Abstract
The transport of nitrate into prokaryotic and eukaryotic cells, of considerable interest to agriculture, ecology, and human health, is carried out by members of a distinct cluster of proteins within the major facilitator superfamily. To obtain structure/function information on this important class of nitrate permeases, a collection of chemically induced mutations in the nrtA gene encoding a 12-transmembrane domain, high-affinity nitrate transporter from the eukaryote Aspergillus nidulans was isolated and characterized. This mutational analysis, coupled with protein alignments, demonstrates the utility of the approach to predicting peptide motifs and individual residues important for the movement of nitrate across the membrane. These include the highly conserved nitrate signature motif (residues 166–173) in Tm 5, the conserved charged residues Arg87 (Tm 2) and Arg368 (Tm 8), as well as the aromatic residue Phe47 (Tm 1), all within transmembrane helices. No mutations were observed in the large central loop (Lp 6/7) between Tm 6 and Tm 7. Finally, the study of a strain with a conversion of Trp481 (Tm 12) to a stop codon suggests that all 12 transmembrane domains and/or the C-terminal tail are required for membrane insertion and/or stability of NrtA.
NITRATE is a key source of nitrogen for a large number of microorganisms and plants. While nitrate limitation frequently confines the growth of these organisms, especially in natural environments, its use in fertilizers to improve plant crop yield may give rise to (i) eutrophication of natural water systems and (ii) human and animal health concerns (reviewed by Crawford and Glass 1998; Daniel-Vedele et al. 1998; Williams and Miller 2001). Futhermore, the central importance of nitrate is highlighted by reports that it is involved in certain plant metabolic and morphogenic processes (Scheible et al. 1997; Zhang and Forde 1998).
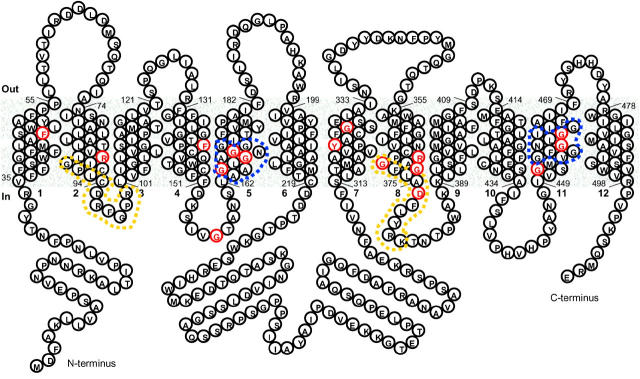
The influx of nitrate into cells is an active process since it occurs against a nitrate gradient (Brownlee and Arst 1983; Forde 2002; Vidmar et al. 2000). At least two classes of nitrate transport systems, high affinity and low affinity, have been identified (Trueman et al. 1996). The Aspergillus nidulans nrtA (formerly crnA) gene (Brownlee and Arst 1983) encodes a membrane protein that belongs to a family of high-affinity nitrate transporters (Unkles et al. 1991, 2001; Forde 2000). A typical secondary structure (Figure 1) was proposed for the 57-kD (507-amino-acid) NrtA protein in which 12 hydrophobic transmembrane domains (Tm's) in α-helical conformation pass through the membrane and connect by hydrophilic loops (Lp). The NrtA homologs belong to a distinct cluster, namely the Nitrate-Nitrite Porter family (TC 2.A.1.8), within the major facilitator superfamily (MFS; TC 2.A.1), which comprises a range of functionally diverse proteins, including mammalian and bacterial sugar transporters (Nelissen et al. 1997; Saier et al. 1999). In common with other MFS transporters, the motif Gly X X X Asp X X Gly X Arg can be observed around Lp 2/3 with a less well-conserved version around Lp 8/9, probably reflecting the proposed duplication of the first six Tm's, which is thought to have occurred early in the evolution of MFS proteins (Pao et al. 1998). One unique feature of high-affinity nitrate transporters is the presence of the consensus sequence Ala Ala Gly X Gly Asn X Gly Gly Gly, residues 163–172 (Figure 1) in Tm 5, described as the nitrate signature with a similar sequence repeated within Tm 11 (Trueman et al. 1996; Forde 2000). Other characteristics are the conserved charged residues within otherwise hydrophobic helices, Arg87 (Tm 2) and Arg368 (Tm 8), or aromatic residue Phe47 (Tm 1), which can be observed within NrtA (Forde 2000). A final feature of A. nidulans NrtA (although common among MFS transporters) is its substantial (∼96 residues) central loop (Lp 6/7), which, although present, is much reduced in the corresponding plant proteins (maximum length of 32 residues). Instead, a long hydrophilic C terminus of 69 residues is observed in plant NrtA-like proteins (Trueman et al. 1996; Forde 2000).
Figure 1.—
Provisional secondary structure model of the high-affinity nitrate transporter NtrA of A. nidulans. Model predicted through assessing the distribution and pattern of charged and hydrophobic amino acids using the TopPred program of Claros and von Heijne (1994) as implemented by Deveaud and Schuerer (Pasteur Institute: http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html). Predicted Tm segments of NrtA were refined by reference to the 52-sequence multiple alignment. Residues indicated in red denote very high conservation (>95%) observed by comparison of the 52 amino acid sequences from bacteria (21), fungi (5), algae and plants (26). Yellow dashes enclose the MFS motifs and blue dashes surround the nitrate signatures. The accession numbers of the 52 sequences used in alignments from which the data in Figure 1 are compiled are T51836, CAC05338, AAC35884, AB015472, AAF00053, AAF00054, Y08210, AY038800, AF0047718, AJ292342, AAC49531, AF091115, AF288688, AF332214, AF091116, AY053452, U34290, AB008519, ABO15472, T48982, AAF78499, AAK59570, Z25438, AY026523, AF135038, AF135039, AJ238664, Z69783, P22152, AF453778, B8G12-170, AAG45172, NP_241478, X15996, P37758, Z70792, CAB72205, AE004604, AAD22549, AAK22599, P42432, AAC06542, Z81360, AF149772, T37042, BAB58550, U40014, P46907, YI5252, AAG34371, CAC48822, and CAB65479.
Complex hydrophobic membrane transporters are inherently difficult to purify and crystallize, clearly an obstacle to the study of their structure. Notwithstanding, three high-resolution MFS protein structures have recently been solved for the oxalate-formate transporter (Hirai et al. 2002), the glycerol-3-phosphate inorganic phosphate transporter (GlpT; Huang et al. 2003), and the lactose transporter (LacY; Abramson et al. 2003). A striking feature of these structures is that the overall architecture of each of the proteins representing different MFS families is very similar, even superimposable (Abramson et al. 2004; Hirai and Subramaniam 2004). A general theme is the presence of a central substrate binding site within a hydrophilic pore, access to which alternates from outside to inside and vice versa by the flexible movement of Tm's. Details of the substrate binding site define the substrate specificity. In this respect and together with the fact that virtually nothing is known about NrtA function, we have used chemical mutagenesis to change NrtA residues and study their effect on protein expression and transport. Such a strategy has yielded valuable information on the Escherichia coli LacY permease (Bailey and Manoil 1998). The NrtA permease from the lower eukaryote A. nidulans is a particularly useful model for studying nitrate transport structure/function relationships due to the amenability of this organism to combined genetic and biochemical approaches. Furthermore, as MFS transporters make up a considerable proportion of membrane proteins in eukaryotes (Ward 2001), information on the NrtA branch of this important family should be of wider significance in our understanding of eukaryotic transport proteins.
MATERIALS AND METHODS
A. nidulans strains and media:
Standard wild-type (with regard to nitrogen regulation) strains with various color markers used for the isolation of nrtA mutants by chemical mutagenesis in this study were (i) biA1, (ii) yA2 pyroA2, (iii) fwA1, and (iv) chA1. A multi-copy nrtA strain, designated SS1, was identified (in an attempt to generate a transformant overexpressing the nitrate transporter) on the basis of supersensitivity to chlorate toxicity at a concentration of chlorate (10 mm) not toxic to the wild type with urea (10 mm) as the sole nitrogen source (S. E. Unkles, unpublished results). Strain nrtAgfp8 contains NrtA fused at the C terminus to green fluorescent protein (GFP; Cormack et al. 1997) encoded by a single-copy construct integrated at the argB locus (D. A. Rouch and S. E. Unkles, unpublished results). Routine Aspergillus growth media and handling techniques were as described before (Clutterbuck 1974).
E. coli strains, plasmids, and media:
Standard procedures were used for propagation of plasmids as well as for subcloning in E. coli strain DH5α. E. coli strain BL21 (DE3; Novagen, Madison, WI) was used to express a Lp 6/7 protein (residues 219–317) from a PCR-generated product ligated into pET21a (Novagen; E. R. DaSilva, unpublished results).
Molecular methods:
DNA was isolated using a Nucleon BACC2 kit (Amersham Pharmacia Biotech, Little Chalfont, UK). The conditions used during Southern blot analysis were as described previously (MacCabe et al. 1990). Following PCR amplification of overlapping fragments using primer pairs (nrtA1.1 and nrtA1.2, nrtA2.1 and nrtA2.2, nrtA3.1 and nrtA3.2, nrtA4.1 and nrtA4.2, nrtA5.1 and nrtA5.2, and nrtA6.1 and nrtA6.2) shown in Table 1, the entire nucleotide sequence of the A. nidulans nrtA mutants was determined by automated sequence analysis as described before (Unkles et al. 1997).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| nrtA1.1 and nrtA1.2 | TTCTACGAACTGCAGTTCC and TCCTTCAGTCCGGTTGTC |
| nrtA2.1 and nrtA2.2 | TGCTTGCATTCCTCTCATG and TGAGGTAACGAGGCCG |
| nrtA3.1 and nrtA3.2 | AGTCTTTATCGGCCTACTG and GCAAGTGAAGAGCATGCC |
| nrtA4.1 and nrtA4.2 | GTTGGGACAGCCAACTC and AGCAGGCGTAGGGGACT |
| nrtA5.1 and nrtA5.2 | CTCCCGCAAGGAGGCTT and AGTAAGACCAAACATAGTCG |
| nrtA6.1 and nrtA6.2 | CAATGGGTTTCTCAGATCC and CCCAAACGCCTCTTGAG |
Sequences are shown 5′ to 3′.
Isolation of chemically induced nrtA mutants:
Wild-type strains (with regard to nitrate assimilation) were treated with chemical mutagens, which preferentially induce single mutations in GC nucleotide base pairs. After mutagenesis with N-methyl-N′-nitro-N-nitrosoguanidine (NTG; Adelberg et al. 1965) or 4-nitroquinoline-1-oxide (NQO; Bal et al. 1977), mutants were isolated on the basis of resistance to chlorate toxicity (200 mm) with uric acid as the sole source of nitrogen (Cove 1976a). This approach takes advantage of the fact that mutations in the untranscribed strand are much less efficiently repaired than those in the transcribed strand, thereby favoring changes, for example, in glycine (G-rich codons) rather than proline (C-rich) residues. In most mutant isolation experiments, sodium deoxycholate (0.08%) was included in the medium to reduce colony size and to increase the number of colonies screened per petri dish. nrtA mutants were distinguished from other chlorate-resistant nitrate-assimilation-defective strains by their ability to utilize nitrate (Cove 1976b).
The generation of nrtA nrtB double mutants:
A number of nrtA mutant strains were crossed to nrtB110, a mutant strain that contains a deletion in the gene encoding the other nitrate transporter, NrtB, in A. nidulans (Unkles et al. 2001). Similar to nrtA loss-of-function mutants, strain nrtB110 grows normally on 10 mm nitrate as the sole sourse of nitrogen. Putative nrtA nrtB double mutants (i.e., strains that grew poorly on nitrate) were verified by Southern blot analysis of the nrtB to detect the deletion mutation nrtB110 (Unkles et al. 2001) and PCR amplification followed by DNA sequencing of the nrtA mutant gene to ensure the presence of the nrtA mutation.
Net nitrate transport assays:
Strains were grown for 6.5–7.5 hr at 37° in liquid minimal medium with 5 mm urea as the sole source of nitrogen (Cove 1966). The inducer of nrtA expression, sodium nitrate (10 mm), was added 100 min prior to harvesting by filtration. Assays were carried out as described by Brownlee and Arst (1983).
Western blotting:
Growth and induction conditions were as above. The resulting spore germlings were washed with cold sterile distilled water and frozen in liquid nitrogen. Crude plasma membrane preparations were made by grinding ∼300 mg pressed wet weight of the spore germlings in liquid nitrogen and suspending the powder in 10 ml extraction buffer [250 mm sucrose, 5% (v/v) glycerol, 1 mm magnesium chloride, 1 mm EDTA, 25 mm MOPS, pH 7.2] at 4° containing 100 μm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and one complete mini protease inhibitor cocktail tablet (Roche Diagnostics, Mannheim, Germany) per 10 ml extraction buffer. The suspension was centrifuged at 2000 × g for 10 min at 4°, and the supernatant was collected and centrifuged at 18,000 × g for 30 min at 4°. The gelatinous pellet was resuspended in 150 μl extraction buffer and stored at −80°. Protein samples (50 μg) were electrophoresed on 10% acrylamide gels (Laemmli 1970) and transferred to nitrocellulose membrane (Towbin et al. 1979). Blots were blocked by incubation overnight at 4° in TBS (137 mm sodium chloride, 20 mm Tris, pH 7.5) containing 5% (w/v) membrane blocking agent (Amersham Pharmacia Biotech). NrtA was detected by incubation of the blot with 1:500 anti-NrtA antibody (Unkles et al. 2004) in TBS containing 0.5% (w/v) membrane blocking agent for 2 hr at room temperature and after washing with TBST [TBS containing 0.1% (v/v) Tween 20] with 1:40,000 horseradish-peroxidase-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hr at room temperature. After further washing with TBST, peroxidase activity was visualized using ECL plus reagents (Amersham Pharmacia Biotech) and Hyperfilm ECL (Amersham Pharmacia Biotech). For GFP detection, peroxidase-conjugated polyclonal anti-GFP antibodies were purchased from Santa Cruz Biotechnology and used at a concentration of 1:500.
RESULTS
Conserved residues:
An alignment of 52 available NrtA sequences was analyzed using Clustal W (Thompson et al. 1994) and refined by eye. These sequences included proteins from a diverse range of organisms, ranging from eubacteria and archaebacteria to algae, fungi, and plants. Sixteen very highly conserved residues (defined on the basis of 50 of 52 conserved, i.e., >95%) are indicated in red (Figure 1). These residues are Phe47 (50/52), Arg87 (52/52), Phe140 (52/52), Gly157 (52/52), Gly165 (51/52), Gly167 (51/52), Gly170 (52/52), Tyr323 (52/52), Gly328 (52/52), Arg368 (52/52), Gly371(52/52), Gly372 (52/52), Asp376 (52/52), Gly452 (52/52), Gly458 (51/52), and Gly461 (51/52).
nrtA mutant isolation:
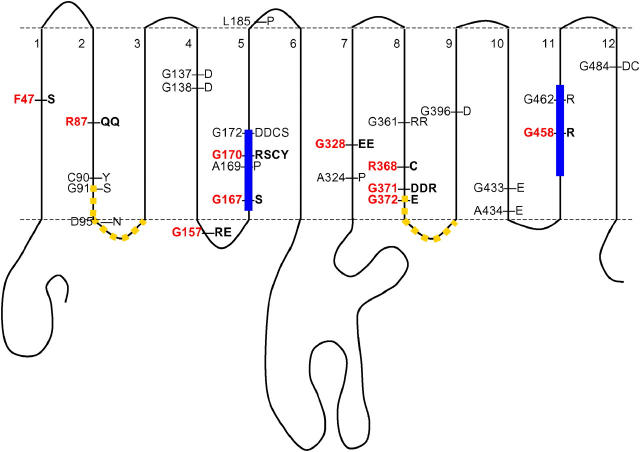
The nrtA gene from 57 nrtA mutants was completely sequenced and, of these, 38 were found to be missense mutants; the remainder were represented by eight deletions, six chain termination, two intron boundary, and three insertions. The location and distribution of residues altered in the 38 missense mutants are presented in Table 2 and Figure 2. Amino acid replacements in all Tm domains except Tm 3 and Tm 6 were observed and, as predicted (see materials and methods), a substantial number of glycine and arginine alterations (33/38) were observed. A large proportion of missense mutations (i.e., 10) occurred within Tm 5 and all these (26% of the entire collection of missense mutants) were found to locate within the nitrate signature motif. Nine of these replacements were found in just three glycine residues, including residues Gly167 and Gly170, which are very highly conserved in diverse organisms (Figure 1). Interestingly, Gly172, which is conserved among the eukaryotes only, is altered in four mutants (cysteine, serine, or aspartate in two independent mutants). There are 46 glycine residues in the entire protein, and so with 28 glycine-altering mutations, it would be expected that three glycine residues would be “hit” with a combined frequency of 1.8 on a random basis (or less than one mutation in each of the three codons). This estimation is based on the assumption that mutation events are entirely random (i.e., independent of DNA sequence conservation) and that only those resulting in a phenotype are recognized.
TABLE 2.
Characteristics ofnrtA mutants
| Allele | Mutagen | Nucleotide change |
Residue substitution |
Positiona | Type of change | Net nitrate uptakeb |
Expressionc |
|---|---|---|---|---|---|---|---|
| nrtA348P | NTG | TTC → TCC | F47S | Tm 1 | Nonpolar → polar | 3.50 ± 1.06 | + |
| nrtA26P, (nrtA19F)c | NTG (NQO) | CGA → CAA | R87Q | Tm 2 | Loss of charge | 4.02 ± 0.35 | + |
| nrtA334P | NTG | TGC → TAC | C90Y | Tm 2 | SH → polar | 3.26 ± 1.38 | + |
| nrtA2015F | NQO | GGC → AGC | G91S | Tm 2 | Conservative | 5.13 ± 1.20 | + |
| nrtA2036F | NQO | GAT → AAT | D95N | Lp 2/3 | Loss of charge | 6.48 ± 0.90 | + |
| nrtA303P | NTG | GGC → GAC | G137D | Tm 4 | Gain of charge | 3.97 ± 0.58 | + |
| nrtA94P | NTG | GGC → GAC | G138D | Tm 4 | Gain of charge | 3.66 ± 0.66 | + |
| nrtA2020F | NQO | GGG → CGG | G157R | Lp 4/5 | Gain of charge | 2.29 ± 0.38 | + |
| nrtA62P | NTG | GGG → GAG | G157E | Lp 4/5 | Gain of charge | 3.62 ± 0.59 | + |
| nrtA7P | NTG | GGT → AGT | G167S | Tm 5 | Conservative | 6.89 ± 0.78 | + |
| nrtA2107F | NQO | GCT → CCT | A169P | Tm 5 | Conservative | 3.36 ± 1.28 | + |
| nrtA13 | NTG | GGT → CGT | G170R | Tm 5 | Gain of charge | 3.66 ± 0.55 | + |
| nrtA337P | NTG | GGT → AGT | G170S | Tm 5 | Conservative | 3.03 ± 0.45 | + |
| nrtA2049F | NQO | GGT → TGT | G170C | Tm 5 | Polar → SH | 5.28 ± 1.75 | + |
| nrtA2046F | NQO | GGT → TAT | G170Y | Tm 5 | Conservative | 3.72 ± 1.26 | + |
| nrtA946, (nrtA55P)d | NTG (NTG) | GGT → GAT | G172D | Tm 5 | Gain of charge | 5.73 ± 0.80 | + |
| nrtA2003F | NQO | GGT → TGT | G172C | Tm 5 | Polar → SH | 4.22 ± 0.03 | + |
| nrtA2082F | NQO | GGT → AGT | G172S | Tm 5 | Conservative | 3.34 ± 0.61 | + |
| nrtA2 | NTG | CTC → CCC | L185P | Lp 5/6 | Conservative | 3.69 ± 0.24 | + |
| nrtA2002F | NQO | GCC → CCC | A324P | Tm 7 | Conservative | 4.97 ± 0.11 | + |
| nrtA14P, (nrtA1087)d | NTG (NTG) | GGG → GAG | G328E | Tm 7 | Gain of charge | 5.94 ± 0.10 | + |
| nrtA2044 (nrtA45P)d | NQO (NTG) | GGG → AGG | G361R | Tm 8 | Gain of charge | 4.60 ± 0.65 | + |
| nrtA8 | NTG | CGT → TGT | R368C | Tm 8 | Loss of charge | 3.79 ± 0.79 | + |
| nrtA301P (nrtA9M)d | NTG | GGT → GAT | G371D | Tm 8 | Gain of charge | 2.85 ± 0.67 | + |
| nrtA2031F | NQO | GGT → CGT | G371R | Tm 8 | Gain of charge | 4.03 ± 1.10 | + |
| nrtA335P | NTG | GGA → GAA | G372E | Tm 8 | Gain of charge | 3.28 ± 0.80 | + |
| nrtA14M | NTG | GGT → GAT | G396D | Tm 9 | Gain of charge | 5.03 ± 1.90 | + |
| nrtA2104 | NQO | GGG → GAG | G433E | Tm 10 | Gain of charge | 3.90 ± 0.79 | + |
| nrtA14F | NQO | GCA → GAA | A434E | Tm 10 | Gain of charge | 4.03 ± 0.30 | + |
| nrtA23P | NTG | GGG → AGG | G458R | Tm 11 | Gain of charge | 7.06 ± 0.03 | + |
| nrtA2043 | NQO | GGT → CGT | G462R | Tm 11 | Gain of charge | 3.97 ± 1.17 | + |
| nrtA19P | NTG | GGT → GAT | G484D | Tm 12 | Gain of charge | 5.13 ± 0.60 | + |
| nrtA2059F | NQO | GGT → TGT | G484C | Tm 12 | Polar → SH | 4.98 ± 1.50 | + |
| Wild type | NA | NA | NA | NA | NA | 10.31 ± 0.60 | + |
| nrtA1 | NTG | GGA → ▪GAe | 125 | Lp 3/4 | NA | 4.15 ± 0.05 | − |
| nrtA747 | NTG | CTA → ▪TAe | 85 | Tm 2 | NA | 3.50 ± 0.20 | − |
| nrtA40P | NTG | TGG → TGA | Y481*f | Tm 12 | NA | ND | − |
| nrtA52P | NTG | GCC → GCCC | A489 →g | Tm 12 | NA | ND | − |
SH, sulfhydryl group of cysteine; NA or ND, not applicable or not done.
Positions are predicted as described in the legend to Figure 1.
The basal level in a loss-of-function nrtA mutant is ∼3–4 nmol min/min/mg under the standard employed for net nitrate uptake (Brownlee and Arst 1983). The results are the mean of three independent experiments ± standard deviation.
The presence or absence of NrtA protein estimated by Western blot is indicated by + or −, respectively.
Two independently isolated mutants with identical changes. The first is the strain for which transport activity was determined.
The black box represents a single nucleotide deletion.
The asterisk represents a stop codon.
A base-pair addition (C:G) at residue 489 results in an altered amino acid sequence after residue Ala489. This altered sequence reads RVYLCLLGSACAEKSDEGVVDRVIGFSRGVWVRFCSVLFSDMIP*.
Figure 2.—
Position of altered residues. Red denotes residues that are very highly conserved (see Figure 1). Yellow dashes represent the position of MFS motifs and blue bars the position of nitrate signatures.
One missense mutation was found to affect Tm 1, and four were located within Tm 2, including two in the highly conserved residue Arg87, two were located in Tm 4, and three were located in Tm 7. Seven mutations occurred within the nucleotides encoding residues of Tm 8, including the highly conserved Arg368. The other six mutants were represented by alterations to glycine residues, Gly361 and the highly conserved Gly371 and Gly372 residues, with Gly371 being changed in three independent mutants. Of the remaining mutants, one mutation was located within Tm 9 and two each were observed in Tm's 10, 11 (including highly conserved Gly458), and 12. With regard to the loops, one missense mutation was observed to affect Lp2/3 (a MFS motif), two mutations altered the highly conserved Gly157 in Lp 4/5, and one change affected Lp 5/6 while none was observed within the large central Lp 6/7.
Six chain termination mutants were observed in our collection of mutants. These were distributed somewhat randomly across the gene and included a mutant, nrtA40P, in which a stop codon replaced Trp481 in Tm 12. This strain was the subject of expression analysis (see below).
Finally, 12 deletions or insertions were identified and included strains used in previous studies vis-à-vis nrtA1, formerly crnA1 (Tomsett and Cove 1979; Brownlee and Arst 1983; Unkles et al. 1991), and nrtA747 (Unkles et al. 2001), both of which were found to have a single base-pair deletion—nrtA1 at nucleotide position 481, codon 125 (Lp 3/4), and nrtA747 at nucleotide position 361, codon 85 (Tm 2). Other deletions included nrtA1009, nrtA1010, nrtA17, nrtA65P, nrtA343P, nrtA2039F, and nrtA24Y, while insertions occurred in mutants nrtA3 and nrtA97P. Noteworthy is mutant nrtA52P, which has a single-base-pair addition near the 3′-end of the coding region (described in Table 2, footnote g) and was used in antibody studies.
Resistance to chlorate toxicity:
Missense mutants isolated on the basis of chlorate resistance with urea as the nitrogen source were tested for resistance to 200 mm chlorate on other nitrogen sources. All mutants remained similarly resistant as the loss-of-function strains nrtA1 and nrtA747 with proline and urea and equally sensitive as the wild-type strain (and as sensitive as nrtA1 and nrtA747) on 10 mm glutamate or 10 mm arginine (Unkles et al. 2001).
Net nitrate uptake:
Most of the missense mutants showed similar net transport basal levels (Table 2) as the deletion strains nrtA1 and nrtA747 (i.e., ∼3–4 nmol/min/mg compared with ∼10–12 nmol/min/mg in the wild-type strain), this basal level in loss-of-function nrtA mutants being due to the nitrate uptake contribution by the NrtB transporter (Unkles et al. 2001). Consequently, such missense mutations were regarded as NrtA loss of function. In addition, a number of mutants (namely nrtA2015F, nrtA2036F, nrtA7P, nrtA2049F, nrtA946, nrtA2002F, nrtA14P, nrtA2044, nrtA14M, nrtA23P, nrtA19P, and nrtA2059F) possessed net nitrate uptake above the basal level (Table 2).
nrtA nrtB double mutants:
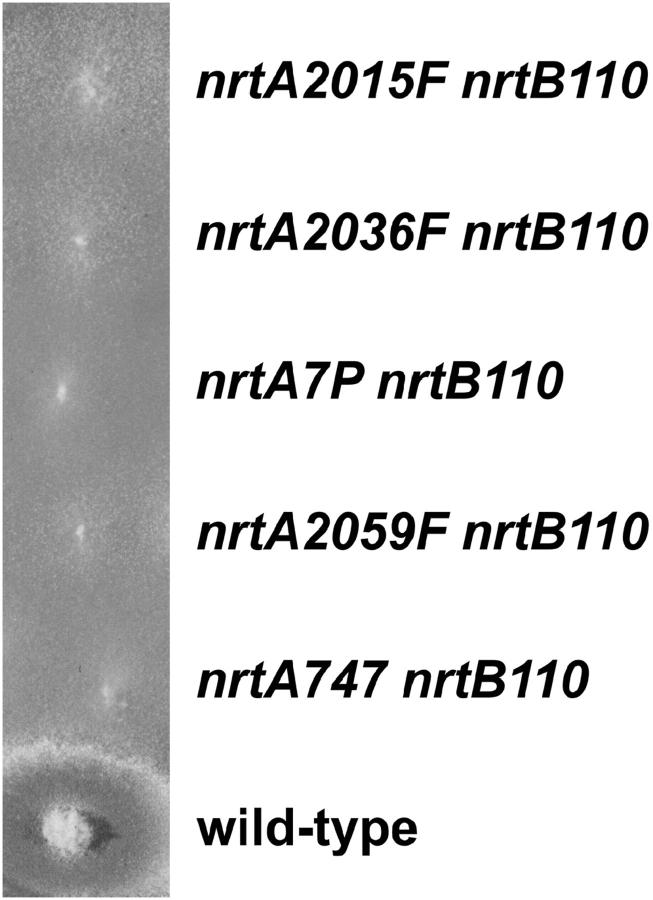
To determine if the higher level of net nitrate uptake in nrtA single-mutant strains nrtA2015F, nrtA2036F, nrtA7P, nrtA2049F, nrtA946, nrtA2002F, nrtA14P, nrtA2044, nrtA14M, nrtA23P, nrtA19P, and nrtA2059F was due to NrtA activity per se, the mutants were crossed one by one to a deletion mutant (nrtB110) in the other A. nidulans nitrate transporter, NrtB (Unkles et al. 2001). Double mutants were identified and verified by molecular technology (see materials and methods). These failed to grow on 10 mm or even 100 mm nitrate, similar to double-deletion mutant strain nrtA747 nrtB110 (Figure 3). Furthermore, negligible net nitrate uptake (i.e., 0 nmol/min/mg values) was observed in these double-mutant strains, similar to double-deletion mutant strain nrtA747 nrtB110.
Figure 3.—
Mutant growth tests. nrtA nrtB double-mutant strains were compared with the wild-type strain for their ability to grow on minimal medium containing 100 mm nitrate as sole nitrogen source. Strains representative of the growth response of all the double mutants are shown.
NrtA expression:
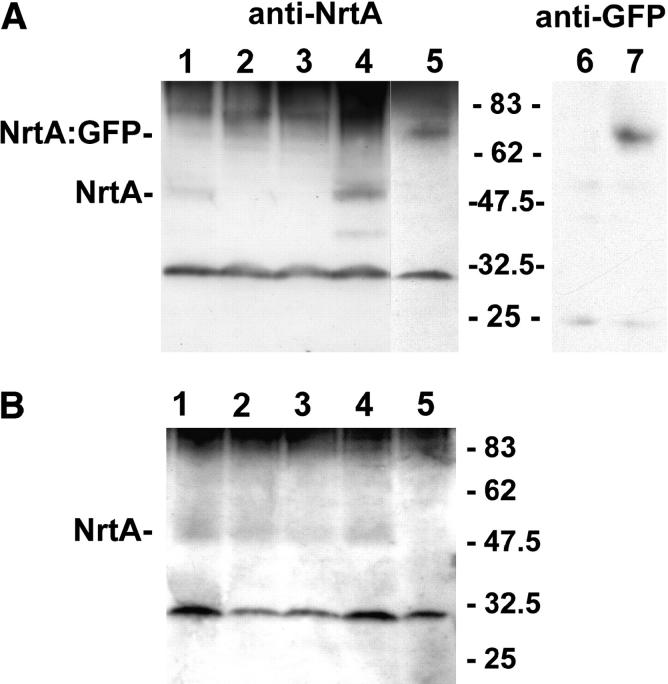
To determine if mutant proteins were stably expressed, we assayed, by Western blotting, steady-state levels in germlings (induced by nitrate) grown 6.5 hr (Figure 4). Low levels of NrtA protein (∼50 kD instead of the predicted 57 kD) could be observed using anti-NrtA antibodies in the wild type (Figure 4A, lane 1) but none were observed in chain termination mutants nrtA747 (lane 2) or nrtA40P (lane 3). The anti-NrtA antibodies also detected Lp 6/7 protein expressed in E. coli (S. E. Unkles and E. R. DaSilva, unpublished results). In contrast, the chain termination mutants nrtA747 (Figure 4A, lane 2) and nrtA1 (S. E. Unkles, unpublished results) lacked the 50-kD signal, as might be expected since these proteins, if expressed, would be devoid of the antibody recognition sites. However, neither chain termination mutant nrtA40P (Figure 4A, lane 3) nor mutant nrtA52P (S. E. Unkles, unpublished results), with an addition of 44 codons following codon 489 at the 3′-end, possessed detectable NrtA protein. In a preparation from strain nrtAgfp8, which contains a single copy of the NrtA:GFP fusion construct at the argB locus, a protein of ∼72 kD (instead of the expected 81 kD) was revealed (Figure 4A, lane 5). Anti-GFP antibodies also detected this 72-kD protein in the preparation from nrtAgfp8 (Figure 4A, lane 7) but not, as expected, in the wild-type protein sample (Figure 4A, lane 6). Confirmation that the 50-kD band was correctly identified as NrtA was obtained with the observation of a high-intensity signal of that molecular size in transformed strain SS1 containing multiple copies of nrtA (Figure 4A, lane 4). Using these anti-NrtA antibodies, all missense mutants such as nrtA8 (Figure 4B, lane 1), nrtA2 (lane 2), and nrtA2082 (lane 3) were shown to have similar low but observable levels of NrtA protein as in the wild type (lane 4). Again, no 50-kD protein was observed in the negative control sample for this blot, i.e., in nrtA747 (lane 5).
Figure 4.—
Analyses of NrtA expression of by Western blot. (A) Lane 1, wild-type biA1; lane 2, nrtA747; lane 3, nrtA40P; lane 4, SS1; lane 5, nrtAgfp8; lane 6, wild-type biA1; lane 7, nrtAgfp8. (B) Lane 1, nrtA8; lane 2, nrtA2; lane 3, nrtA2082F; lane 4, wild-type biA1; lane 5, nrtA747. Growth of strains is described in materials and methods. A band of 30 kD was observed with the anti-NrtA antibody preparation in all strains and was also present when preimmune serum was used in Western blots. The positions of molecular size markers (in kilodaltons) are shown.
DISCUSSION
A mutational survey of NrtA giving clues to the residues that may play a role in nitrate transporter function not only for fungi but also for higher plants, which are less amenable to such genetical approaches, has been accomplished. Chemical mutagens were employed to target GC base pairs yielding mutant strains with a recognizable phenotype, i.e., chlorate resistance, thus identifying possibly important residue positions for activity of NrtA.
One clear hotspot for mutations was observed within the first nitrate signature of Tm 5, where residues Gly167, Ala169, Gly170, and Gly172 were represented by a total of 10 substitutions. This relatively high frequency of change, together with the very high level of conservation of Gly167 and Gly170, suggests that this motif is crucial for function, and even conservative substitutions, for example, glycine to serine or cysteine, result in loss of function. The preponderance of such compact residues with small side chains and intolerance of even moderate increase in side-chain bulk suggests a requirement for particularly tight helix packing in the region of the nitrate signature of Tm 5.
Two arginine residues, Arg87 and Arg368, are found within the otherwise hydrophobic helices of Tm 2 and Tm 8, respectively. These are the only positively charged residues conserved within Tm regions in all nitrate transporters and both were represented by mutations in this study, two resulting in Arg87 conversion to Gln and one in which Arg368 was changed to Cys. Since such mutations involved a loss of charge, it would appear that a positive charge at these positions is necessary for transport function.
Interestingly, helical wheel analysis of Tm's in which several mutations have been identified (in Tm 2, Tm 5, Tm 7, and Tm 8) showed that all of the mutations in Tm's 2, 7, and 8 locate on one face of the Tm (S. E. Unkles, unpublished results). This distribution of mutations fits well within the context of known MFS transporter structures where Tm's 1, 2, 4, and 5 of the N-terminal half of the protein, along with Tm's 7, 8, 10, and 11 of the C-terminal portion, form the substrate translocation pore. Thus in NrtA also, Tm's 2, 7, and 8 appear to have a specific orientation with those residues altered in this study, probably facing the substrate channel. Of key interest among the conserved residues targeted by our chemical mutagenesis are Arg87 in Tm 2 and Arg368 in Tm 8 since their positively charged side chains have the potential to interact directly with the nitrate anion. Unlike Tm's 2, 7, and 8, however, residue positions of Tm 5 mutations are scattered throughout the helix, supporting a structural role for these glycine and alanine residues by virtue of their small side-chain volume, rather than a specific function in nitrate translocation. Finally, it is noteworthy that although Tm 3 possesses three glycine residues, no mutants in this helix were recovered, probably reflecting the nonessentiality of these residues. Indeed, in terms of known MFS structures, Tm 3 along with Tm's 6, 9, and 12 are embedded within the membrane. As such they are unlikely to participate directly in transport and perhaps therefore are more tolerant of minor or localized conformational changes, which would not have been recognized by our selection for loss of function.
The aromatic residue Phe47 is the only Tm 1 residue conserved in all species studied thus far and the only Tm 1 change (to Ser) present in our missense mutant collection. Interestingly, Tm 1 has a high proportion of aromatic residues (10/21), several of which are highly conserved within eukaryotic nitrate transporters, forming the motif Phe X X X Trp X X Phe X X X Phe X X X Phe/Tyr from position 36 to 51. Helical wheel analysis again places Phe47 on the same face as the other aromatic residues in this motif, suggesting that these bulky side chains may have a function equivalent to aromatic residues of GlpT whose role is to close the translocation pore following binding of substrate (Huang et al. 2003). Alternatively, Phe47 (and its symmetrical equivalent, the highly conserved aromatic amino acid Tyr323 in Tm 7) may be positioned in such a way as to constrain the flexibility of the long side chains of Arg87 and Arg368 within the translocation pore. Either way, alteration of the bulky Phe47 to a compact residue such as serine might be expected to lead to loss of function.
With regard to the functionality of loops, not unexpectedly perhaps, alterations were observed in the sequence Gly X X X Asp X X Gly X Arg (residues 91–100 in A. nidulans) of Lp 2/3 and vicinity, a well-conserved motif in the MFS superfamily. A conservative polar alteration to Ser of the highly conserved residue Gly91 (predicted to lie near the border of Tm 2 and the first residue of the conserved Lp2/3 MFS motif) results in concomitant reduction of transport activity, suggesting that this residue is essential in NrtA. In the lactose transporter of E. coli, replacement of the equivalent glycine residue with amino acids of increased bulk (other than alanine) resulted in marked reduction of activity (Jessen-Marshall et al. 1995). A similar sensitivity to bulk could account for the mutant phenotype observed with Gly372 (to Asp) within the repeated MFS motif at Lp 8/9 and possibly also for the adjacent Gly371 (represented by three mutations to Glu or Asp) although the effect may be due to charge introduction. In addition, a mutant with an Asp95-to-Asn modification was isolated in this study. This aspartate residue is thought to be critical for the function of this MFS motif as a conformationally versatile region (Pazdernik et al. 2000). Within Lp 4/5, Gly157 is conserved (among a peptide stretch of lower similarity) in all nitrate transporters studied and modification of this residue (two independent changes to Arg or Glu) was noted. Although six glycine and four arginine residues are present in the poorly conserved large central loop (Lp 6/7), the fact that no missense mutations were found in the stretch of DNA encoding this loop might suggest that individual residues within Lp 6/7 play no crucial functional role.
Of the remaining mutants, alterations were observed in two of the glycine residues (Gly137 and Gly138) of Tm 4, which compose a motif somewhat loosely conserved in many MFS proteins including sugar transporters (Henderson 1991). Further glycine residues altered within Tm's included the conserved residues Gly328 in Tm 7, Gly361 in Tm 8 (represented by two independent mutations), Gly433 in Tm 10, two glycines of the repeated nitrate signature motif of Tm 11 (Gly458 and Gly462 with two mutations), and Gly484 in Tm 12 as well as the nonconserved Gly396. However, all of these changes introduce a bulkier residue with a positive or negative charge into the Tm, which may cause major local structural perturbation, and so it cannot be deduced whether these glycine residues per se are necessary for transport. The exception is Gly484 for which one of the changes was to Cys, which might be expected to be tolerated.
The NrtA protein was expressed in all missense mutants (but not in chain termination mutants; see below), as detected by Western blots, at approximately similar levels although it is difficult to be precise due to the low levels of NrtA expression. Extensive in vitro mutagenesis of the E. coli LacY protein has revealed that only 5% of missense mutations result in lack of expression of the lacY protein (Bailey and Manoil 1998; Frillingos et al. 1998) and so expression of all the mutant proteins generated in this study, representing just 5% of NrtA residues, might be expected. Among the six chain termination mutants observed, the mutant phenotype obtained by alteration of Trp481 within Tm 12 (mutant nrtA40P) was of particular interest. Together with nrtA52P in which the C-terminal 18 residues were replaced by 44 residues, these were the only mutants encoding epitopes recognized by our anti-NrtA antibodies that had no detectable protein in Western blots. This suggests that all 12 transmembrane domains and/or the short C-terminal tail are required for NrtA insertion into the membrane and/or protein stability.
While most of the missense mutants showed net transport basal levels of ∼3–4 nmol/min/mg, similar to deletion strains nrtA1 or nrtA747 (compared with ∼10–12 nmol/min/mg in the wild-type strain), certain missense mutants possessed activity above the basal level. Surprisingly, double mutants of these and strain nrtB110 (a deletion mutation within the other nitrate transporter NrtB) showed neither growth on even high concentrations of nitrate nor significant detectable net nitrate transport. Clearly, therefore, these nrtA mutants are NrtA loss-of-function, enhanced levels of activity observed in the single missense mutants perhaps due to increased NrtB expression or possibly to a gain-of-function mutation in another transporter, alternatives which are currently being investigated. The results suggest that the selection of nitrate transport mutants on the basis of resistance to chlorate toxicity (at least at a concentration of 200 mm chlorate with uric acid as the sole source of nitrogen) yields nrtA mutants, which all appear to be loss of function. This is in contrast to nrtB mutants, which are sensitive to chlorate toxicity at a range of concentrations and with various single nitrogen sources.
Acknowledgments
We appreciated the strains sent to us by J. Clutterbuck, University of Glasgow, United Kingdom. S.E.U. gratefully acknowledges the Australian Research Council for financial support. J.R.K. thanks The Royal Society (London) for funds to travel to Australia.
References
- Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback et al., 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301: 610–615. [DOI] [PubMed] [Google Scholar]
- Abramson, J., H. R. Kaback and S. Iwata, 2004. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: members of the major facilitator superfamily. Curr. Opin. Struct. Biol. 14: 413–419. [DOI] [PubMed] [Google Scholar]
- Adelberg, E. A., M. Mandel and G. C. C. Chen, 1965. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem. Biophys. Res. Commun. 18: 788–795. [Google Scholar]
- Bailey, J., and C. Manoil, 1998. Missense mutations that inactivate Escherichia coli lac permease. J. Mol. Biol. 277: 199–213. [DOI] [PubMed] [Google Scholar]
- Bal, J., E. M. Kajtaniak and N. J. Pieniazek, 1977. 4-Nitroquinolone-1-oxide: a good mutagen for Aspergillus nidulans. Mutat. Res. 56: 153–156. [Google Scholar]
- Brownlee, A. G., and H. N. Arst, Jr., 1983. Nitrate uptake in Aspergillus nidulans and involvement of the third gene of the nitrate assimilation gene cluster. J. Bacteriol. 155: 1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M. G., and G. von Heijne, 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10: 685–686. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A. J., 1974 Aspergillus nidulans, pp. 447–510 in Handbook of Genetics, Vol. 1, edited by R. C. King. Plenum Press, New York.
- Cormack, B. P., G. Bertram, M. Egerton, N. A. R. Gow, S. Falkow et al., 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143: 303–311. [DOI] [PubMed] [Google Scholar]
- Cove, D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56. [DOI] [PubMed] [Google Scholar]
- Cove, D. J., 1976. a Chlorate toxicity in Aspergillus nidulans: the selection and characterisation of chlorate resistant mutants. Heredity 36: 191–203. [DOI] [PubMed] [Google Scholar]
- Cove, D. J., 1976. b Chlorate toxicity in Aspergillus nidulans. Studies of mutants altered in nitrate assimilation. Mol. Gen. Genet. 146: 147–159. [DOI] [PubMed] [Google Scholar]
- Crawford, N. M., and A. D. M. Glass, 1998. Molecular and physiological aspects of nitrate uptake in plants. Plant Sci. 3: 389–395. [Google Scholar]
- Daniel-Vedele, F., S. Filleur and M. Caboche, 1998. Nitrate transport: a key step in nitrate assimilation. Curr. Opin. Plant Biol. 1: 235–239. [DOI] [PubMed] [Google Scholar]
- Forde, B. G., 2000. Nitrate transporters in plants: structure, function and regulation. Biochim. Biophys. Acta 1465: 219–235. [DOI] [PubMed] [Google Scholar]
- Forde, B. G., 2002. The role of long-distance signaling in plant responses to nitrate and other nutrients. J. Exp. Bot. 53: 39–43. [PubMed] [Google Scholar]
- Frillingos, S., M. Sahin-Toth, J. Wu and H. R. Kaback, 1998. Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J. 12: 1281–1299. [DOI] [PubMed] [Google Scholar]
- Henderson, P. J. F., 1991. Sugar transport proteins. Curr. Opin. Struct. Biol. 1: 590–601. [Google Scholar]
- Hirai, T., and S. Subramaniam, 2004. Structure and transport mechanism of the bacterial oxalate transporter OxlT. Biophys. J. 87: 3600–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, T., J. A. Heymann, D. Shi, R. Sarker, P. C. Maloney et al., 2002. Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9: 597–600. [DOI] [PubMed] [Google Scholar]
- Huang, Y., M. J. Lemieux, J. Song, M. Auer and D. N. Wang, 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301: 616–620. [DOI] [PubMed] [Google Scholar]
- Jessen-Marshall, A. E., N. J. Paul and R. J. Brooker, 1995. The conserved motif, GXXX(D/E)(R/K)XG[X](R/K)(R/K), in hydrophilic loop 2/3 of the lactose permease. J. Biol. Chem. 270: 16251–16257. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- MacCabe, A. P., M. B. R. Riach, S. E. Unkles and J. R. Kinghorn, 1990. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 9: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, B., R. Dewachter and A. Goffeau, 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21: 113–134. [DOI] [PubMed] [Google Scholar]
- Pao, S. S., I. T. Paulsen and M. H., Saier, Jr., 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazdernik, N. J., E. A. Matzke, A. E. Jessen-Marshall and R. J. Brooker, 2000. Roles of charged residues in the conserved motif, G-X-X-X-D/E-R/K-X-G-[X]-R/K-R/K, of the lactose permease of Escherichia coli. J. Membr. Biol. 174: 31–40. [DOI] [PubMed] [Google Scholar]
- Saier, M. H., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. M. Heijne et al., 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1: 257–279. [PubMed] [Google Scholar]
- Scheible, W. R., A. Gonzales-Fontes, M. Laurer, B. Muller-Rober, M. Caboche et al., 1997. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsett, A. B., and D. J. Cove, 1979. Deletion mapping of the niiA niaD gene region of Aspergillus nidulans. Genet. Res. 34: 19–32. [DOI] [PubMed] [Google Scholar]
- Towbin, H., T. Staehelin and J. Gordon, 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman, L. J., A. Richardson and B. G. Forde, 1996. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene 175: 223–231. [DOI] [PubMed] [Google Scholar]
- Unkles, S. E., K. L. Hawker, C. Grieve, E. I. Campbell, P. Montague et al., 1991. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 88: 204–208 (erratum: Proc. Natl. Acad. Sci. USA 92: 3076). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles, S. E., J. Smith, G. J. M. Kana'n, L. J. Millar, I. S. Heck et al., 1997. The Aspergillus nidulans cnxABC locus is a single gene encoding two catalytic domains required for synthesis of precursor Z, an intermediate in molybdenum cofactor biosynthesis. J. Biol. Chem. 272: 28381–28389. [DOI] [PubMed] [Google Scholar]
- Unkles, S. E., D. Zhou, M. Y. Siddiqi, J. R. Kinghorn and A. D. M. Glass, 2001. Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J. 20: 6246–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles, S. E., R. Wang, Y. Wang, A. D. M. Glass, N. M. Crawford et al., 2004. Nitrate reductase is required for nitrate uptake into fungal but not plant cells. J. Biol. Chem. 270: 28182–28186. [DOI] [PubMed] [Google Scholar]
- Vidmar, J. J., D. Zhuo, M. Y. Siddiqi, J. K. Schjoerring, B. Touraine et al., 2000. Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol. 123: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J. M., 2001. Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 17: 560–563. [DOI] [PubMed] [Google Scholar]
- Williams, L. E., and A. J. Miller, 2001. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 659–688. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and B. G. Forde, 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. [DOI] [PubMed] [Google Scholar]