Abstract
Double-strand breaks (DSBs) initiate meiotic recombination. The DSB repair model predicts that both genetic markers spanning the DSB should be included in heteroduplex DNA and be detectable as non-Mendelian segregations (NMS). In experiments testing this, a significant fraction of events do not conform to this prediction, as only one of the markers displays NMS (one-sided events). Two explanations have been proposed to account for the discrepancies between the predictions and experimental observations. One suggests that two-sided events are the norm but are “hidden” as heteroduplex repair frequently restores the parental configuration of one of the markers. Another explanation posits that one-sided events reflect events in which heteroduplex is formed predominantly on only one side of the DSB. In the absence of heteroduplex repair, the first model predicts that two-sided events would be revealed at the expense of one-sided events, while the second predicts no effect on the distribution of events when heteroduplex repair is lost. We tested these predictions by deleting the DNA mismatch repair genes MSH2 or MLH1 and analyzing the proportion of two-sided events. Unexpectedly, the results do not match the predictions of either model. In both mlh1Δ and msh2Δ, the proportion of two-sided events is significantly decreased relative to wild type. These observations can be explained in one of two ways. Either Msh2p/Mlh1p-independent mispair removal leads to restoration of one of the markers flanking the DSB site or Msh2p/Mlh1p actively promote two-sided events.
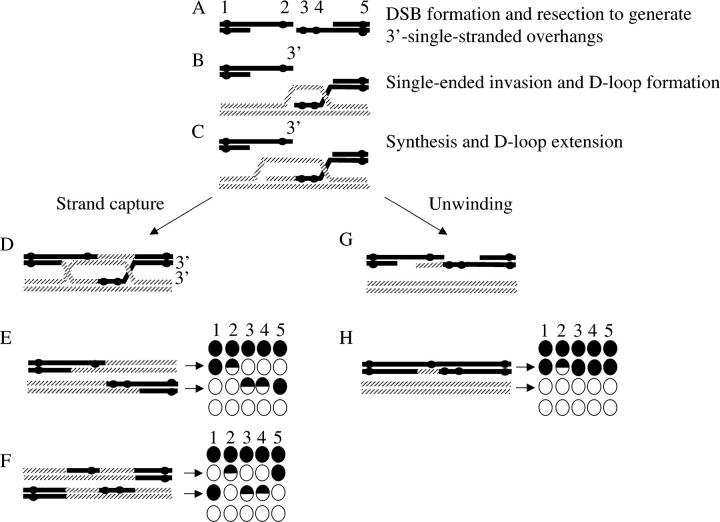
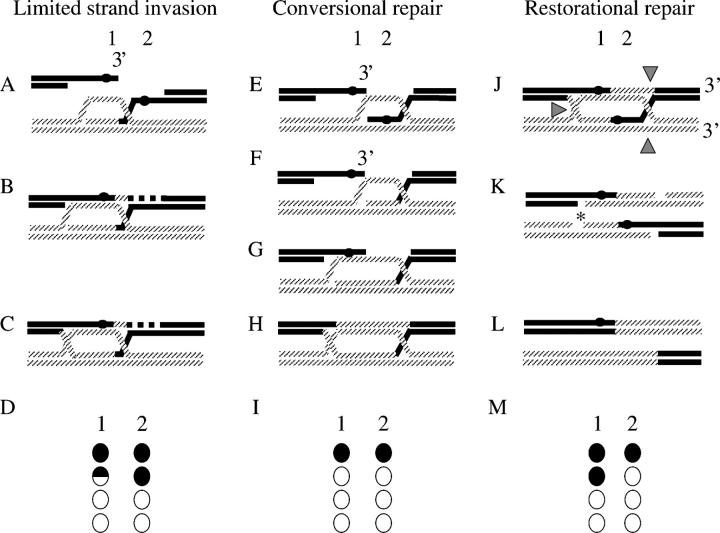
THE double-strand break (DSB) repair model proposed by Szostak et al. (1983) predicts that heteroduplex DNA is formed on both sides of the DSB (Figure 1). Thus, when resection, strand invasion, and strand capture include markers placed on opposite sides of the DSB, both markers should show non-Mendelian segregation (NMS; Schultes and Szostak 1990; Figure 1). Such events have been termed “two-sided” events. However, other genetic evidence suggests that “one-sided” events are common and that the proportion thereof may be hotspot specific (Schultes and Szostak 1990; Porter et al. 1993; Gilbertson and Stahl 1996; Merker et al. 2003; Jessop et al. 2005). Although there is general agreement that DSB repair (DSBR) events are inherently two sided, the extent of resection and/or heteroduplex formed on each side of the DSB is not clear. Petes and colleagues (Porter et al. 1993; Merker et al. 2003) have suggested two physical models whereby heteroduplex DNA is formed on only one side of the break. Porter et al. (1993) suggest that one-sided events might derive from substantial resection of one side of the DSB but not the other, while Merker et al. (2003) suggest that the extent of heteroduplex formed upon invasion is short, while that of strand capture is long (Figure 2). Either of these mechanisms leads to extensive heteroduplex formation on only one side of the DSB. The model of Merker et al. (2003) is illustrated in Figure 2, A–D. As an alternative mechanism, Foss et al. (1999) propose that two-sided events are processed to one-sided events by heteroduplex repair of one of the markers to the parental configuration (also known as restoration; Figure 2, J and K). All of the above experiments require that the interacting DNA strands are identifiable. Therefore, heteroduplex DNA must remain unrepaired (Figure 1). To accomplish this, palindromes have been the preferred genetic markers for these studies. Heteroduplex that contains palindromes is poorly repaired (i.e., fewer 6:2 or 2:6 full conversions), due to the fact that palindromes are partially refractory to mismatch repair by the Msh2p/Mlh1p mismatch repair system (Nag et al. 1989). This is presumably due to the failure of mismatch repair proteins to process the palindrome when found in heteroduplex DNA (Alani 1996). The failure to remove the palindrome allows the monitoring of heteroduplex DNA in the end products of the meiotic recombination event (i.e., as 5:3 or 3:5 half conversions/postmeiotic segregation in the tetrads).
Figure 1.—
A modified double-strand break repair model and a synthesis-dependent single-strand annealing (SDSA) for recombination. Five markers, labeled 1–5 and spanning the DSB, are shown as solid circles. These correspond to HPH, BIK1-939, his4-ATC, HIS4-1605, and NAT, as illustrated in Figure 3. Markers 2, 3, and 4 are included in heteroduplex DNA and are left unrepaired so that the origin of the DNA strands is clear. Markers 1 and 5 are flanking markers used to assess crossovers. For simplicity, only the two interacting chromatids are shown. (A) DSB formation and subsequent resection initiates meiotic recombination and generates 3′-single-stranded overhangs. (B) Invasion of the intact homologous chromosome by only one of the 3′-single-stranded overhang generates a D-loop. This is the first stage during which heteroduplex DNA may be formed. Synthesis (C) and subsequent capture of the D-loop by the second end lead to formation of the joint molecule (double Holliday junction; D). Double Holliday junctions may give rise to crossovers (E and F). The resulting tetrads are shown to the right. The spores arising from the interacting DNA molecules are indicated by arrows. The parental chromatids are shown above (all markers solid) and below (all markers open). Markers are as given in A. (G) The D-loop, shown in C, may also be disassembled, leading to SDSA. In the absence of heteroduplex repair, SDSA will generate a one-sided event (marker 2) that will not be associated with a crossover.
Figure 2.—
Models for the origin of sidedness. Markers 1 and 2, corresponding to BIK1-939 and his4-ATC, flank the double-strand break. The tetrads resulting from the DNA interactions are illustrated below, as in Figure 1. Only the two interacting DNA strands are illustrated, but the top and bottom line of the tetrad show the parental configuration of the two uninvolved strands. (A) Limited strand invasion. Invasion of the 3′-single-strand overhang does not include marker 2, shown as a solid circle. D-loop capture by the left-hand side of the DSB (B) and DSB repair synthesis will generate a double Holliday junction (C; redrawn from Merker et al. 2003). Marker 2 is not included into heteroduplex DNA at any stage and therefore shows Mendelian segregation (D). When marker 1 remains unrepaired or is converted, a one-sided event results. Conversional repair: The single-end invasion generates heteroduplex DNA containing marker 2 (E). Early mismatch repair of marker 2 (F) leads to a full gene conversion. For marker 1, placed on the left-hand side of the DSB, heteroduplex is formed upon capture of the second end (G). Mismatch repair directed by this end (H) results in this genetic marker undergoing a full conversion as well, thereby generating a two-sided event (I). Nick-directed restorational repair. If early heteroduplex repair is inefficient, as has been suggested for palindromic markers (see text), the heteroduplex DNA persists while the double Holliday junction is formed (J). Cutting of the double Holliday junction to yield a crossover (configuration in Figure 1E), indicated by shaded arrowheads, generates nicks that can be used to direct the mispair removal. If the nicks from only one Holliday junction are used, indicated by an asterisk in K, to direct mispair removal, marker 1 would be restored and marker 2 would be converted (L). Consequently, a one-sided event would result (M). Use of both nicks can lead to both one- and two-sided events (not illustrated).
The mismatch repair proteins of Saccharomyces cerevisiae are orthologs of the Escherichia coli long-patch repair proteins MutS and MutL. Heterodimers of Msh2p and either Msh3p or Msh6p recognize insertion/deletion loops as well as base mispairs. Msh2p/Msh3p and Msh2p/Msh6p heterodimers subsequently form a tetramer with a heterodimer consisting of Mlh1p with Pms1p, Mlh2p, or Mlh3p that is presumed to recruit repair enzymes (reviewed in Surtees et al. 2004). We hypothesized that if the model of Foss et al. (1999) was correct, and if restorational repair is dependent on Msh2p and/or Mlh1p, then deleting MSH2 or MLH1 should allow all two-sided events to be detected (Figures 1 and 2). On the other hand, if, as is implicit in the Foss et al. (1999) model, restorational repair is at least partially independent of conventional mismatch repair, then abolishing mismatch repair should mimic the results obtained with palindromic markers. Furthermore, neither of the models put forth by Petes and colleagues predict an effect of abolishing Msh2p/Mlh1p-dependent mismatch repair. We tested these predictions by deleting MLH1 or MSH2 in appropriately marked strains. When selecting tetrads in which one marker had undergone a NMS, we found a decrease in the proportion of two-sided events in the msh2Δ and mlh1Δ strains compared to wild type. In other words, DSB repair events are initially two sided but are processed to one-sided events in the absence of Msh2p/Mlh1p. We suggest that in the absence of Msh2p and Mlh1p restorational repair can occur and/or that Msh2p and Mlh1p actively promote two-sided events.
MATERIALS AND METHODS
Genetic analysis:
Standard genetic procedures and omission media were used as described previously (Abdullah and Borts 2001). The alleles used to study meiotic segregation at the HIS4 hotspot (his4-ATC, BIK1-939, and HIS4-1605) are described below. The ade1-1, met13-2, trp5-1, leu2-r, and CYH2 alleles have all been described previously (Abdullah and Borts 2001; Abdullah et al. 2004) as have the HphR/HYG and NouR/NAT genes (Goldstein and McCusker 1999).
Double-strand break analysis:
Diploid strains were induced to undergo sporulation in liquid as described previously (Goyon and Lichten 1993) and samples were collected for DSB analysis at 0 and 24 hr. Genomic DNA was isolated using standard procedures (Borts et al. 1986). Genomic DNA concentrations were determined using the KODAK 2000 Image Station with the KODAK 1D v3.5 software to determine the integrity and concentration of DNA. Plasmid DNA (pEH12) and λBstEII ladder (New England Biolabs, Beverly, MA) were labeled using the genes images random labeling module (Amersham Pharmacia) following the instructions of the supplier. To visualize the HIS4 double-strand break hotspot, 1 μg of genomic DNA was digested with XbaI (New England Biolabs) and the DNA fragments were separated on a 1.1% agarose gel. DNA was transferred to a solid membrane (Southern 1975; Sambrook et al. 1989) and hybridized with labeled probes detected following the manufacturer's instructions (Amersham Pharmacia).
Quantification of DSBs:
Chemiluminescence was quantified using a CCD camera (Kodak Image Station 2000; see above). Similar to quantification using radioactivity, chemiluminescence is proportional to the amount of probe hybridized and therefore the amount of homologous DNA on the filter when probe is in excess. The emitted signal was linear with time and with amount of DNA over the experimental ranges utilized (data not shown). Chemiluminescence was also detected by exposure to film. For bands too faint for the camera to detect, film was digitized and quantified. The amount of signal detected on film was also linear with concentration of DNA and time of exposure. The intensity of DSBs was quantified using the KODAK 1D v3.5 software.
Strains:
All of the strains were derivatives of S. cerevisiae strain Y55 and have been described previously (Hoffmann et al. 2003). All of the mutant strains were isogenic derivatives of EY97 and EY128 (Table 1). The spore viability of the wild-type strains was 95%. ERY102 had a spore viability of 86% and ERY112 of 83%. The spore viabilities of all of these strains agree with those observed previously (Hoffmann et al. 2003).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| Haploids | |
| EY97 |
MATα, LEU2, FUS1-HPH; his4-ATC; RRP7-NAT; ade1-1; trp5-1; cyh2-R; lys2-c; ura3-1 |
| EY128 |
BIK1-939; HIS4-1605; leu2-r; met13-2; lys2-d; ura3-1 |
| EY111 | As EY97 but msh2Δ::KANMX4 |
| EY129 | As EY128 but msh2Δ::KANMX4 |
| EY130 | As EY97 but mlh1Δ::KANMX4 |
| EY131 | As EY128 but mlh1Δ::KANMX4 |
| EY201 | As EY97 but com1Δ::KANMX4 |
| EY203 | As EY128 but com1Δ::KANMX4 |
| EY255 | As EY97 but MATa |
| Diploids | |
| ERY102 | EY111 × EY129 (msh2Δ/msh2Δ) |
| ERY103 | EY97 × EY128 |
| ERY112 | EY130 × EY131 (mlh1Δ/mlh1Δ) |
| ERY188 | EY201 × EY203 (com1Δ/com1Δ) |
Plasmids:
The HIS4 (pEH24) and BIK1 (pEH13) ORFs and the surrounding sequences were PCR amplified from S. cerevisiae Y55 using Pfu polymerase (Stratagene, La Jolla, CA) and cloned into pMOSBlue (Amersham Pharmacia; Table 2). KlURA3 was PCR amplified from pWJ716 (Erdeniz et al. 1997) and cloned into the SmaI site of pEH24 to yield pEH26. Mutations were introduced into HIS4 and BIK1 using the quick change site-directed mutagenesis kit (Stratagene, Cambridge, UK) following the manufacturer's instructions. pEH27, containing the his4-ATC allele, and pEH28, containing the HIS4-1605 mutation, were constructed from pEH26 using primer sets HIS4.g3c.F plus HIS4.g3c.R and HIS4.c1605g.F plus HIS4.c1605g.R, respectively (Table 3). HIS4-1605 has a silent guanosine-to-cytosine change that deletes a HhaI site. Similarly, BIK1-939 was created in pEH13 using the BIK1.g939a.F and BIK1.g939a.R primers, resulting in pEH19. This change results in a silent guanisine-to-adenosine change that deletes a PvuII site in the BIK1-939 allele.
TABLE 2.
Plasmids
| Plasmid | Description |
|---|---|
| PMOSBlue (Stratagene) | |
| pEH12 | HIS4 ORF into pMOSBlue |
| pEH13 | BIK1 ORF including the upstream 500 and downstream 223 into pMOSBlue |
| pEH19 | pEH13 but change BIK1-939 g → a (ΔPvuII site) |
| pEH24 | HIS4 ORF and upstream 1000 bp into pMOSBlue |
| pEH26 | pEH24, but KlURA3 PCR into SmaI site |
| pEH27 | pEH26 but change HIS4-1605 c → g (ΔHhaI site) |
| pEH28 | pEH26 but change HIS4-3 g → c |
| pAG25 and pAG32 | HPHMX4 and NATMX4 plasmids (Goldstein and McCusker 1999) |
| pRS416 | KANMX4 plasmid (Wach et al. 1994) |
All plasmids were verified by sequencing the relevant parts of the insertion.
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| Mutagenesis | |
| HIS4-g3c.F | TTTTTTCTGAATAATCGTTTTGCCGATTCTACCG |
| HIS4-g3c.R | CGGTAGAATCGGCAAAACGATTATTCAGAAAAAA |
| HIS4.g3c-adaptamerA.F | AATTCCAGCTGACCACCATGTATGACTATGAACAGTAG |
| HIS4.g3c-adaptamerB.R | GATCCCCGGGAATTGCCATGACCAACCCTAGACAACGCTC |
| HIS4.c1605g.F | CCAGCTAGAACAATCTTGGAAGCCCCAACTTTTTCTGCGAC |
| HIS4.c1605g.R | GTCGCAGAAAAAGTTGGGGCTTCCAAGATTGTTCTAGCTGG |
| HIS4-1605-MX4.F | TGTCTTGTGTTCCAGATTCCCTCGTCCTATTGAAAAAGTTGGCGTACGCTGCAGGTCGAC |
| HIS4-1605-MX4.R | CCAAAACTTCACTTGGGCCAGCTGGCATATCAATGGAACATAGAGCATCGATGAATTCGAGCTCG |
| BIK1.g939a.F | CGAACTCCTGAAAAAACAGCTAGAACAATTACGCAAC |
| BIK1.g939a.R | GTTGCGTAATTGTTCTAGCTGTTTTTTCAGGAGTTCG |
| BIK1.g939a-adaptamerA.F | GAACTGACTCTAATAGTGACTCCGGTAAATTAGTTAATTAATTGCGATGATGTAGTTTCTGGTT |
| BIK1.g939a-adaptamerB.R | ATGTATGTACAACACACATCGGAGGTGAATATAACGTTCCGTGATTCTGGGTAGAAGATCG |
| Cloning | |
| HIS4-1000 upstream.F | GAATTACGAGAAGCCCAATTGACCATCGAA |
| HIS4-ORF.F | ATGGTTTTGCCGATTCTACCGTTAATTGAT |
| HIS4-ORF.R | CTACTGGAAATCCTTTGGGAACAACCCAAGC |
| BIK1-500 upstream.F | AGTTGCGTTTGGGAAGAA |
| BIK1-223 downstream.R | ACAGCGCAGTTGTGCTATGATAT |
| Insertion | |
| FUS1-HPH.F | TTGTCATGCACATCATCATACTAAACTTACACGAATAGGAATCGATGAATTCGAGCTCG |
| FUS1-HPH.R | TTGTCATGCACATCATCATACTAAACTTACACGAATAGGAATCGATGAATTCGAGCTCG |
| FUS1-A1 | CATTGCCGCTTACTCCAAAC |
| FUS1-A4 | CATTGCCGCTTACTCCAAAC |
| RRP7-NAT.F | TTTAAAGGCATCAATAATTTTTTTCTTTCATATATATTCTGCGTACGCTGCAGGTCGAC |
| RRP7-NAT.R | TTGTCATGCACATCATCATACTAAACTTACACGAATAGGAATCGATGAATTCGAGCTCG |
| RRP7-A1 | GTGGATGAGGATGGATTCAC |
| RRP7-A4 | CATAACCGACAATCACCGTC |
| Allele detection | |
| HIS4.1200-1220.fwd | TCTGTTTTTTTGGAGTACAC |
| HIS4.1700-1681.rev | GGACCCAAGATCTTATCCAC |
| BIK1.751-771.fwd | AAGCAACAATTGGAGCTCGAACGC |
| BIK1.1323-1300.R | CTAGAAGAACTGCTGGTTGTCAGG |
Disruption oligonucleotide primers for MLH1, MSH2, and COM1 were designed using the immediate 40–45 bp upstream and downstream and linking these to the KANMX4 primer sequence. Verification primers were designed upstream and downstream as described previously (Wach et al. 1994). Sequences are available upon request.
Construction of alleles:
The his4-ATC and BIK1-939 mutant alleles were verified by sequencing around the mutations and introduced into the genome using a cloning-free method described previously (Erdeniz et al. 1997). The primers labeled “.adaptermer” in Table 3 were used for this purpose. HIS4-1605 was introduced by replacing 250 bp to each side of the HhaI site with KANMX4 (primers HIS4-1605MX4.F and .R; Table 3). The KANMX4 cassette was then replaced by transformation with the mutated pEH28 fragment, selecting for histidine prototrophy. All of the introduced alleles were verified by sequencing.
The HIS4 locus was flanked by the hygromycin B resistance gene (HPHMX4, or HPH) upstream of FUS1 (inserted 5130 bp upstream of the start of the HIS4 ORF, deleting 7 bp), and the nourseothricin resistance gene (NATMX4, or NAT) was inserted 3804 bp downstream of HIS4 (the oligonucleotides are listed in Table 3 under “Insertion”). PCR, Southern blotting, and genetic linkage to HIS4 were used to check both insertions.
Colony PCR:
The silent alleles at BIK1-939 and HIS4-1605 were analyzed by colony PCR. The entire colony (∼107 cells) was resuspended in 20 μl 0.02 m NaOH. These resuspended colonies can be used for PCR for at least 6 months if stored at 4°. From this, 2 μl was added to a standard PCR reaction (Jeffreys et al. 1990) containing either the primer set for BIK1 or HIS4 amplification in a total volume of 10 μl. The allele present in the PCR product was determined by restriction digestion with either PvuII or HhaI (New England Biolabs), according to the manufacturer's instructions, followed by electrophoresis. Wild-type information at HIS4-1605 yields two bands of 100 and 400 bp, whereas the mutation at HhaI results in a single 500-bp band. Similarly, by destroying the PvuII site in BIK1-939, the two wild-type bands of 200 and 600 bp now result in a single 800-bp band. The PCRs yielded results with a range of cells (from <10 to 107) and were able to detect the minority band when cells containing the majority information were in 104-fold excess (data not shown).
Reconstruction experiment:
To determine the probability of detecting a half conversion of either HIS4-1605 or BIK1-939, we performed a reconstruction experiment as follows: Spore colonies mimicking a half-conversion at BIK1-939, his4-ATC, and HIS4-1605 were “constructed” by placing a single cell from each parent adjacent to each other on a YEPD plate. For this purpose, it was necessary to change the mating type of one of the parents to prevent the two haploid cells from mating (EY255 was derived from EY97, but expressed the MATa allele rather than MATa; Table 1). In total, 83 reconstructed colonies were analyzed, all of which contained half conversions at his4-ATC as detected genetically by sectoring for His+. Segregations of HIS4-1605 and BIK1-939 were analyzed by colony PCR as described above. At HIS4-1605, all of the 83 reconstructed colonies contained both parental bands in approximately equal proportions. This indicates that the HIS4-1605 allele does not influence the growth rate compared to the wild-type HIS4. Similarly for BIK1-939, 82 of the colonies contained all three bands in approximately equal proportions, again suggesting that the single-base change does not influence the growth rate. However, one of the colonies showed only mutant information; hence the failure rate of detecting a half conversion at BIK1-939 was 1.2% (1/83). On the basis of this experiment we are 95% certain of detecting heteroduplex DNA in ≥95% of the cases encompassing his4-ATC, HIS4-1605, and BIK-939. These frequencies are similar to those observed previously (Porter et al. 1993; Hillers and Stahl 1999).
Statistical analysis:
Statistical tests were applied as indicated throughout (Sokal and Rohlf 1995; http://faculty.vassar.edu/lowry/VassarStats.html). P < 0.05 was considered significant, except when multiple data sets were analyzed using pairwise comparisons. In such cases, the Dunn-Sidak adjustment of the P-value was used (Sokal and Rohlf 1995) to avoid a type I error, as applied previously (Hoffmann et al. 2003).
RESULTS
Rationale and terminology:
To test predictions regarding the deposition of heteroduplex DNA and crossovers that can made from the models discussed above, we flanked the HIS4 hotspot with genetic markers. The proportion of events that flanked the DSB (two-sided events) was then determined in wild-type and mismatch-repair-defective strains. According to Merker et al. (2003), the absence of mismatch repair proteins should have no effect on the proportion of two-sided events, whereas the model by Foss et al. (1999) predicts a decrease in two-sided events in the mismatch-repair-defective strains. Furthermore, the Foss et al. (1999) model predicts that crossovers associated with one-sided events should map to a specific interval.
The terminology used to describe tetrads in which more than one genetic marker segregates is as follows: When two markers segregate independently of each other because the heteroduplex DNA that contained them arose from two different DSB repair events, we term this a “co-event.” When the two alleles show non-Mendelian segregation consistent with heteroduplex formation and/or repair of a single DSB, the two markers are said to display co-conversion. Conversions could be either full conversions (6:2 or 2:6) or half conversions (5:3 or 3:5, also known as postmeiotic segregations). When reporting ratios of NMS (e.g., 6:2), the genotype of EY128 is given first. This corresponds to wild type for his4-ATC, but not for BIK1-939 and HIS4-1605, and to growth on nourseothricin and hygromycin-B-containing medium. DSBR refers to any mechanism that involves strand invasion. Restorational repair refers to removal of a mispair from heteroduplex DNA such that a 4:4 segregation was obtained. Similarly, conversional repair leads to 6:2 or 2:6 segregation. Heteroduplex repair can refer to either. MMR is used only in the context of mismatch removal by Msh2p/Mlh1p. Mispair removal is used when the repair system has not been experimentally established.
S. cerevisiae Y55 contains a strong HIS4 double-strand break hotspot:
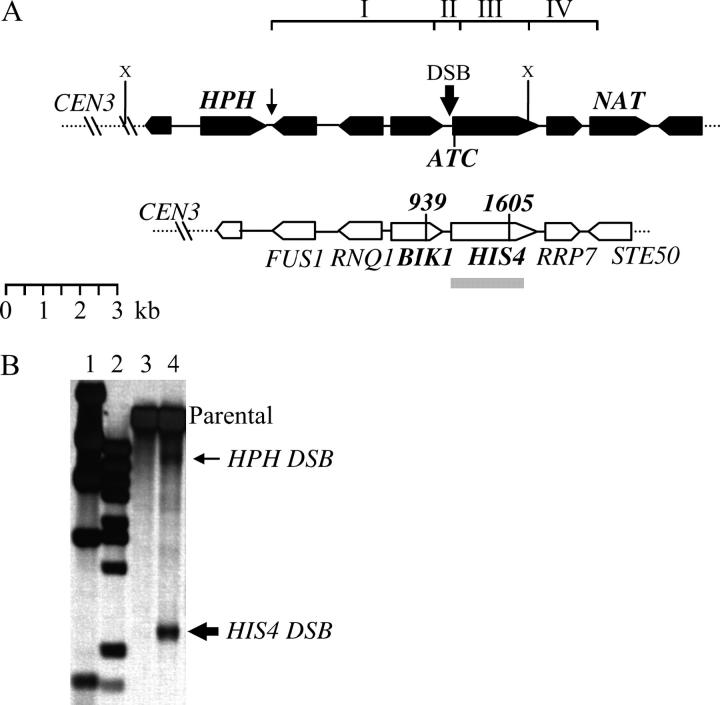
Two strains differing in intensity and in the distribution of DSBs within the HIS4 promoter have been characterized (Fan et al. 1995; Baudat and Nicolas 1997; Gerton et al. 2000). To demonstrate that the S. cerevisiae Y55 strain also contains a HIS4 DSB hotspot, we analyzed genomic DNA from a strain (ERY188; Table 1) that accumulates unprocessed DSBs (McKee and Kleckner 1997; Prinz et al. 1997; see materials and methods). Upon XbaI digestion of the genomic DNA and probing with the HIS4 ORF, a DSB within the HIS4 promoter was expected to give rise to an ∼2.4- to 2.6-kb fragment (Figure 3B). The fragment size suggested that the HIS4 DSB hotspot is placed ∼300 bp upstream of the HIS4 start codon. This places BIK1-939 and his4-ATC 350 and ∼300 bp, respectively, on opposite sides of the HIS4 DSB. Quantification indicated that ∼4–5% of the total DNA was cut at the HIS4 hotspot. Other, less strong DSBs were observed at HPH (Figure 2B; <0.5%) and NAT (data not shown); however, no other DSBs were detected within ∼4 kb spanning the HIS4 DSB.
Figure 3.—
The HIS4 region on the left arm of chromosome III (Crick strand). The chromosome is drawn opposite to the conventional orientation. (A) The two haploid parents EY97 (top chromatid; solid) and EY128 (bottom chromatid; open) are shown. The direction of transcription is indicated by the tapered end. The boxes labeled NAT and HPH represent insertions introduced in EY97 but not in EY128. EY97 is auxotrophic for histidine synthesis as the start codon of HIS4 has been changed from ATG to ATC (his4-ATC allele). HIS4 and BIK1 of parent EY128 contain silent single nucleotide changes at HIS4-1605 and BIK1-939. The regions between HPH and BIK1-939, BIK1-939 and his4-ATC, his4-ATC and HIS4-1605, and HIS4-1605 and NAT to which crossovers could be mapped are labeled interval I, II, III, and IV, respectively. (B) Mapping of DSBs within the HIS4 region. Lanes 1 and 2 contain the size standards λHindIII and λBstEII, respectively. Meiotic time course DNA from strain ERY188 was extracted after 0 hr in sporulation medium (lane 3) or after 24 hr (lane 4). The DNA was digested with XbaI indicated by an “X” in A and the Southern blot was probed with the HIS4 ORF (shaded bar in A). Arrows to the right indicate the position and relative intensities of the two DSBs in the region.
Deletion of MSH2 or MLH1 leads to an increase in non-Mendelian segregation and a decrease in heteroduplex repair:
As observed previously, the msh2Δ and mlh1Δ strains showed an increase in non-Mendelian segregation (Alani et al. 1994; Prolla et al. 1994). The NMS frequencies of the his4-ATC, HIS4-1605, BIK1-939, leu2-R1, ade1-1, trp5-1, cyh2, and met13-2 alleles were all increased in the mlh1Δ strain and at seven of the eight loci in the msh2Δ strain (Table 4). The probabilities for such directional increases in aberrant segregation frequencies were 0.0039 and 0.03, respectively (exact binomial probabilities). Such an effect has been observed previously in the absence of functional mismatch repair (Alani et al. 1994; Prolla et al. 1994; Hunter and Borts 1997; Hoffmann et al. 2003). Moreover, heteroduplex intermediates at all of the genetic markers were removed less efficiently (increased half conversions/postmeiotic segregations) in the mlh1Δ and msh2Δ strains compared to the wild-type strain (Table 5; data not shown).
TABLE 4.
Non-Mendelian segregation of several loci
| Allele
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | his4-ATC | HIS4-1605a | BIK1-939a | leu2-R1 | ade1-1 | met13-2 | cyh2 | trp5-1 |
| Wild-type | 14 (243/1731) | 2.8 (8/289) | 9.3 (10/107) | 1.7 (29/1731) | 2.0 (36/1731) | 4.5 (78/1731) | 0.6 (11/1731) | 0.9 (16/1731) |
| mlh1Δ | 20b (116/585) | 5.6 (6/106) | 13 (14/106) | 2.7 (16/585) | 3.6 (21/585) | 5.5 (32/585) | 1.4 (8/585) | 1.4 (8/585) |
| msh2Δ | 18 (96/545) | 4.0 (4/101) | 8.9 (9/101) | 2.2 (12/545) | 2.8 (15/545) | 2.6 (14/545) | 0.9 (5/545) | 2.9 (16/545) |
NMS is given as the percentage of total half-conversion and full-conversion events divided by the total number of tetrads analyzed. The actual number of tetrads with an NMS event and the total number of tetrads are given in parentheses.
Randomly selected tetrads were analyzed for the wild-type, mlh1Δ, and msh2Δ strains to estimate the overall frequency of NMS at HIS4-1605 and BIK1-939.
Values significantly different from the wild-type strain using a G-test. P-values <0.05 were considered significant. None of the values from the subsets were different from the corresponding main data set (G-test). Therefore, the subsets are representative of the main data sets.
TABLE 5.
Non-Mendelian segregation ofhis4-ATC andBIK1-939
| NMS class
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Relevant genotype | 6:2 | 2:6 | 5:3 | 3:5 | 8:0 | 0:8 | 7:1 | 1:7 | Ab4:4 | Othersa | Total NMS |
Total tetrads |
NMS (%) |
FC/NMS (%) |
| his4-ATC | Wild type | 96 | 111 | 14 | 15 | 1 | 3 | 1 | 2 | 0 | 0 | 243 | 1731 | 14 | 87 |
| mlh1Δ | 5 | 7 | 35 | 65 | 0 | 0 | 0 | 0 | 2 | 2 | 116 | 585 | 20b | 12 | |
| msh2Δ | 15 | 18 | 17 | 36 | 0 | 1 | 0 | 1 | 3 | 3 | 96 | 545 | 18 | 37 | |
| Wild-type subsetc | 1 | 7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 107 | 9.3 | 80 | |
| mlh1Δ subsetc | 2 | 0 | 9 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 106 | 21 | 9 | |
| msh2Δ subsetc | 2 | 3 | 3 | 11 | 0 | 0 | 0 | 0 | 1 | 1 | 21 | 101 | 21 | 24 | |
| BIK1-939 | Wild-type subsetc | 0 | 9 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 10 | 107 | 9.3 | 100 |
| mlh1Δ subsetc | 0 | 1 | 6 | 4 | 0 | 2 | 0 | 0 | 1 | 0 | 14 | 106 | 13 | 21 | |
| msh2Δ subsetc | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 101 | 8.9 | 0 | |
| Wild type selectedd | 38 | 51 | 2 | 1 | 1 | 6 | 0 | 1 | 0 | 0 | 100 | 144 | 69 | 96 | |
| mlh1Δ selectedd | 0 | 3 | 23 | 25 | 1 | 0 | 0 | 0 | 2 | 1 | 51 | 116 | 44b | 10 | |
| msh2Δ selectedd | 0 | 4 | 14 | 23 | 0 | 0 | 0 | 0 | 2 | 0 | 43 | 96 | 45b | 10 | |
FC, full conversion.
Mainly Ab6:2 and Ab2:6.
Statistically significant compared to the equivalent wild-type proportion (P < 0.025; G-test for homogeneity).
Randomly chosen tetrads were analyzed by PCR to estimate the NMS frequencies of BIK1-939 and HIS4-1605.
Tetrads that were selected for an NMS at his4-ATC and were analyzed further for NMS at BIK1-939 and HIS4-1605.
The total NMS frequency of his4-ATC was increased compared to the wild-type strain in both the mlh1Δ and msh2Δ strains (compare 14% to 20% and 18%, respectively; Table 5). The frequencies of NMS of BIK1-939 were less than that of his4-ATC (9.3, 13, and 8.9% in the wild-type, mlh1Δ, and msh2Δ strains, respectively; Table 5) in all of the three strains. This may be because BIK1-939 is placed ∼50 bp farther from the HIS4 DSB and is less likely to be included in heteroduplex DNA or because BIK1-939 mispairs are more readily restored compared to his4-ATC mispairs in all three strains. Finally, the repair of heteroduplex DNA containing either allele was decreased in the mismatch repair mutants. The repair rate containing BIK1-939 was decreased from 100% to 0% in the mlh1Δ strain and to 21% in the msh2Δ strain (Table 5; mlh1Δ subset and msh2Δ subset). The repair rate of his4-ATC was decreased from 87% in the wild-type strain to 12% and 37% in the mlh1Δ and msh2Δ strains, respectively. The repair rate of his4-ATC in the msh2Δ strain was higher than that in the mlh1Δ strain (P < 0.05; z-test for proportions). The NMS frequencies and heteroduplex repair of the NAT and HPH markers were not affected in the mlh1Δ and msh2Δ strains. Since it is known that the removal of large insertion/deletions such as NAT and HPH present in meiotic heteroduplex DNA is independent of the mismatch repair system (Kearney et al. 2001), NAT and HPH were excluded from the analysis above.
The proportions of co-events and co-conversions were decreased in mlh1Δ and msh2Δ strains:
Tetrads in which his4-ATC showed a NMS were identified (“selected” in Table 5) and analyzed for NMS at BIK1-939 and HIS4-1605 (the marker configurations of all these tetrads are illustrated in the supplementary appendix at http://www.genetics.org/supplemental/). In the wild-type strain, 144 of the 243 tetrads showing NMS at his4-ATC were analyzed. In the wild-type strain, 69% of events were co-events of his4-ATC and BIK1-939, whereas in the mlh1Δ and msh2Δ strains there were significantly fewer (44% and 45%, respectively; P < 0.025; z-test for proportions).
Identification and analysis of complex events:
Co-events may not be an accurate measure of two sidedness as “complex events,” consisting of more than one DSB repair event, may obscure the actual proportion of two-sided events. Tetrads that contained more than two recombinant chromatids in the HIS4 interval (Figure 4; supplementary material section 1A at http://www.genetics.org/supplemental/) most likely arose from multiple initiations. Finally, tetrads that contained two recombinant spores in which the NMS of BIK-939 and his4-ATC had arisen from repair of more than a single DSB were also considered complex (supplementary material section 1B at http://www.genetics.org/supplemental/; Figure 4D). All three strains contained a similar proportion of such complex events. This resulted in 38%, 30%, and 36% of the tetrads that showed non-Mendelian segregation for his4-ATC being excluded from further analysis in the wild-type, mlh1Δ, and msh2Δ strains, respectively (Table 6; supplementary material section 1A at http://www.genetics.org/supplemental/). These values are similar to those observed previously at HIS4 (Hillers and Stahl 1999; Merker et al. 2003). The majority of these complex events (>70%) can be explained in terms of two independent DSB repair events initiated from the HIS4 promoter. This is consistent with no other DSBs having been detected within the HIS4 region.
Figure 4.—
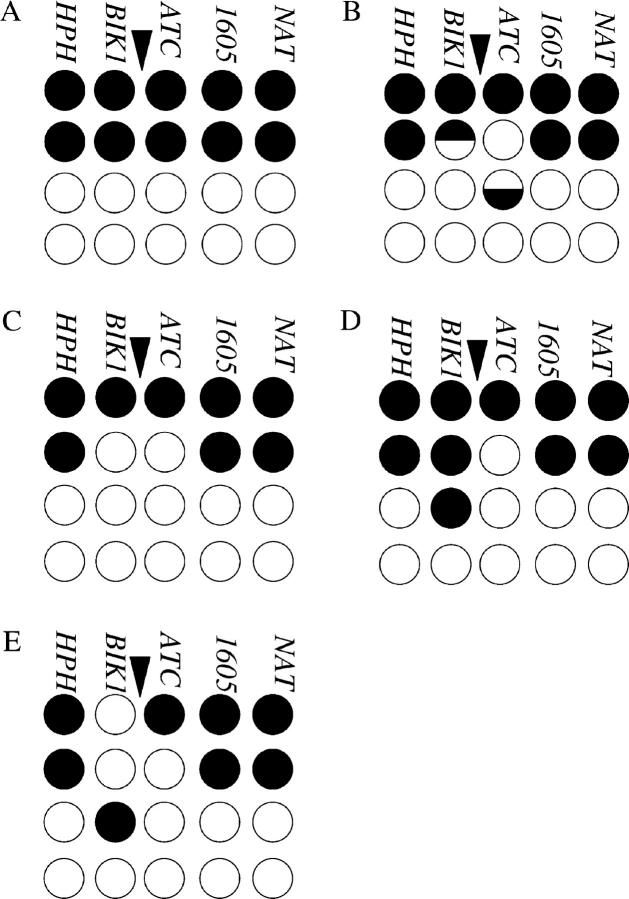
Co-conversion and co-event tetrads. A tetrad with the parental configuration of the genetic markers is given in A. Each tetrad is shown by four rows and five columns. Each row represents one chromatid and each column represents one marker [HPH, BIK1-939 (BIK1), his4-ATC (ATC), HIS4-1605 (1605), and NAT)]. The position of the HIS4 DSB is indicated by an arrowhead. Open circles signify the EY128 parental information (Figure 3), and solid circles show EY97 parental information (Figure 3). A NMS event is represented as the number of white to the number of black strands. (B) The tetrad illustrated represents a co-half conversion of BIK1-939 and his4-ATC. Such tetrads may arise from unrepaired heteroduplex DNA from a single DSB repair event. The half conversions are placed on two different recombinant chromatids as predicted by the DSB repair model (Figure 1, E and F). The half-conversions occur in the 5:3 and 5:3 orientation for BIK1-939 and his4-ATC as expected for a co-half conversion of markers on opposite sides of the same DSB. (C) Represents bona fide full co-conversion events arising from the removal of both mispairs within the heteroduplex DNA from a single DSB repair event as illustrated in Figure 2 (E–I). Other bona fide co-conversions arising from the heteroduplex repair of B could have a half conversion at BIK1-939 accompanied by a full conversion at his4-ATC or vice versa (for example, see class 5a and class 677a in the supplementary appendix at http://www.genetics.org/supplemental/). (D) A co-event where the non-Mendelian segregation of the two markers (2:6 and 6:2) is most likely caused by two independent DSB repair events. Mispair removal of the heteroduplex in B cannot account for this type of tetrad. An example of a tetrad that contained three recombinant chromatids is shown in E. Such tetrads, as well as those that contained four recombinant chromatids, were deemed to have arisen from multiple DSB repair events and were excluded from further analysis. Classifications of all tetrads analyzed (supplementary appendix) are given in the supplementary tables at http://www.genetics.org/supplemental/.
TABLE 6.
One- and two-sided events at theHIS4 recombination hotspot
| Complexb
|
Single eventsf
|
||||||
|---|---|---|---|---|---|---|---|
| Strain | Total ATCa |
One sidedc | Two sidedd | Two DSBRe | One sided | Two sidedg | Potentialh |
| Wild-type | 144 | 28 (15/54) | 65 (35/54) | 7.0 (4/54) | 32 (29/90) | 63 (57/90) | 4.4 (4/90) |
| mlh1Δ/mlh1Δ | 116 | 26 (9/35) | 46 (16/35) | 28 (10/35) | 64i (52/81) | 31i (25/81) | 4.9 (4/81) |
| msh2Δ/msh2Δ | 96 | 34 (12/35) | 60 (21/35) | 5.7 (2/35) | 67i (41/61) | 28i (17/61) | 4.9 (3/61) |
The total number of NMS events at his4-ATC (abbreviated ATC) that were analyzed for NMS of BIK1-939.
Tetrads in which more than two spores were recombinant for the five genetic markers (e.g., Figure 4E).
The proportion of complex events in which his4-ATC but not BIK1-939 showed NMS.
The proportion of complex events in which both his4-ATC and BIK1-939 showed NMS.
Tetrads in which only two spores were recombinant and in which both his4-ATC and BIK1-939 showed NMS but not explicable as coming from a single DSB repair event (Figure 4D).
Tetrads in which no more than two spores were recombinant for the five genetic markers.
The proportion of single events in which both his4-ATC and BIK1-939 showed co-conversion (two-sided events).
Two-sided events in which both his4-ATC and BIK1-939 showed NMS according to the DSBR model, although, one additional marker also showed NMS.
Statistically significantly different from the wild-type strain (P < 0.025, z-test for proportions).
It was possible that the wild-type strain only appeared to contain more two-sided events compared to the mutant strains. This would be the case if a greater proportion of two-sided events in the mutants were complex events and therefore excluded from the analysis. In the wild type, approximately two-thirds of the complex events were two sided. Since the proportions of two-sided complex events in the mlh1Δ and msh2Δ strains were similar (Table 6), we conclude that eliminating complex events could not account for the decreased proportion of two-sided events in the mlh1Δ and msh2Δ strains.
Identification of true co-conversion events:
To identify true co-conversions, and thus two-sided events arising from a single DSB repair event, only tetrads in which no more than two spores were recombinant were analyzed. These tetrads were placed into the classes given in Table 6. Tetrads in which his4-ATC, but not BIK1-939, showed NMS were considered to be one-sided events. When his4-ATC and BIK1-939 segregated in a fashion predicted by the DSBR model (Figures 1 and 4), the DSBR event was considered two sided. If other markers showed a co-event, the DSBR event was considered potentially two sided. Similarly, when his4-ATC and BIK1-939 showed a co-event not predicted by the DSBR models, the co-event was considered two sided but not due to a single DSBR event. Using only the co-conversions to estimate the proportion of two-sided events, 63% of the events in the wild-type strain were two sided. In contrast, only 31% of events were two sided in the mlh1Δ and 28% were two sided in the msh2Δ strains (P < 0.025; z-test for proportions; Table 6). Thus, the decrease in the proportion of co-events in the two mutant strains was also reflected in the decrease in two-sided co-conversion events arising from a single DSB repair event.
Potential sources of error:
One reason for the apparent decrease in the proportion of co-conversions in the mutant strains may be the erroneous classification of events as two sided in the wild-type strain, since the wild type contained very few informative tetrads (half conversions at BIK1-939 as well as at his4-ATC). For example, the apparent two-sided events might have been the result of two independent DSB repair events that by chance looked like a co-conversion. However, from the number of events that unambiguously arose from multiple initiations (two-strand complex, Figure 4D and Table 6), we estimated the number of co-conversions of BIK1-939 and his4-ATC that were the consequence of two different meiotic DSBs and arose by chance. If two non-Mendelian segregation events arose from two independent DSB repair events, then a 6:2 (6 white strands:2 black strands) non-Mendelian segregation of his4-ATC should be equally likely to be associated with a 6:2 or a 2:6 non-Mendelian segregation of BIK1-939 (Figure 4, C or D). For example, tetrads in which his4-ATC shows a 6:2 NMS while BIK1-939 shows a 2:6 NMS (Figure 4D) can have come only from two initiations. Therefore, a number of his4-ATC and BIK1-939 co-conversions, where both markers converted in the same direction, for example, 6:2/6:2's (Figure 4C), could have arisen from two independent initiations. This number of apparent co-conversions is equivalent to the number of obvious co-events (Figure 4D). Since the wild-type strain had four such events, the 57 co-conversion events (Table 6) contained an equivalent number of co-events that were in the correct pattern by chance (and thus would mimic co-conversion events). Therefore, we estimated that only 53 of the 57 co-conversions were real co-conversions. This is still a significantly greater proportion than that predicted by chance (P < 0.05, χ2 goodness-of-fit test). In addition, the wild-type adjusted co-conversion frequency was significantly greater than that of the Mlh1p- and Msh2p-deficient mutant strains (compare 53/90 to 25/81 and to 17/61; P < 0.025, z-test for proportions; pairwise comparisons). The values for the mutant strains were not adjusted since their adjustment would only exacerbate the differences between wild type and mutants.
G-G mismatches are restored in the mlh1Δ and msh2Δ strains:
Since DSBs occur with equal frequency on both parental strands (data not shown), the frequency of 5:3/6:2 non-Mendelian segregations should equal the frequency of 3:5/2:6 non-Mendelian segregations, as was observed in the wild-type strain. However, in the mlh1Δ and msh2Δ strains there was a deficit of 5:3 half conversions compared to 3:5 half conversions at his4-ATC that were not compensated for by an increase in 6:2 full conversions (compare 35 to 65 and 17 to 36 in the mlh1Δ and msh2Δ strain, respectively, Table 5; P < 0.05, χ2 goodness-of-fit test, for both strains). This deficit of 5:3 half conversions suggested that G-G mismatches were restored in the mlh1Δ and msh2Δ strains. Assuming that we recovered all C-C mispairs as either 2:6 or 3:5's, then the total number of G-G mismatches formed (number of 5:3 + number of 6:2) should equal the total number of C-C mismatches formed (number of 3:5 + number of 2:6). From this we can estimate the rate of restoration of G-G pairs by dividing the number of “missing” G-G mismatches by the total number of G-G mismatches, estimated from the rate of C-C mismatches. In the mlh1Δ strain there were 40 observed G-G mismatches (35 tetrads in which his4-ATC segregated 5:3 and 5 in which they segregated 6:2; Table 5). Similarly, there were 72 C-C mismatches formed (65 and 7 tetrads, respectively, in which his4-ATC segregated 3:5 and 2:6). Thus, 32 G-G mismatches were missing, resulting in a restoration rate of 44% (32/72). Similarly, 41% of the G-G mismatches were potentially restored in the msh2Δ strain. Alternatively, if there is some restoration of C-C mispairs in the mutants, then the restoration rates of G-G are underestimated. It is impossible to determine whether C-C mispairs were restored from the genetic data. However, the apparent restoration of G-G mispairs raises the possibility that some of the two-sided events may have been processed to one-sided events in an Msh2p/Mlh1p-independent fashion (see discussion). A similar disparity has also been observed when other poorly repaired markers, such as palindromes, are used (Porter et al. 1993; Gilbertson and Stahl 1996; Merker et al. 2003; Jessop et al. 2005).
Deletion of MLH1 affects crossing over:
Tetrads containing two-sided or one-sided events were analyzed for crossing over (Table 7; see supplementary material at http://www.genetics.org/supplemental/, section 3, for classes included). If the crossover was not immediately adjacent to the genetic marker that showed an aberrant segregation, it was deemed incidental and excluded from the analysis (Merker et al. 2003). The map distance in the mlh1Δ strain was decreased compared to the wild type. This is in agreement with previous observations that mlh1Δ shows decreased crossing over (Hunter and Borts 1997). To ask whether mlh1Δ affects the frequency of crossing over associated with one- and/or two-sided events, we divided the events into four classes: one-sided events associated with crossovers, one-sided events associated with noncrossovers, two-sided events associated with crossovers, and two-sided associated with noncrossovers (Table 7). Both MMR mutant strains showed a difference in the distribution of events into those four classes compared to the wild-type strain (P < 0.05; G-test of homogeneity), reflecting that the mlh1Δ and msh2Δ strains contain more one-sided events. When we compared the distribution of mlh1Δ to that of the msh2Δ strain, we did not observe a significant difference. However, given the size of the data sets presented here, we may not have been able to detect any differences.
TABLE 7.
Distribution of crossovers and noncrossovers associated with NMS ofhis4-ATC
| % two sided
|
% one sided
|
|||||
|---|---|---|---|---|---|---|
| CO | NCO | CO | NCO | Total events | COa | |
| Wild type | 49 (35) | 27 (19) | 13 (9) | 11 (8) | 71 | 0.68 |
| mlh1Δ/mlh1Δb | 24 (15) | 14 (9) | 16 (10) | 46 (29) | 63 | 0.40c |
| msh2Δ/msh2Δb | 19 (9) | 13 (6) | 36 (17) | 32 (15) | 47 | 0.55 |
Events were classified as two sided with a crossover, two sided with a noncrossover, one sided with a crossover, or one sided with a noncrossover (for classification see supplementary section 3 at http://www.genetics.org/supplemental/). Incidental crossovers (supplementary Table 2 at http://www.genetics.org/supplemental/) were not included in this analysis. Events in which HPH or NAT co-converted were excluded as the crossover could not be mapped. CO, crossovers; NCO, noncrossovers.
Calculated as the number of two- and one-sided events associated with a crossover divided by the number of total events.
Distribution of events significantly different from the distribution of the wild-type strain (P < 0.025; G-test for homogeneity).
Frequency significantly decreased compared to the wild-type strain (P < 0.025; z-test for proportions).
Crossovers map to either side of the non-Mendelian segregation:
Foss et al. (1999) suggest that two-sided events are processed to a one-sided event (for example, an apparently simple NMS of his4-ATC) due to restoration using the nicks generated from Holliday junction cleavage (Figure 2, J–L). If this is correct, then crossovers associated with the apparent one-sided event should map between his4-ATC and BIK1-939 (interval II). As a test of the model, we analyzed the position of the crossovers associated with one-sided events. Of the 19 informative one-sided events in the two mutant strains, seven crossovers appeared to be incidental, mapping in either interval I or IV (supplementary Table 2 at http://www.genetics.org/supplemental/). Of the remaining crossovers, three mapped to interval II whereas nine mapped to interval III (between his4-ATC and HIS4-1605). Thus, the crossovers associated with one-sided events did not show a significant bias toward mapping in interval II. We conclude that the one-sided events associated with crossovers do not arise from Holliday junction resolution that was associated with restoration of BIK1-939.
Gene conversion tracts are short:
Previous studies at HIS4 have found evidence for long gene conversion tracts and break-induced replication (Merker et al. 2003 and references therein). To investigate whether this was also the case in this system, we analyzed co-events between HPH and BIK1-939 as well as NAT and HIS4-1605. In both cases, co-events were rare. Of all of the unselected tetrads analyzed in the three strains, only 2 tetrads of 314 showed co-events of BIK1-939 with HPH. In these 2 tetrads, NAT and HIS4-1605 also were co-events. Moreover, all of these segregations were 8:0 and thus clearly identifiable as multiple events. Therefore, we did not observe any co-conversions involving HPH-BIK1-939 and NAT-HIS4-1605. Hence, we did not observe any evidence of break-induced replication or very long gene conversion tracts. We also assessed the frequency of co-events between his4-ATC and HIS4-1605. Fewer than 10% of tracts extended as far as HIS4-1605 (1.6 kb from his4-ATC and ∼1.9 kb from the DSB). Thus, most gene conversion tracts at HIS4 were short.
DISCUSSION
MSH2 and MLH1 mutants have fewer two-sided events:
The loss of Msh2p/Mlh1p-mediated mismatch repair could have one of three possible outcomes, depending on its role(s) in recombinational DNA transactions. First, the proportion of two-sided events could be increased, as all heteroduplex DNA would be recovered (Figures 1 and 2) due to the absence of any mispair removal. Second, if the extent of 3′-tail invasion/assimilation were the sole determinant of sidedness, then deletion of Msh2p/Mlh1p should have no effect (Figure 2, A–D). Third, the proportion of two-sided events would be decreased if Msh2p/Mlh1p-directed mispair removal from the invading/captured strand's end “fixed” heteroduplex DNA as full conversions and in its absence they became subject to restorational repair (Figure 2, E–H). Finally, a similar decrease in two-sided events is expected if mismatch repair proteins actively promote two-sided events. In the absence of Msh2p/Mlh2p, we found that the proportion of two-sided events was significantly decreased, indicating that the latter possibilities are more likely.
Models for Msh2p/Mlh1p functions:
We can envision a number of mechanisms by which Msh2p/Mlh1p might promote two-sided events. In the first, Mlh1p and Msh2p modulate strand invasion such that a greater proportion of the 3′-tail invades the homolog and/or a greater proportion of heteroduplex DNA is formed upon strand capture. The Mer3p helicase has recently been demonstrated to carry out strand assimilation (Mazina et al. 2004) and has been proposed to promote crossovers. If Msh2p/M1h1p were to facilitate this function, one-sided events might be extended into two-sided events. However, this mechanism is inconsistent with previous proposals for the influence of mismatch repair proteins on heteroduplex formation (reviewed in Borts et al. 2000) as well as the lack of a crossover defect in msh2Δ strains. It is also inconsistent with the in vitro experiments demonstrating that the MutL/S complex disrupts RecA-mediated filament formation in the presence of mismatches (Worth et al. 1994, 1998).
A mechanism more consistent with the known activities of mismatch repair proteins is one in which Msh2p/Mlh1p promote two-sided events via mismatch removal from the invading strand by excision. This could lead to destabilization of the strand invasion structure. Such an event might necessitate that the other side of the DSB invades and more extensive heteroduplex DNA than normal is formed. In a wild-type strain, this will always lead to full conversion of any markers involved. In addition, when multiple mismatches are contained in the same heteroduplex, as might occur during SDSA, a second round of recombination extending the conversion tract might occur (Borts and Haber 1987). As these processes are dependent on mismatch repair proteins (Borts et al. 1990, 2000), the absence of Msh2p/Mlh1p would cause the proportion of two-sided events to be decreased. No physical evidence addresses these issues.
Foss et al. (1999) proposed that markers remaining in heteroduplex until resolution are subject to restoration. This implies a hierarchical removal of mispairs within heteroduplex DNA. During 3′-tail invasion and/or annealing, mispairs are removed using the invading end to direct the removal of the mispair, thus fixing the event as a conversion. When MMR is efficient, a two-sided event can occur (Figure 2, E–I). In contrast, should early removal of the mispair fail—for example, if MMR is absent or inactive—the ends generated from double Holliday junction resolution (Figure 2, J–L) or SDSA (Figure 1G) can be used to direct removal of the mispair. This “late” mispair removal, however, is likely to cause both restorations and conversions due to the positions of the ends. This model, based on data generated using poorly removed palindromes as genetic markers, requires that early and late mispair removal have different properties. Most importantly, the “early” mispair removal must be less able to recognize/remove a palindrome-containing mispair such that it is not converted. The late mispair removal must then be able to remove the palindrome such that restoration or conversion occurs. Both restorational and conversional mispair removal in the absence of Msh2p have been observed (Coic et al. 2000), and the data presented here provide additional, albeit indirect, evidence for restorational mispair removal.
The model of Foss et al. (1999) also predicts that for one-sided events associated with crossovers, the crossover should be positioned between his4-ATC and BIK1-939 for the nicks generated by Holliday junction resolution to have promoted a restoration of the BIK1-939 marker. This was clearly not the case. Thus, while the model by Foss et al. (1999) is formally possible, we suggest a model that involves restorational repair without assuming that nicks generated from Holliday junction resolution direct mispair removal. In addition, we suggest that Msh2p/Mlh1p-dependent repair is nick directed and is active both during strand invasion/assimilation (early) and after Holliday junction resolution (late: Figure 1, E and F, and Figure 2, J and K) and D-loop disassembly during SDSA (Figure 1G). This would lead predominantly to full conversions but also some restorations, depending on which late nicks are used to direct mispair removal (e.g., Figure 2, J–L). If SDSA prevails, some of the one-sided events may also be restored, thus leaving no sign of a DSB repair event. In the absence of the Msh2p/Mlh1p nick-directed repair pathway, a less efficient repair pathway may be able to remove some, but not all, of the mispairs in the heteroduplex DNA. However, rather than using nicks, this repair complex may create short, discontinuous heteroduplex repair tracts with no strand bias, as have been observed previously in msh2Δ (Coic et al. 2000). Without strand bias, there would be a 50% chance of a restoration. This is in good agreement with the observation that the mlh1Δ and msh2Δ strains contained approximately half the number of two-sided events compared to the wild-type strain (Table 6). This also agrees with the estimate that ∼40% of the G-G mispairs are restored.
Palindromes and mismatch repair mutants:
The proportion of two-sided events differs markedly between wild-type strains utilizing poorly repaired palindromes (Porter et al. 1993; Gilbertson and Stahl 1996; Jessop et al. 2005) and those using single-base mismatches as markers for heteroduplex formation (this study and Schultes and Szostak 1990). However, the proportion of two-sided events obtained in the mlh1Δ and msh2Δ strains with single-base-pair markers was similar to that obtained in wild-type strains using palindromes. There may be several explanations for this. First, palindromes and single nucleotide polymorphisms may have different effects on heteroduplex DNA formation. For example, palindromes may be less likely to be included in heteroduplex DNA. Thus, palindromes may cause events to be one-sided by limiting strand invasion. However, no experimental data have addressed this question. On the other hand, assuming that palindromes are indeed incorporated into heteroduplex DNA as frequently as single nucleotide mispairs, the heteroduplex repair of single nucleotide mispairs in MLH1 and MSH2 mutants may be analogous to the failure to remove palindromes in wild-type cells. If this were true, one would predict that for base-pair mismatches, the absence of Mlh1p/Msh2p should mimic the effect of palindromes and therefore have a greater proportion of one-sided events. This is indeed the case (Table 6). Whether palindromes are less frequently incorporated into heteroduplex DNA or subject to alternative repair remains to be determined.
In conclusion, we suggest that in wild-type cells the initial DSB repair event is two sided. The absence of MMR, by either mutation or use of poorly repaired palindromes, allows a second, unbiased mispair removal pathway to restore a proportion of heteroduplex, leading to apparent one-sided events. This is consistent with the observed disparity in the recovery of conversions of palindromes.
Acknowledgments
We thank Craig Griffin, Jette Foss, Frank Stahl, Michael Lichten, Lea Jessop, Victoria Cotton, and Ed Loius for critical reading of various manuscripts. We thank Jette Foss, Frank Stahl, Lea Jessop, Tom Petes, Ian Hickson, and the anonymous reviewers for helpful comments and discussions. This work was supported by the Wellcome Trust.
References
- Abdullah, M. F., and R. H. Borts, 2001. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah, M. F., E. R. Hoffmann, V. E. Cotton and R. H. Borts, 2004. A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet. Genome Res. 107: 180–190. [DOI] [PubMed] [Google Scholar]
- Alani, E., 1996. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 16: 5604–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani, A., R. A. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., and J. E. Haber, 1987. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science 237: 1459–1463. [DOI] [PubMed] [Google Scholar]
- Borts, R. H., M. Lichten and J. E. Haber, 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113: 551–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., W.-Y. Leung, K. Kramer, B. Kramer, M. S. Williamson et al., 1990. Mismatch repair-induced meiotic recombination requires the PMS1 gene product. Genetics 124: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Coic, E., L. Gluck and F. Fabre, 2000. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 19: 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz, N., U. H. Mortensen and R. Rothstein, 1997. Cloning-free PCR-based allele replacement methods. Genome Res. 7: 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Q., F. Xu and T. D. Petes, 1995. Meiosis-specific double-strand breaks at the HIS4 recombination hotspot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15: 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss, G. M., K. J. Hillers and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. II. A role for mismatch repair directed by biased resolution of the recombinational intermediate. Genetics 153: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson, L. A., and F. W. Stahl, 1996. A test of the double-strand break model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Goyon, C., and M. Lichten, 1993. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex is formed late in meiotic prophase. Mol. Cell. Biol. 13: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. I. Rejection and restoration of Mendelian segregation. Genetics 153: 555–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, E. R., P. V. Shcherbakova, T. A. Kunkel and R. H. Borts, 2003. MLH1 mutations differentially affect meiotic functions in Saccharomyces cerevisiae. Genetics 163: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1p is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., R. Neumann and V. Wilson, 1990. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell 60: 473–485. [DOI] [PubMed] [Google Scholar]
- Jessop, L., T. Allers and M. Lichten, 2005. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton and T. D. Petes, 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina, O. M., A. V. Mazin, T. Nakagawa, R. D. Kolodner and S. C. Kowalczykowski, 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51: implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- McKee, A. H., and N. Kleckner, 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker, J. D., M. Dominska and T. D. Petes, 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, K. K., M. A. White and T. D. Petes, 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340: 318–320. [DOI] [PubMed] [Google Scholar]
- Porter, S. E., M. A. White and T. D. Petes, 1993. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, S., A. Amon and F. Klein, 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla, T. A., D.-M. Christie and R. M. Liskay, 1994. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 14: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schultes, N. P., and J. W. Szostak, 1990. Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics 126: 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995 Biometrics. W. H. Freeman, San Francisco.
- Southern, E. M., 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98: 503–517. [DOI] [PubMed] [Google Scholar]
- Surtees, J., J. Argueso and E. Alani, 2004. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107: 146–159. [DOI] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Worth, L., Jr., S. Clark, M. Radman and P. Modrich, 1994. Mismatch repair proteins MutS and MutL inhibit RecA-catalysed strand transfer between diverged DNAs. Proc. Natl. Acad. Sci. USA 91: 3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth, L., Jr., T. Bader, J. Yang and S. Clark, 1998. Role of MutS ATPase activity in MutS, L-dependent block of in vitro strand transfer. J. Biol. Chem. 273: 23176–23182. [DOI] [PubMed] [Google Scholar]