Abstract
A screen of random, autosomal, homozygous-viable P-element insertions in D. melanogaster found small effects on wing shape in 11 of 50 lines. The effects were due to single insertions and remained stable and significant for over 5 years, in repeated, high-resolution measurements. All 11 insertions were within or near protein-coding transcription units, none of which were previously known to affect wing shape. Many sites in the genome can affect wing shape.
FOR every quantitative trait some set of genomic sites can yield mutational variation. The full distribution of potential effects is unknowable, since it is a function of all possible modifications; but any mutational screen can help to identify quantitative trait loci (QTL). Here we report the provisional identification of 11 loci affecting wing shape in Drosophila melanogaster, based on effects produced with high frequency by random P-element insertions.
Wing shape is the third morphological trait in Drosophila to be mapped for natural QTL (Weber et al. 1999), after bristle number (Long et al. 1995) and features of male genitalia (Liu et al. 1996). Studies of natural genetic variation in wing shape in D. melanogaster find evidence of numerous small, mainly additive effects (Weber et al. 1999, 2001), which are largely independent of sex and body size (Weber 1990; Birdsall et al. 2000; Zimmerman et al. 2000) and sometimes act in small regions of the wing (Weber 1992).
Common natural alleles can be quite different from new mutations. Their effects can arise from multiple sequence differences (Haenlin et al. 1994; Stam and Laurie 1996; Robin et al. 2002), unlike the new alleles that show up in mutational screens. Natural alleles may evolve piecemeal over long periods (Kreitman and Hudson 1991; Phillips 1999). Moreover, the natural genetic variance for a quantitative trait can arise mainly from a few major loci (Robertson 1967; Mackay 2001a,b), whereas mutational genetic variation for the same trait could have a broader potential base.
Common alleles in nature do not necessarily represent loci that would contribute to major evolutionary changes. Genes often affect particular traits through secondary pleiotropic pathways, as the classical trait of bristle number in Drosophila has shown. For example, much of the standing genetic variance for bristle number appears to arise from genes with primary roles in the development of the peripheral nervous system (PNS; Nuzhdin 1999; Norga et al. 2003). Similarly, one of the first quantitative trait genes to be identified was the bobbed (bb) locus, which codes for ribosomal RNA but also happens to affect bristle number (Frankham et al. 1978). Despite these findings, one would think that natural selection could produce macroevolutionary changes in bristle number without the inevitable derangement of either ribosome production or the PNS. Thus, during a major adaptive change, the common alleles first available to selection might sometimes present genetic conflicts, rather than a reserve of evolutionary potential. New mutational effects should provide a more complete picture of the genetic basis of traits and of their potential to evolve. A key question in evolutionary genetics is the ultimate mutational target size for typical traits.
Most P-element insertion screens have focused on qualitative effects that can be scored in a few individuals. Other screens have studied quantitative effects by measuring the increase in phenotypic variance among P-element lines compared to controls or by quantifying effects in individual insertion lines as deviations from the control mean. However, the creation and extraction of P-element insertion lines can cause various types of new genetic variation, and small effects of insertions are hard to separate from these and other residual variations in the genetic background. This study employs new tactics to reduce both genetic and environmental variation to help ensure that insertion lines differ from controls only in the effect of the insertion.
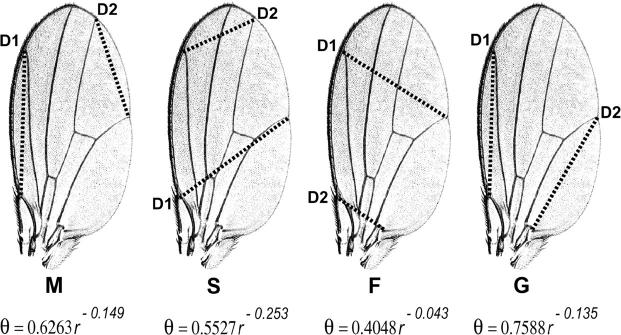
The shape metrics employed in these studies are angular offsets (Weber 1990). Angular offsets are sometimes inadvertently described as ratios (Klingenberg et al. 2001; Houle et al. 2003), but ratios are not valid metrics of allometric shapes. Angular offsets resemble ratios in that each value is a function of two numbers (dimensions D1 and D2) with opposite effects on its magnitude, but unlike ratios they are size independent in allometric forms. Angular offsets provide the most reductionistic shape metrics possible on the basis of the simplest aspect of shape, which is the allometric relation between two dimensions.
MATERIALS AND METHODS
Creation of insertion lines:
The P-element construct P{lacW} contains a functional allele of white (w+mC), providing a visible marker (Bier et al. 1989). P{lacW} retains the left and right P-element terminal sequences required for transposition, but lacks the transposase gene. We crossed a line with a single P{lacW} inserted on the X chromosome, in a background marked by yellow (y) and w, to another M-cytotype line, also marked by y and w. Thereafter, red-eyed virgin flies, heterozygous for the insertion, were mated to their white-eyed brothers lacking the insertion for 20 generations of single-pair matings. This created an inbred line in which this single P-element insertion was still segregating in an otherwise nearly isogenic y w background. This line was the P-element source and the target genome in all mobilizations.
We mobilized P{lacW} in the standard way, by crossing red-eyed virgin heterozygotes, from the inbred stock just described, to males with dominantly marked and balanced second and third chromosomes, carrying the defective P-element “jumpstart” or “Δ2-3,” which produces transposase but cannot jump (Robertson et al. 1988; Bier et al. 1989). Sons containing both P elements, and with red sectored eyes showing mobilization of P{lacW}, were backcrossed to virgin white-eyed cousins segregating from the inbred P-element source line. From this mating, new autosomal insertions of P{lacW} were picked up as heterozygous red-eyed males lacking their grandfather's dominant markers. By this procedure, 60 random autosomal insertions were collected, each from a different vial to avoid duplicate insertions.
Each new heterozygous insertion male was crossed to one virgin white-eyed sib, also lacking dominant markers. Thereafter, each insertion was maintained in a segregating line by single-pair matings of red-eyed insertion-heterozygote virgins to their white-eyed brothers for 10 more generations of inbreeding, leading up to the first screen.
Screening for wing-shape effects:
During the entire experiment each insertion was maintained continuously in its own inbred segregating line. Insertions were screened three times, and for each screen new, temporary, homozygous insertion-bearing and insertion-free lines were extracted from the segregating line and compared. Insertions were always made homozygous without using balancers to avoid the chance of small effects entering the inbred lines from balancer stocks. Wherever possible, we used the slight differences in the depth of red eye color to distinguish between insertion homozygotes and heterozygotes. Where eye colors were not distinguishable, we relied on the inevitability of success in large numbers of single-pair matings between red-eyed flies and the fact that eye colors would not segregate in fixed lines. Of the 60 insertions, 10 could not be made homozygous.
To eliminate effects of environmental differences among vials, flies of paired homozygous insertion and control lines were cultured together in vials containing five pregnant females from each line. From the progeny of each vial, 20 red-eyed males and 20 white-eyed males were compared. Paired samples of insertion and control were always measured by the same operator. Five vials per insertion were pooled, so that 100 insertion-homozygote males were compared to 100 insertion-free males. This measurement protocol was standard for all lines in all screens, except for line 41 in the third screen, when an extra comparison was done using pooled sample sizes of 300. Flies were cultured in plastic 75-ml vials on commercial potato-flake medium (Carolina Biological Supply, http://www.carolina.com) with yeast at 26°.
First screen:
Both homozygous insertion and noninsertion control lines were extracted from the same inbred segregating line for each insertion that could be made homozygous. Wing shapes between each homozygous insertion line and its paired control line were compared. Insertions that showed significant differences were maintained by propagating the original lines in which the insertions were still segregating. Insertions that failed the screen were abandoned.
Second screen:
New homozygous insertion and noninsertion lines were extracted from each remaining segregating line, for the second time, at least 15 generations (depending on the line) after the completion of the first screen. During the interval between the first and second screens, the segregating lines were maintained by mass mating in single-vial cultures. The second extraction was again done without balancers, as described above. Once again, flies of both homozygous types were cultured together in the same vials to eliminate effects of environmental differences between vials. Again, some insertions failed the screen and were abandoned.
Third screen:
Insertions that showed the same pattern of effects in both the first and second screens were perpetuated in their segregating lines for 5 more years after the second screen on an ∼2-week generation cycle. During this time, for ∼50 consecutive generations, the lines were maintained by strict single-pair mating of red-eyed virgins to their white-eyed brothers. Finally, new homozygous insertion and noninsertion lines were extracted for the third time. Again, homozygous lines were extracted without using balancers, and flies of both homozygous types were cultured together in the same vials for measurement.
Angular offsets as a shape metric:
Vein intersection landmarks were digitized, and interlandmark dimensions were used to quantify angular offsets for four wing-shape indexes, designated M, S, F, and G. These are calculated from the four pairs of wing dimensions (D1 and D2) shown in Figure 1.The method is fully explained and illustrated in Weber (1990). Briefly, to create each index, a curve was passed through a scatterplot of points (D1, D2) for wild-type flies by regressing log θ on log r, where θ = arctan (D2/D1), and 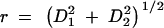 . This yields a polar equation for each trait that expresses the mean allometric relation between dimensions D1 and D2 over all body sizes in wild-type flies and serves as a baseline. The trait value or angular offset of the point (D1, D2) for any individual wing is its rotation about the origin from the baseline in radians. Selection on angular offset produces a rotation of this line, i.e., a quantified change in allometry between D1 and D2 (Weber 1990). The baselines for traits M, S, F, and G were derived in Weber (1990) and are still the same. This method gives a simple metric of shape that is independent of body size and most environmental influences.
. This yields a polar equation for each trait that expresses the mean allometric relation between dimensions D1 and D2 over all body sizes in wild-type flies and serves as a baseline. The trait value or angular offset of the point (D1, D2) for any individual wing is its rotation about the origin from the baseline in radians. Selection on angular offset produces a rotation of this line, i.e., a quantified change in allometry between D1 and D2 (Weber 1990). The baselines for traits M, S, F, and G were derived in Weber (1990) and are still the same. This method gives a simple metric of shape that is independent of body size and most environmental influences.
Figure 1.—
The four size-independent shape indexes (M, S, F, and G) are defined by polar equations that express the allometric relation between paired dimensions (D1 and D2) in wild-type male flies.
Plasmid rescue and sequencing:
A fragment of flanking genomic DNA to the right of each P{lacW} insertion site was retrieved by plasmid rescue (Pirotta 1986). On its right end, P{lacW} includes a bacterial plasmid sequence with replication origin and ampicillin resistance to the right of an EcoRI restriction site. Whole genomic DNA of male flies was digested with EcoRI, ligated, and used to transform Escherichia coli. Clones surviving ampicillin were screened by electrophoresis. The flanking genomic DNA in typical clones was sequenced using the right end of P{lacW} as the primer: 5′-CGACGGGACCACCTTATGTTATTTCATCATG-3′ (Ballinger and Benzer 1989; Lindsley and Zimm 1992). Most clones were subsequently sequenced with a second primer starting 111 bases to the left of the right end, as a check on the exact insertion site: 5′-GGGTTAATCAACAATCATATCGCTGTCTCAC-3′.
Chromosome labeling:
Polytene chromosomes from larval salivary glands were labeled using biotinylated DNA probes, including both P-element and rescued plasmid sequences in separate assays, by a standard in situ protocol (cf. Long et al. 1995). The treatments (separated by appropriate transitional baths) were 20 min 2× SSC at 65°, 3 min 0.14 m NaOH, air drying, hybridization with biotinylated probe overnight, 20 min streptavidin/biotin treatment (Vectastain, Vector Laboratories, Burlingame, CA), 30 min in diaminobenzidine/H2O2 solution, and light Giemsa counterstaining.
RESULTS
Phenotypic screening of insertions:
Each insertion was maintained in a segregating line with inbreeding arranged to reduce genetic variation other than the insertion. To quantify insertion effects, paired homozygous lines with and without the insertion were extracted from each segregating line and compared. New homozygous lines were extracted from the same segregating lines and compared three times in consecutive screens, as inbreeding proceeded.
First screen:
Of 60 original insertions, 10 could not be made homozygous and were compared to controls as heterozygotes. In t-test comparisons (not corrected for the number of tests), we found differences in wing shape in one or more of the four traits, which were significant at P ≤ 0.05 in 29 of the 50 homozygous insertion lines and in 7 of the 10 still-heterozygous lines. Our criterion to retain a line in this initial screen was a difference with an individual P ≤ 0.01 in at least one of the four shape traits (M, S, F, and G). Of the 50 homozygous lines, 17 passed this criterion. Of the 10 heterozygous lines, 3 passed. Only the 17 homozygous-viable insertions that passed were maintained for long-term study. No insertions had any obvious visible effects on the size, shape, or venation of wings, except that one insertion (insertion 36) sometimes showed a small gap at the distal end of vein L5.
Table 1 shows the grand means of line means and the variances among line means for all 50 homozygous-viable insertions and for their controls. The difference between insertion and control grand means is not significant in t-tests for any trait (d.f. = 98). Thus the insertion of P{lacW} seems to act randomly with no consistent directional effect. This was also true in later comparisons involving only insertions that do show significant individual effects.
TABLE 1.
Means and variances of homozygous control and insertion line means in first screen
| 50 control line means
|
50 insertion line means
|
|||
|---|---|---|---|---|
| Trait | Grand mean | Variance | Grand mean | Variance |
| M | +8.0 × 10−3 | 2.4 × 10−5 | +9.0 × 10−3 | 3.4 × 10−5 |
| S | −2.1 × 10−3 | 3.5 × 10−5 | −1.4 × 10−3 | 3.4 × 10−5 |
| F | −7.3 × 10−3 | 1.6 × 10−5 | −8.0 × 10−3 | 2.3 × 10−5 |
| G | −1.9 × 10−2 | 8.6 × 10−6 | −1.8 × 10−2 | 1.3 × 10−5 |
Means are in radians of angular offset from wild-type baseline.
Model II ANOVAs, with line as a random, main effect and insertion (a random effect) nested within line, show a moderate variance component due to line for each trait (M—19.2% of total variance, P < 0.0001; S—23.0%, P < 0.0001; F—8.9%, P = 0.0006; G—9.4%, P < 0.0001) and a small, but significant variance component due to differences between insertion and noninsertion lines (M—6.3% of total variance, P < 0.0001; S—4.2%, P < 0.0001; F—10.5%, P < 0.0001; G—3.7%, P < 0.0001) for all four traits. Each insertion line sample and its paired control were measured by the same operator, but many different operators measured the different lines. (The data set includes 10,000 wings.) The among-line variance, then, reflects both (1) the effect of the mutagenic transposition process in 50 crosses on a common genetic background followed by inbreeding in 50 separate segregating lines and (2) among- and within-operator error. Variance due to insertions vs. controls reflects only insertion effects and within-operator error.
Although the variance due to insertions is a significant component of total variance in these ANOVAs in all four traits, a direct comparison between the among-line variances of insertion line means and control line means is not significant in any of the four traits. As seen in Table 1, the variances among the 50 lines with the insertion are larger in three of four cases, but none of the differences among variances are significant in terms of F-ratio (d.f. = 49, 49).
Second screen:
At least 15 generations after the first screen, we extracted new homozygous insertion and noninsertion lines from the segregating lines for the 17 insertions remaining from the first screen. These were tested again in the same way. Our criterion in the second screen was not only a significant trait difference, but also consistency between the first and second screens in the pattern of values of the four traits. Of the 17 lines, 2 (lines 6 and 37) showed no significant trait differences in the second screen. Three of the lines (lines 8, 50, and 52) showed significant differences, but in patterns that were not consistent with the first screen. The most contradictory of these (line 50) was immediately reextracted and compared again and showed no significant differences in the third comparison. These 5 problematic lines were dropped without further study, as type I errors, unstable or multiple insertions, or lines with persistent residual variation affecting the trait. The remaining 12 lines all still showed the same approximate patterns of relative values in the four traits and still showed significant trait differences. Eleven of these were still significant at P ≤ 0.01 in at least one trait. One (line 47) was only significant at P = 0.036, but was retained for study.
Third screen:
The 12 insertions retained after the second screen were maintained in their original segregating lines for ∼120 more generations in small vial cultures. During this time, for ∼50 consecutive generations, these lines were propagated by single-pair matings of red-eyed, heterozygous virgins to white-eyed, insertion-free brothers. Finally, new homozygous insertion and noninsertion lines were extracted and compared a third time. In this screen, all insertions showed patterns of differentiation consistent with previous measurements, but only 10 of the 12 insertions passed initially, with P ≤ 0.01 in at least one shape index. The two insertions that failed this third screen (insertions 41 and 51) still showed some evidence of effects.
We immediately retested line 41 with sample sizes of N = 300 for both insertions and controls. This was the fourth test on line 41 on the third pair of extracted homozygous lines. In this test, line 41 showed P ≤ 0.01 in one trait and the same profile of trait differences as before. Unfortunately, line 51 could not be retested because the fixed white-eyed control was lost, and the segregating stock was contaminated by wild type. This insertion survives only in a fixed line.
Leaving aside the doubtful line 51, we conclude that 11 of these 12 insertions still show effects of the P{lacW} insertion. Table 2 shows the mean effects of these 11 insertions, calculated as the mean absolute difference between insertion and control lines for all three assays, expressed as a fraction of the wild-type phenotypic standard deviation for each trait (Weber 1990). These differences are all too small to be reliably scored by eye.
TABLE 2.
Mean absolute trait differences from three screens in SD of wild flies
| Insertion no. | M | S | F | G |
|---|---|---|---|---|
| 7 | 0.37* | 0.11 | 0.50* | 0.15 |
| 12 | 0.16 | 0.00 | 0.33* | 0.52* |
| 16 | 0.06 | 0.30* | 0.37* | 0.16 |
| 18 | 0.66* | 1.02* | 0.93* | 0.15 |
| 24 | 0.24 | 0.15 | 0.60* | 0.48* |
| 25 | 0.74* | 0.42* | 0.30 | 0.10 |
| 27 | 0.21 | 0.15 | 0.22 | 0.39* |
| 36 | 0.22 | 0.54* | 0.15 | 0.05 |
| 41 | 0.18* | 0.18 | 0.10 | 0.05 |
| 45 | 0.37* | 0.35* | 0.58* | 0.14 |
| 47 | 0.21 | 0.01 | 0.23 | 0.23* |
SD from Weber (1990).
Difference had P < 0.01 in at least two screens.
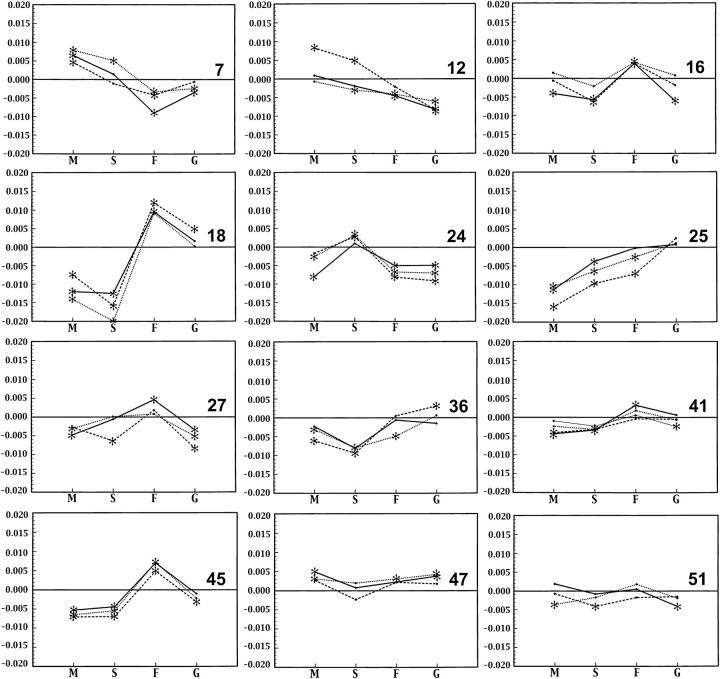
Figure 2 shows the results of all three screens for the 12 insertions in the third screen. Even where individual trait differences are not significant, they tend to preserve a recognizable profile across the four traits, although measured by different teams in each screen. Line 12 shows significant deviations in opposite directions in index S, but otherwise retains the same overall pattern. Line 51 shows only P < 0.05 in one trait in the third screen and only weak consistency between screens. Measurement error would explain inconsistencies in the profiles in Figure 2, but some changes may reflect background mutations that modify the effects of the insertion.
Figure 2.—
Differences between insertion homozygotes and controls in three separate extractions from the same inbred lines for four shape indexes (M, S, F, and G) in males. Solid, dashed, and dotted lines correspond to first, second, and third extractions. Asterisks mark all differences with individual P < 0.05. Insertion line numbers are shown at the right. Line 41 was tested a fourth time on the third extraction (narrow solid line).
Gene identification and confirmation:
Flanking DNA on the right-hand (3′) side of each insertion was retrieved by plasmid rescue, for all 12 insertions that passed the second screen. Significant unique hits were obtained for all sequences with BLAST searches of the D. melanogaster genome (http://www.fruitfly.org/blast/). Polytene chromosomes of each insertion line were twice labeled in situ, using as probes a P-element sequence and also the retrieved plasmid. Table 3 shows the approximate chromosome band locations assigned to each insertion by in situ labeling and also the band locations assigned by subsequent BLAST searches using flanking DNA. In each case, the apparent in situ site was reasonably consistent with the computed or known band assignment according to FlyBase (http://flybase.bio.indiana.edu). No secondary insertion sites were noted.
TABLE 3.
Insertion site identifications byin situ labeling and by BLAST search
| Insertion | Gene(s) | Band: in situ/FlyBase | DNA site |
|---|---|---|---|
| 7 (+) | hairy (h) (+) | 66D/66D10 | 301 bp upstream |
| 12 (−) | tribbles (trbl) (−) | 77B/77C1 | 31 bp upstream |
| 16 (−) | CG31605 or gelded (gel) (+) | 28D/28E3-E5 | In 16-kb intron |
| 18 (−) | hephaestus (heph) (−) | 100F/100D3-E1 | In 76-kb intron |
| 24 (+) | foxo (+) | 88A/88A6-A8 | In short intron |
| 25 (+) | seven in absentia (sina) (+) | 73C/73D2-D3 | In exon (5′-UTR) |
| Rhodopsin 4 (Rh4) (−) | In 8.8-kb intron | ||
| 27 (+) | sugarless (sgl) (−) | 65D/65D5 | In 2.4-kb intron |
| 36 (+) | stem cell tumor (stet) (−) | 62A/62A2 | Downstream enda |
| 41 (+) | yippee interacting protein 2 (yip2) (+) | 30C/30E4 | 90 bp upstream |
| Srp54 (+) | 140 bp downstream | ||
| 45 (+) | out at first (oaf) (+) | 22F/22F3 | In 2.1-kb intron |
| 47 (+) | Alhambra (−) | 84B/84B-C | In intron |
| 51 (−) | Glycerol-3-phosph. dehyd. (Gpdh) (+) | 26A/26A | In exon (5′-UTR) |
Polarity of insertions and genes is indicated by + or −. All data according to FlyBase Release 3.
The 3′-UTR of stet (no. 36) contains an insertion of Doc, 140 bp to right of P{lacW}.
Genes near the insertions:
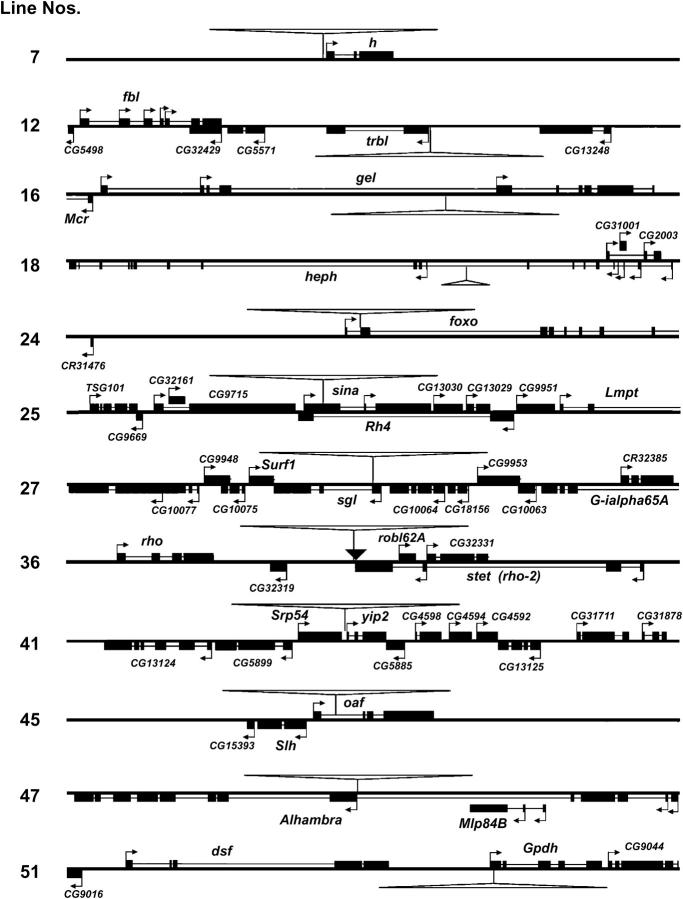
Figure 3 shows each insertion in its genomic context of recognized transcription units. All insertions are shown at the same scale, including 30 kb of flanking DNA, except for line 18 (heph). In many cases one gene is the obvious candidate for the source of the effects, but not in all. Insertions may fall in a short interval between two genes or in an intron containing other genes. In some cases the gene affected is uncertain because of the density of small genes near the insertion. The following summaries, with reference to Figure 3, describe the genes most likely to be affected by each insertion. All information about gene functions, locations, exons and introns, alternative transcripts, etc. is based on the FlyBase annotation under Release 3, if not attributed to other sources.
Figure 3.—
P{lacW} insertions in genomic context. Each diagram shows an identical-sized region of 30 kb, except for no. 18, which includes 145 kb. Items above or below the central lines are in plus or minus orientation, respectively. Arrows show transcription start sites, including multiple start sites for some genes. Exons are thick; introns are thin. The diagrams do not show some very short introns. Gene symbols are given according to FlyBase. Large open triangles show the location and orientation of P{lacW} and are proportional to its length of 10.7 kb. The small solid triangle in no. 36 marks the site of an insertion of Doc of unknown length.
Insertion 7:
The insertion site is 301 bp upstream of the 5′-end of the gene hairy (h). The closest other genes are 25.2 kb upstream and 42.4 kb downstream. h codes for a transcription factor of the basic helix-loop-helix type. It acts as a pair-rule gene in embryonic segmentation (Ingham et al. 1985) and is involved in patterning the nervous system (Carroll and Whyte 1989). It has widespread effects on sensory bristle patterning and represses bristle formation by negative regulation of the achaete-scute complex (Moscoso Del Prado and García-Bellido 1984). h has been implicated as an important quantitative trait gene for sternopleural and abdominal bristle number (Shrimpton and Robertson 1988; Long et al. 1995; Gurganus et al. 1999; Robin et al. 2002). Mutant alleles of h show effects on the placement of sensory bristles on the wing (Rushlow et al. 1989; Thompson and Preston 1992) and modulate the effects of vestigial (vg) and Notch (N) on the wing (Abu-Issa and Cavicchi 1996). Aside from this, h has not previously been connected to wing morphology.
Insertion 12:
The insertion is ∼30 bp upstream of the tribbles (trbl) transcription start site. The closest other genes are 5.5 kb upstream (CG13248) and 8 kb downstream (CG5571). trbl codes for a cell-cycle regulatory protein with an amino acid sequence that indicates protein serine/threonine kinase activity. This protein appears to act as a cell-cycle brake in G2-phase (Mata et al. 2000). Mutations of trbl have many effects on morphogenesis through their influence on the coordination of mitosis (Mata et al. 2000; Seher and Leptin 2000). Overexpression of trbl in the posterior wing compartment causes cells inside the compartment to be fewer and larger than normal, without visibly changing the size of the compartment (Mata et al. 2000). Among genes known to interact with trbl, several show independent effects on the wing in some of their mutant alleles. These include slow border cells (slbo), a transcription factor (Rørth et al. 2000); string (stg), another cell-cycle control gene (Milan et al. 1996a,b; Verheyen et al. 1996; Salzberg et al. 1997; Toba et al. 1999); wee, a protein kinase that may be a cell-cycle control gene (Price et al. 2002); and Notch (N; de Celis and Bray 1997; Lawrence et al. 2000).
Insertion 16:
This insertion falls in an intron of the gene gelded (gel), also designated as CG31605. Known alleles of gel include numerous recessive lethals (Roch 1998) and one allele that causes male sterility by a recessive effect on spermatid development (Castrillon et al. 1993). This gene has no previously reported effects on the wing, nor any reported interactions with other genes that affect the wing. The complete transcribed region of gel is 26 kb long, with many exons, at least nine known transcripts, and three transcription start sites. The insertion site falls in the central region of the gene, within the longest intron (16 kb in some transcripts), and 1.8 kb upstream of the start site for the shortest transcript. The closest promoters of other genes are 18 kb to the left of the insertion at Macroglobulin complement-related (Mcr) and 16 kb to the right at CG31756 (not included in Figure 3).
Insertion 18:
The insertion is in an intron of the unusually long (142.3 kb) gene hephaestus (heph). This gene has at least 15 exons, 11 known transcripts, and five different transcription start sites. Just as in the case of insertion 16, this insertion occurs in a long central intron (76 kb in most transcripts), and the transcription start site of the shortest transcript falls in the same intron as the insertion, downstream (2380 bp) of the insertion site.
heph appears to be a spliceosome component. Its amino acid sequence contains an RNA polypyrimidine tract-binding region of four domains, already characterized in several vertebrates (Dansereau et al. 2002). In Drosophila, this gene has been implicated in the regulation of the Notch signaling pathway, appearing to suppress peripheral activity of Notch within the broad regions where Notch is first expressed during wing-pattern formation. In homozygous clones, recessive lethal mutations in heph eliminate wing-vein formation and also induce ectopic wing margin tissue, producing both effects by interaction of heph with Notch (Dansereau et al. 2002). heph also interacts with fringe (fng; Dansereau et al. 2002), a gene expressed in dorsal cells of the wing (Irvine and Wieschaus 1994), which can independently induce ectopic wing-vein material in hypomorphic alleles (Correia et al. 2003). Norga et al. (2003) reported significant bristle number effects from a P-element insertion in heph.
Insertion 24:
This insertion is located in the first intron of foxo, a long (30.4 kb) gene with one known transcript. The site of the insertion is 31.8 kb from the promoter of the next gene to the right (CG3153, not shown), and 13.1 kb from the promoter of the next gene to the left (CR31476, a tRNA gene). The exact location of the insertion is between the last two bases (A and G) of the intron. The right terminal base of P{lacW} would not reconstitute the AG signal. However, another AG site occurs six bases downstream, and, with an eight-base target-site duplication (O'Hare and Rubin 1983), this and the original AG site may remain to the left of the insertion. Thus the insertion may be spliced out with the intron, or the intron may be spliced out leaving the insertion. Only the 5′-UTR would be affected by this insertion, since the protein-coding sequence begins after the second intron. This insertion could affect either transcription or translation rates of foxo and might activate unknown alternative transcription start sites. foxo appears to have 9–14 exons and numerous expressed sequence tags, suggesting that variable start sites generate transcript diversity.
foxo codes for an apparent transcription factor with a forkhead (or winged helix) DNA-binding domain. In other organisms, proteins with this domain are involved in cell determination in early embryogenesis. Only one allele of foxo (wild type) is known in Drosophila. Seven other transcription factors in Drosophila with forkhead domains are known. All are expressed in small, highly localized regions of embryos early in development (Häcker et al. 1992).
Insertion 25:
This insertion involves two genes as the most likely targets: seven in absentia (sina) on the positive strand and Rhodopsin 4 (Rh4) on the negative strand. Rh4 has a single long intron (8.8 kb), and sina is mainly contained within this intron, although its first transcription start site appears to overlap with the second exon of Rh4. The insertion site is near the middle of the 5′-UTR of sina (base position 996 of 1902 bases) and 630 bp from the left end of the intron of Rh4. The closest flanking loci are all on the positive strand. Upstream, they include the nested genes CG9715 and CG32161, with their promoters 8 kb to the left of the insertion site. Downstream, they include two more genes, both of which are nested tightly together with sina within the same intron of Rh4: CG13030, with its promoter 5.3 kb to the right of the insertion, and CG13029.
Rh4 functions in the eye in phototransduction and is expressed exclusively in photoreceptor cells, so it is unlikely to mediate any effect on the wing. sina codes for a protein with a single-ring-type zinc-finger domain, which is expressed in various tissues during all phases of development, particularly in sense organ development. Mutations in sina affect the formation of receptor cells in ommatidia and also in sensory bristles. One mutant allele of sina causes outstretched wings (Lindsley and Zimm 1992), and another allele reduces the numbers of bristles along the anterior wing margin (Carthew and Rubin 1990). sina also interacts with many other genes, including at least six that are normally expressed in the wing and that can show incidental effects on the wing. These include, for example, tramtrack (ttk) and phyllopod (phyl). ttk is expressed everywhere in the wing disk except in proneural clusters (Lehembre et al. 2000). When overexpressed, it ablates almost all sensory bristle growth and severely reduces the size of the wing (Badenhorst et al. 2002). phyl and sina antagonize ttk, and overexpression of phyl in the wing causes ectopic bristle formation on the third wing vein (Pi et al. 2001). The products of sina, ttk, and phyl appear to act together as a protein complex in both photoreceptor and sensory bristle differentiation (Pi et al. 2001). musashi (msi) interacts with both sina and ttk in eye development (Hirota et al. 1999) and causes variation in the number of bristle support cells in sensory bristles, notably along the anterior wing margin (Nakamura et al. 1994). GTPase-inactivating protein I (Gap1) is another gene of interest that interacts with sina. Gap1 is involved in differentiation of ommatidia and bristles and affects chromosome segregation, and Gap1 mutants also show effects on wing-vein formation, including extra veinlets attached to the posterior crossvein parallel to the long veins (Gaul et al. 1992). Again, echinoid (ed) interacts with sina and also has direct effects on the differentiation of photoreceptor cells, and in one reported mutant genotype causes extra wing-vein growth and enlarged wings (Bai et al. 2001). Finally, small wing (sl) has effects on the differentiation of ommatidia and also produces a short, blunted wing with extra wing-vein material in two characteristic locations (Thackeray et al. 1998). A rather consistent pattern emerges for the genes in this group. They interact with sina, and, like sina, can affect the differentiation of both photoreceptors (specifically, the R7 cell of the ommatidium) and sensory bristles, including bristles on the notum and wing; and usually they also show some effects on wing shape, size, or venation.
Insertion 27:
The insertion site is in the first intron of the gene sugarless (sgl). The intron is ∼2.4 kb long, and the insertion is at the very beginning of the intron between the fourth and fifth bases. This location is not far from the promoter region of the gene, because the first exon is short (406 bp). A short interval of 458 bp separates the 5′-end of sgl and the 3′-end of the next upstream gene (CG10064). The surrounding region has a high density of genes, and the promoters of five other genes are located 3.6–6.3 kb from the insertion site.
sgl encodes a UDP-glucose dehydrogenase. This enzyme is essential in the production of UDP-glucuronate, which is utilized in the biosynthesis of several glycosaminoglycans. Glycosaminoglycans have many structural and metabolic roles and are somehow involved in modulating the signaling of wingless (wg; Binari et al. 1997), a segment polarity gene, which interacts with sgl. Mutations in sgl produce a pattern of defects in embryonic cuticle that is similar to the effects of nonlethal mutations of wg (Haerry et al. 1997). This phenotype in sgl mutants can be rescued by overexpression of wg (Häcker et al. 1997) and also by embryonic microinjection of exogenous heparan sulfate, a glycosaminoglycan (Binari et al. 1997).
Insertion 36:
This is the least conclusive case in regard to the identity of the affected gene. The insertion is not located within any gene or promoter region, but is 3–4 kb from the transcription start sites of four different genes. Another complication of this case is a natural transposable element (Doc) that was discovered in our line adjacent to the P{lacW} insertion.
The closest gene is stem cell tumor (stet), also known as rhomboid-2 (rho-2). The insertion is 118 bases to the left of the end of transcript A (which begins at the second start site of stet) and 137 bases to the left of the end of transcript B (which begins at the first start site). The retrieved flanking sequence, to the right of P{lacW}, includes part of this 3′-UTR and reveals the presence of an insertion of the transposable element Doc, just 22 bases inside the 3′-UTR of transcript A and three bases inside the 3′-UTR of transcript B. Thus, in our original inbred P{lacW} source line, stet most probably already carried this insertion of Doc inside the transcription unit before P{lacW} was inserted just outside it. The retrieved flanking sequence includes ∼500 bases matching the left end of Doc. Complete Doc sequences are ∼5 kb long (Lindsley and Zimm 1992).
The product of stet belongs to the rhomboid-like group of proteins with seven transmembrane domains. The stet protein functions as a membrane-bound serine-peptidase in epidermal growth factor signaling (Klämbt 2000). Mutations in stet affect male and female germline cells and cause defects in gonadal development (Schulz et al. 2002). Normal expression of stet appears to be limited to cells within male and female gonads at an early stage of differentiation. However, misexpression of stet in the wings of transgenic flies has direct effects on wing morphology, including thickening of veins and formation of ectopic vein material (Guichard et al. 2000; Urban et al. 2002). Thus stet could be the source of wing-shape effects, perhaps through effects on its second transcription start site. But the transcription start sites of two other genes—CG32319 and robl62A—are closer to the insertion.
The interpolation of a complete sequence of Doc to the right of P{lacW} would make CG32319 the closest transcription start site, and robl62A the next closest. Neither gene has a reported mutant phenotype but both have been classified by homology as to molecular function. CG32319 encodes a protein with N-acetyltransferase activity, which is involved in acetylation of amino acids in proteins. robl62A encodes a dynein subunit protein with ATPase activity, which should be involved in microtubule movement (Goldstein and Gunawardena 2000).
Insertion 41:
This insertion falls in a short interval of 230 bases between two adjacent genes that both run left to right. The insertion is 140 bases downstream of the gene Srp54 and 90 bases upstream of the gene yippee interacting protein 2 (yip2) within the promoter region of yip2. Aside from yip2, three other genes have their promoters within 3 kb of the insertion site, including Srp54, CG5899, and CG5885. yip2 codes for an acetyl-CoA C-acyltransferase, which is found in the mitochondrion. Srp54 codes for a protein that includes an RNA-binding sequence. CG5899 codes for a DNA helicase. CG5885 codes for a signal sequence receptor component. No mutant effects have been described for any of these four loci.
Insertion 45:
This insertion is inside a 2.1-kb intron, following the first exon of out at first (oaf), ∼768 bp from the left end of the intron. oaf has three introns and four known transcripts, all with the same start site. The oaf gene codes for a protein of unknown functional type involved in neurogenesis. It is transcribed in the embryonic central nervous system in segmental clusters and in gonads of both sexes throughout development and adulthood (Bergstrom et al. 1995). Some reported mutations in oaf are viable and have no obvious phenotypic effect; others are recessive lethal and affect the nervous system (Bergstrom et al. 1995). oaf has no reported effects on the wing, but does appear to be expressed uniformly throughout the wing disk at low levels (Merli et al. 1996).
Two other genes, SLY-1 homologous (Slh) and CG15393, are close to oaf but code in the opposite direction. The promoters of Slh and CG15393 are 1.4 and 2.0 kb from the insertion. Slh appears to be involved in membrane trafficking (Littleton 2000) and is not expressed in the wing (Merli et al. 1996). CG15393, to the left of Slh, is a small gene coding for 121 amino acids with no recognized functional motif. The tight three-gene cluster of oaf, Slh, and CG15393 is isolated from neighboring genes by ∼30 kb on the left and ∼20 kb on the right.
The next gene upstream of oaf is decapentaplegic (dpp), which affects many aspects of development (Spencer et al. 1982) and is strongly expressed in the wing. Although the coding regions of oaf and dpp are separated by 33.5 kb, most of this interval (30.0 kb) is occupied by an array of enhancers that control dpp (Blackman et al. 1991; Bergstrom et al. 1995). Perhaps insertion of the 10.7-kb P{lacW} in the oaf intron could have weak remote effects on the regulation of dpp, given the size of the region devoted to dpp transcriptional control and the proximity of this region to the insertion site. A regulatory site for human α-globin is also located in an intron of an adjacent gene at a similar distance (Vyas et al. 1992).
Insertion 47:
This insertion is in the Alhambra gene, which has four known transcripts. Two are long and include a 17-kb central intron, and the other two are short and start inside this central intron, beginning with an exon that is spliced out of the long transcripts. The insertion site is near the middle of this gene, just 17 bases upstream of the common start site of the two short transcripts. About 5.6 kb upstream of the site of the insertion, and within the same long intron of Alhambra, is the small included gene Muscle LIM protein at 84B (Mlp84B), a gene that also runs right to left in the same direction as Alhambra. (LIM stands for three homeodomain proteins with a shared motif.) The whole Alhambra transcription unit occupies 29.6 kb, so the central insertion site is rather isolated from other genes.
Alhambra codes for a protein that is thought to incorporate two zinc ions in a domain resembling a plant homeodomain finger, which is predicted to be involved in transcriptional regulation (Bahri et al. 2001). Reported mutations in Alhambra have recessive effects on the larval nervous system and development rate. Mlp84B codes for a protein with a glucocorticoid-receptor-like DNA-binding domain (again, a zinc-bound feature). Proteins with the LIM domain regulate cell growth and differentiation (Dawid et al. 1998). The product of Mlp84B belongs to a group of LIM proteins that regulate muscle differentiation, and the gene is expressed during differentiation at sites of muscle attachment (Stronach et al. 1996, 1999).
Insertion 51:
The insertion falls within the 5′-UTR of the gene Glycerol 3 phosphate dehydrogenase (Gpdh), ∼285 bp after the transcription start site and ∼145 bp before the first codon. Gpdh encodes an enzyme important in flight-muscle metabolism (Wojtas et al. 1997). Numerous recessive lethal and sublethal mutations of Gpdh have been reported, as well as a few mutations causing flightlessness (Kotarski et al. 1983). There are no reports of morphological effects. Insertion 51 may have had some variable effect on wing shape.
DISCUSSION
The use of paired insertions and controls, extracted from the same segregating, long-inbred stocks, with three repeated screens and multi-trait comparisons, leads to high confidence in the detection of small genetic effects. Our protocol was not designed to confirm every minor effect that may have been present in the first screen, but to select only the more significant and stable effects. We found such effects in 11 of 50 random homozygous-viable insertions.
Effects are strongly associated with the insertion of P{lacW}:
No other cause would be likely to produce trait profiles that consistently segregate with the insertion in three independent extractions, separated by many generations of recombination and inbreeding. Initial variation was already low due to preliminary inbreeding of the source line. In the transposition generation, variation could have arisen by male recombination in mobilized hybrid males or from mobilization-induced mutations. After transposition, much residual variation would have been eliminated before the first screen by the first 10 generations of single-pair matings between insertion-heterozygote virgins and their insertion-free brothers. Further recombination and inbreeding leading to the second and third screens would gradually homogenize and isogenize the genetic background even more. By never extracting insertions and controls except at the times of measurement, we avoided any chance for background divergence to accumulate. Balancers were never used in extractions so genetic variation could not arise from rare recombination with balancers. Some apparent effects in the first screen were not repeatable, due perhaps to unstable effects, multiple insertions, or residual variation. Elimination of these problematic lines left 11 that were still stable and consistent 5 years later in the third extraction and screen. During these 5 years, ∼50 generations were single-pair matings between sisters heterozygous for the insertion and brothers lacking it, increasing the likelihood that by the time of the third extraction, trait differences were caused only by the presence or absence of the insertion.
Effects arise from gene disruption, not from P{lacW} itself:
In the region surrounding each insertion, nearby loci might be affected by interactions with specific parts of P{lacW} or simply by the insertion of 10.7 kb. But the shape changes are not an autonomous effect of P{lacW} itself, because each insertion causes a unique pattern. Each trait was increased or decreased at roughly equal frequencies among lines. The grand means of the 50 control and 50 insertion line means from the first screen are not significantly different in any of the four traits (Table 1). In their screen of the effects of P{lArB} insertions on bristle number, Lyman et al. (1996) detected a significant mean directional effect that was due to the insertion itself, relative to ry− controls, and this was attributed to rescue of ry− by ry+ in the insertion.
Although ANOVAs detected a significant contribution of insertion and control differences to total variance, F-ratios did not indicate significant increases in variance among insertion line means compared to controls. This is not surprising, considering the relatively small number of lines in our screen and the absence of large effects. In larger screens of P-element insertion lines, others have reported significant effects on variance in bristle number that were attributed mainly to a few insertions with large effects (Mackay et al. 1992; Lyman et al. 1996).
No wing-shape effects have been reported for these genes:
All the genes closest to or including each insertion have been discussed in at least one publication. Six have previously known effects on wing veins, wing bristles, or wing posture, but none were known to affect wing shape. FlyBase lists ∼1900 loci (∼14% of the genome) with known effects on wing morphology, including effects on size, shape, veins, bristles, etc. This entire list includes only four genes encountered in our screen (h, trbl, sgl, sina). Two other implicated genes not on the FlyBase list are known to affect wing morphology in special circumstances: heph (a recessive lethal affecting the wing only in homozygous clones; Dansereau et al. 2002) and stet (affecting the wing only by ectopic expression; Urban et al. 2002). Thus some of these genes do have known effects on the wing, but none have been reported to affect wing shape. Butler et al. (2003) recently reported 56 genes with at least twofold higher expression in the blade and hinge of wing imaginal disks than in the body-wall region. These are likely to include some genes that can affect wing shape, but include none of our 11.
Diverse genetic pathways appear to affect wing shape:
The genes most likely affected by these insertions include some with prominent developmental roles, mainly outside the wing. These include four transcription factors (h, foxo, sina, Alhambra), two genes involved in signaling (sgl, stet), and one gene involved in cell-cycle control (trbl). Also included are a putative spliceosome component (heph), a mitochondrial protein (yip2), and two genes of unknown types (gel, oaf). It is interesting that two genes previously implicated as quantitative trait genes for bristle number—hairy (Robin et al. 2002), and heph Norga et al. (2003)—now turn out to be potential genes for wing shape as well.
Mutations affecting wing morphogenesis have been reported in two other nucleus-encoded mitochondrial genes: colt (Hartenstein et al. 1997) and Gart (Tiong and Nash 1990). Thus it is not implausible that a third such gene may affect wing shape, as suggested by our results for yip2. Although results for Gpdh were inconsistent, it is also not implausible that a basic metabolic enzyme could affect wing shape. For example, the rudimentary (r) locus codes for an enzyme involved in pyrimidine synthesis, and mutations at r cause variation in wing shape (Fausto-Sterling and Hsieh 1976).
No effects are due to amino-acid-coding interruptions:
Seven insertions are in introns, four are in flanking regions, and two are in exons—with one in two categories at once (insertion 25). The two insertions in exons (sina and Gpdh) are both in the 5′-UTR of the first exon, where they could influence either transcription or translation. Of the four insertions in flanking regions, two are just upstream of a gene (h, trbl) in the promoter region, one is near the downstream end (stet), and one falls in a short interval between two genes, upstream of one (yip2) and downstream of the other (Srp54). Insertions near either end of a gene could influence transcription. Of the seven insertions in introns, one is in an intron of a gene (Alhambra) that contains another gene (Mlp84B). Another is in a gene (sina) that is inside an intron of another gene (Rh4). The remaining five insertions are all in introns without such complications (gel, heph, foxo, sgl, and oaf). Insertions in introns could affect transcription rates, alternative transcription start or stop sites, or the frequencies of different splicing patterns. Various P-element insertions have been reported to increase or decrease transcription rates or to change the timing or the location of expression (reviewed in Engels 1989). Insertions might alter gene regulation in other ways. Alteration of the 5′-UTR might affect mRNA stability or change the tertiary structure of mRNA in a way that affects gene expression (Parsch et al. 1997). Insertions near promoters could activate secondary promoter sites.
It is still not clear which genes produce the effects:
Eight insertions are inside genes and four are within a few hundred base pairs of one. In most cases one gene is an obvious primary candidate. However, the effect on wing shape could actually arise from some other nearby gene. Small regulatory influences can extend over local neighborhoods. For example, transgenes with transcription start sites embedded in large P-element vectors are not isolated from surrounding DNA, but show many position effects on the transgene, ranging from strong enhancement of weak promoters to complete suppression of strong promoters (Horn et al. 2003), and other effects such as new expression patterns within normally expressing tissues (Sun et al. 1995). In this study, the distances from some of the insertion sites to other nearby loci are well within the distances of other cis regulatory sites with major effects, reported for various loci with complex regulation (Blackman et al. 1991; Dorsett 1993; Bachmann and Knust 1998; Berman et al. 2002). Maroni (1994) found correlations in length among major functional gene regions, such as the 5′- and 3′-UTRs, the coding region, and the first intron in Drosophila; therefore, longer transcription units might also be influenced by more far-flung regions, even when they seem to be packed between other genes. Where genes are close together, major regulatory effects may be efficiently separated in various ways (Vazquez and Schedl 2000; Bell et al. 2001; Burgess-Beusse et al. 2002; Levine and Tjian 2003). However, even if regulatory compartmentalization is qualitatively complete, minor changes in expression might occur as a kind of leakage through regulatory barriers. Figure 3 shows that some of these insertions are surrounded by many genes. The identity of the gene (or genes) causing the wing-shape effect is especially uncertain in these cases.
The percentage of insertions with effects is unusually high:
Some recent screens have looked for genes with gain-of-function effects in various targets by using a gene that normally turns on in the target to express Gal4, so as to drive overexpression or misexpression in a collection of other genes that carry insertions with a Gal4-driven transcriptional activator (Rørth 1996). Rørth et al. (1998) found various wing effects in 7% of 2300 such insertion lines, driven by a Gal4 source covering most of the wing blade. Abdelilah-Seyfried et al. (2000) found effects on sensory bristles in 5% of 2293 insertion lines, driven by a Gal4 source in the scabrous gene. Kraut et al. (2001) found effects on growth patterns of larval motor neurons in 5% of 2293 lines, driven by a neuron-specific Gal4 source. Peña-Rangel et al. (2002) found thorax modifications in 9% of 2100 lines, driven by a thorax-specific Gal4 source. Finally, Tseng and Hariharan (2002) found phenotypic effects on the eye in 2.3% of 2296 insertion lines, driven by a Gal4 source in the eye disk.
Other screens have looked for direct effects of P-element insertions on specific quantitative traits. In a screen of 379 insertions, ∼4% showed effects on avoidance of an odorant, benzaldehyde (Anholt et al. 1996). Two screens totaling 2825 insertions were analyzed in Norga et al. (2003) for P-element effects on bristle number. When insertion line means for abdominal and sternopleural bristle number were compared to the phenotypic distribution in control lines, >20% had phenotypes outside the 95% confidence limits, and 4–10% had phenotypes outside the 99.9% confidence limits.
A screen of metabolic effects of P-element insertions by Clark et al. (1995) also yielded high percentages. In a screen of 263 random, single, autosomal, homozygous-viable P-element insertions, they found 153 with a significant difference from controls in 1 or more of 16 different traits, of which 1 was body weight, 3 were biochemical fractions, and 12 were enzyme activities. Many insertions caused effects in >1 trait. From the published data, the average number of lines affected per trait would have been ∼24 of 263, or ∼9.1%. Since this is an average, the frequency of effects must have exceeded this in some traits.
We report positive results in 11 of our 50 lines, or 22%, supported by repeated high-resolution tests of individual lines. On the basis of our small sample of lines, the percentage of all random, autosomal, homozygous-viable insertions of P{lacW} that affect wing shape is, with 95% confidence, at least 11.5% (Rohlf and Sokal 1981).
According to Miklos and Rubin (1996), 65–75% of all genes in Drosophila have no obvious loss-of-function phenotype. However, none of the small effects reported here are obvious phenotypes. Perhaps most genes have small loss-of-function phenotypes, if the right features are measured with sufficient precision.
The genome must have a large potential for such variability:
P elements do not insert into DNA sites randomly, and perhaps are not even random with respect to traits (Engels 1989; Spradling et al. 1995). Still, it seems clear from these results that the total number of genes that could affect wing shape must be much higher than the number segregating in any wild population. The latter number was estimated for one trait and population sample as at least 20 by QTL analysis of selected lines (Weber et al. 2001) and at least 140 by the Castle-Wright method (Weber 1990). The number of possible effects must have some inverse relation to their magnitude, but empirical studies are only beginning to quantify how large the mutational target size of the genome is at different magnitudes of effect.
Transposable elements must generate similar variation in nature. P elements can create quantitative genetic variation (Mackay et al. 1992; Keightley et al. 1993), which is selectable (Mackay 1985; Torkamanzehi et al. 1992). In D. melanogaster, ∼10% of the genome consists of transposable elements, and their activity causes at least half of all spontaneous mutations (Finnegan 1992). The long-term evolutionary importance of transposable elements has been recognized in their ability to rearrange bits of the genome and to generate small length variations (Shapiro 1999; Makalowski 2003) and partly depends on how they are usually eliminated from the genome, which is still an issue (Nuzhdin 1999). Any other source of local variation in DNA length—such as imprecise repair of double-strand breaks (Liang et al. 1998)—could probably produce genetic variability in the same sites. Recent detailed comparisons between whole genomes of related organisms show that genomes are constantly undergoing localized reduction, expansion, and rearrangement (Waterston et al. 2002). This may create large amounts of minor regulatory variation.
These effects are all definitely microevolutionary:
None of the effects are clearly visible or qualitative except for the small occasional vein L5 gap noted in line 36. Yet we would almost certainly have detected additional, even smaller effects if we had measured more wings per line. We would probably also have found some large effects, as in other P-element screens (Clark et al. 1995; Lyman et al. 1996), if we had looked at many lines. Our limited study can show only an intermediate region of the distribution.
The primary motivation for this study was to explore the latent evolutionary potential of shape traits in the wing. As P-element insertion screens are applied to more and more traits, with increasing resolution, results show that very large portions of the genome can contribute small effects. What is the evolutionary and adaptive significance of such findings?
If many genes can affect a trait, adaptive flexibility greatly increases. Selection will tend to utilize the most suitable genes with the fewest pleiotropic complications. Moreover, smaller effects allow more precise control of detail, as if one were building with grains of sand instead of stones. But adaptive potential may not reside only in small effects that can accumulate at many loci. It may also depend importantly on particular small effects that have the potential to become large.
Immediate effect is not ultimate adaptive potential:
A gene for a mitochondrial protein might affect wing shape, but would be unlikely to play a role in wing-shape evolution. By contrast, a gene involved in genetic regulation or signaling might have the potential for a large role, even if its immediate mutational effects are small. The effect of an allele can increase by selection of interacting alleles or by incremental changes of the allele itself (Stam and Laurie 1996). Such incremental changes could include multiple regulatory effects on timing, location, or rate of expression, as well as amino acid changes.
Because alleles can evolve, no analysis of quantitative trait genes in a trait that is assumed to have diverged by selection can establish with certainty the original distribution of selected effects by measuring the present distribution of fixed differences in effect, as is often assumed when major trait genes are identified. Because alleles can evolve, a small mutational effect could be seen as the tip of an unknown wedge. Selected effects may start out small, but a bit part might evolve into a starring role in a future adaptation and perhaps do so by steps that are all (in the words of Darwin 1859) “insensibly small.”
Acknowledgments
We thank two anonymous reviewers and Jeff Walker for comments. This project was supported by National Science Foundation grant DEB-9407005 and a grant from the Bioscience Research Institute of Southern Maine to K.W. and National Institutes of Health grant R15GM65116-01 to D.C.
References
- Abdelilah-Seyfried, S., Y.-M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Issa, R., and S. Cavicchi, 1996. Genetic interactions among vestigial, hairy, and Notch suggest a role of vestigial in the differentiation of epidermal and neural cells of the wing and haltere of Drosophila melanogaster. J. Neurogenet. 10: 239–246. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., F. L. Lyman and T. F. C. Mackay, 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann, A., and E. Knust, 1998. Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev. Genes Evol. 208: 346–351. [DOI] [PubMed] [Google Scholar]
- Badenhorst, P., J. T. Finch and A. A. Travers, 2002. Tramtrack co-operates to prevent inappropriate neural development in Drosophila. Mech. Dev. 117: 87–101. [DOI] [PubMed] [Google Scholar]
- Bahri, S. M., W. Chia and X. Yang, 2001. The Drosophila homolog of human AF10/AF17 leukemia fusion genes (Dalf) encodes a zinc finger/leucine zipper nuclear protein required in the nervous system for maintaining EVE expression and normal growth. Mech. Dev. 100: 291–301. [DOI] [PubMed] [Google Scholar]
- Bai, J., W. Chiu, J. Wang, T. Tzeng, N. Perrimon et al., 2001. The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development 128: 591–601. [DOI] [PubMed] [Google Scholar]
- Ballinger, D. G., and S. Benzer, 1989. Targeted gene mutations in Drosophila. Proc. Natl. Acad. Sci. USA 86: 9402–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A. C., A. G. West and G. Felsenfeld, 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291: 447–450. [DOI] [PubMed] [Google Scholar]
- Bergstrom, D. E., C. A. Merli, J. A. Cygan, R. Shelby and R. K. Blackman, 1995. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics 139: 1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker et al., 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier, E., H. Vaessin, S. Shepherd, K. Lee, K. McCall et al., 1989. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3: 1273–1287. [DOI] [PubMed] [Google Scholar]
- Binari, R. C., B. E. Staveley, W. A. Johnson, R. Godavarti, R. Sasisekharan et al., 1997. Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development 124: 2623–2632. [DOI] [PubMed] [Google Scholar]
- Birdsall, K., E. Zimmerman, K. Teeter and G. Gibson, 2000. Genetic variation for the positioning of wing veins in Drosophila melanogaster. Evol. Dev. 2: 16–24. [DOI] [PubMed] [Google Scholar]
- Blackman, R. K., M. Sanicola, L. A. Raftery, T. Gillevet and W. M. Gelbart, 1991. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111: 657–666. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse, B., C. Farrell, M. Glaszner, M. Litt, V. Mutskov et al., 2002. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA 99: 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M. J., T. L. Jacobsen, D. M. Cain, M. G. Jarman, M. Hubank et al., 2003. Discovery of genes with highly restricted expression patterns in the Drosophila wing disc using DNA oligonucleotide microarrays. Development 130: 659–670. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., and J. S. Whyte, 1989. The role of the hairy gene during Drosophila morphogenesis—stripes in imaginal discs. Genes Dev. 3: 905–916. [Google Scholar]
- Carthew, R. W., and G. M. Rubin, 1990. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63: 561–577. [DOI] [PubMed] [Google Scholar]
- Castrillon, D. H., P. Gonczy, S. Alexander, R. Rawson, C. G. Eberhart et al., 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., L. Wang and T. Hulleberg, 1995. P-element-induced variation in metabolic regulation in Drosophila. Genetics 139: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia, T., V. Papayannopoulos, V. Panin, P. Woronoff, J. Jiang et al., 2003. Molecular genetic analysis of the glycosyltransferase Fringe in Drosophila. Proc. Natl. Acad. Sci. USA 100: 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansereau, D. A., M. D. Lunke, A. Finkielsztein, M. A. Russell and W. J. Brook, 2002. hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development 129: 5553–5566. [DOI] [PubMed] [Google Scholar]
- Darwin, C., 1859 On the Origin of Species by Means of Natural Selection, or Preservation of Favoured Races in the Struggle for Life. Murray, London. [PMC free article] [PubMed]
- Dawid, I. B., J. J. Breen and R. Toyama, 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14: 156–162. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., and S. Bray, 1997. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124: 3241–3251. [DOI] [PubMed] [Google Scholar]
- Dorsett, D., 1993. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134: 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., 1989 P elements in Drosophila melanogaster, pp. 437–484 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, DC.
- Fausto-Sterling, A., and L. Hsieh, 1976. Studies on the female-sterile mutant rudimentary of Drosophila melanogaster. Dev. Biol. 51: 269–281. [DOI] [PubMed] [Google Scholar]
- Finnegan, D. J., 1992 Transposable elements, pp. 1096–1107 in The Genome of Drosophila melanogaster, edited by D. L. Lindsley and G. G. Zimm. Academic Press, San Diego/New York/London.
- Frankham, R., D. A. Briscoe and R. K. Nurthen, 1978. Unequal crossing over at the rRNA locus as a source of quantitative genetic variation. Nature 272: 80–81. [DOI] [PubMed] [Google Scholar]
- Gaul, U., G. Mardon and G. M. Rubin, 1992. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell 68: 1007–1019. [DOI] [PubMed] [Google Scholar]
- Goldstein, L. S. B., and S. Gunawardena, 2000. Flying through the Drosophila cytoskeletal genome. J. Cell Biol. 150: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard, A., M. Roark, M. Ronshaugen and E. Bier, 2000. brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogeneisis, promotes EGF-R/MAPK signalling. Dev. Biol. 226: 255–266. [DOI] [PubMed] [Google Scholar]
- Gurganus, M. C., S. V. Nuzhdin, J. W. Leips and T. F. C. Mackay, 1999. High-resolution mapping of quantitative trait loci for sternopleural bristle number in Drosophila melanogaster. Genetics 152: 1585–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker, U., U. Grossniklaus, W. J. Gehring and H. Jäckle, 1992. Developmentally regulated Drosophila gene family encoding the fork head domain. Proc. Natl. Acad. Sci. USA 89: 8754–8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker, U., X. Lin and N. Perrimon, 1997. The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development 124: 3565–3573. [DOI] [PubMed] [Google Scholar]
- Haenlin, M., M. Kunisch, B. Kramatschek and J. A. Campos-Ortega, 1994. Genomic regions regulating early embryonic expression of the Drosophila neurogenic gene Delta. Mech. Dev. 47: 99–110. [DOI] [PubMed] [Google Scholar]
- Haerry, T., E., T. R. Heslip, J. L. Marsh, and M. B. O'Connor, 1997. Defects in glucuronate biosynthesis disrupt Wingless signaling in Drosophila. Development 124: 3055–3064. [DOI] [PubMed] [Google Scholar]
- Hartenstein, K., P. Sinha, A. Mishra, H. Schenkel, I. Török et al., 1997. The congested-like tracheae gene of Drosophila melanogaster encodes a member of the mitochondrial carrier family required for gas-filling of the tracheal system and expansion of the wings after eclosion. Genetics 147: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, Y., M. Okabe, T. Imai, M. Kurusu, A. Yamamoto et al., 1999. Musashi and Seven in absentia downregulate Tramtrack through distinct mechanisms in Drosophila eye development. Mech. Dev. 87: 93–101. [DOI] [PubMed] [Google Scholar]
- Horn, C., N. Offen, S. Nystedt, U. Haecker and E. A. Wimmer, 2003. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 163: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle, D., J. Mezey, P. Galpern and A. Carter, 2003. Automated measurement of Drosophila wings. BMC Evol. Biol. 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham, P. W., K. R. Howard and D. Ish-Horowicz, 1985. Transcription of the Drosophila segmentation gene hairy. Nature 318: 439–444. [Google Scholar]
- Irvine, K. D., and E. Wieschaus, 1994. Fringe, a boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79: 595–606. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., M. J. Evans and W. G. Hill, 1993. Effects of multiple retrovirus insertions on quantitative traits of mice. Genetics 135: 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt, C., 2000. EGF receptor signaling: the importance of presentation. Curr. Biol. 10: 388–391. [DOI] [PubMed] [Google Scholar]
- Klingenberg, C. P., L. J. Leamy, E. J. Routman and J. M. Cheverud, 2001. Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157: 785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotarski, M. A., S. Pickert, D. A. Leonard, G. J. Larosa and R. J. Macintyre, 1983. The characterization of α-glycerophosphate dehydrogenase mutants in Drosophila melanogaster. Genetics 105: 387–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. [DOI] [PubMed] [Google Scholar]
- Kreitman, M., and R. R. Hudson, 1991. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics 127: 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, N., T. Klein, K. Brennan and A. M. Arias, 2000. Structural requirements for Notch signaling with Delta and Serrate during the development and patterning of the wing disc of Drosophila. Development 127: 3185–3195. [DOI] [PubMed] [Google Scholar]
- Lehembre, F., P. Badenhorst, S. Muller, A. Travers, F. Schweisguth et al., 2000. Covalent modification of the transcriptional repressor tramtrack by the ubiquitin related protein Smt3 in Drosophila flies. Mol. Cell. Biol. 20: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M., and R. Tjian, 2003. Transcription regulation and animal diversity. Nature 424: 147–151. [DOI] [PubMed] [Google Scholar]
- Liang, F., M. Han, P. J. Romanienko and M. Jasin, 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95: 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, San Diego/New York/London.
- Littleton, J. T., 2000. A genomic analysis of membrane trafficking and neurotransmitter release in Drosophila. J. Cell Biol. 150: 77–82. [DOI] [PubMed] [Google Scholar]
- Liu, J., J. M. Mercer, L. F. Stam, G. C. Gibson, Z-B. Zeng et al., 1996. Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics 142: 1129–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., S. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, R. F., F. Lawrence, S. V. Nuzhdin and T. F. C. Mackay, 1996. Effects of single P-element insertions on bristle number and viability in Drosophila melanogaster. Genetics 143: 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F. C., 1985. Transposable element-induced response to artificial selection in Drosophila melanogaster. Genetics 111: 351–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. a The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. b Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2: 11–20. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., R. F. Lyman and M. S. Jackson, 1992. Effects of P element insertions on quantitative traits in Drosophila melanogaster. Genetics 130: 315–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W., 2003. Not junk after all. Science 300: 1246–1247. [DOI] [PubMed] [Google Scholar]
- Maroni, G., 1994. The organization of Drosophila genes. DNA Sci. 4: 347–354. [DOI] [PubMed] [Google Scholar]
- Mata, J., S. Curado, A. Ephrussi and P. Rorth, 2000. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101: 511–522. [DOI] [PubMed] [Google Scholar]
- Merli, C., D. E. Bergstrom, J. A. Cygan and R. K. Blackman, 1996. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Miklos, G., and G. Rubin, 1996. The role of the genome project in determining gene function: insights from model organisms. Cell 86: 521–529. [DOI] [PubMed] [Google Scholar]
- Milan, M., S. Campuzano and A. García-Bellido, 1996. a Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc. Natl. Acad. Sci. USA 93: 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan, M., S. Campuzano and A. García-Bellido, 1996. b Cell cycling and patterned cell proliferation in the Drosophila wing during metamorphosis. Proc. Natl. Acad. Sci. USA 93: 11687–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso Del Prado, J., and A. García-Bellido, 1984. Cell interactions in the generation of chaetae pattern in Drosophila. Roux's Arch. Dev. Biol. 193: 246–251. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., H. Okano, J. A. Blendy and C. Montell, 1994. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13: 67–81. [DOI] [PubMed] [Google Scholar]
- Norga, K. K., M. C. Gurganus, C. L. Dilda, A. Yamamoto, R. F. Lyman et al., 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13: 1388–1397. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., 1999. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica 107: 129–137. [PubMed] [Google Scholar]
- O'Hare, K., and G. M. Rubin, 1983. Structure of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34: 25–35. [DOI] [PubMed] [Google Scholar]
- Parsch, J., S. Tanda and W. Stephan, 1997. Site-directed mutations reveal long-range compensatory interactions in the Adh gene of Drosophila melanogaster Proc. Natl. Acad. Sci. USA 94: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Rangel, M. T., I. Rodriguez and J. R. Riesgo-Escovar, 2002. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 160: 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, P. C., 1999. From complex traits to complex alleles. Trends Genet. 15: 6–8. [DOI] [PubMed] [Google Scholar]
- Pi, H., H.-J. Wu and C.-T. Chien, 2001. A dual function of phyllopod in Drosophila external sensory organ development: cell fate specification of sensory organ precursor and its progeny. Development 128: 2699–2710. [DOI] [PubMed] [Google Scholar]
- Pirotta, V., 1986 Cloning Drosophila genes, pp. 83–109 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL Press, Oxford.
- Price, D. M., Z. Jin, S. Rabinovitch and S. D. Campbell, 2002. Ectopic expression of the Drosophila Cdk1 inhibitory kinases, Wee1 and Myt1, interferes with the second mitotic wave and disrupts pattern formation during eye development. Genetics 161: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A., 1967 The nature of quantitative genetic variation, pp. 265–280 in Heritage From Mendel, edited by R. A. Brink and E. D. Styles. University of Wisconsin Press, Madison, WI.
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. Johnson-Schlitz, W. K. Benz et al., 1988. A stable source of P-element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, C., R. F. Lyman, A. D. Long, C. H. Langley and T. F. C. Mackay, 2002. hairy: a quantitative trait locus for Drosophila sensory bristle number. Genetics 162: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch, F., F. Serras, F. J. Cifuentes, M. Corominas, B. Alsina et al., 1998. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: clonal analysis and morphology of imaginal discs. Mol. Gen. Genet. 257: 103–112. [DOI] [PubMed] [Google Scholar]
- Rohlf, F. J., and R. R. Sokal, 1981 Statistical Tables, Ed. 2. W. H. Freeman, San Francisco.
- Rørth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Rørth, P., K. Szabo and G. Texido, 2000. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell 6: 23–30. [DOI] [PubMed] [Google Scholar]
- Rushlow, C. A., A. Hogan, S. M. Pinchin, K. M. Howe, M. Lardelli et al., 1989. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 8: 3095–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg, A., S. N. Prokopenko, Y. He, P. Tsai, M. Pal et al., 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: mutations affecting embryonic PNS development. Genetics 147: 1723–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, C., C. G. Wood, D. L. Jones, S. I. Tazuke and M. T. Fuller, 2002. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129: 4523–4534. [DOI] [PubMed] [Google Scholar]
- Seher, T. C., and M. Leptin, 2000. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr. Biol. 10: 623–629. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1999. Transposable elements as the key to a 21st century view of evolution. Genetica 107: 171–179. [PubMed] [Google Scholar]
- Shrimpton, A. E., and A. Robertson, 1988. The isolation of polygenic factors controlling bristle score in Drosophila melanogaster. II. Distribution of third chromosome bristle effects within chromosome sections. Genetics 118: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, F. A., F. M. Hoffmann and W. M. Gelbart, 1982. Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell 28: 451–461. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, I. Kiss, J. Roote, T. Laverty et al., 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92: 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, L. F., and C. C. Laurie, 1996. Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 144: 1559–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronach, B. E., S. E. Siegrist and M. C. Beckerle, 1996. Two muscle-specific LIM proteins in Drosophila. J. Cell Biol. 134: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronach, B. E., P. J. Renfranz, B. Lilly and M. C. Beckerle, 1999. Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol. Biol. Cell 10: 2329–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. H., C.-J. Tsai, M. M. Green, J.-L. Chao, C.-T. Yu et al., 1995. white as a reporter gene to detect transcriptional silencers specifying position-specific gene expression during Drosophila melanogaster eye development. Genetics 141: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray, J. R., P. C. W. Gaines, P. Ebert and J. R. Carlson, 1998. small wing encodes a phospholipase C-(γ) that acts as a negative regulator of R7 development in Drosophila. Development 125: 5033–5042. [DOI] [PubMed] [Google Scholar]
- Thompson, J. N., and A. D. Preston, 1992. The Drosophila segmentation gene hairy responds to a late postembryonic gradient in the wing. Dros. Inf. Serv. 71: 247–249. [Google Scholar]
- Tiong, S. Y., and D. Nash, 1990. Genetic analysis of the adenosine3 (Gart) region of the second chromosome of Drosophila melanogaster. Genetics 124: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba, G., T. Ohsako, N. Miyata, T. Ohtsuka, K.-H. Seong et al., 1999. The gene search system: a method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamanzehi, A., C. Moran and F. W. Nicholas, 1992. P element transposition contributes substantial new variation for a quantitative trait in Drosophila melanogaster. Genetics 131: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, A.-S. K., and I. K. Hariharan, 2002. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, S., J. R. Lee and M. Freeman, 2002. A family of rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 21: 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, J., and P. Schedl, 2000. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics 155: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen, E. M., K. J. Purcell, M. E. Fortini and S. Artavanis-Tsakonas, 1996. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics 144: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas, P., M. A. Vickers, D. L. Simmons, H. Ayyub, C. F. Craddock et al., 1992. Cis-acting sequences regulating expression of human α-globin cluster lie within constitutively open chromatin. Cell 69: 781–793. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–561. [DOI] [PubMed] [Google Scholar]
- Weber, K. E., 1990. Selection on wing allometry in Drosophila melanogaster. Genetics 126: 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, K. E., 1992. How small are the smallest selectable domains of form? Genetics 130: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, K. E., R. Eisman, L. Morey, A. Patty, J. Sparks et al., 1999. An analysis of polygenes affecting wing shape on chromosome 3 in Drosophila melanogaster. Genetics 153: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, K. E., R. Eisman, S. Higgins, L. Morey, A. Patty et al., 2001. An analysis of polygenes affecting wing shape on chromosome 2 in Drosophila melanogaster. Genetics 159: 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas, K., N. Slepecky, L. Von Kalm and D. Sullivan, 1997. Flight muscle function in Drosophila requires colocalization of glycolytic enzymes. Mol. Biol. Cell 8: 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, E., A. Palsson and G. Gibson, 2000. Quantitative trait loci affecting components of wing shape in Drosophila melanogaster. Genetics 155: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]