatggcttgt-AGCTT. No full match of the 15 bp to S. pombe or Escherichia coli genomic DNA could be detected.
Figure 1.—
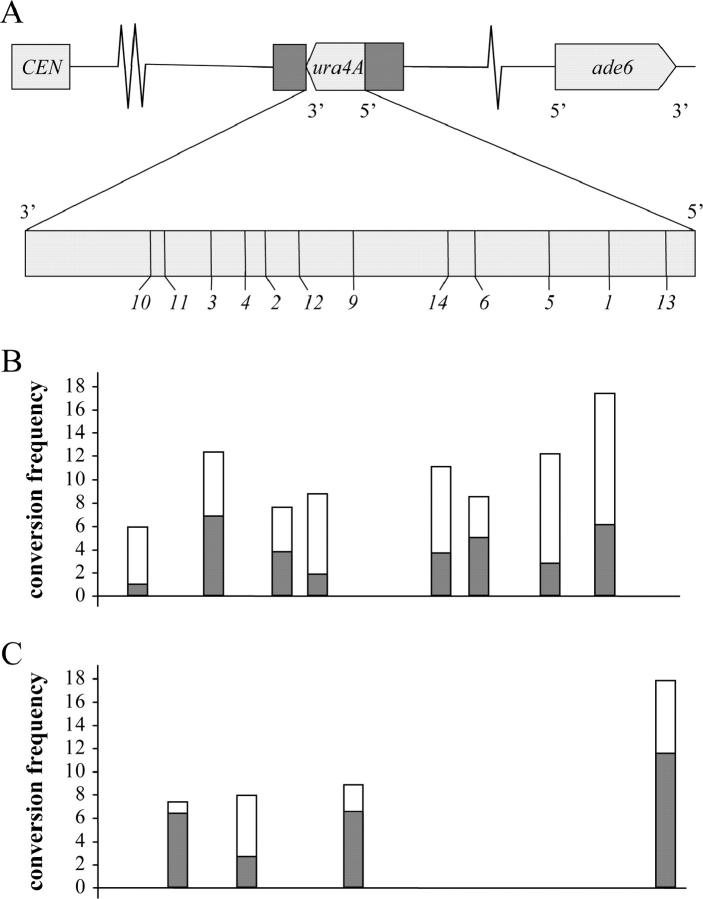
The ura4A hot spot. (A, top) The location and orientation of the ura4A insertion on chromosome III. (A, bottom) The locations of the sequenced mutations in the open reading frame (see Table 2 for details). In B and C, a gradient of conversion frequencies (percentage of NMS of tetrads examined) is apparent with the high frequencies for the mutations at the 5′ end. (B) Tetrad analysis of type I crosses (h+ ura4A+ × h− ura4A−) yielded overall more 2:6 (open bars) than 6:2 conversions (shaded bars). (C) Type II crosses (h− ura4A+ × h+ ura4A−) yielded more 6:2 than 2:6 conversions. For further explanations see the text.
The ura4A-x mutations are of spontaneous origin. Their positions in the gene and the physical alterations are given in Table 2 and Figure 1. Most are single-base-pair changes. ura4A-6 is a G-to-C transversion, which is known to show strongly enhanced prototroph frequencies in intragenic two-factor crosses (Schär and Kohli 1993). This is also true for this mutation (data not shown). Yet conversion frequency of ura4A-6 in single-point crosses is comparable to the frequencies obtained with neighboring markers. ura4A-11 is mitotically highly unstable (data not shown). This is understandable, considering its molecular nature (22-bp repeat). It cannot be used in random spore analysis of intragenic two-factor crosses. No problems arise with tetrads because the numerous matings between revertant and wild-type cells give 8+:0− segregations, which are recognized and excluded. ura4A-11 yields a conversion frequency similar to the mutations to its left and right.
TABLE 2.
Conversion frequencies ofura4A mutations in one-point crosses
| Conversionsc
|
|||||||
|---|---|---|---|---|---|---|---|
| Mutation | Positiona | Sequence change | Cross typeb |
6:2 | 2:6 | Tetrads analyzed |
Conversion (%) |
| 13 | 562 | C to A | II | 24 | 13 | 208 | 17.8 |
| 1 | 634 | A to G | I | 13 | 24 | 213 | 17.4 |
| 5 | 709 | G to T | I | 6 | 20 | 214 | 12.1 |
| 6 | 793 | G to C | I | 10 | 7 | 200 | 8.5 |
| 14 | 823 | C to A | I | 8 | 16 | 217 | 11.1 |
| 9 | 934 | G deletion | II | 14 | 5 | 216 | 8.8 |
| 12 | 1001 | G insertion | I | 4 | 15 | 217 | 8.8 |
| 2 | 1043 | G to A | I | 8 | 8 | 211 | 7.6 |
| 4 | 1066 | G to A | II | 4 | 8 | 153 | 7.8 |
| 3 | 1104 | C to T | I | 15 | 12 | 220 | 12.3 |
| 11 | 1163 | 22-bp repeat | II | 13 | 2 | 205 | 7.3 |
| 10 | 1167 | C to T | I | 2 | 10 | 204 | 5.9 |
Numbering according to Grimm et al. (1988). First base of start codon is 534; first base of stop codon is 1326.
Cross type I is h− ura4A− × h+ ura4A+; cross type II is h− ura4A+ × h+ ura4A−.
In this tetrad run it was not possible to detect PMS. A conversion of 6:2 means three ura4A+ spore colonies (prototrophs) and one ura4A− colony (auxotroph); 2:6 conversion tetrads have only one ura4A+ spore colony.
Hot spot, polarity, and mat bias as revealed by tetrad analysis:
The data on tetrad analyses of monofactorial crosses are given in Table 2 and Figure 1. The overall conversion frequencies of the extreme markers to the 5′ and 3′ sides are 18% (ura4A-13) and 6% (ura4A-10). Values of intervening mutations are in between these two values. In S. pombe 17 multifactorial crosses producing a total of 12,119 segregations of allelic differences showed only 17 3:1 and 15 1:3 conversion tetrads, giving an average NMS frequency of 0.26% (P. Munz, unpublished results). The 20- to 60-fold higher conversion frequencies of the tested alleles clearly show that ura4A is a hot spot of meiotic recombination.
Figure 1 shows that conversion frequencies gradually decline from the 5′ to the 3′ side of the gene. This reveals a conversion gradient, as is also seen in other organisms (Nicolas and Petes 1994; Paques and Haber 1999). The decrease is not strictly monotonic. Alleles ura4A-6 and ura4A-3 show departures. This might be attributed to statistical fluctuations or, in the case of ura4A-6, to the marker effect of G-to-C transversions (Schär and Kohli 1993).
Before describing the novel phenomenon called mating-type bias (mat bias), some conventions applied in this article will be introduced.
Allele ratios in tetrads are stated according to the mode used for eight-spored asci (each single-strand is counted in the four-spored asci).
Plus (+) is always stated first, followed by minus (−), and the wild-type allele + at ura4A promotes growth on uracil-free media.
Type I and type II crosses are defined as follows: In type I crosses, the h− parent contributes the ura4A mutant allele, whereas in type II crosses, the wild-type allele is associated with h−.
The mat bias is documented in Table 2 and Figure 1. The two conversion classes do not occur with equal frequencies. In type I, 6:2 conversions are less frequent (shaded frequency bars in Figure 1B) than 2:6 (open bars). In type II, the situation is reversed; 6:2 tetrads are in the majority (Figure 1C). This rule is not strict. Of the 12 markers analyzed, 8 conform to the scheme, 3 violate it, and 1 shows equality (ura4A-2). When all tetrad numbers were pooled within types, type I showed a total of 66 6:2 tetrads and 112 2:6 tetrads. In type II, the numbers were 55 (6:2) and 28 (2:6). The departure from equality is significant in both cases (χ2 test, P < 0.005). It appears that information carried by the allele entering with the h− parent was preferably donated for conversion, while the allele coming with the h+ parent acted as acceptor more often than parity would predict.
Polarity, mat bias, and map expansion as revealed by random spore analysis:
Random spores from a number of two-factor crosses have been analyzed. For ease of discussion, we define type III crosses as h− a + × h+ + b. The 5′ mutation a enters with the h− parent, and the 3′ mutation b with the h+ parent. The reciprocal type IV crosses were h− + b × h+ a +. The data are given in Table 3 [one determination per cross, prototrophs per million (ppm) spores rounded to two figures]. A series of preliminary experiments (data not shown) with a number of mutants led to the recognition that polarity, mat bias, and map expansion are detectable by this approach. Then the seven mutations, 13, 1, 5, 14, 9, 2, and 10 in 5′–3′ order, were chosen (low reversion frequency, suitable location) for crosses carried out under rigorously standardized conditions. The data on six mutations are shown in Figure 2. The observations of mutant ura4A-9 are treated in the next section. All pairwise combinations have been analyzed. In control selfings of the mutants, no prototrophs were observed in ∼2 × 107 spores, with exception of ura4A-9 (2 × 10−5). The concentrations of prototrophic recombinants and of total spores in spore suspensions were determined as described in materials and methods. Prototrophs per million spores were calculated by the division of the prototroph concentration by the concentration of total spores and then multiplied by 106. The recombination density (D) is defined as the quotient of the ppm value and the number of base pairs separating the two mutations involved. D is a measure of hotness for recombination of a given interval.
TABLE 3.
Random spore analysis of two-point crosses
| Type III cross (low)c
|
Type IV cross (high)c
|
|||||
|---|---|---|---|---|---|---|
| Mutationsa | Distanceb (bp) | ppmd | Densitye | ppmd | Densitye | Quotientf |
| 13 × 1 | 72 | 620 | 8.6 | 880 | 12.3 | 0.70 |
| 13 × 5 | 147 | 960 | 6.5 | 2,200 | 14.8 | 0.44 |
| 13 × 14 | 261 | 2100 | 7.9 | 4,600 | 17.5 | 0.45 |
| 13 × 9 | 371 | 6900 | 18.5 | 7,900 | 21.2 | 0.87 |
| 13 × 2 | 481 | 6800 | 14.1 | 9,500 | 19.8 | 0.71 |
| 13 × 10 | 605 | 8600 | 14.2 | 12,000 | 20.2 | 0.71 |
| 1 × 5 | 75 | 540 | 7.1 | 670 | 9.0 | 0.80 |
| 1 × 14 | 189 | 1700 | 8.9 | 2,300 | 12.0 | 0.74 |
| 1 × 9 | 299 | 5500 | 18.3 | 5,100 | 17.1 | 1.07 |
| 1 × 2 | 409 | 5000 | 12.2 | 5,800 | 14.3 | 0.86 |
| 1 × 10 | 533 | 5700 | 10.8 | 9,400 | 17.7 | 0.61 |
| 5 × 14 | 114 | 950 | 8.3 | 1,100 | 10.1 | 0.83 |
| 5 × 9 | 224 | 3700 | 16.6 | 3,500 | 15.8 | 1.05 |
| 5 × 2 | 334 | 4800 | 14.3 | 4,100 | 12.4 | 1.16 |
| 5 × 10 | 458 | 6000 | 13.2 | 5,900 | 12.8 | 1.03 |
| 14 × 9 | 110 | 1100 | 9.9 | 1,400 | 12.8 | 0.78 |
| 14 × 2 | 220 | 1600 | 7.3 | 2,000 | 9.3 | 0.79 |
| 14 × 10 | 344 | 2400 | 6.9 | 4,000 | 11.6 | 0.59 |
| 9 × 2 | 110 | 930 | 8.4 | 1,100 | 10.2 | 0.83 |
| 9 × 10 | 234 | 1900 | 8.2 | 2,400 | 10.3 | 0.80 |
| 2 × 10 | 124 | 480 | 3.9 | 750 | 6.0 | 0.64 |
The mutation stated first is the one closer to the 5′ end of the gene.
The distance between the mutation sites in base pairs.
In type III crosses, the mutation closer to the 5′ end is in the h− strain; in type IV crosses, it is in the h+ strain.
Prototrophic recombinants per million spores (one experiment per cross).
Prototrophs per million divided by base pairs (distance between mutations).
Prototrophs per million of the type III cross divided by ppm of the type IV cross.
Figure 2.—
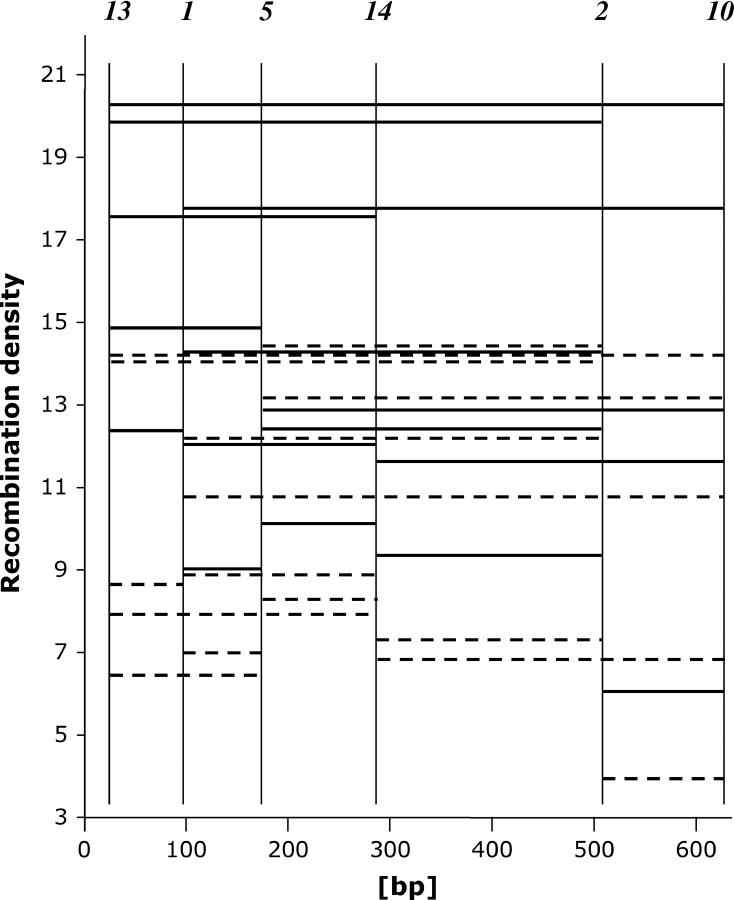
Intragenic two-factor crosses at ura4A. The abscissa shows the open reading frame of ura4A (5′ end to the left) and the physical position of six selected mutations. The ordinate portrays the recombination density D (ppm per base pair). The horizontal lines connect the two mutations involved in a given cross. Type III crosses show lower densities (dashed lines) than the corresponding type IV crosses (solid lines). The detailed data are shown in Table 3. For further explanations, see the text.
In Figure 2 intragenic recombination is plotted in an unconventional way by jointly using genetic and physical information. The abscissa represents the physical map of ura4A in base pairs with the six mutations located at their sites. The recombination densities are shown as horizontal bars whose endpoints connect the mutations involved and whose heights indicate the density value.
The hot spot nature of ura4A is clearly revealed by the two-factor crosses: Whereas in non-hotspot genes ppm values obtained with intragenic markers farthest apart rarely exceed 1000 (Reymond et al. 1992; Grishchuk and Kohli 2003), the value for the extreme mutations ura4A-13 and ura4A-10 is >10,000 in the type IV cross.
Inspection of the data in Table 3 reveals differences between the prototroph frequencies (ppm) and the recombination densities (D) for the mating-type reciprocal crosses. Prototrophs are mainly produced by events starting at one end of the gene and ending between the two mutations. Like the tetrad data, the random spore data can be explained by assuming that the 5′ part of ura4A entering with the h+ parent is the preferred acceptor of information. Thus, in type III crosses there is a preponderance of the proximal a mutation replacing + by conversion. This produces not a prototroph but rather the double mutant (a b). On the other hand, in type IV, + replaces a, generating a wild-type allele.
In Figure 2 the recombination densities are plotted for type III (dashed lines) and type IV crosses (solid lines). In both series the highest D-values were generally obtained for intervals bordered by alleles close to the 5′ end of the gene. The D-values gradually fall for intervals toward the 3′ end. Estimates for intervals of approximately equal physical length decline from left to right. The lowest values were obtained for the interval defined by the mutations 2 and 10 closest to the 3′ end. This gradient of D-values reflects the 5′–3′ polarity of NMS already observed in the one-factor crosses (Figure 1).
In the two-factor crosses the mat bias first described by tetrad analysis (Figure 1) reveals itself for most intervals by comparison of the D-value from the type III cross with that from the type IV cross. The corresponding data are given in Table 3 and plotted in Figure 2 (cross III, dashed lines; cross IV, solid lines). With four exceptions, the D-values from type IV crosses are higher than those from type III crosses. The quotients (Q) of types III and IV ppm values are shown in Table 3. Under the assumption that mating-type configuration has no effect on prototroph formation, one expects an equal number of Q-values below and above unity, yet the observed ratio is 17:4, clearly deviating from 1:1. The range of the 21 Q-values is 0.44–1.16, the median 0.79, and the mean 0.78 with an associated standard deviation of 0.19. In addition to polarity and mat bias, map expansion is visible (Holliday 1964). This cannot be seen in monofactorial crosses. Map expansion is the term for the observation that intragenic recombinant frequencies (ppm) for a given interval are higher than the sum of the frequencies obtained for subintervals. If map expansion were absent in ura4A, the density of a compound interval could not be larger than the largest value for the subintervals. Inspection of Figure 2 shows that this is never the case for the type IV crosses. In any family of intervals chosen, the compound value always surpasses the subinterval values. There are a few exceptions for the type III crosses, however, but basically the same conclusion can be reached for the series yielding low ppm values. It is evident that mixing the two data sets would confuse the situation.
A novel marker effect:
The data on ura4A-9 are given in Table 3 but not in Figure 2. The mutations included in Figure 2 behave in a regular way in the sense that in families of intervals with the same left-hand marker the densities monotonically increase as the right-hand partner allele moves toward the 3′ end. ura4A-9, located between 14 and 2, is an exception. Its D-values are too large to fit into the pattern of the other mutations. This, we think, reflects a mild marker effect. Mutation 9, a deletion of a G, enhances prototroph formation independent of polarity and map expansion effects. For this reason, it was excluded from Figure 2 and the analysis of mat bias and map expansion. For the same reason, mutation 6, a G-to-C transversion, was also excluded. G-to-C transversion marker effects are strong and have been described in detail (Schär and Kohli 1993).
Hybrid DNA, conversion, and restoration at ura4A:
The high conversion frequency at ura4A (in contrast to the forbidding quarter percentage generally seen) makes it possible to obtain some information on the nature of recombination events. Two crosses, X1119 and MX1124, have been analyzed. With respect to ura4A, they are identical type IV crosses: h− ura4A-10 × h+ ura4A-13. The difference is in the genetic background. In X1119, the mismatch repair genes pms1 and swi10 (Fleck et al. 1999) were wild type; in MX1124, they were disrupted.
Table 4 sums up the tetrad analysis of X1119. The overall frequency of NMS is 51/290 (18%). Most events involve both sites: 35 co-conversions (tetrad types D and E) and two co-PMS (F and G), totaling 37/51 (73%). Single-site PMS was not detected. A total of 14 single-site conversions (B and C) were found at the 5′ mutation 13. In contrast, the 3′ mutation 10 was never involved alone. The asymmetries within the single conversion and the co-conversion classes are attributed to the mat bias: The allele entering the cross with h+ acts predominantly as acceptor.
TABLE 4.
Tetrad analysis of two-point crosses
| Tetrad types | Spore 1 | Spore 2 | Spore 3 | Spore 4 | No.c |
|---|---|---|---|---|---|
| Cross X1119a: h+ ura4A-13 × h− ura4A-10 | |||||
| A | + − | + − | − + | − + | 239 |
| + − | + − | − + | − + | ||
| B | + − | + − | + + | − + | 10 |
| + − | + − | + + | − + | ||
| C | + − | − − | − + | − + | 4 |
| + − | − − | − + | − + | ||
| D | + − | + − | + − | − + | 25 |
| + − | + − | + − | − + | ||
| E | + − | − + | − + | − + | 10 |
| + − | − + | − + | − + | ||
| F | + − | + − | + − | − + | 1 |
| + − | + − | − + | − + | ||
| G | + − | + − | − + | − + | 1 |
| + − | − + | − + | − + | ||
| Total: | 290 | ||||
| Cross MX1124b: h+ ura4A-13 pms1− swi10− × h− ura4A-10 pms1− swi10− | |||||
| A | + − | + − | − + | − + | 32, 8, 13 |
| + − | + − | − + | − + | ||
| B | + − | + − | + + | − + | 1, 1, 0 |
| + − | + − | + + | − + | ||
| D | + − | + − | + − | − + | 1, 0, 0 |
| + − | + − | + − | − + | ||
| F | + − | + − | + − | − + | 3, 0, 2 |
| + − | + − | − + | − + | ||
| G | + − | + − | − + | − + | 2, 1, 2 |
| + − | − + | − + | − + | ||
| H | + − | + − | + + | − + | 3, 0, 0 |
| + − | + − | − + | − + | ||
| I | + − | + − | − + | − + | 0, 2, 3 |
| + − | + + | − + | − + | ||
| K | + − | + − | − + | − + | 0, 0, 2 |
| + − | − − | − + | − + | ||
| L | + − | + − | + − | − + | 1, 0, 0 |
| + − | + + | − + | − + | ||
| M | + − | + − | + − | − + | 0, 0, 1 |
| + − | + + | − − | − + | ||
| Total: | 43, 12, 23 | ||||
For each spore of the tetrads the genotype is given for both DNA strands (+, wild type; −, mutant), first for the 5′ site (13), followed by the 3′ site (10).
Octad analysis was performed on this cross. Thus, the + and − symbols refer to homoduplex DNA derived from the first mitotic division of spores.
Only complete tetrads were considered for cross X1119. For cross MX1124, the numbers of complete octads for a specific type are given first, followed by the numbers of octads with one or two missing colonies (see text).
The data are interpreted as follows: All or most hDNA starts upstream of the gene and extends to the 3′ end of the gene. It may terminate within the gene or, more frequently, beyond it. Repair of base mismatches in hDNA is efficient (35 co-conversions) but not absolute (2 co-PMS). If hDNA were exclusively symmetrical, one would expect some apparent reciprocal recombination between 13 and 10, due to coverage of 13 alone combined with conversion-type repair in both hDNA chromatids. Since this was not found, we prefer the view that hDNA is mostly asymmetrical at ura4A.
The results from cross MX1124 (mismatch repair defective) are also presented in Table 4. In this case, the first postmeiotic cells deriving from the spores have been separated to yield octads composed of four pairs of colonies deriving from a single meiosis. The inactivation of mismatch repair led to a marked increase of PMS events. Yet mismatch repair apparently was not completely abolished. For instance, octad B was classified as a single-site conversion of 13 and octad D as a co-conversion.
The analysis suffered from considerable spore lethality, likely due to the repair mutant background. Disruption of pms1 has a deleterious effect on spore viability (Schar et al. 1997). In addition to the complete octads, we tentatively incorporated information from the incomplete octads, when they were missing only one colony, or from the two sister colonies deriving from a single spore (as indicated in Table 4). The genotype of missing colonies was always assumed to be one of the parentals and chosen to avoid creation of an NMS event.
First, the complete octads are considered alone. The overall frequency of NMS is 11/43 (26%). Ten events are easily explained by asymmetrical hDNA, and one (L) shows evidence for symmetrical hDNA. Mutation 13 is involved alone four times (B and H). In the remaining seven NMS octads, both markers participate. No conversion or PMS of 10 alone was found. Thus, the same conclusions as with X1119 are reached: hDNA enters predominantly from the 5′ side, covering in a minority of cases 13 alone, and in the majority of events 13 and 10 jointly. Furthermore, hDNA is mostly asymmetrical.
Two interpretations are possible for octad L.
hDNA is fully symmetrical over the entire gene, and one 13/+ mismatch experiences restoration-type repair. Thus, the repair deficiency might not only reduce the repair frequency, but also shorten the tracts in the remaining cases of repair. Single-site repair would then occur at the expense of corepair.
hDNA formation starts asymmetrically in one chromatid and shifts to a symmetrical phase between the markers. Absence of repair of the three resulting mismatches then results directly in octad L.
In what follows, all octads of cross MX1124 (shown in Table 4) are considered. The frequency of NMS is 32% (25/78). All but two (L and M) can be explained by asymmetrical hDNA formation. The 5′ site 13 was involved alone five times (B and H). Both sites were involved in 15 octads (D, F, G, K, L, M). The 3′ site 10 was involved alone in five octads (I). The data indicate that hDNA may cover marker 10 alone. This could occur if an event started within the gene and proceeded toward the 3′ end, and, alternatively, started in the 3′ flanking region and terminated between the markers. These possibilities cannot be disproved at this stage. On the other hand, on the basis of X1119 and the sample of complete octads with no sign of NMS of 10 alone, we prefer the interpretation that the five octads under discussion are the result of hDNA spanning both sites with restoration repair limited to the 13/+ mismatch.
In considering 13 strictly on its own, we find support for the occurrence of restoration at the 5′ end of ura4A. In the repair-proficient cross X1119, there are two PMS and 49 conversion events in 290 tetrads, resulting in a conversion frequency of 17% (49/290). In the repair-deficient cross MX1124, there are 17 PMS and three conversion events. We imagine that almost all of the 17 PMS events would be repaired in a repair-proficient cross. If one assumes that 16 of the 17 PMS events yielded eight conversions and eight restorations, the conversion frequency would be 14% (11/78). If, on the other hand, all 16 PMS events were repaired into conversions, the frequency would be 24% (19/78). In conclusion, the assumption that 13/+ mismatches are about half converted and half restored in repair-proficient crosses leads to a better agreement with the 17% conversion observed at 13 in the X1119 cross.
DISCUSSION
The frequency of meiotic recombination at the ura4A gene is markedly elevated compared with other S. pombe genes analyzed. The ura4A hot spot originated from insertion of the ura4 gene 15 kb away from ade6 on chromosome III (Grimm et al. 1994). At its original locus ura4 shows low recombination frequencies: the maximal prototroph frequency obtained in crosses between mutants was 1400 × 10−6 (data not shown). Not all relocations of genes lead to hot spot activity in S. pombe. The insertion of arg3 on chromosome III 20 kb from ura4A does not result in a hot spot (B. Sakem and J. Kohli, unpublished results). Relocation of the ade6-M26 hot spot to various places in the genome resulted in strong variation, including loss, of hot-spot activity (Virgin and Bailey 1998).
The well-characterized hot spot ade6-M26 (Gutz 1971; Davis and Smith 2001) derives from a single base substitution in the open reading frame of ade6. It leads to the formation of an oligonucleotide sequence that is required for hot spot activity (Schuchert et al. 1991; Steiner et al. 2002). ura4A is only the second meiotic hot spot that has been characterized in detail in S. pombe. Its maximal frequency of NMS events (18% in wild type, 26% in mismatch-repair-deficient strains) is higher than that of the homozygous ade6-M26 hot spot: 7.5% in wild type (Schär and Kohli 1994), and 9.5% in repair-deficient background (Fleck et al. 1999). Many hot spots of recombination have been characterized in S. cerevisiae (Lichten and Goldman 1995; Paques and Haber 1999) and also in higher eukaryotes (de Massy 2003). It remains to be elucidated to what extent the mechanisms leading to hot-spot activity are conserved in different organisms. In addition, hot spots like ura4A render studies of recombination mechanisms feasible, which are not possible elsewhere in the genomes.
Polarity and double-strand breaks:
A conversion gradient is seen in ura4A ranging from 18% NMS at the 5′ end to 6% at the 3′ end (Figure 1). This polarity is also apparent in prototroph frequency analysis of two-factor crosses (Figure 2). ura4A is the first S. pombe gene with a clearly described polarity. In S. cerevisiae and other fungi, this pattern is frequent and indicates initiation of recombination by DSB formation in the promoter region (Lichten and Goldman 1995; Liu et al. 1995; Paques and Haber 1999; Petes 2001). Recently, a meiotic DSB has been detected in the region upstream of ura4A (B. Sakem and J. Kohli, unpublished results). But there are exceptions to initiation in the 5′ region: In the HIS2 gene of S. cerevisiae higher frequencies of conversion are observed for mutations at the 3′ end where two DSBs also have been located (Malone et al. 1992; Haring et al. 2003).
For the ade6-M26 hot spot, DSB formation has been shown to move through the ade6 gene in register with movement of the M26 oligonucleotide sequence. Also, the amount of DNA breakage correlated with intragenic recombination frequency (Steiner et al. 2002). It has been shown that Atf1/Pcr1 transcription-factor binding to the M26 sequence is required for M26 hot-spot activity (Kon et al. 1997). Thus, the M26 hot spot may correspond to α-type hot spots of S. cerevisiae that require transcription factor binding for efficient DSB formation (Petes 2001). In S. cerevisiae, DSB formation is also promoted by sequences refractive to nucleosome formation (β-type hot spots) and by sequences high in G + C content γ-type hot spots (Petes 2001). A large number of meiotic DSBs were visualized throughout the S. pombe genome by pulse-field gel electrophoresis and proposed to be involved in meiotic recombination (Cervantes et al. 2000; Young et al. 2002). Additional experiments are required for determination of the features that lead to hot spot activity at ura4A.
Hybrid DNA, conversion, and restoration:
Hybrid DNA is a central intermediate in homologous recombination (Paques and Haber 1999). It has been shown to occur with high frequency across the ade6-M26 hot spot sequence (Schär and Kohli 1994). This work demonstrates frequent coverage of the whole ura4A gene by hDNA as concluded from the observed co-conversion and PMS events found in tetrad analysis of two-factor crosses (Table 4). hDNA extends predominantly from the 5′ side and is likely to terminate to some extent in the middle of the gene, since conversion of a 5′ mutation alone was observed (Table 4). Conversion of the 3′ allele alone was not observed, indicating that extension of hDNA from the 3′ side into the gene is rare. The hDNA formation at ura4A mostly occurs in only one chromatid (asymmetrical), but two cases of symmetrical hDNA have been demonstrated (Table 4), one in the wild-type (3% of NMS tetrads) and the other in the repair-deficient cross (4% of NMS tetrads). The corresponding values for ade6-M26 are: no aberrant 4:4 tetrads in wild type and 23% in the repair-deficient cross (Fleck et al. 1999). Lower frequency of symmetrical hDNA formation may distinguish the ura4A hot spot from the M26 hot spot.
Mismatches in hDNA are repaired to different extents, depending on the types of cells and base mismatches formed. In S. pombe, repair is efficient for all mismatches (95% repair or more), with the notable exception of C/C mismatches with 70% repair (Schär et al. 1993; Schär and Kohli 1993). With the exception of the marker-effect mutations (see below), the ura4A mutations used in this study produced mismatches that were corrected with high efficiency. Few PMS events were found with the exception of cross MX1124, in which two different pathways of mismatch repair were inactivated (Fleck et al. 1999).
There are different models for the explanation of gradients of NMS events across genes. One model, the restoration/conversion model, explains such gradients assuming that 5′ markers are mostly converted, whereas mismatches at the 3′ end are half converted and half restored (Detloff et al. 1992; Kirkpatrick et al. 1998). This scenario assumes that hDNA practically always covers the entire gene. If hDNA enters from one side and is allowed to terminate within the gene, the gradient is explained. Then there is no need to assume different frequencies of conversion and restoration in function of the site within the gene.
For ura4A we prefer the termination-within-gene hypothesis to the restoration/conversion model. First, ura4A-13, the extreme 5′ mutation, is apparently not only converted, but also restored (see the corresponding section in results). Second, frequencies of prototrophic recombinants in segments of equal physical size drop from the 5′ to the 3′ region of the gene (Figure 2). This is explained by more frequent termination of hDNA between mutations at the 5′ end than at the 3′ end. In the restoration/conversion model, hDNA is assumed to span the whole gene, and recombinants must arise by independent repair of the two mismatches. Depending on the chromatid involved, one mismatch must experience conversion-type repair, the other restoration-type repair. If chromatids of different parentage are equally used, and no bias in repair direction exists, no drop of recombination is expected. If 5′ markers are preferentially converted, again no drop of recombination is expected, since gain in one chromatid is balanced by loss in the other. Finally, the mat bias to be described below has no impact on this matter: crosses of both types, III (low) and IV (high), show the same polar decrease of recombination density.
Marker effects:
The ura4A-6 mutation is a G-to-C transversion (Table 2, Figure 1). It shows an overall frequency of NMS in one-factor crosses expected from its location within the gene, but strongly enhanced prototroph frequencies in two-factor crosses with other ura4A mutants (data not shown). This feature of G-to-C transversions has been investigated (Schär et al. 1993; Schär and Kohli 1993; Fleck et al. 1999). It is based on the inability of the MutLS long-patch mismatch-repair system to correct C/C mismatches. Instead, C/Cs are partially repaired by the short-patch nucleotide excision repair system and remain partially unrepaired, resulting in PMS. Both the short repair tracts and the failure of repair contribute to higher prototroph frequencies, when two mutations involved in a two-factor cross are included in the same hDNA tract.
The mutation ura4A-9, deletion of a single G-C pair, showed normal behavior in one-factor crosses (Table 2, Figure 1). In the two-factor crosses, it yielded generally higher prototroph frequencies than expected from its position in the middle of the gene. The average recombination density calculated from the six crosses shown in Table 3 is 14.5, while the averages for the flanking mutations ura4A-14 and ura4A-2 are 12.2 and 12, respectively. This novel marker effect is comparably weak. It may originate from inefficient repair of the single nucleotide loops (G and C) in hDNA by the MutLS system (PMS as consequence) or by recognition of the single base loops by an alternative, short-patch repair system.
The two examples of marker-effect alleles demonstrate that prototroph frequencies, as determined in two-factor crosses, can be affected by special features of the different mismatch repair pathways. For this reason mutations showing marker effects must be excluded when overall features of intragenic recombination are studied (Figure 2). The phenomena of mat bias and map expansion discussed below could not be clearly described with inclusion of the data from the two marker-effect alleles.
Mat bias:
This phenomenon, to our knowledge, has not been described before. The ura4A allele entering the cross with the h+ parent acts as information acceptor almost twice as often as the allele being contributed by the h− parent (167: 94, Q = 0.56), as seen by summing up of the tetrad analysis of all mutants (Table 2). In the random spore analysis of two-factor crosses (Table 3), the mean bias (Q = 0.78) is lower. This may derive from the lack of detection of events that do not result in prototrophic recombinants (co-conversions, Table 4). Since the h+ and h− mating types of S. pombe carry different DNA rearrangements at the mating-type locus on chromosome II (Beach et al. 1982), we wondered whether the structural differences between the h+N and h−S mating types used in our experiments could account for the mat bias. From the h+N ura4A-13 and h+N ura4A-10 strains spontaneous h−U derivatives were isolated (Egel 1989), and random spores of the two reciprocal crosses 13 × 10 with isogenic backgrounds were analyzed (data not shown). The asymmetry in prototroph frequencies persisted, again with the chromosome from the h+ parent as the preferred acceptor. Since h+N and h−U are structurally equivalent, it seems that mating-type function, rather than mating-type structure, is responsible for the mat bias. h+ mating-type function derives from the expression of the two genes Pc and Pi from the mat1P allele at mat1 (Kelly et al. 1988). That the mat bias is caused by a genetic factor unrelated to mating type and residing elsewhere in the genome is unlikely, when all the reported experiments are considered.
A number of questions about the mat bias require additional experiments. Is it specific for the ura4A hot spot, or does it affect other genes as well? Recent experiments with two other genes indicate that the mat bias is a general phenomenon in S. pombe (E. Parvanov and J. Kohli, unpublished results). Obviously the chromosomes contributed by the h+ parent maintain a memory, probably a feature of chromatin structure not present in chromosomes of h− cells, through cell mating, karyogamy, and premeiotic DNA replication. An obvious question is whether the mat bias is absent in azygotic meiosis after homologous chromosomes have coexisted in diploid cells for many mitotic divisions. Recent experiments indicate loss of the mat bias in azygotic meiosis (E. Parvanov and J. Kohli, unpublished results).
Map expansion:
This phenomenon was described on the basis of fine-structure mapping of genes (Holliday 1964). The maps were constructed from prototroph frequencies measured in two-factor crosses of auxotrophic mutants. The prototroph frequency measured for an interval defined by mutations a and b was often larger than the sum of the prototroph frequencies obtained from two subintervals. The subintervals were defined by additional mutations mapped between a and b. The longer the a-b intervals were, the more pronounced was map expansion. Models for the explanation of map expansion are founded on variations of parameters of hDNA formation, of mismatch repair, and of mismatch-induced rejection of hDNA (Fincham and Holliday 1970; Fujitani and Kobayashi 1997). The extreme map expansion found in S. pombe tRNA genes (Fincham and Holliday 1970) was later attributed to marker effects of G-to-C transversion mutations (Hofer et al. 1979; Schär and Kohli 1993). Since physical ordering of mutations was not possible at the time, the description of map expansion was hampered by mistakes in mapping and by marker-effect mutations that could not be identified easily. Considering these difficulties, the very existence of map expansion was questioned (Stahl 1979).
The distribution of physically mapped mutations along ura4A, the exclusion of marker-effect alleles, the frequent coverage of the whole gene by hDNA, and the frequent long repair tracts covering the 605 bp between the outer markers 13 and 10 (co-conversion) were ideal for a reevaluation of the existence of map expansion. As shown in Figure 2 and explained in the results, map expansion was clearly demonstrated in ura4A. On the basis of the results obtained from tetrad and octad analysis of the two-factor cross 13 × 10 (Table 4), we offer the following explanation. Prototrophic recombinants appear to be generated mainly by hDNA ending between the mutation sites in ura4A. A minor contribution is likely to derive from hDNA covering both sites, coupled with independent repair of the two mismatches. Map expansion could then be the result of this second component gaining overproportionally in importance as markers become farther apart. Alternatively, hDNA spanning two markers may be destabilized more frequently, when the markers are close, resulting in a relatively greater loss of potential recombinants for close markers.
In conclusion, the detailed analysis of intragenic recombination at ura4A has revealed the novel phenomenon of mat bias and established the existence of map expansion and will serve as the basis for the investigation of the molecular mechanisms of recombination.
Acknowledgments
We acknowledge support from grants of the Swiss National Science Foundation. K. Ludin was supported by a Marie-Heim-Vögtlin fellowship. Part of this work is contained in the Ph.D. thesis of M. Baur, submitted to the Faculty of Science, University of Berne.
References
- Beach, D., P. Nurse and R. Egel, 1982. Molecular rearrangement of mating-type genes in fission yeast. Nature 296: 682–683. [DOI] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2001. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98: 8395–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy, B., 2003. Distribution of meiotic recombination sites. Trends Genet. 19: 514–522. [DOI] [PubMed] [Google Scholar]
- de Massy, B., V. Rocco and A. Nicolas, 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14: 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., M. A. White and T. D. Petes, 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel, R., 1989 Mating-type genes, meiosis and sporulation, pp. 31–73 in Molecular Biology of the Fission Yeast, edited by P. Y. A. Nasim and B. F. Johnson. Academic Press, San Diego.
- Fincham, J. R., and R. Holliday, 1970. An explanation of fine structure map expansion in terms of excision repair. Mol. Gen. Genet. 109: 309–322. [DOI] [PubMed] [Google Scholar]
- Fleck, O., E. Lehmann, P. Schar and J. Kohli, 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21: 314–317. [DOI] [PubMed] [Google Scholar]
- Fujitani, Y., and I. Kobayashi, 1997. Mismatch-stimulated destruction of intermediates as an explanation for map expansion in genetic recombination. J. Theor. Biol. 189: 443–447. [DOI] [PubMed] [Google Scholar]
- Grimm, C., J. Kohli, J. Murray and K. Maundrell, 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215: 81–86. [DOI] [PubMed] [Google Scholar]
- Grimm, C., J. Bahler and J. Kohli, 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 136: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, A. L., and J. Kohli, 2003. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69: 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974 Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Haring, S. J., G. R. Halley, A. J. Jones and R. E. Malone, 2003. Properties of natural double-strand-break sites at a recombination hotspot in Saccharomyces cerevisiae. Genetics 165: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer, F., H. Hollenstein, F. Janner, M. Minet, P. Thuriaux et al., 1979. The genetic fine structure of nonsense suppressors in Schizosaccharomyces pombe. Curr. Genet. 1: 45–61. [DOI] [PubMed] [Google Scholar]
- Holliday, R., 1964. A mechanism for gene conversion in fungi. Genet. Res. 5: 282–304. [DOI] [PubMed] [Google Scholar]
- Kelly, M., J. Burke, M. Smith, A. Klar and D. Beach, 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., M. Dominska and T. D. Petes, 1998. Conversion-type and restoration-type repair of DNA mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Genetics 149: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith and W. P. Wahls, 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94: 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M., and A. S. Goldman, 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Liu, J., T. C. Wu and M. Lichten, 1995. The localization and structure of double-strand breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, R. E., S. Bullard, S. Lundquist, S. Kim and T. Tarkowski, 1992. A meiotic gene conversion gradient opposite to the direction of transcription. Nature 359: 154–155. [DOI] [PubMed] [Google Scholar]
- Nicolas, A., and T. D. Petes, 1994. Polarity of meiotic gene conversion in fungi: contrasting views. Experientia 50: 242–252. [DOI] [PubMed] [Google Scholar]
- Ohta, K., T. Shibata and A. Nicolas, 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 13: 5754–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Reymond, A., S. Schmidt and V. Simanis, 1992. Mutations in the cdc10 start gene of Schizosaccharomyces pombe implicate the region of homology between cdc10 and SWI6 as important for p85cdc10 function. Mol. Gen. Genet. 234: 449–456. [DOI] [PubMed] [Google Scholar]
- Schär, P., and J. Kohli, 1993. Marker effects of G to C transversions on intragenic recombination and mismatch repair in Schizosaccharomyces pombe. Genetics 133: 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schär, P., and J. Kohli, 1994. Preferential strand transfer and hybrid DNA formation at the recombination hotspot ade6–M26 of Schizosaccharomyces pombe. EMBO J. 13: 5212–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schär, P., P. Munz and J. Kohli, 1993. Meiotic mismatch repair quantified on the basis of segregation patterns in Schizosaccharomyces pombe. Genetics 133: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar, P., M. Baur, C. Schneider and J. Kohli, 1997. Mismatch repair in Schizosaccharomyces pombe requires the mutL homologous gene pms1: molecular cloning and functional analysis. Genetics 146: 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert, P., M. Langsford, E. Kaeslin and J. Kohli, 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10: 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35: 243–274. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., 1979 Genetic Recombination: Thinking About It in Phage and Fungi. W. H. Freeman, San Francisco.
- Steiner, W. W., R. W. Schreckhise and G. R. Smith, 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9: 847–855. [DOI] [PubMed] [Google Scholar]
- Virgin, J. B., and J. P. Bailey, 1998. The M26 hotspot of Schizosaccharomyces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics 149: 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A. P. H., K. Maundrell and S. Shall, 1986. Transformation of Schizosaccharomyces pombe by non-homologous unstable integration of plasmids in the genome. Curr. Genet. 10: 503–508. [DOI] [PubMed] [Google Scholar]
- Wu, T. C., and M. Lichten, 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518. [DOI] [PubMed] [Google Scholar]
- Young, J. A., R. W. Schreckhise, W. W. Steiner and G. R. Smith, 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Zahn-Zabal, M., E. Lehmann and J. Kohli, 1995. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics 140: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]