Abstract
Candida albicans, which is diploid, possesses a single mating-type (MTL) locus on chromosome 5, which is normally heterozygous (a/α). To mate, C. albicans must undergo MTL homozygosis to a/a or α/α. Three possible mechanisms may be used in this process, mitotic recombination, gene conversion, or loss of one chromosome 5 homolog, followed by duplication of the retained homolog. To distinguish among these mechanisms, 16 spontaneous a/a and α/α derivatives were cloned from four natural a/α strains, P37037, P37039, P75063, and P34048, grown on nutrient agar. Eighteen polymorphic (heterozygous) markers were identified on chromosome 5, 6 to the left and 12 to the right of the MTL locus. These markers were then analyzed in MTL-homozygous derivatives of the four natural a/α strains to distinguish among the three mechanisms of homozygosis. An analysis of polymorphisms on chromosomes 1, 2, and R excluded meiosis as a mechanism of MTL homozygosis. The results demonstrate that while mitotic recombination was the mechanism for homozygosis in one offspring, loss of one chromosome 5 homolog followed by duplication of the retained homolog was the mechanism in the remaining 15 offspring, indicating that the latter mechanism is the most common in the spontaneous generation of MTL homozygotes in natural strains of C. albicans in culture.
IN haploid Saccharomyces cerevisiae, mating type is regulated by a cassette system consisting of two loci, HML and HMR, which contain unexpressed copies of the MATa and MATα locus, respectively, and a third, MAT, which contains one expressed copy of either the MATa or MATα locus (Herskowitz and Oshima 1981; Haber 1998). The MATa locus contains the mating-type gene MATa1, and the MATα locus contains the mating-type genes MATα1 and MATα2. Mating type is dictated by the MAT locus genotype. Haploid S. cerevisiae can change its mating type by site-specific recombination at the MAT locus with a copy of the alternative silent locus (Butler et al. 2004). This represents a conserved system, since no mating-type information is lost when cells switch mating type. Although the yeast pathogen Candida albicans possesses mating-type genes similar to those in S. cerevisiae, C. albicans, which is diploid, possesses a single mating-type (MTL) locus situated on chromosome 5 (Hull and Johnson 1999), which is normally heterozygous (a/α) in natural strains (Lockhart et al. 2002). The MTL locus of one chromosome 5 homolog contains the genes MTLa1 and MTLa2, while the MTL locus on the other homolog contains the genes MTLα1 and MTLα2 (Hull and Johnson 1999; Tsong et al. 2003). MTLa2 plays a unique role as a positive regulator of a-specific genes in C. albicans (Tsong et al. 2003). For C. albicans to express a mating type, the MTL heterozygote must undergo homozygosis to either a/a or α/α (Hull et al. 2000; Magee and Magee 2000; Lockhart et al. 2003). Hence, in contrast to S. cerevisiae, C. albicans loses the alternative mating-type information to be mating competent. An analysis of the MTL genotypes of a collection of 220 natural C. albicans strains revealed that ∼97% were MTL heterozygous, while only 3% were MTL homozygous (Lockhart et al. 2002). Of the MTL-heterozygous strains, ∼4% underwent spontaneous homozygosis in culture (Lockhart et al. 2002; Pujol et al. 2003). MTL zygosity has been shown to regulate not only mating competency, but also white-opaque switching. Cells can switch from white to opaque, a requirement for mating, only when they have undergone homozygosis (Lockhart et al. 2002; Miller and Johnson 2002). Hence, understanding how a/α cells become a/a or α/α is important not only for understanding mating, but also in understanding phenotypic switching.
C. albicans might employ three possible mechanisms to achieve homozygosis at the MTL locus. First, mitotic recombination (Whelan and Soll 1982) may occur between the chromosome 5 homologs at a site between the MTL locus and centromere, as depicted in Figure 1A, resulting in cosegregation of two MTLa or two MTLα loci. Second, one MTL locus could undergo precise gene conversion to the alternative mating type (i.e., a to α or α to a), as occurs at the MAT locus in S. cerevisiae (Butler et al. 2004), resulting in cosegregation of two MTLa or two MTLα loci, as depicted in Figure 1B. Third, one chromosome 5 homolog may be lost and the retained homolog duplicated, as depicted in Figure 1C. This last mechanism (Figure 1C) has been demonstrated to occur when C. albicans is grown in medium in which sorbose is the sole carbon source (Janbon et al. 1998; Magee and Magee 2000).
Figure 1.—
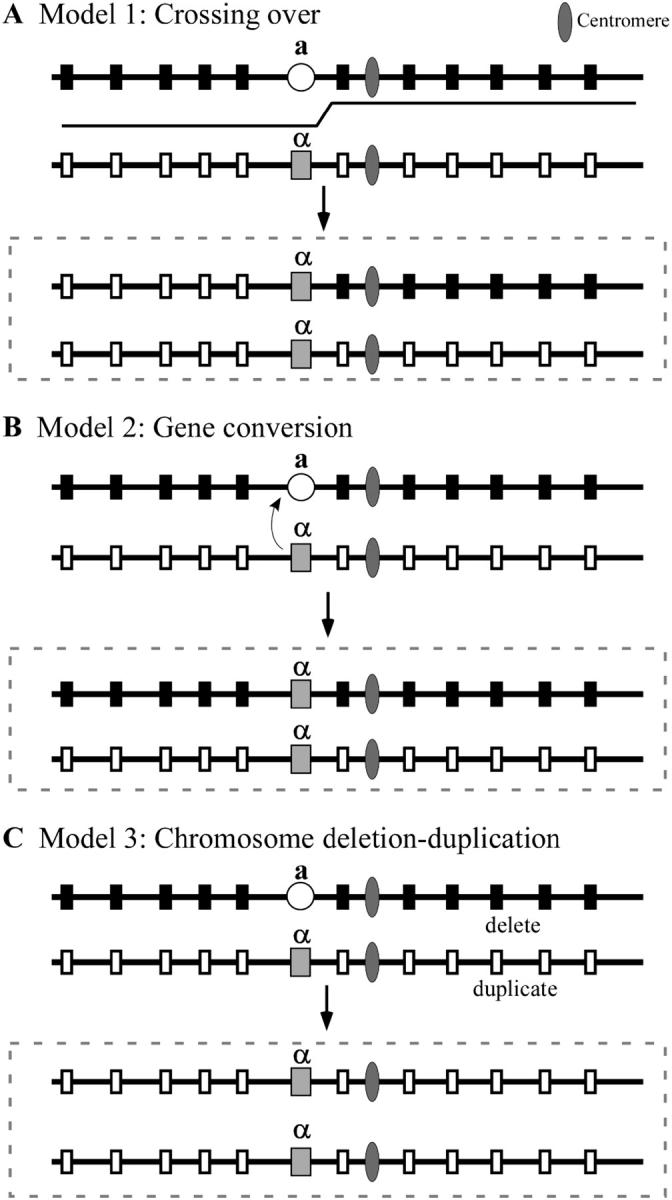
Models for the three possible mechanisms of mating-type locus (MTL) homozygosis in Candida albicans, mitotic recombination (A), gene conversion (B), and deletion of one homolog of chromosome 5 followed by duplication of the retained homolog (C). Hypothetical polymorphic alleles on the chromosome 5 homolog harboring MTLa (a) are represented as solid boxes and those on the chromosome 5 homolog harboring MTLα (α) are represented as open boxes. The centromere is represented as a solid ellipsoid. Dashed boxes represent outcome of homozygosis for each possible mechanism. The step in the line between the a and α homologs in A is the position of a crossover. The small curved arrow from α to a in B represents a gene conversion. The a homolog is deleted and the α homolog duplicates in C.
To test which mechanism(s) is responsible for spontaneous MTL homozygosis in natural strains of C. albicans in culture, we first identified polymorphic (heterozygous) sequences on either side of the MTL locus on chromosome 5 in four C. albicans a/α strains that were previously demonstrated to undergo spontaneous MTL homozygosis in culture (Lockhart et al. 2002; Pujol et al. 2003; our unpublished observations). We then analyzed the zygosity of these markers in spontaneous MTL-homozygous derivatives of these strains. If MTL homozygosis results from mitotic recombination, then the MTL-homozygous derivative would contain a combination of chromosome segments containing homozygous and heterozygous markers, and the MTL locus would be harbored in a segment containing other homozygous markers, as in Figure 1A. If MTL homozygosis results from precise gene conversion, as in S. cerevisiae, all markers in the MTL-homozygous derivative should be heterozygous except for the MTL locus, as in Figure 1B. Finally, if MTL homozygosis results from chromosome deletion, all markers in the MTL-homozygous derivatives should be homozygous, including the MTL locus, as in Figure 1C. In this scenario, polymorphisms on chromosomes other than chromosome 5 should remain polymorphic. The results we have obtained demonstrate that spontaneous MTL homozygosis in C. albicans is achieved both by mitotic recombination along chromosome 5 and by deletion of one chromosome 5 homolog followed by duplication of the retained homolog. The latter proved to be by far the most frequent mechanism.
MATERIALS AND METHODS
Strain origin, maintenance, and growth:
The four C. albicans MTL-heterozygous (a/α) strains P37037, P37039, P75063, and P34048, selected for analysis, have been demonstrated in earlier studies to undergo spontaneous MTL homozygosis in culture (Lockhart et al. 2002; Pujol et al. 2003; unpublished observations). To obtain MTL-homozygous derivatives, cells were plated at low density (∼50 colony-forming units per 10-cm-diameter petri dish) on nutrient agar medium (Lee et al. 1975) supplemented with zinc and arginine (“modified Lee's medium”), according to Bedell and Soll (1979), and 5 μg/ml of phloxine-B, which differentially stains opaque phase sectors and colonies red (Anderson and Soll 1987). Since only MTL-homozygous cells switch from white to opaque (Lockhart et al. 2002; Miller and Johnson 2002), MTL-homozygous derivatives were readily obtained by cloning cells from red sectors or colonies and verifying MTL homozygosity by polymerase chain reaction or Southern analysis (Lockhart et al. 2002; see later section). Six MTL-homozygous derivatives were obtained from a/α strain P37037 (α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, and a/a-1), one from a/α strain P37039 (α/α-1), two from a/α strain P75063 (a/a-1, a/a-2), and seven from a/α strain P34048 (α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, α/α-6, and a/a-1). The minority of MTL-homozygous offspring were obtained independently from colonies generated by different clones of the a/α parent strains. The majority were from sectors. Hence, all MTL-homozygous offspring originated from independent homozygosis events. All original a/α and derivative a/a or α/α strains were stored in 25% glycerol at −80°. For experimental purposes, stored cells were plated and grown on modified Lee's medium agar.
Polymorphisms identified by sequence analysis:
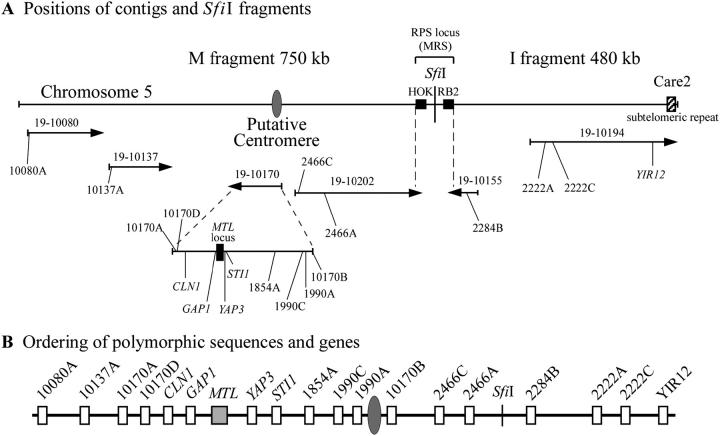
Fifty intergenic regions 500–600 bp in length along chromosome 5 were randomly selected from a contig map created from data provided by the Stanford Genome Technology Center C. albicans sequencing project (http://www-sequence.stanford.edu/group/candida/) and sequenced in the four MTL-heterozygous test strains. Of these, 13 intergenic regions on chromosome 5 were found to be polymorphic in one or more of the four a/α strains. The sites on chromosome 5 are listed as “target regions” for primers in Table 1 and diagrammed in Figure 2. The placement of polymorphic sequences on chromosome 5 in relation to the MTL locus, the putative centromeric region (Sanyal et al. 2004), the SfiI restriction sites (Chu et al. 1993; Chibana et al. 1998), and the subtelomeric Care2 site on chromosome 5 is presented in Figure 2A. Their placement was in agreement with the partial physical map of P. T. Magee and colleagues (http://alces.med.umn.edu/Candida.html; Forche et al. 2004) and Whiteway and colleagues (http://cbr-rbc.nrc-cnrc.gc.ca/biovis/candida/). Intergenic regions along chromosomes 1, 2, and R were similarly selected and sequenced. One polymorphic region was selected on each chromosome from contigs 19-10205, 19-10196, and 19-10057, respectively (Table 1).
TABLE 1.
Oligonucleotides used in this study and their positions in contigs
| Primer | Target regionsa | Oligonucleotide sequences |
|---|---|---|
| Contig19-10080 | ||
| 10080A-F | 1845–1862 | 5′-CAAAAGTAGACCCCTTCCTC-3′ |
| 10080A-R | 2379–2397 | 5′-CCTGCACATTGTCAATCTCG-3′ |
| Contig19-10137 | ||
| 10137A-F | 5130–5151 | 5′-GCAGGCATCGATACATATTCTC-3′ |
| 10137A-R | 5682–5701 | 5′-ACCAATGCAGCCAAACCAAG-3′ |
| Contig19-10170 | ||
| 10170B-F | 1–21 | 5′-CATGCCAAGTCTGTACACGCT-3′ |
| 10170B-R | 650–670 | 5′-GTTGTGGTAGCCATAGTGTGG-3′ |
| 1990A-F | 5212–5232 | 5′-TGGAGACATTTAGAGAACTTA-3′ |
| 1990A-R | 5829–5849 | 5′-CCGGTGACAACTCCACTCAAC-3′ |
| 1990C-F | 6792–6802 | 5′-GTCATCATCATTTCTTCTGAA-3′ |
| 1990C-R | 7515–7535 | 5′-CGGCAAATATTGTATGTTAAT-3′ |
| 1854A-F | 25288–25306 | 5′-TGACTACTTTTAGCTAGT-3′ |
| 1854A-R | 26737–26755 | 5′-CCACTCACCTTCACGGTC-3′ |
| STI1-F | 61258–61278 | 5′-GGATTACCCGAAGCAGTG-3′ |
| STI1-R | 60689–60719 | 5′-CAGGAGTTCACCCTTCAA-3′ |
| YAP3-F | 62976–62996 | 5′-CCTGTGAATATACTCAATTGG-3′ |
| YAP3-R | 63584–63604 | 5′-TCCGAGAACGTAAAGAATCTA-3′ |
| GAP1-F | 69153–69173 | 5′-CAGCATAGTTCCCCTTGG-3′ |
| GAP1-R | 69657–69677 | 5′-GAGTCTATATGCACATAC-3′ |
| CLN1-F | 92354–92374 | 5′-GAAGCAAATAGCCAACCTCAA-3′ |
| CLN1-R | 92954–92974 | 5′-GGATCCATTAGTTGATATTGACT-3′ |
| 10170D-F | 96001–96021 | 5′-CATCAATACTATAATTGATTT-3′ |
| 10170D-R | 96675–96695 | 5′-ATATTGAAGATGGTAAGAAC-3′ |
| 10170A-F | 97726–97746 | 5′-ATGCATCTACATGTAGTATTT-3′ |
| 10170A-R | 98400–98420 | 5′-ACG ATTAATCTATATTGTTAT-3′ |
| Contig19-10202 | ||
| 2466C-F | 11481–11501 | 5′-GAAGATGATCAACAACTCGCG-3′ |
| 2466C-R | 10810–10830 | 5′-TCGTTCATGACGCTATCAACA-3′ |
| 2466A-F | 64543–64523 | 5′-CTAAGTACCCATTCAAACTAT-3′ |
| 2466A-R | 63889–63869 | 5′-CCTACTCAGTATTAAGAAGCA-3′ |
| Contig19-10155 | ||
| 2284B-F | 23818–23798 | 5′-GTGATAACCCATCACCATCAC-3′ |
| 2284B-R | 23274–23254 | 5′-GTACAGAGTTGTCTCAGTGCG-3′ |
| Contig19-10194 | ||
| 2222C-F | 84429–84449 | 5′-TCAGATTGTTTAACAACCACT-3′ |
| 2222C-R | 84388–84408 | 5′-TCAACTGGTTTAGTACATCGA-3′ |
| 2222A-F | 70577–70597 | 5′-AGCTCGTGAAATGGTCAGGTT-3′ |
| 2222A-R | 71176–71195 | 5′-TTCAACCCATATAGATTCTCT-3′ |
| YIR12-F | 238693–238713 | 5′-AGATATAACAGACTCTTTATG-3′ |
| YIR12-R | 238308–238328 | 5′-TTTGGAAGGTTCTTCCCAGAT-3′ |
| Contig19-10205 | ||
| 10205A-F | 8016–8036 | 5′-TGCTTGTCTCTTCAATTGTAA-3′ |
| 10205A-R | 8662–8642 | 5′-CCCAACTTTAATTCCTTCCTT-3′ |
| Contig19-10196 | ||
| 10196A-F | 82083–82103 | 5′-AAGGAGCTTCATTTGTTAACA-3′ |
| 10196A-R | 82603–82583 | 5′-TAGTACCATTAGTGGTGCTGT-3′ |
| Contig19-10057 | ||
| 10057A-F | 83015–83035 | 5′-TGCTTGTCTCTTCAATTGTAA-3′ |
| 10057A-R | 83498–83478 | 5′-CCCAACTTTAATTCCTTCCTTT-3′ |
Position of the primer on the contig sequences obtained from the Stanford Genome Technology Center Candida albicans sequencing project. Contig19-10205, contig19-10196, and contig19-10057 are located on chromosomes 1, 2, and R, respectively.
Figure 2.—
Development of a partial contig map for chromosome 5 and placement of the polymorphic genes and intergenic sequences used in this study in their respective contigs. (A) the placements of contigs with associated genes along chromosome 5 were interpreted from data obtained from the Stanford Genome Technology Center C. albicans sequencing project database (http://www-sequence.stanford.edu/group/candida/), the partial physical map posted by P. T. Magee and colleagues (http://alces.med.umn.edu/Candida.html; Forche et al. 2004) and Whiteway and colleagues (http://cbr-rbc.nrc-cnrc.gc.ca/biovis/candida/). An explanation of the logic behind the map is presented in results. The 5′-3′ orientations of the contigs are indicated by arrow direction. The positions of the putative centromere (Sanyal et al. 2004), the RPS locus, and the CARE2 locus are noted. (B) Tentative ordering of polymorphic genes and intergenic sequences along chromosome 5. Note that the orientation of the markers 1990C and 1990A could be either on the right or left of the centromere, but have been arbitrarily ordered as in the sequence of contig 19-10170 for simplicity. A change in orientation changes none of the interpretations or conclusions in this article. The distances between markers are arbitrary.
The polymorphic regions were then sequenced in MTL-homozygous derivatives of the four original MTL heterozygous strains. For sequencing, primers were designed from the genome sequences (Table 1). These regions were then amplified as follows. One nanogram of genomic DNA isolated by methods previously described (Soll et al. 1996) was used in each 50-μl amplification reaction. Amplification with Taq DNA polymerase was performed as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Polymerase chain reaction (PCR) mixtures were denatured by incubating them for 10 min at 94°. PCR reactions were then run through 40 cycles for 1 min at 94°, for 1 min at 47°, and for 1 min at 68° and then elongated for 8 min at 68°. Sequencing was performed in both directions with an ABI sequencing apparatus (PE-ABI, Foster City, CA), using the same primers as those used for PCR amplification.
Polymorphisms identified by Southern analysis:
Primers were generated from the open reading frame sequences of 20 genes along chromosome 5 obtained from the Stanford Genome Technology Center C. albicans sequencing project database and the sequences were synthesized by PCR. The PCR products were labeled and used as probes in Southern analysis of the four MTL-heterozygous strains. Southern analysis was performed according to methods previously described (Srikantha et al. 2000, 2001). Five of the genes (CLN1, GAP1, YAP3, STI1, and YIR12) proved polymorphic in one or more of the a/α strains. The primers used to synthesize these gene sequences are presented in Table 1. The amplified probes were then used to assess polymorphisms in the MTL-homozygous derivatives primarily by Southern analysis or sequence analysis of the PCR products.
DNA fingerprinting:
To verify that MTL-homozygous strains were indeed derivatives of the four MTL-heterozygous strains, cells were DNA fingerprinted with the complex DNA fingerprinting probe Ca3 by methods previously described (Schmid et al. 1990; Soll 2000). All derivatives were genetically similar to the presumed strains of origin.
RESULTS
Isolation of MTL-homozygous derivatives:
Because switching occurs only in MTL-homozygous cells (Lockhart et al. 2002; Miller and Johnson 2002), MTL-homozygous derivatives of the four original MTL-heterozygous strains (P37037, P37039, P75063, and P34048) were isolated by screening for opaque phase sectors and colonies, which are selectively stained red by phloxine B in the supporting agar (Anderson and Soll 1987). To be sure that each MTL-homozygous derivative was the result of an independent homozygosis event, selected derivatives were from colony sectors or colonies from different clones of the a/α strain. MTL homozygosity of the selected offspring was verified by Southern blot hybridization with labeled MTLa1 and MTLα2 probes (Hull and Johnson 1999). While the original strain in each case possessed copies of both MTLa1 and MTLα2, all derivative clones contained copies of either MTLa1 or MTLα2, but not both (Figure 3). In each case, the hybridization signal of MTLa1 or MTLα2 in the MTL-homozygous derivative was approximately twice that of either MTLa1 or MTLα2 of the MTL-heterozygous strain from which it was derived (Figure 3), indicating that two copies of either MTLa1 or MTLα2 were in the derivative clone. DNA fingerprinting with the complex probe Ca3 (Schmid et al. 1990; Soll 2000) verified that all of the identified MTL-homozygous strains were true derivatives of the MTL-heterozygous strain of origin (data not shown).
Figure 3.—
Southern blot analysis of MTLa1 and MTLα2 for three of the MTL-heterozygous strains and one MTL-homozygous derivative from each. Note that the intensity of MTLa1 and MTLα2 bands in the MTL-heterozygous strains is roughly half that of the derivative MTL-homozygous strain. Similar results were obtained for the remaining MTL-heterozygous strain and additional MTL-homozygous derivatives not shown.
Identification of polymorphic sequences along chromosome 5:
To assess the mechanism of MTL homozygosis (Figure 1), polymorphic sequences were identified in the four MTL-heterozygous strains along chromosome 5 by sequence analysis of 50 random intergenic regions and 5 genes. In addition, polymorphic genes were identified in three strains (P37037, P37039, and P75063) by Southern analysis of 20 genes distributed throughout the chromosome. A combined 13 intergenic polymorphisms and 5 gene polymorphisms were identified (Figure 2). Six polymorphic genes and intergenic sequences were identified to the left of the MTL locus and 12 to the right (Figure 2).
Development of the contig map for chromosome 5:
The sequences of the contigs were obtained from assembly 19 of the Stanford Genome Technology Center C. albicans sequencing project database (http://www.sequence.stanford.edu/group/candida/). A single RPS locus, also known as MRS (major repeat sequence), on chromosome 5 contained the repeat sequence HOK at one end and the tandemly repeated RPS sequences and the repeat sequence RB2 at the opposite end (Iwaguchi et al. 1992; Anderson et al. 1993; Lockhart et al. 1995; Chindamporn et al. 1998; Pujol et al. 1999). The RPS locus contains the SfiI sites that separate chromosome 5 into fragments I and M (Figure 2). The orientation of contig 19-10202 was based on the fact that it contained the HOK sequence at one end. The orientation of contig 19-10155 was based on the position of the RB2 sequence at one end. Their locations on the chromosome were in agreement with the partial physical map generated by P. T. Magee and colleagues (http://alces.med.umn.edu/Candida.html; Forche et al. 2004). Contigs 19-10194 and 19-10170 were assigned to fragments I and M, respectively, according to the partial physical map and Forche et al. (2004). The orientation of contig 19-10194 was based upon the fact that it contains CARE2-homologous sequences at one end. CARE2 has been shown to be subtelomeric on chromosome 7 and has been suggested to be subtelomeric on other chromosomes (Chibana et al. 1998). This orientation is in agreement with that in Forche et al. (2004). The orientation of contig 19-10170, 19-10080, and 19-10137 follows that of Forche et al. (2004). The location of a putative centromere on chromosome 5 was based on Cse4p binding studies recently reported by Sanyal et al. (2004). This sequence is located in contig 19-10170 between positions 1 kb and 3.2 kb. This latter region is between two inverted repeat sequences (Figure 2). The intergenic marker 10170B is located ∼1 kb downstream of the putative centromeric region (Figure 2). The markers 1990A and 1990C are located ∼2 and 3.6 kb, respectively, from the putative centromeric region, but because of the inverted repeat sequences surrounding the putative centromeric region, the orientation (to the left or right) has not yet been resolved. We have placed these markers to the left, as in the latest assembly (assembly 19) of the C. albicans genome (http://www-sequence.stanford.edu/group/candida/).
The analysis of homozygous derivatives of strains P37037, P37039, P37063, and P34048 was made under the assumption that the integrity of chromosome 5 was not disrupted by chromosomal rearrangements, specifically at the RPS locus (Chu et al. 1993; Pujol et al. 1999; Chibana et al. 2000; Joly et al. 2002). The integrity of chromosome 5 was verified in the majority of derivative strains by Southern blot hybridization of contour-clamped homogeneous electric field (CHEF) gels with the markers 1854A and 2466C on fragment M and markers 2284B and YIR12 to the right of the RPS locus on fragment I (data not shown).
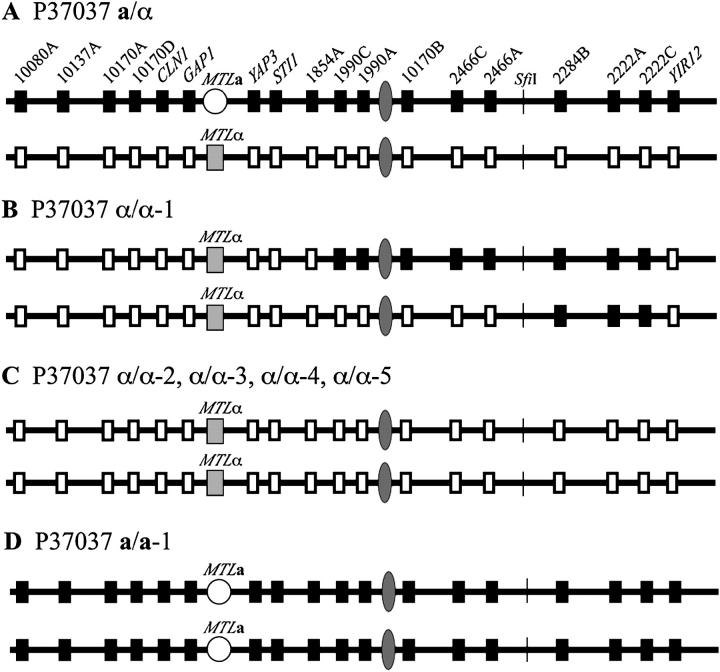
MTL homozygosis in strain P37037:
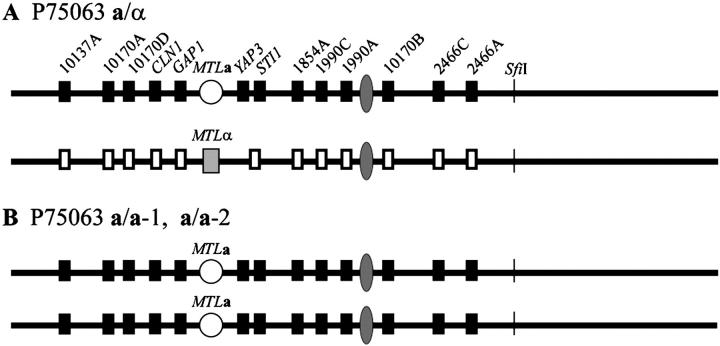
In the MTL-heterozygous (a/α) strain P37037, 6 markers to the left and 12 to the right of the MTL locus were heterozygous (Figure 4A). The nucleotide polymorphisms in a/α P37037 of the intergenic sites, and genes YAP3 and YIR12, are presented in Table 2, and the polymorphisms identified by Southern analysis for the genes STI1, CLN1, GAP1, and YAP3 are presented in Figure 5. Of the six MTL-homozygous derivatives of a/α P37037 identified in the screen for white-opaque switching, five were α/α derivatives (α/α-1, α/α-2, α/α-3, α/α-4, and α/α-5) and one an a/a derivative (a/a-1). In derivative α/α-1 markers in the portion of chromosome 5 beginning between markers 1854A and 1990C and ending between markers 2466A and 2284B remained heterozygous (Figure 4B). This region included the putative centromeric sequence. All markers to the left of this heterozygous region, however, were homozygous, containing the polymorphisms associated with the original MTLα homolog of chromosome 5. Three markers to the right of the heterozygous region, beginning between markers 2466A and 2284 B and ending between markers 2222C and YIR12, were homozygous, containing the polymorphisms associated with the original MTLa homolog of chromosome 5, and the last marker to the right, YIR12, was homozygous for the polymorphism associated with the original MTLα homolog of chromosome 5. These results indicate that at least three crossovers occurred in the genesis of α/α-1, one between markers 1854A and 1990C, one between markers 2466A and 2284B, and one between markers 2222C and YIR12 (Figure 4, A and B). The region between markers 2644A and 2284B harbors the RPS locus (Figure 2). These results indicate that homozygosity in α/α-1 arose by mitotic recombination and exclude a mechanism of precise gene conversion at the MTL locus, as occurs in S. cerevisiae, or a mechanism of chromosome deletion.
Figure 4.—
MTL homozygosis in the a/α strain P37037 generating six MTL-homozygous derivatives occurred by either mitotic recombination along chromosome 5 in one case or by loss of one chromosome 5 homolog followed by duplication of the retained homolog in five cases. (A) Diagram of polymorphic sites along the homologs of chromosome 5 in the original a/α strain. The solid and open boxes represent gene and intergenic sequence polymorphisms. Diagrams of sites along the homologs of chromosome 5 are presented for the α/α-1 derivative (B); the α/α-2, α/α-3, α/α-4, and α/α-5 derivatives (C); and the a/a-1 derivative (D). The interpretive mechanisms of MTL homozygosis for the derivatives are mitotic recombination for α/α-1 and loss of one chromosome 5 homolog followed by duplication of the retained homolog for α/α-2, α/α-3, α/α-4, α/α-5, and a/a-1.
TABLE 2.
Sequence analysis of chromosome 5 intergenic regions and the genesYAP3 andYIR12 for strain P37037 a/α and theMTL-homozygous derivatives α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, and a/a-1
| Marker | Strain | Polymorphic sites | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 43 | 158 | 170 | 180 | 200 | 213 | 217 | 223 | 228 | 234 | 236 | 238 | 248 | 342 | ||
| 10080A | a/α | C/T | A/G | A/G | A/G | C/T | G/T | C/T | C/T | G/T | A/C | A/T | A/T | C/T | A/G | |
| α/α-1 | C/C | A/A | A/A | G/G | T/T | G/G | C/C | T/T | G/G | C/C | T/T | A/A | C/C | A/A | ||
| α/α-2, -3, -4, -5 | C/C | A/A | A/A | G/G | T/T | G/G | C/C | T/T | G/G | C/C | T/T | A/A | C/C | A/A | ||
| a/a-1 | T/T | G/G | G/G | A/A | C/C | T/T | T/T | C/C | T/T | A/A | A/A | T/T | T/T | G/G | ||
| bp: | 218 | 234 | 255 | 324 | 340 | 377 | ||||||||||
| 10137A | a/α | C/T | A/G | A/G | C/T | C/T | C/T | |||||||||
| α/α-1 | C/C | A/A | A/A | T/T | C/C | T/T | ||||||||||
| α/α-2, -3, -4, -5 | C/C | A/A | A/A | T/T | C/C | T/T | ||||||||||
| a/a-1 | T/T | G/G | G/G | C/C | T/T | C/C | ||||||||||
| bp: | 75 | |||||||||||||||
| 10170A | a/α | C/T | ||||||||||||||
| α/α-1 | T/T | |||||||||||||||
| α/α-2, -3, -4, -5 | T/T | |||||||||||||||
| a/a-1 | C/C | |||||||||||||||
| bp: | 137 | 265 | ||||||||||||||
| 10170D | a/α | A/T | C/T | |||||||||||||
| α/α-1 | A/A | T/T | ||||||||||||||
| α/α-2, -3, -4, -5 | A/A | T/T | ||||||||||||||
| a/a-1 | T/T | C/C | ||||||||||||||
| bp: | 130 | 214 | 268 | 442 | 539 | 555 | ||||||||||
| YAP3 | a/α | A/T | A/G | A/T | A/G | C/T | A/G | |||||||||
| α/α-1 | A/A | G/G | T/T | G/G | T/T | A/A | ||||||||||
| α/α-2, -3, -4, -5 | A/A | G/G | T/T | G/G | T/T | A/A | ||||||||||
| a/a-1 | T/T | A/A | A/A | A/A | C/C | G/G | ||||||||||
| bp: | 196 | |||||||||||||||
| 1854A | a/α | T/− | ||||||||||||||
| α/α-1 | −/− | |||||||||||||||
| α/α-2, -3, -4, -5 | −/− | |||||||||||||||
| a/a-1 | T/T | |||||||||||||||
| bp: | 112 | 177 | 221 | 431 | ||||||||||||
| 1990C | a/α | A/G | C/T | A/G | C/T | |||||||||||
| α/α-1 | A/G | C/T | A/G | C/T | ||||||||||||
| α/α-2, -3, -4, -5 | G/G | C/C | G/G | C/C | ||||||||||||
| a/a-1 | A/A | T/T | A/A | T/T | ||||||||||||
| bp: | 255 | 460 | ||||||||||||||
| 1990A | a/α | A/T | A/G | |||||||||||||
| α/α-1 | A/T | A/G | ||||||||||||||
| α/α-2, -3, -4, -5 | T/T | G/G | ||||||||||||||
| a/a-1 | A/A | A/A | ||||||||||||||
| bp: | 74 | 125 | 190 | 326 | 370 | 375 | 393 | 403 | 426 | 435 | ||||||
| 10170B | a/α | A/G | C/G | A/G | C/T | C/G | C/T | C/T | C/T | C/T | C/T | |||||
| α/α-1 | A/G | C/G | A/G | C/T | C/G | C/T | C/T | C/T | C/T | C/T | ||||||
| α/α-2, -3, -4, -5 | G/G | G/G | A/A | C/C | C/C | T/T | C/C | C/C | C/C | C/C | ||||||
| a/a-1 | A/A | C/C | G/G | T/T | G/G | C/C | T/T | T/T | T/T | T/T | ||||||
| bp: | 138 | 144 | 423 | 448 | 449 | 567 | 579 | 603 | ||||||||
| 2466C | a/α | A/G | A/T | A/G | C/T | A/T | C/T | A/G | A/G | |||||||
| α/α-1 | A/G | A/T | A/G | C/T | A/T | C/T | A/G | A/G | ||||||||
| α/α-2, -3, -4, -5 | G/G | A/A | G/G | T/T | T/T | T/T | G/G | G/G | ||||||||
| a/a-1 | A/A | T/T | A/A | C/C | A/A | C/C | A/A | A/A | ||||||||
| bp: | 380 | 436 | ||||||||||||||
| 2466A | a/α | C/T | C/T | |||||||||||||
| α/α-1 | C/T | C/T | ||||||||||||||
| α/α-2, -3, -4, -5 | T/T | T/T | ||||||||||||||
| a/a-1 | C/C | C/C | ||||||||||||||
| bp: | 152 | 349 | 391 | 406 | 438 | 443 | ||||||||||
| 2284B | a/α | C/T | A/G | A/C | C/T | A/G | C/T | |||||||||
| α/α-1 | T/T | A/A | A/A | T/T | A/A | T/T | ||||||||||
| α/α-2, -3, -4, -5 | C/C | G/G | C/C | C/C | G/G | C/C | ||||||||||
| a/a-1 | T/T | A/A | A/A | T/T | A/A | T/T | ||||||||||
| bp: | 59 | 60 | 74 | 93 | 95 | 114 | 135 | 139 | 214 | 220 | 223 | 232 | 335 | 544 | ||
| 2222A | a/α | A/G | C/G | A/C | A/G | G/T | A/C | C/T | C/T | A/G | C/T | C/T | C/G | C/G | A/G | |
| α/α-1 | A/A | G/G | C/C | G/G | T/T | C/C | T/T | G/G | G/G | C/C | C/C | C/C | C/C | A/A | ||
| α/α-2, -3, -4, -5 | G/G | C/C | A/A | A/A | G/G | A/A | C/C | C/C | A/A | T/T | T/T | G/G | G/G | G/G | ||
| a/a-1 | A/A | G/G | C/C | G/G | T/T | C/C | T/T | G/G | G/G | C/C | C/C | C/C | C/C | A/A | ||
| bp: | 290 | 300 | 307 | 321 | 334 | 336 | 347 | 360 | 372 | |||||||
| 2222C | a/α | A/C | A/G | C/T | A/C | C/− | A/G | A/T | C/T | A/G | ||||||
| α/α-1 | C/C | A/A | T/T | C/C | −/− | G/G | A/A | T/T | G/G | |||||||
| α/α-2, -3, -4, -5 | A/A | G/G | C/C | A/A | C/C | A/A | T/T | C/C | A/A | |||||||
| a/a-1 | C/C | A/A | T/T | C/C | −/− | G/G | A/A | T/T | G/G | |||||||
| bp: | 312 | |||||||||||||||
| YIR12 | a/α | A/G | ||||||||||||||
| α/α-1 | G/G | |||||||||||||||
| α/α-2, -3, -4, -5 | G/G | |||||||||||||||
| a/a-1 | A/A | |||||||||||||||
Consensus sequences were generated between the sequences obtained for P37037, P37039, P75063, P34048, and their derivatives. Base pair positions are given for the polymorphic sites as a function of the consensus sequences. Position 1 corresponds to the 5′ end of the forward primers described in Table 1, except for the sequences of genes YAP3 and YIR12, where position 1 corresponds to the 5′ end of the reverse primer due to the presence of a 6-bp insertion/deletion near the beginning of the sequence obtained with the forward primer. Dashes in the sequence indicate single-base-pair deletions.
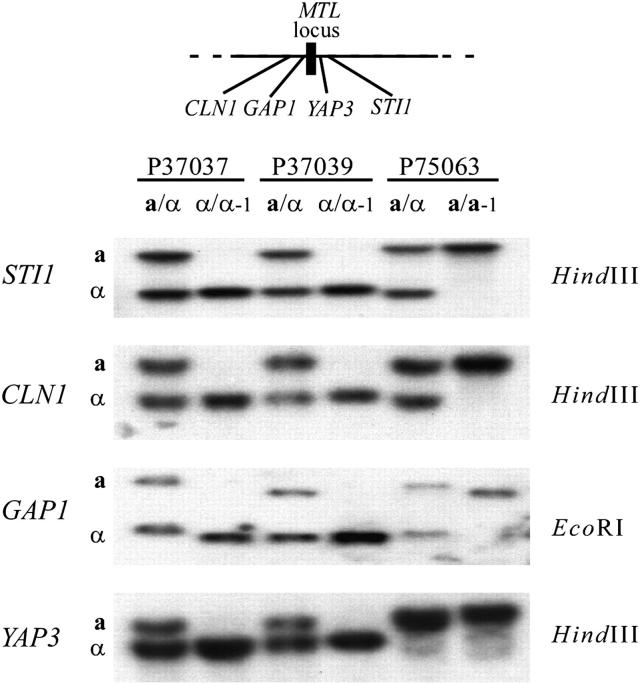
Figure 5.—
Southern blot hybridization analysis of four polymorphic genes positioned within a 30-kb region harboring the mating-type locus (MTL). The relative positions of the genes are shown in the schematic above the Southern blots. Restriction enzymes used to show the polymorphism for each gene are indicated to the right. The primers used to generate the open reading frame of each gene are presented in Table 1.
In the MTL-homozygous derivatives α/α-2, α/α-3, α/α-4, and α/α-5, all of the markers, including 6 to the left and 12 to the right of the MTL locus, were homozygous (Figure 4C). Most notably, the markers 1990A, 1990C, and 10170B, located close to the putative centromeric region, were homozygous. The nucleotide sequences of intergenic markers and the genes YAP3 and YIR12 are presented in Table 2. These results indicate that the chromosome 5 homolog harboring the MTLa locus was deleted in the genesis of the α/α-2, α/α-3, α/α-4, and α/α-5 derivatives of P37037. Similarly, in the MTL-homozygous derivative a/a-1, the 6 markers to the left and the 12 markers to the right of the MTL locus were homozygous (Figure 4D). The nucleotide sequences of intergenic markers and genes YAP3 and YIR12 are presented in Table 2. These results indicate that the chromosome 5 homolog harboring the MTLα locus was lost, and the retained chromosome 5 homolog duplicated in the genesis of the a/a-1 derivative of P37037.
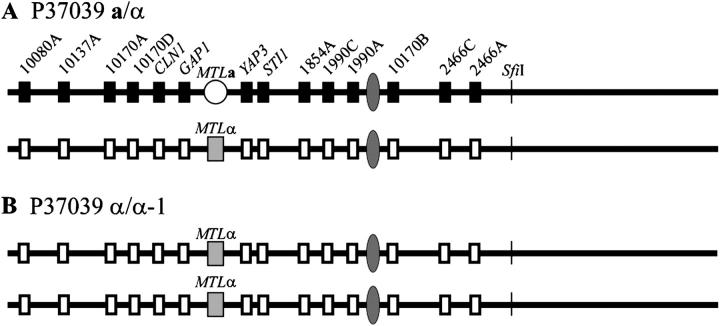
MTL homozygosis in strain P37039:
In the MTL-heterozygous (a/α) strain P37039, six markers to the left and eight to the right were polymorphic (Figure 6A). The polymorphic nucleotide sequences of the intergenic sites and the gene YAP3 are presented for a/α P37039 in Table 3, and the polymorphisms identified by Southern analysis of STI1, CLN1, GAP1, and YAP3 are presented in Figure 5. In the MTL-homozygous derivative α/α-1 of strain P37039, all markers were homozygous, including those in close proximity to the putative centromeric region (Figure 6B). A comparison of the polymorphic sequences between the original a/α and α/α-1 derivative of strain P37039 is presented in Table 3, and Southern analysis of the polymorphic genes is shown in Figure 5. These results indicate that the chromosome 5 homolog harboring the MTLa locus was deleted in the genesis of the α/α-1 derivative of P37039. The intensity of the MTLα2 band in the Southern blot of the P37039 α/α-1 derivative was approximately twice that of either the a or α band of the parent MTL-heterozygous strain (Figure 3), suggesting that loss of the chromosome 5 homolog harboring the MTLa locus was followed by duplication of the homolog harboring the MTLα locus. Similar results were obtained in Southern analyses of the genes CLN1, GAP1, STI1, and YAP3 (Figure 5), supporting this scenario of homolog duplication after loss.
Figure 6.—
MTL homozygosis in the a/α strain P37039 generating an MTL-homozygous derivative occurred by loss of one chromosome 5 homolog followed by duplication of the retained homolog. (A) Diagram of polymorphic sites along the homologs of chromosome 5 in the original a/α strain. The solid and open boxes represent the gene and intergenic sequence polymorphisms. (B) Diagram of sites is presented for the α/α-1 derivative.
TABLE 3.
Sequence analysis of chromosome 5 intergenic regions and the geneYAP3 for strain a/α p37039 and theMTL-homozygous derivative α/α-1
| Marker | Strain | Polymorphic sites | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 43 | 158 | 170 | 180 | 200 | 213 | 217 | 223 | 228 | 234 | 236 | 238 | 248 | 342 | ||
| 10080A | a/α | C/T | A/G | A/G | A/G | C/T | G/T | C/T | C/T | G/T | A/C | A/T | A/T | C/T | A/G | |
| α/α-1 | C/C | A/A | A/A | G/G | T/T | G/G | C/C | T/T | G/G | C/C | T/T | A/A | C/C | A/A | ||
| bp: | 218 | 234 | 255 | 324 | 340 | 377 | ||||||||||
| 10137A | a/α | C/T | A/G | A/G | C/T | C/T | C/T | |||||||||
| α/α-1 | C/C | A/A | A/A | T/T | C/C | T/T | ||||||||||
| bp: | 75 | |||||||||||||||
| 10170A | a/α | C/T | ||||||||||||||
| α/α-1 | C/C | |||||||||||||||
| bp: | 137 | 265 | ||||||||||||||
| 10170D | a/α | A/T | C/T | |||||||||||||
| α/α-1 | A/A | T/T | ||||||||||||||
| 130 | 214 | 268 | 442 | 539 | 555 | |||||||||||
| YAP3 | a/α | A/T | A/G | A/T | A/G | C/T | A/G | |||||||||
| α/α-1 | A/A | G/G | T/T | G/G | T/T | A/A | ||||||||||
| bp: | 196 | |||||||||||||||
| 1854A | a/α | T/− | ||||||||||||||
| α/α-1 | −/− | |||||||||||||||
| bp: | 112 | 177 | 221 | 431 | ||||||||||||
| 1990C | a/α | A/G | C/T | A/G | C/T | |||||||||||
| α/α-1 | G/G | C/C | G/G | C/C | ||||||||||||
| bp: | 255 | 460 | ||||||||||||||
| 1990A | a/α | A/T | A/G | |||||||||||||
| α/α-1 | T/T | G/G | ||||||||||||||
| bp: | 74 | 125 | 190 | 326 | 370 | 375 | 393 | 403 | 426 | 435 | ||||||
| 10170B | a/α | A/G | C/G | A/G | C/T | C/G | C/T | C/T | C/T | C/T | C/T | |||||
| α/α-1 | G/G | G/G | A/A | C/C | C/C | T/T | C/C | C/C | C/C | C/C | ||||||
| bp: | 138 | 144 | 423 | 448 | 449 | 567 | 579 | 603 | ||||||||
| 2466C | a/α | A/G | A/T | A/G | C/T | A/T | C/T | A/G | A/G | |||||||
| α/α-1 | G/G | A/A | G/G | T/T | T/T | T/T | G/G | G/G | ||||||||
| bp: | 380 | |||||||||||||||
| 2466A | a/α | C/T | ||||||||||||||
| α/α-1 | T/T | |||||||||||||||
See Table 2 footnote.
MTL homozygosis in strain P75063:
In the MTL-heterozygous (a/α) strain P75063, five markers to the left and eight to the right of the MTL locus were heterozygous (Figure 7A). In each of the two independently isolated MTL-homozygous derivatives a/a-1 and a/a-2, all markers were also homozygous, including those in close proximity to the putative centromere (Figure 7B). A comparison of a/a-1 and a/a-2 revealed that all homozygous alleles were identical, suggesting that no additional recombination event had occurred. A comparison of the nucleotide sequences of polymorphic intergenic and YAP3 sequences between the original a/α strain, a/a-1 derivative, and a/a-2 derivative is presented in Table 4, and Southern analysis of the polymorphic genes STI1, CLN1, and GAP1 in the a/α P75063 strain, and the a/a-1 derivative, is presented in Figure 5. YAP3 exhibited no polymorphisms in Southern blots. These results indicate that the chromosome 5 homolog harboring the MTLα locus in strain P75063 was deleted in the independent genesis of both the a/a-1 and a/a-2 derivatives of P75063. In addition, the intensity of the MTLa1 band in the Southern blot of the P75063 a/a-1 derivative was approximately twice that of either the a or α bands of the parent MTL heterozygote (Figure 3), again suggesting that the loss of the homolog of chromosome 5 harboring the MTLα locus was followed by duplication of the homolog that was retained. Similar results were obtained in Southern analyses of the genes STI1, CLN1, and GAP1 (Figure 5), further suggesting that duplication followed chromosome loss.
Figure 7.—
MTL homozygosis in the a/α strain P75063 generating two independent MTL-homozygous derivatives occurred by loss of one chromosome 5 homolog followed by duplication of the retained homolog. (A) Diagram of the polymorphic sites along the homologs of chromosome 5 in the original a/α strain. The solid and open boxes represent the gene and intergenic sequence polymorphisms. (B) Diagrams of the tested sites along the homologs of chromosome 5 are presented for the P75063 derivatives a/a-1 and a/a-2.
TABLE 4.
Sequence analysis of chromosome 5 intergenic regions and the geneYAP3 for a/α strain P75063 and theMTL-homozygous derivatives a/a-1 and a/a-2
| Marker | Strain | Polymorphic sites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 545 | |||||||||
| 10137A | a/α | C/T | ||||||||
| a/a-1, a/a-2 | C/C | |||||||||
| bp: | 70 | 80 | 161 | 173 | ||||||
| 10170A | a/α | G/T | C/T | C/T | A/G | |||||
| a/a-1, a/a-2 | G/G | C/C | T/T | A/A | ||||||
| bp: | 587 | |||||||||
| 10170D | a/α | A/G | ||||||||
| a/a-1, a/a-2 | G/G | |||||||||
| bp: | 540 | 555 | 559 | |||||||
| YAP3 | a/α | C/T | A/G | A/G | ||||||
| a/a-1, a/a-2 | T/T | A/A | A/A | |||||||
| bp: | 67 | 116 | 196 | |||||||
| 1854A | a/α | A/G | A/G | T/− | ||||||
| a/a-1, a/a-2 | A/A | G/G | T/T | |||||||
| bp: | 112 | 167 | 170 | 177 | ||||||
| 1990C | a/α | A/G | C/T | A/C | C/T | |||||
| a/a-1, a/a-2 | A/A | T/T | C/C | T/T | ||||||
| bp: | 160 | 253 | 255 | 316 | 460 | |||||
| 1990A | a/α | A/G | C/T | A/T | C/T | A/G | ||||
| a/a-1 | A/A | T/T | A/A | C/C | A/A | |||||
| bp: | 72 | 190 | 252 | 326 | 375 | 426 | 435 | 455 | ||
| 10170B | a/α | C/T | A/G | C/T | C/T | C/T | C/T | C/T | C/T | |
| a/a-1, a/a-2 | T/T | G/G | C/C | T/T | C/C | T/T | T/T | C/C | ||
| bp: | 96 | 99 | 546 | 567 | ||||||
| 2466C | a/α | A/G | A/T | C/T | C/T | |||||
| a/a-1, a/a-2 | G/G | T/T | C/C | C/C | ||||||
| bp: | 122 | 130 | 133 | 231 | 345 | 459 | 462 | 602 | ||
| 2466A | a/α | A/G | A/G | C/T | A/G | A/G | A/G | A/T | A/G | |
| a/a-1, a/a-2 | G/G | A/A | C/C | G/G | G/G | A/A | A/A | G/G | ||
See Table 2 footnote.
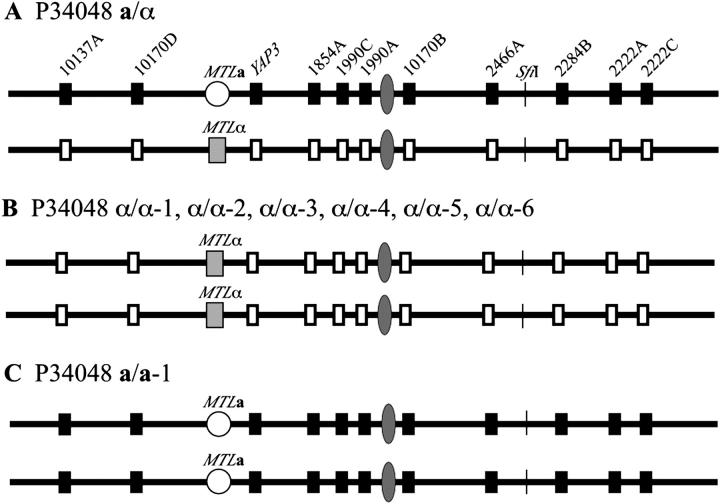
MTL homozygosis in strain P34048:
In the MTL-heterozygous (a/α) strain P34048, two markers to the left and nine markers to the right of the MTL locus were heterozygous (Figure 8A). The nucleotide sequences are presented in Table 5. In the MTL-homozygous derivatives α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, and α/α-6, all markers were homozygous (Figure 8B; Table 5). In the MTL-homozygous derivative a/a-1, all markers were also homozygous (Figure 8C; Table 5). Hence, all MTL-homozygous derivatives of P34048a/α were generated by loss of a chromosome 5 homolog.
Figure 8.—
MTL homozygosis in the a/α strain P34048 generating seven MTL-homozygous derivatives occurred by loss of one chromosome 5 homolog followed by duplication of the retained homolog in all cases. (A) Diagram of polymorphic sites along the homologs of chromosome 5 in the original a/α strain. The solid and open boxes represent gene and intergenic sequence polymorphisms. Diagrams of sites along the homologs of chromosome 5 are presented for the α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, and α/α-6 derivatives (B) and the a/a-1 derivative (C).
TABLE 5.
Sequence analysis of chromosome 5 intergenic regions and the geneYAP3 for strain P34048 a/α and theMTL-homozygous derivatives α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, α/α-6, and a/a-1
| Marker | Strain | Polymorphic sites | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 96 | 251 | 327 | 490 | |||||||
| 10137A | a/α | C/T | G/T | A/T | A/G | ||||||
| α/α-1, -2, -3, -4, -5, -6 | T/T | G/G | A/A | G/G | |||||||
| a/a-1 | C/C | T/T | T/T | A/A | |||||||
| bp: | 126 | 180 | 275 | 321 | 372 | ||||||
| 10170D | a/α | C/T | C/T | C/T | C/T | A/G | |||||
| α/α-1, -2, -3, -4, -5, -6 | C/C | T/T | T/T | T/T | A/A | ||||||
| a/a-1 | T/T | C/C | C/C | C/C | G/G | ||||||
| bp: | 292 | 540 | 559 | ||||||||
| YAP3 | a/α | A/G | C/T | A/G | |||||||
| α/α-1, -2, -3, -4, -5, -6 | A/A | T/T | A/A | ||||||||
| a/a-1 | G/G | C/C | G/G | ||||||||
| bp: | 541 | 608 | 609 | ||||||||
| 1854A | a/α | A/G | A/C | A/G | |||||||
| α/α-1, -2, -3, -4, -5, -6 | A/A | C/C | A/A | ||||||||
| a/a-1 | G/G | A/A | G/G | ||||||||
| bp: | 125 | 193 | 221 | ||||||||
| 1990C | a/α | A/C | C/T | A/G | |||||||
| α/α-1, -2, -3, -4, -5, -6 | C/C | T/T | A/A | ||||||||
| a/a-1 | A/A | C/C | G/G | ||||||||
| bp: | 590 | ||||||||||
| 1990A | a/α | A/G | |||||||||
| α/α-1, -2, -3, -4, -5, -6 | A/A | ||||||||||
| a/a-1 | G/G | ||||||||||
| bp: | 192 | 240 | 297 | 385 | 405 | ||||||
| 10170B | a/α | A/G | C/T | C/T | A/G | C/T | |||||
| α/α-1, -2, -3, -4, -5, -6 | G/G | T/T | C/C | A/A | T/T | ||||||
| a/a-1 | A/A | C/C | T/T | G/G | C/C | ||||||
| bp: | 122 | 130 | 133 | 151 | |||||||
| 2466A | a/α | A/G | A/G | C/T | C/T | ||||||
| α/α-1, -2, -3, -4, -5, -6 | A/A | G/G | T/T | C/C | |||||||
| a/a-1 | G/G | A/A | C/C | T/T | |||||||
| bp: | 176 | 180 | 349 | 406 | |||||||
| 2284B | a/α | G/T | C/T | A/G | C/T | ||||||
| α/α-1, -2, -3, -4, -5, -6 | T/T | T/T | G/G | C/C | |||||||
| a/a-1 | G/G | C/C | A/A | T/T | |||||||
| bp: | 59 | 300 | |||||||||
| 2222A | a/α | A/G | C/T | ||||||||
| α/α-1, -2, -3, -4, -5, -6 | G/G | C/C | |||||||||
| a/a-1 | A/A | T/T | |||||||||
| bp: | 290 | 300 | 307 | 321 | 334 | 336 | 347 | 360 | 372 | ||
| 2222C | a/α | A/C | A/G | C/T | A/C | C/− | A/G | A/T | C/T | A/G | |
| α/α-1, -2, -3, -4, -5, -6 | A/A | G/G | C/C | A/A | C/C | A/A | T/T | C/C | A/A | ||
| a/a-1 | C/C | A/A | T/T | C/C | −/− | G/G | A/A | T/T | G/G | ||
See Table 2 footnote.
Chromosome 5 homolog-specific polymorphisms:
In comparing the polymorphic sites along chromosome 5 in MTL-homozygous offspring from the different a/α strains, we noted similar linkage of specific polymorphisms associated with either MTLa or MTLα. Hence, when a/α strains shared the base polymorphism X/Y at a particular site, X segregated with MTLa and Y segregated with MTLα in the majority of cases for unrelated strains (Table 6).
TABLE 6.
Linkage of specific polymorphisms withMTLa andMTLα in chromosome 5 homologs
| Marker | Strain | Polymorphic sitesa | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 43 | 158 | 170 | 180 | 200 | 213 | 217 | 223 | 228 | 234 | 236 | 238 | 248 | 342 | ||||||||||||||||
| 10080A | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | ||
| P37037 | T | C | G | A | G | A | A | G | C | T | T | G | T | C | C | T | T | G | A | C | A | T | T | A | T | C | G | A | ||
| P37039 | C | A | A | G | T | G | C | T | G | C | T | A | C | A | ||||||||||||||||
| 218 | 234 | 255 | 324 | 340 | 377 | |||||||||||||||||||||||||
| 10137A | bp: | a | α | a | α | a | α | a | α | a | α | a | α | |||||||||||||||||
| P37037 | T | C | G | A | G | A | C | T | T | C | C | T | ||||||||||||||||||
| P37039 | C | A | A | T | C | T | ||||||||||||||||||||||||
| 75 | ||||||||||||||||||||||||||||||
| 10170A | bp: | a | α | |||||||||||||||||||||||||||
| P37037 | C | T | ||||||||||||||||||||||||||||
| P37039 | C | |||||||||||||||||||||||||||||
| 137 | 265 | |||||||||||||||||||||||||||||
| 10170D | bp: | a | α | a | α | |||||||||||||||||||||||||
| P37037 | T | A | C | T | ||||||||||||||||||||||||||
| P37039 | A | T | ||||||||||||||||||||||||||||
| 130 | 214 | 268 | 442 | 539 | 540 | 555 | 559 | |||||||||||||||||||||||
| YAP3 | bp: | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | |||||||||||||
| P37037 | T | A | A | G | A | T | A | G | C | T | NPb | G | A | NPb | ||||||||||||||||
| P37039 | A | G | T | G | T | NPb | A | NPb | ||||||||||||||||||||||
| P75063 | NPb | NPb | NPb | NPb | NPb | T | A | A | ||||||||||||||||||||||
| P34048 | NPb | NPb | NPb | NPb | NPb | C | T | NPb | G | A | ||||||||||||||||||||
| bp: | 196 | |||||||||||||||||||||||||||||
| 1854A | a | α | ||||||||||||||||||||||||||||
| P37037 | T | — | ||||||||||||||||||||||||||||
| P37039 | — | |||||||||||||||||||||||||||||
| P75063 | T | |||||||||||||||||||||||||||||
| 112 | 177 | 221 | 431 | |||||||||||||||||||||||||||
| 1990C | bp: | a | α | a | α | a | α | a | α | |||||||||||||||||||||
| P37037 | A | G | T | C | A | G | T | C | ||||||||||||||||||||||
| P37039 | G | C | G | C | ||||||||||||||||||||||||||
| P75063 | A | T | NPb | NPb | ||||||||||||||||||||||||||
| P34048 | NPb | NPb | G | A | NPb | |||||||||||||||||||||||||
| bp: | 255 | 460 | ||||||||||||||||||||||||||||
| 1990A | a | α | a | α | ||||||||||||||||||||||||||
| P37037 | A | T | A | G | ||||||||||||||||||||||||||
| P37039 | T | G | ||||||||||||||||||||||||||||
| P75063 | A | A | ||||||||||||||||||||||||||||
| bp: | 74 | 125 | 190 | 326 | 370 | 375 | 393 | 403 | 426 | 435 | ||||||||||||||||||||
| 10170B | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | ||||||||||
| P37037 | A | G | C | G | G | A | T | C | G | C | C | T | T | C | T | C | T | C | T | C | ||||||||||
| P37039 | G | G | A | C | C | T | C | C | C | C | ||||||||||||||||||||
| P75063 | NPb | NPb | G | T | NPb | C | NPb | NPb | T | T | ||||||||||||||||||||
| 138 | 144 | 423 | 448 | 449 | 567 | 579 | 603 | |||||||||||||||||||||||
| 2466C | bp: | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | |||||||||||||
| P37037 | A | G | T | A | A | G | C | T | A | T | C | T | A | G | A | G | ||||||||||||||
| P37039 | G | A | G | T | T | T | G | G | ||||||||||||||||||||||
| P75063 | NPb | NPb | NPb | NPb | NPb | C | NPb | NPb | ||||||||||||||||||||||
| bp: | 122 | 130 | 133 | 380 | ||||||||||||||||||||||||||
| 2466A | a | α | a | α | a | α | a | α | ||||||||||||||||||||||
| P37037 | NPb | NPb | NPb | C | T | |||||||||||||||||||||||||
| P37039 | NPb | NPb | NPb | T | ||||||||||||||||||||||||||
| P75063 | G | A | C | NPb | ||||||||||||||||||||||||||
| P34048 | G | A | A | G | C | T | NPb | |||||||||||||||||||||||
| 349 | 406 | |||||||||||||||||||||||||||||
| 2284B | bp: | a | α | a | α | |||||||||||||||||||||||||
| P37037 | A | G | T | C | ||||||||||||||||||||||||||
| P34048 | A | G | T | C | ||||||||||||||||||||||||||
| bp: | 59 | |||||||||||||||||||||||||||||
| 2222A | a | α | ||||||||||||||||||||||||||||
| P37037 | A | G | ||||||||||||||||||||||||||||
| P34048 | A | G | ||||||||||||||||||||||||||||
| 290 | 300 | 307 | 321 | 334 | 336 | 347 | 360 | 372 | ||||||||||||||||||||||
| 2222C | bp: | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | a | α | |||||||||||
| P37037 | C | A | A | G | T | C | C | A | — | C | G | A | A | T | T | C | G | A | ||||||||||||
| P34048 | C | A | A | G | T | C | C | A | — | C | G | A | A | T | T | C | G | A | ||||||||||||
In addition to these shared polymorphisms obtained through sequencing, polymorphisms obtained by Southern analyses in strains P37037, P37039, and P75063 and their derivatives showed additional polymorphisms specific to MTLa and MTLα.
Polymorphic sites that were shared by at least two of the four a/α strains used in this study are represented for each marker. The nucleotides associated to MTLa (a) and (or) MTLα (α) are represented for each strain. In strains P37037 and P34048 for which MTLa and MTLα derivatives were obtained, nucleotides are given for both. In strain P37039, nucleotides are given for the lone MTLα derivative obtained. In strain P75063, nucleotides are given for the MTLa derivatives obtained.
No polymorphism was found for these strains at these positions.
Ruling out meiosis:
Although we proposed three possible mechanisms for homozygosis that could be discriminated by analyzing polymorphic markers along chromosome 5, there was also a fourth improbable mechanism that would give the same chromosome 5 results as chromosome loss followed by duplication, namely meiosis. If meiosis occurred, all genes in the genome would become monomorphic. To test this possibility, we identified polymorphic intergenic sequences on contig 19-10205 of chromosome 1, 19-10196 of chromosome 2, and 19-10057 of chromosome R and analyzed these sequences in all of the MTL-homozygous offspring of strain a/α P37037, a/α P37039, a/α P75063, and a/α P34048 (Table 7). Polymorphisms on chromosomes other than chromosome 5 were maintained in all 16 MTL-homozygous offspring (Table 7). In Table 2, P37037α/α-2 and P37037α/α-4, loss of heterozygosity occurred on the chromosome 1 marker, but polymorphisms were conserved on chromosomes 2 and R (Table 6), suggesting that these offspring, which had become α/α through independent homozygosis events, had both undergone parallel homozygosis at the chromosome 1 marker analyzed, but not meiosis. Together, these results support the conclusion that meiosis can be excluded as the mechanism for MTL homozygosis in all 16 MTL-homozygous offspring analyzed in this study.
TABLE 7.
Heterozygosity of independent genetic markers eliminates meiosis as a mechanism ofMTL homozygosis
| Marker | Strains | Polymorphic sites | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp: | 59 | 60 | 135 | 147 | 150 | 165 | 241 | 383 | 483 | 543 | 561 | 567 | 576 | ||
| 10205A on chromosome 1 |
P37037 a/α, α/α-1, α/α-3, α/α-5, a/a-1 |
A/C | A/T | C/T | A/C | C/T | A/G | A/C | C/T | C/T | A/G | A/G | A/G | A/G | |
| P37037 α/α-2, α/α-4 | C/C | T/T | C/C | C/C | T/T | G/G | A/A | C/C | T/T | G/G | G/G | A/A | G/G | ||
| P37039 a/α, α/α-1 | A/C | A/T | C/T | A/C | C/T | A/G | A/C | C/T | C/T | A/G | A/G | A/G | A/G | ||
| P75063 a/α, a/a-1, a/a-2 | C/C | T/T | C/T | A/C | C/T | A/G | C/C | T/T | T/T | A/A | A/A | G/G | A/A | ||
|
P34048 a/α, α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, α/α-6, a/a-1 |
A/A | A/A | T/T | A/A | C/C | A/G | A/C | C/T | C/T | A/G | A/G | A/G | A/G | ||
| bp: | 72 | 140 | 266 | 336 | 343 | ||||||||||
| 10196A on chromosome 2 |
P37037 a/α, α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, a/a-1 |
C/T | C/T | C/T | G/T | A/G | |||||||||
| P37039 a/α, α/α-1 | C/T | C/T | C/T | G/T | A/G | ||||||||||
| P75063 a/α, a/a-1, a/a-2 | C/C | C/C | C/C | G/T | A/G | ||||||||||
|
P34048 a/α, α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, α/α-6, a/a-1 |
C/C | C/C | C/C | G/G | A/A | ||||||||||
| bp: | 78 | 119b | |||||||||||||
| 10057A on chromosome R |
P37037 a/α, α/α- 1, α/α-2, α/α-3, α/α-4, α/α-5, a/a-1 |
T/T | A/− | ||||||||||||
| P37039 a/α, α/α-1 | C/T | A/− | |||||||||||||
| P75063 a/α, a/a-1, a/a-2 | C/C | A/− | |||||||||||||
|
P34048 a/α, α/α-1, α/α-2, α/α-3, α/α-4, α/α-5, α/α-6, a/a-1 |
C/C | A/− | |||||||||||||
a Heterozygous nucleotide positions are noted in boldface type.
b Heterozygosity due to a single-base-pair insertion/deletion (dashes denote a deletion).
DISCUSSION
MTL zygosity regulates not only mating (Hull et al. 2000; Magee and Magee 2000; Lockhart et al. 2003) but also phenotypic switching (Lockhart et al. 2002; Miller and Johnson 2002). Here, we have examined the mechanism of spontaneous MTL homozygosis in natural strains of C. albicans grown in culture. Sixteen spontaneous MTL-homozygous offspring, both a/a and α/α, that were derived from four natural a/α strains were obtained by screening colonies for the opaque phenotype. Care was taken to be sure that each MTL-homozygous derivative was the product of an independent homozygosis event. By analyzing markers distributed along the entire length of chromosome 5, which harbors the MTL locus, we were able to deduce the mechanism of homozygosis for each of the 16 MTL-homozygous derivatives. We proposed three possible mechanisms for homozygosis, mitotic recombination, precise gene conversion, as in S. cerevisiae, and loss of a chromosome 5 homolog followed by duplication of the retained homolog. We found that only 1 of the 16 derivatives arose through mitotic recombination, while 15 derivatives arose by loss of one chromosome 5 homolog followed by duplication of the retained homolog. We found no evidence for precise gene conversion, although it cannot be ruled out as a minor mechanism, such as mitotic recombination. However, there was also the unlikely possibility that homozygosis could arise by meiosis. An analysis of polymorphic markers on chromosomes 1, 2, and R, however, revealed that all 16 MTL-homozygous derivatives maintained heterozygosity in chromosomes other than chromosome 5 homologs, excluding meiosis as a mechanism of MTL homozygosis in vitro. Two independent derivatives of P37037, P37037α/α-2 and P37037α/α-4, appeared to have similarly undergone the loss of one chromosome 1 homolog as well as one chromosome 5 homolog. Both exhibited polymorphisms on chromosomes 2 and R, indicating that they had undergone chromosome loss, not meiosis.
In the one documented case of mitotic recombination (P37037α/α-1), the parent strain was observed to undergo minor but frequent changes in its DNA fingerprint patterns identified by Southern blot hybridization with the complex DNA fingerprinting probe Ca3 (data not shown). Changes in the Ca3 fingerprinting pattern can be due to unequal crossing over at RPS loci (Pujol et al. 1999; Joly et al. 2002). P37037 was also the only strain that exhibited homozygosis in a second chromosome, in this case in chromosome 1, consistent with the conclusion that this a/α strain is genetically less stable than the other three that were analyzed.
In two of the four a/α strains analyzed (P37037 and P34048), we isolated both a/a and α/α offspring that arose through loss of one chromosome 5 homolog followed by duplication of the retained homolog. In a previous study of mitotic recombination, the presence of recessive lethal alleles was demonstrated (Whelan and Soll 1982). Since offspring from the same strains possessing duplicate copies of either homolog of chromosome 5 were viable, we can conclude that chromosome 5 does not harbor recessive lethal alleles. However, it is curious that in these same strains (P37037 and P34048), from which we isolated the most MTL-homozygous offspring, the majority of offspring were α/α, 5 out of 6 in the case of P37037 and 6 out of 7 in the case of P34048. Indeed, of the 16 MTL-homozygous offspring isolated from the four natural a/α strains in this study, 12 (75%) were α/α. The probability of this happening by chance is 0.02, suggesting that there may be a bias toward the loss of the chromosome 5 homolog harboring the MTLa locus. This bias may be the result of the culture conditions used in our study, since it has been demonstrated that the sugar source can affect loss of a chromosome 5 homolog (Janbon et al. 1998; Magee and Magee 2000). Hence, it is possible that the amino acid-rich medium used in our study may have biased loss of the homolog harboring the MTLa locus. Alternatively, the bias may be due to deleterious, but not necessarily lethal, alleles.
Our results also indicate that the chromosome 5 homolog that harbors the MTLa locus also harbors alleles of other genes that are specific to that homolog and that the homolog harboring MTLα also harbors specific alleles. This held true for shared polymorphic sites between the four unrelated a/α strains, which represented three clades (Soll and Pujol 2003), suggesting that there is strong selection pressure for these allelic linkages on the chromosome 5 homologs.
Acknowledgments
The authors thank Karla Daniels for her help in generating tables and figures; Julie Collins for helping assemble the article; and Kaustuv Sanyal, Mary Baum, and John Carbon of the University of California, Santa Barbara, for sharing their data on the location of the putative centromere in chromosome 5 prior to publication. This research was funded by National Institutes of Health grant AI2392.
References
- Anderson, J. M., and D. R. Soll, 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169: 5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., T. Srikantha, B. Morrow, S. H. Miyasaki, T. C. White et al., 1993. Characterization and partial sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J. Clin. Microbiol. 31: 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, G., and D. R. Soll, 1979. The effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc resistant and zinc sensitive pathways for mycelium formation. Infect. Immun. 26: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin et al., 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101: 1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana, H., B. B. Magee, S. Griddle, Y. Ran, S. Scherer et al., 1998. A physical map of chromosome 7 of Candida albicans. Genetics 149: 1739–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana, H., J. L. Beckerman and P. T. Magee, 2000. Fine resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10: 1865–1877. [DOI] [PubMed] [Google Scholar]
- Chindamporn, A., Y. Nakagawa, I. Nizuguchi, H. Chibana, M. Doi et al., 1998. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology 144: 849–857. [DOI] [PubMed] [Google Scholar]
- Chu, W.-S., B. B. Magee and P. T. Magee, 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J. Bacteriol. 175: 6637–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., P. T. Magee, B. B. Magee and G. May, 2004. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32: 561–599. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I., and Y. Oshima, 1981 Control of cell type in Saccharomyces cerevisiae: mating type and mating-type interconversion, pp. 181–210 in The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, edited by J. N. Strathern, E. W. Jones and J. R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hull, C. M., and A. D. Johnson, 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285: 1271–1275. [DOI] [PubMed] [Google Scholar]
- Hull, C. M., R. M. Raisner and A. D. Johnson, 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Iwaguchi, S. I., M. Homma, H. Chibana and K. Tanaka, 1992. Isolation and characterization of a repeated sequence (RPS1) of Candida albicans. J. Gen. Microbiol. 138: 1893–1900. [DOI] [PubMed] [Google Scholar]
- Janbon, G., F. Sherman and E. Rustchenko, 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95: 5150–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, S., C. Pujol and D. R. Soll, 2002. Microevolutionary changes and chromosomal translocations are more frequent at RPS loci in Candida dubliniensis than in Candida albicans. Infect. Genet. Evol. 2: 19–37. [DOI] [PubMed] [Google Scholar]
- Lee, K. L., H. R. Buckley and H. R. Campbell, 1975. An amino acid liquid synthetic medium for development of mycelia and yeast forms of Candida albicans. Sabouraudia 13: 148–153. [DOI] [PubMed] [Google Scholar]
- Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schröppel, T. Srikantha et al., 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., C. Pujol, K. Daniels, M. Miller, A. Johnson et al., 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels and D. R. Soll, 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, B. B., and P. T. Magee, 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Miller, M. G., and A. D. Johnson, 2002. White-opaque switching in Candida albicans is controlled by the mating type (MTL) locus and allows efficient mating. Cell 110: 293–302. [DOI] [PubMed] [Google Scholar]
- Pujol, C., S. Joly, B. Nolan, T. Srikantha and D. R. Soll, 1999. Microevolutionary changes in Candida albicans identified by the complex Ca3 fingerprinting probe involve insertions and deletions of the full-length repetitive sequence RPS at specific genomic sites. Microbiology 145: 2635–2646. [DOI] [PubMed] [Google Scholar]
- Pujol, C., S. A. Messer, M. Pfaller and D. R. Soll, 2003. Drug resistance is not directly affected by mating type locus zygosity in Candida albicans. Antimicrob. Agents Chemother. 47: 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, K., M. Baum and J. Carbon, 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 101: 11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, J., E. Voss and D. R. Soll, 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28: 1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll, D. R., 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13: 332–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll, D. R., and C. Pujol, 2003. DNA fingerprinting Candida albicans clades. FEMS Immunol. Med. Microbiol. 39: 1–7. [DOI] [PubMed] [Google Scholar]
- Soll, D. R., T. Srikantha and S. Lockhart, 1996 Cloning and characterizing developmentally regulated genes in Candida albicans, pp. 17–37 in Microbial Genome Methods, edited by K. W. Adolph. CRC Press, Boca Raton, FL.
- Srikantha, T., L. Tsai, K. Daniels and D. R. Soll, 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 182: 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha, T., L. K. Tsai, A. Klar and D. R. Soll, 2001. The histone deacetylase HDA1 and RPD3 play distinct roles in the regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183: 4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong, A. E., M. G. Miller, R. M. Raisner and A. D. Johnson, 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115: 389–399. [DOI] [PubMed] [Google Scholar]
- Whelan, W. L., and D. R. Soll, 1982. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol. Gen. Genet. 187: 477–485. [DOI] [PubMed] [Google Scholar]