Abstract
The population structure of domesticated species is influenced by the natural history of the populations of predomesticated ancestors, as well as by the breeding system and complexity of the breeding practices exercised by humans. Within Oryza sativa, there is an ancient and well-established divergence between the two major subspecies, indica and japonica, but finer levels of genetic structure are suggested by the breeding history. In this study, a sample of 234 accessions of rice was genotyped at 169 nuclear SSRs and two chloroplast loci. The data were analyzed to resolve the genetic structure and to interpret the evolutionary relationships between groups. Five distinct groups were detected, corresponding to indica, aus, aromatic, temperate japonica, and tropical japonica rices. Nuclear and chloroplast data support a closer evolutionary relationship between the indica and the aus and among the tropical japonica, temperate japonica, and aromatic groups. Group differences can be explained through contrasting demographic histories. With the availability of rice genome sequence, coupled with a large collection of publicly available genetic resources, it is of interest to develop a population-based framework for the molecular analysis of diversity in O. sativa.
ASIAN cultivated rice (Oryza sativa L.) holds a unique position among domesticated crop species in that it is both a critical food staple and the first fully sequenced crop genome. Rice is consumed as a grain almost exclusively by humans, supplying 20% of daily calories for the world population (World Rice Statistics, http://www.irri.org; FAOSTAT, http://apps.fao.org). As a model organism with a fully sequenced genome, rice affords unique opportunities to use genomic approaches to study its domestication, adaptive diversity, and the history of crop improvement.
Archeological evidence supports a similar time of domestication for rice, wheat (Triticum aestivum), and maize (Zea mays ssp. mays), 5–10,000 years ago, but the evolutionary histories of these cereals differ in several significant ways (Solheim 1972; Sharma and Manda 1980; Zohary and Hopf 2000; Piperno and Flannery 2001). Recent studies tracing the molecular evolution of maize offer several points of comparison that help illuminate the genetic history of rice. Unlike maize, rice is predominantly autogamous and, hence, gene flow is restricted. As a result, geographically or ecologically distinct groups of rice are expected to show greater genetic differentiation than would be the case in an outcrossing species. Because of fewer opportunities for cross-pollination, the structure of landraces in rice and maize is also predicted to be fundamentally different. A greater proportion of diversity is expected to reside in differences between homozygous lines within a heterogenous landrace in rice (Olufowote et al. 1997) compared to the distribution of diversity among heterozygous individuals within a landrace of maize (Labate et al. 2003). In addition, evidence suggests that the two primary subspecies of rice, indica and japonica, are the products of separate domestication events from the ancestral species, O. rufipogon, a hypothesis initially based on studies of biochemical traits (Second 1982) and hybrid sterility (Kato et al. 1928) and subsequently supported by molecular analyses (Doi et al. 2002; Cheng et al. 2003). This is in contrast to the single domestication event that led to the evolution of modern maize (Matsuoka et al. 2002).
At all levels of analysis, the differences between the indica and japonica subspecies are very apparent. Differences between nonsticky (indica) and sticky (japonica) rices are documented in Chinese literature as early as 100 AD (Matsuo et al. 1997). In ecogeographical terms, indica are primarily known as lowland rices that are grown throughout tropical Asia, while japonica are typically found in temperate East Asia, upland areas of Southeast Asia, and high elevations in South Asia. The traits that have been used to classify indica and japonica have included grain shape, phenol reaction, sensitivity to potassium chlorate, leaf color, and apiculus hair length, although the spectra of variation for any of these individual traits overlap in the two subspecies (Oka 1988).
Using RFLPs, the indica-japonica division was very clear (Wang and Tanksley 1989; Nakano et al. 1992; Zhang et al. 1992) but additional population structure consisting of the six varietal groups indica, japonica, aus, aromatic, rayada, and ashina was discerned using 15 isozyme loci (Glaszmann 1987). The aus, rayada, and ashina are minor groups that have generally been considered to be ssp. indica ecotypes, and all have a comparatively small geographic distribution along the Himalayan foothills. The drought-tolerant, early maturing aus rices are grown in Bangladesh during the summer season from March to June. Rayada and ashina are floating rices of Bangladesh and India, respectively. Aromatic rices such as basmati from Pakistan, Nepal, and India and sadri from Iran have a distinctive popcorn-like aroma and are highly prized for their quality. Because there has been no reliable way to distinguish ecotypes on the basis of phenotypic evaluation and because information about the varietal groupings is rarely available from genetic resource collections, a genetically based identification of groups is required to fully utilize these resources.
The purpose of this study is (1) to establish a population genetics framework for the evaluation of rice by characterizing the intraspecific divergence within a set of 234 rice accessions using simple sequence repeats (SSRs) and chloroplast sequence and (2) to address the evolutionary relationships among groups within the species. Intraspecific classification of rice has been of importance to rice geneticists and breeders, but with the advent of population genetics approaches, it is now feasible to examine the genetic basis of domestication, adaptation, plant development, and agricultural performance. SSR loci are particularly useful for the study of population structure and demographic history of domesticated species because their high level of allelic diversity facilitates the detection of the fine structure of diversity more efficiently than an equal number of RFLP, AFLP, or SNP loci. The specific goals of this study are to characterize population structure within O. sativa, to examine the differences between and relationships among genetically defined groups, and to analyze aspects of demographic history that may explain them. The resulting framework will be used to pose questions about the origin and diversity of gene pools that exist within cultivated Asian rice and to lay the foundation for characterizing the genes that distinguish them.
MATERIALS AND METHODS
Plant material:
We sampled 234 rice accessions representing the geographic range of O. sativa. The sample included accessions collected in Asia (187), the Americas (27), Africa (14), Europe (3), and Oceania (2). Information about the accessions used (accession name, accession number, seed source, country of origin, membership in one of the five model-based populations, accession number cited in supplemental Figure S1, and choloroplast haplotype) is listed in supplemental Table S1 at http://www.genetics.org/supplemental/. Aroma of rice leaves was evaluated using the protocol of Pinson (1994), modified to include warming the samples in a 67° water bath for 10 min prior to analysis.
Genomic DNA extraction and SSR genotyping:
DNA was extracted using a modified potassium acetate-SDS protocol (Dellaporta et al. 1983). The 169 nuclear SSRs employed to analyze population structure are published in supplemental Table S2 at http://www.genetics.org/supplemental/ (Chen et al. 1997; Temnykh et al. 2000, 2001; Coburn et al. 2002). PCR was performed as in Coburn et al. (2002) except that mixtures contained 20 ng template DNA, 4 pmol of forward and reverse primers, and 1 unit of Taq polymerase. Pooled PCR products, diluted to equalize signal strength, were size separated by capillary electrophoresis using an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA). SSRs were analyzed with GenScan 3.1.2 software (Applied Biosystems) and scored with Genotyper 2.5 software (Applied Biosystems). Genotype data for all accessions are available at http://ricelab.plbr.cornell.edu/publications/2005/garris/Genotype_Data/.
Chloroplast sequencing:
The plastid subtype-identity (PS-ID) sequence, which captures linker sequences between plastid genes rp116 and rp114, was amplified as reported by Nakamura et al. (1998). A second fragment, ORF100, is known to harbor length variation in rice and was amplified as in Nakamura et al. (1998), except that a new forward primer was designed to amplify a smaller fragment (5′ CAACCCACCCCATAAAATTG 3′). Quantified PCR product (10 μl) was treated with 10 units exonuclease I and 2 units shrimp alkaline phosphatase and incubated at 37° for 15 min followed by 80° for 15 min. Single-pass sequencing was performed by automated sequencing using an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA) at the Cornell BioResource Center (Ithaca, NY). Direct sequencing of PCR products resulted in a homozygous sequence. Sequences were aligned using Sequencher 4.0.5 (Gene Codes, Ann Arbor, MI) for base calling and CLUSTAL W (Thompson et al. 1994) with manual quality control for insertion/deletions. The ends of fragments were trimmed to remove low-quality sequence.
Statistical analysis:
Genetic distance was calculated using the C.S. Chord distance (Cavalli-Sforza and Edwards 1967) because it has been shown by analysis of simulations to generate correct tree topologies regardless of the microsatellite mutation model (Takezaki and Nei 1996). Phylogenetic reconstruction was based on the neighbor-joining method implemented in PowerMarker version 2.7 (Liu and Muse 2004; http://www.powermarker.net) In addition, the model-based program STRUCTURE (Pritchard et al. 2000; Falush et al. 2003) was used to infer population structure using a burn-in of 10,000, run length of 100,000, and a model allowing for admixture and correlated allele frequencies. Five independent runs yielded consistent results. Inferred ancestry for each accession and the key for identifying the accessions shown in the neighbor-joining tree are given in supplemental Table S1 and Figure S1 at http://www.genetics.org/supplemental/. The graphical display of the STRUCTURE results was generated using Distruct software (Rosenberg 2002; http://www.cmb.usc.edu/noahr/distruct.html). PowerMarker was used to calculate the average number of alleles, gene diversity, and polymorphism information content (PIC) values. FST, the correlation of alleles within subpopulations, was calculated using an AMOVA approach in Arlequin V2.000 (Weir 1996; Schneider and Excoffier 1999). To utilize analysis approaches that are based on the stepwise mutation model (SMM), a set of 60 SSR loci that behaved in a stepwise manner (<10% of alleles were at nonstepwise intervals) was identified (as indicated in the list of SSRs published in supplemental Table S2 at http://www.genetics.org/supplemental/). This set of loci was used for analysis of directional evolution and population bottlenecks. Average standardized allele sizes for analysis of directional evolution were calculated as in Vigouroux et al. (2003). Ascertainment bias was assessed by comparing the difference in allele lengths between ssp. indica and japonica when the markers were originally derived from cv. IR36 (indica, 67 markers) or cv. Nipponbare (japonica, 100 markers). Ascertainment bias was nonsignificant (t = 0.24, P-value = 0.83).
The program BOTTLENECK (Cornuet and Luikart 1996) was used to test each group for deviation from mutation-drift equilibrium under the SMM. This program conducts tests for recent (within the past 2Ne to 4Ne generations) population bottlenecks that severely reduce effective population size (Ne) and produce an excess in heterozygosity. Significance was determined by the sign, standardized differences, and Wilcoxon tests.
RESULTS
Genetic structure in rice:
Analysis of genetic distance and population structure provided evidence for significant population structure in rice. Analysis of these data, using STRUCTURE, produced the highest log likelihood scores when the number of populations was set at five, which was consistent with clustering based on genetic distance. Most accessions were classified into one of the five groups, which corresponded to indica (79), aus (20), aromatic (19), temperate japonica (45), and tropical japonica (44; Figure 1). In addition to the accessions that were clearly assigned to a single population, where >80% of their inferred ancestry derived from one of the model-based populations, 24 accessions (10%) in the sample were categorized as having admixed ancestry (Figure 2). While the majority of these were identified as admixture between temperate and tropical japonica groups, other admixture combinations were present as well (Figure 2; supplemental Table S1).
Figure 1.—
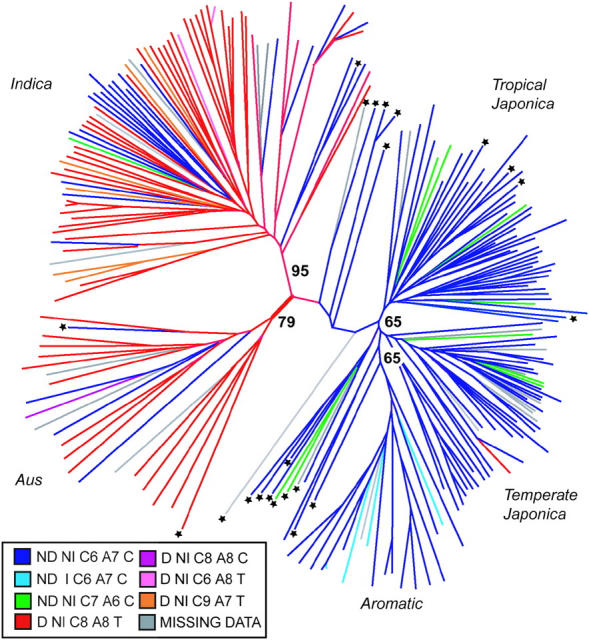
Unrooted neighbor-joining tree based on C.S. Chord (Cavalli-Sforza and Edwards 1967) based on 169 nuclear SSRs. The key relates the color of the line to the chloroplast haplotype based on ORF100 and PS-ID sequences. Admixed individuals are identified with an asterisk. Bootstrap values (out of 100) are indicated at the branch points.
Figure 2.—
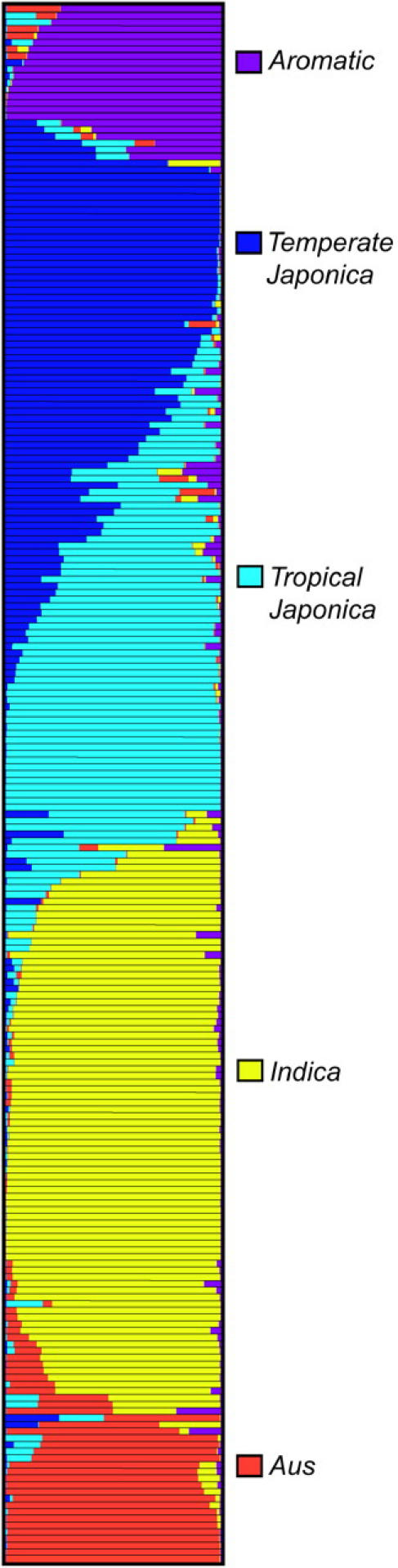
Model-based ancestry for each accession. Color codes are as follows: aromatic, purple; aus, orange; indica, yellow; temperate japonica, dark blue; and tropical japonica, light blue.
The overall AMOVA indicates that 37.5% of the variation was due to differences among groups with the remaining 62.5% due to differences within groups. Pairwise estimates of FST using the AMOVA approach indicated a high degree of differentiation between the five model-based groups with values ranging from 0.20 to 0.42 (Table 1). Lower levels of differentiation were observed in pairwise comparisons of temperate with tropical japonica (0.20) and aus with indica (0.25).
TABLE 1.
PairwiseFST values and AMOVA
| Aromatic | Aus | Indica | Temperate japonica | Tropical japonica | |
|---|---|---|---|---|---|
| Aromatic | 0.34 | 0.39 | 0.37 | 0.31 | |
| Aus | 0.28 | 0.26 | 0.44 | 0.37 | |
| Indica | 0.34 | 0.25 | 0.45 | 0.39 | |
| Temperate japonica | 0.29 | 0.42 | 0.43 | 0.22 | |
| Tropical japonica | 0.23 | 0.35 | 0.36 | 0.20 |
AMOVA-based estimates appear above the diagonal and classical estimates (θ) appear below the diagonal. For AMOVA-based estimates, P < 0.000001 for 110 permutations for all population comparisons.
Chloroplast diversity:
As an alternative method to assess the relationships among populations, two plastid loci were examined. Overall, eight chloroplast haplotypes based on five polymorphic sites (two indels, one SNP, and a polyC/polyA region) were in the PSID and ORF100 fragments (see supplemental Table S1 at http://www.genetics.org/supplemental/; Figure 1). The indica subpopulation contained the most chloroplast diversity, harboring seven of the eight haplotypes and encompassing all the chloroplast diversity found in the temperate and tropical japonica groups. Four of the eight haplotypes were observed in aus chloroplasts, and these represented the most frequent indica haplotype as well as one found in higher frequency in the japonicas. Only two haplotypes were found in the japonica subpopulations and both were shared between the temperate and tropical groups. The aromatic rices share a more recent maternal ancestor with the japonica, consistent with their position based on nuclear SSRs, but 15% of aromatic rices also contained a 4-bp deletion in the ORF100 fragment that was unique to this group.
Nuclear diversity:
The amount and organization of genetic diversity differed among the model-based populations (Table 2). The indica and tropical japonica groups contained a high percentage of polymorphic loci (99%) and an average of 7.26 and 6.09 alleles per locus, respectively. Even with a much smaller sample size, the aus group had very high diversity with 98% of loci polymorphic and an average of 5.1 alleles per locus. These three groups also had the highest heterozygosity values (0.55 for indica, 0.54 for aus, and 0.47 for tropical japonica). The temperate japonica and aromatic groups had lower diversity with 91 and 88% polymorphic loci and 4.9 and 3.4 alleles per locus, respectively, and lower He values (0.39 for both temperate japonica and aromatic). The temperate japonica and aromatic populations also had the highest incidence of monomorphism, with 15 and 21 monomorphic loci, respectively. Interestingly, the alleles at all 15 monomorphic loci in the temperate japonica group were identical in size to the most frequent allele among the tropical japonica. This observation is consistent with the hypothesis that temperate japonica rices were derived from tropical japonica. For 15 of the 21 monomorphic loci in the aromatic sample, the allele was identical in size to the most frequent allele in the tropical japonica, and this was often the most frequent or only allele in the temperate japonica.
TABLE 2.
Summary of polymorphism for total data set and by population
| All | Aus | Indica | Aromatic | Temperate japonica | Tropical japonica | |
|---|---|---|---|---|---|---|
| Sample size | 234a (7.35) | 21 (0.990) | 79 (2.92) | 19 (1.85) | 41 (2.24) | 48 (1.73) |
| Average no. of alleles/locus | 11.8 (7.35) | 5.1 (2.73) | 7.3 (4.44) | 3.4 (2.00) | 4.9 (3.68) | 6.1 (3.90) |
| Average gene diversity | 0.7 (0.16) | 0.54 (0.23) | 0.55 (0.24) | 0.39 (0.26) | 0.39 (0.28) | 0.47 (0.27) |
| Average PIC value | 0.67 (0.18) | 0.52 (0.22) | 0.52 (0.24) | 0.38 (0.24) | 0.37 (0.27) | 0.46 (0.26) |
Standard deviations are indicated in parentheses.
Includes admixed.
Directional evolution in allele length:
It has been proposed that there is an upward bias in the number of repeats responsible for the hypervariability of SSRs that would lead to larger average allele sizes in “derived” groups (Rubinsztein et al. 1995). This has been shown to be true in the comparison of humans and nonhuman primates (Rubinsztein et al. 1995) and in nonancestral populations of maize (Vigouroux et al. 2003). Using the framework established by the population structure analysis, comparisons of allele lengths between groups using the subset of 60 SSR loci that have evolved in a stepwise fashion resulted in statistically significant differences among some populations of rice (Table 3). The average standardized allele lengths in the indica, aus, and aromatic groups were significantly smaller than those in the temperate and tropical japonica groups although the allele lengths in the indica, aus, and aromatic groups were not statistically different from each other. Furthermore, in the comparison between temperate and tropical japonica, the average standardized allele size is greater in the former (t = 9.31, P < 0.0001), supporting the hypothesis that the temperate japonica group is derived from the tropical japonica group.
TABLE 3.
Differences in average standardized allele sizes for pairs of populations
| Aromatic | Aus | Indica | Temperate japonica | Tropical japonica | |
|---|---|---|---|---|---|
| Aromatic | 1.1824 (NS) | 1.0143 (NS) | 13.1320* | 6.3997* | |
| Aus | 0.4801 (NS) | 11.4668* | 6.6454* | ||
| Indica | 6.2148* | 4.0691* | |||
| Temperate japonica | 9.3128* | ||||
| Tropical japonica |
NS, not significant (P > 0.05).
P < 0.0001.
Population bottlenecks:
The observed differences in diversity among the rice populations suggest differences in demographic history that have shaped these patterns. To assess the effect of historical population sizes in the distribution of diversity, we examined the five model-based populations for evidence of recent bottlenecks. A likely cause of differences in the effective population sizes of the rice groups is the proximity, duration, and severity of population bottlenecks. Deviation of allelic diversity and heterozygosity from mutation-drift equilibrium under the SMM was assessed to determine whether any of the genetic populations had recently experienced a bottleneck. Analysis of a set of 60 dinucleotide SSR markers that exhibited stepwise mutation patterns revealed strong evidence of bottlenecks for the aus, aromatic, temperate japonica, and tropical japonica populations. These data did not support a recent bottleneck in the indica population. No estimate is currently available for the mutation rate of SSRs in rice, which would assist in the estimation of the time since the divergence of these groups.
DISCUSSION
Genetic structure has been previously documented in rice (Glaszmann 1987; Parsons et al. 1999; Ni et al. 2002), but this analysis combines a large number of accessions (234) with a large number of loci (169). The O. sativa rice accessions sampled show significant differentiation into five groups: aromatic, aus, indica, temperate japonica, and tropical japonica. This deep genetic structure is, in part, a legacy of structure in ancestral rice populations. Analysis of sequence divergence between cv. GLA4, an indica cultivar and cv. Nipponbare, a temperate japonica, suggests that these two groups diverged ∼440,000 years ago, supporting the hypothesis that indica and japonica are derived from independent domestication events from an ancestral rice that had already differentiated into (at least) two gene pools (Cai and Morishima 2002; Ma and Bennetzen 2004; Yamanaka et al. 2004). Sequence divergence between the chloroplast genomes of cv. 93-11, an indica, and cv. PA64S, an indica-like variety with a japonica chloroplast, yielded a divergence time of 86,000–200,000 years ago (Tang et al. 2004). These results suggest that the divergence between indica and japonica in our sample is in part due to differentiation of ancestral O. rufipogon populations in different locations and at different times. Rice presents a contrast to the history of domestication of maize, which involved a single domestication event with a clear geographic center and expansion to the north and south (Matsuoka et al. 2002). As sequence comparisons in rice are enlarged to include representatives of each subpopulation, the relationships among the groups can be clarified and the times of divergence estimated.
The deep genetic structure in rice may also be an effect of the autogamous breeding system. In self-pollinated species, one would predict a greater partitioning of diversity among rather than within populations in the absence of human-mediated gene flow between populations by breeding. Indeed, the large amount of variation attributable to differences between groups in rice (37.5% in this study) can be compared to results of a comparable sample of maize inbred lines, in which only 8.3% of the variation was due to differences between groups (Liu et al. 2003).
While both breeding system and domestication history have had large effects on the structuring of diversity in rice, the independent population histories of the groups have also shaped the gene pools. For example, the indica is a diverse group relative to the others with no evidence of a genetic bottleneck. The source of this variation could include mitigation of the domestication bottleneck by gene flow with sympatric wild relatives or a historically larger effective population size due to overland dispersal routes.
The aus had high diversity values relative to its sample size and, like the indica, contained several chloroplast haplotypes. Aus rices were traditionally grown in a short summer season in Bangladesh under rainfed conditions (Parsons et al. 1999). Adaptation to flowering under long days required evolution of day length neutrality, fostering temporal reproductive isolation and divergence. Although the aus types have a historically smaller geographical distribution and receive less attention than indica and japonica rices in breeding programs, their drought tolerance and early maturity are adaptive traits that could be usefully targeted in breeding applications.
The temperate and tropical japonica have a very close genetic relationship and have overall lower genetic diversity than indica (Glaszmann 1987; Zhang et al. 1992; Ni et al. 2002) as well as larger standardized allele lengths. In contrast to indica, which was able to utilize land routes for migration, many of the tropical japonica in our sample were collected from the islands of Indonesia and the Philippines where migration via islands could have acted to decrease diversity by a chain of bottlenecks. In addition, the two japonica groups represent an adaptive spectrum of an ancient subpopulation from tropical origins to temperate latitudes, with the necessary adaptations to environmental signals such as day length and temperature. As the only pairwise comparison that embodies such obvious adaptation to a new environment, the temperate and tropical japonica groups offer a valuable tool for studying the genetic basis of adaptation. The statistical significance of the larger allele size in the temperate relative to the tropical japonica group supports the hypothesis that temperate japonica were derived from the tropical japonica group. One explanation for the differences in average allele lengths is a higher mutation rate in the temperate population. Previous observations of enhanced transposable element activity in temperate compared to tropical japonica groups (Jiang et al. 2002) suggest that this hypothesis may be worthy of further investigation.
Previously described as intermediate between indica and japonica rice (Ahuja et al. 1995), aromatic rice forms a distinct subgroup in this and other studies (Jain et al. 2004). Both the nuclear and the chloroplast data demonstrate a close relationship to japonica. The aromatic group had a high proportion of monomorphic loci suggestive of a severe or recent bottleneck (Nagaraju et al. 2002; this study). The genetics of aroma may contribute to the apparent genetic bottleneck in this group (Lorieux et al. 1996; Garland et al. 2000) but this question awaits further research.
In addition to the groups identified by this analysis, 10% of individuals show evidence of mixed population ancestry. In some cases these admixed individuals are likely to be the result of modern breeding; in other cases they may be landraces belonging to groups that were underrepresented in our sample. For example, Ashina and rayada rices (isozyme-based varietal groups III and IV) composed only 1% of all 1688 varieties sampled by Glaszmann (1987), and their adaptation to deep water conditions makes them less amenable to ex situ conservation. The identities of some admixed individuals could perhaps be better resolved through deliberate addition of deep water rices to the data set. The public availability of the genotypic data presented here should facilitate further characterization of rice population structure and diversity and highlights the need for complementary research on the regional and national levels.
Using this framework of genetically defined populations, it may be possible to exploit the rice gene pools more effectively with population genetics-based approaches using the extensive collections of rice genetic resources. In particular, different subpopulations are likely to provide differing levels of resolution for association mapping studies (Garris et al. 2003) as well as different allele frequencies associated with desirable traits for plant improvement. In an evolutionary context, many of the most intriguing questions remain to be answered, such as to what extent allelic distribution in O. sativa is shaped by these populations, whether a predomestication divergence between indica and japonica can be detected in O. rufipogon and O. nivara ancestral groups, and whether comparisons among populations will help identify loci showing footprints of selection. Studies designed to address these and other questions will lead to a better understanding of the processes of domestication and adaptation in this cultivated, inbreeding species.
Acknowledgments
We thank D. Mackill of the International Rice Research Institute in the Philippines, H. Bockelman of the National Small Grains Collection in Aberdeen, Idaho, R. Dilday and J. N. Rutger of the Dale Bumpers National Rice Research Center, and K. Moldenhauer of the Rice Research and Extension Center in Stuttgart, Arkansas, for seeds; E. Septiningsih for developing the RM623 primer pair; S. Harrington for editing spellings and checking accession numbers in supplemental Table S1; and L. Swales for formatting. We also thank J. Edwards and E. Buckler for critical reading of the manuscript prior to submission. This research was supported by U.S. Department of Agriculture (USDA) National Research Initiative Competitive Grants Program 00-35300-9216 (T.H.T. and S.R.M.) and Current Research Information System project 6225-21000-006 (T.H.T.). A.J.G. was supported by USDA/Cooperative State Research Service competitive grant 97-35300-5101, representing Food and Agricultural Sciences National Needs Graduate Fellowship in Plant Biotechnology.
References
- Ahuja, S. C., D. V. S. Panwar, U. Ahuja and K. R. Gupta, 1995 Basmati Rice: The Scented Pearl. Directorate of Publications, C.C.S. Haryana Agricultural University, Hisar, Haryana, India.
- Cai, H. W., and H. Morishima, 2002. QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L. L., and A. W. F. Edwards, 1967. Phylogenetic Analysis: Models and Estimation Procedures. Am. J. Human Genet. 19: 233–257. [PMC free article] [PubMed] [Google Scholar]
- Chen, X., S. Temnykh, Y. Xu, Y. G. Cho and S. R. McCouch, 1997. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95: 553–567. [Google Scholar]
- Cheng, C. Y., R. Motohashi, S. Tsuchimoto, Y. Fukuta, H. Ohtsubo et al., 2003. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20: 67–75. [DOI] [PubMed] [Google Scholar]
- Coburn, J. R., S. V. Temnykh, E. M. Paul and S. R. McCouch, 2002. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L.). Crop Sci. 42: 2092–2099. [Google Scholar]
- Cornuet, J. M., and G. Luikart, 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S. L., H. Wood and J. B. Hicks, 1983. A plant DNA minipreparation. Plant Mol. Biol. Rep. 1: 19–21. [Google Scholar]
- Doi, K., K. Sobrizal, K. Ikeda, P. L. Sanchez, T. Kurakazu et al., 2002 Developing and evaluating rice chromosome segment substitution lines, pp. 275–287 in IRRI Conference September 16–19, 2002. International Rice Research Institute, Beijing, China.
- Falush, D., M. Stephens and J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland, S., L. Lewin, A. Blakeney, R. Reinke and R. Henry, 2000. PCR-based molecular markers for the fragrance gene in rice (Oryza sativa L.). Theor. Appl. Genet. 101: 364–371. [Google Scholar]
- Garris, A., S. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann, J. C., 1987. Isozymes and classification of asian rice varieties. Theor. Appl. Genet. 74: 21–30. [DOI] [PubMed] [Google Scholar]
- Jain, S., R. Jain and S. McCouch, 2004. Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor. Appl. Genet. 109: 965–977. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, S. Temnykh, Z. Cheng, J. Jiang et al., 2002. Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 161: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, S., H. Kosaka and S. Hara, 1928. On the affinity of rice varieties as shown by the fertility of rice plants. Central Agricultural Institute of Kyushu Imperial University 2: 241–276. [Google Scholar]
- Labate, J. A., K. R. Lamkey, S. E. Mitchell, S. Kresovich, H. Sullivan et al., 2003. Molecular and historical aspects of corn belt dent diversity. Crop Sci. 43: 80–91. [Google Scholar]
- Liu, K., and S. Muse, 2004 PowerMarker: New Genetic Data Analysis Software, Version 2.7 (http://www.powermarker.net).
- Liu, K. J., M. Goodman, S. Muse, J. S. Smith, E. Buckler et al., 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M., M. Petrov, N. Huang, E. Guiderdoni and A. Ghesquiere, 1996. Aroma in rice: genetic analysis of a quantitative trait. Theor. Appl. Genet. 93: 1145–1151. [DOI] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T., Y. Futsuhara, F. Kikuchi and H. Yamaguchi, 1997 Science of the Rice Plant. Food and Agriculture Policy Research Center, Tokyo.
- Matsuoka, Y., Y. Vigouroux, M. M. Goodman, G. J. Sanchez, E. Buckler et al., 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju, J., M. Kathirvel, R. R. Kumar, E. A. Siddiq and S. E. Hasnain, 2002. Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc. Natl. Acad. Sci. USA 99: 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, I., H. Urairong, N. Kameya, Y. Fukuta, S. Chitrakon et al., 1998. Six different plastid subtypes were found in O. sativa-O. rufipogon complex. Rice Genet. Newsl. 15: 80–81. [Google Scholar]
- Nakano, M., A. Yoshimura and N. Iwata, 1992. Phylogenetic study of cultivated rice and its wild relatives by RFLP. Rice Genet. Newsl. 9: 132–134. [Google Scholar]
- Ni, J., P. M. Colowit and D. J. Mackill, 2002. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 42: 601–607. [Google Scholar]
- Oka, H. I., 1988 Origin of Cultivated Rice. Elsevier, Tokyo.
- Olufowote, J. O., Y. B. Xu, X. L. Chen, W. D. Park, H. M. Beachell et al., 1997. Comparative evaluation of within-cultivar variation of rice (Oryza sativa L.) using microsatellite and RFLP markers. Genome 40: 370–378. [DOI] [PubMed] [Google Scholar]
- Parsons, B. J., H. J. Newbury, M. T. Jackson and B. V. Ford-Lloyd, 1999. The genetic structure and conservation of aus, aman and boro rices from Bangladesh. Genet. Res. Crop Evol. 46: 587–598. [Google Scholar]
- Pinson, S. R. M., 1994. Inheritance of aroma in 6 rice cultivars. Crop Sci. 34: 1151–1157. [Google Scholar]
- Piperno, D. R., and K. V. Flannery, 2001. The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proc. Natl. Acad. Sci. USA 98: 2101–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A., 2002 Distruct: A Program for the Graphical Display of Structure Results (http://www.cmb.usc.edu/~noahr/distruct.html).
- Rubinsztein, D. C., W. Amos, J. Leggo, S. Goodburn, S. Jain et al., 1995. Microsatellites are generally longer in humans compared to their homologs in nonhuman-primates—evidence for directional evolution at microsatellite loci. Nat. Genet. 10: 337–343. [DOI] [PubMed] [Google Scholar]
- Schneider, S., and L. Excoffier, 1999. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Second, G., 1982. Origin of the genic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isozyme loci. Jpn. J. Genet. 57: 25–57. [Google Scholar]
- Sharma, G. R., and D. Manda, 1980 Excavations at Mahagara 1977–1978: A Neolithic Settlement in Belan Valley (Archaeology of the Vindhyas and Ganga Valley 6). Department of Ancient History, Culture, and Archeology, Allahabad, India.
- Solheim, W. G., 1972. An earlier agricultural revolution. Sci. Am. 266: 34–41. [Google Scholar]
- Takezaki, N., and M. Nei, 1996. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J. B., H. A. Xia, M. L. Cao, X. Q. Zhang, W. Y. Zeng et al., 2004. A comparison of rice chloroplast genomes. Plant Physiol. 135: 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh, S., W. D. Park, N. Ayres, S. Cartinhour, N. Hauck et al., 2000. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100: 697–712. [Google Scholar]
- Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour et al., 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W—Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, Y., Y. Matsuoka and J. Doebley, 2003. Directional evolution for microsatellite size in maize. Mol. Biol. Evol. 20: 1480–1483. [DOI] [PubMed] [Google Scholar]
- Wang, Z. Y., and S. D. Tanksley, 1989. Restriction fragment length polymorphism in Oryza sativa L. Genome 32: 1113–1118. [Google Scholar]
- Weir, B. S., 1996 Genetic Data Analysis II: Materials for Discrete Population Genetic Data. Sinauer Associates, Sunderland, MA.
- Yamanaka, S., I. Nakamura, K. N. Watanabe and Y. I. Sato, 2004. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 108: 1200–1204. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. F., M. A. S. Maroof, T. Y. Lu and B. Z. Shen, 1992. Genetic diversity and differentiation of Indica and Japonica rice detected by Rflp analysis. Theor. Appl. Genet. 83: 495–499. [DOI] [PubMed] [Google Scholar]
- Zohary, D., and M. Hopf, 2000 Domestication of Plants in the Old World. Oxford University Press, Oxford.


