Abstract
DNA palindromes are rare in humans but are associated with meiosis-specific translocations. The conserved Mre11/Rad50/Nbs1 (MRN) complex is likely directly involved in processing palindromes through the homologous recombination pathway of DNA repair. Using the fission yeast Schizosaccharomyces pombe as a model system, we show that a 160-bp palindrome (M-pal) is a meiotic recombination hotspot and is preferentially eliminated by gene conversion. Importantly, this hotspot depends on the MRN complex for full activity and reveals a new pathway for generating meiotic DNA double-strand breaks (DSBs), separately from the Rec12 (ortholog of Spo11) pathway. We show that MRN-dependent DSBs are formed at or near the M-pal in vivo, and in contrast to the Rec12-dependent breaks, they appear early, during premeiotic replication. Analysis of mrn mutants indicates that the early DSBs are generated by the MRN nuclease activity, demonstrating the previously hypothesized MRN-dependent breakage of hairpins during replication. Our studies provide a genetic and physical basis for frequent translocations between palindromes in human meiosis and identify a conserved meiotic process that constantly selects against palindromes in eukaryotic genomes.
MEIOSIS, the hallmark of sexual reproduction, allows eukaryotes to transmit through their germline all the genetic characteristics of the species to the next generation. In most species, meiosis is characterized by a high level of homologous recombination during prophase of the first meiotic division. Meiotic recombination creates new combinations of alleles, thereby increasing genetic diversity, and also generates the connection between homologs that allows their proper alignment and segregation by the meiotic spindle. Cells undergoing the meiotic program must therefore allow genome-wide recombination yet at the same time keep this recombination under control to avoid deleterious outcomes such as ectopic recombination and translocations. Cells have solved this problem by allowing recombination to happen in a timely and spatially defined manner after premeiotic DNA replication, when the recombining homologs are closely juxtaposed or synapsed (Roeder 1997).
Recombination is stimulated by lesions in DNA. Accidental DNA lesions can be introduced into the genome from exogenous sources, such as exposure to UV and ionizing radiation or radiomimetic chemicals, or from endogenous sources, such as free radicals generated during normal cellular metabolism. These lesions threaten genome integrity but can be repaired with efficiencies that depend on the stage of the cell cycle during which they happen (Lee et al. 1997). Much more dangerous to cells are genomic DNA elements that can adopt secondary structures and be cleaved by nucleases. Such elements can be faithfully repaired by recombination with a sister or a homolog. But they can also be hotspots of DNA breakage with a high potential to recombine with ectopic sequences and thereby generate translocations. This may be one reason why DNA palindromes are underrepresented in genomes, including that of humans (Lobachev et al. 2000).
Cells have protein complexes to deal with such deleterious DNA elements. The best studied of these complexes is the Mre11/Rad50/Nbs1 (MRN) complex (in Saccharomyces cerevisiae Xrs2 replaces Nbs1 and the complex is denoted MRX). This complex is conserved from bacteriophages to mammals and is involved in many aspects of DNA metabolism (Haber 1998). The MRN complex associates with S-phase chromatin and is required for the intra-S-phase checkpoint (D'Amours and Jackson 2001; Maser et al. 2001; Chahwan et al. 2003). Xenopus extracts depleted of Mre11 accumulate doublestrand breaks (DSBs) during DNA replication (Costanzo et al. 2001). This accumulation of DSBs may be the basis of MRN being essential in vertebrates (Luo et al. 1999; Zhu et al. 2001).
The involvement of the MRN complex in palindrome elimination is revealed by both in vitro and in vivo studies. MRN or MRX complexes isolated from human cells or budding yeast cells, respectively, cleave hairpin structures, thus supporting the notion that the MRN complex can process palindromes (Paull and Gellert 1999; Trujillo and Sung 2001; Trujillo et al. 2003). Palindromes introduced into both fission yeast and budding yeast behave as MRN-dependent mitotic recombination hotspots (Farah et al. 2002; Lobachev et al. 2002). Most notably, in fission yeast, palindrome elimination through gene conversion is dependent on a wild-type MRN complex (Farah et al. 2002). In a model that integrates the above results, we proposed that during replication the MRN complex cleaves an extruded palindrome, thus generating a DSB that is further processed by the recombination machinery (Farah et al. 2002). However, no MRN-dependent in vivo DSBs have been reported at any palindrome studied during the mitotic cell cycle.
Meiosis is probably the ideal stage at which to eliminate palindromes because recombination between homologs is strongly stimulated at the expense of recombination between sisters, where recombination would regenerate a palindrome (Collins and Newlon 1994; Schwacha and Kleckner 1994). This is the case in S. cerevisiae, where a 140-bp palindrome is a meiotic recombination hotspot and is eliminated through homologous recombination entirely dependent on Spo11 (Nag and Kurst 1997; Nasar et al. 2000). In mice and humans, DNA palindromes promote genomic instability during germline development (Kurahashi and Emanuel 2001a; Zhou et al. 2001). The MRN (or MRX) complex has not been reported to be specifically involved in initiating any of the above meiotic processes.
Using Schizosaccharomyces pombe as a model system, we show here that a 160-bp palindrome is a meiotic recombination hotspot. Importantly, we show that the hotspot is dependent on the MRN complex, in addition to Rec12 (ortholog of human Spo11), for full activity. We show for the first time that MRN-dependent DSBs are formed in vivo during premeiotic DNA replication, lending support to the model that the MRN complex eliminates palindromes during S phase. We show that the MRN-dependent pathway generates crossovers and needs the Rad16/Swi10 (ortholog of human XPF/ERCC1) complex to efficiently eliminate the palindrome. We discuss the implications of this discovery of a new MRN-dependent meiotic recombination pathway on the stability of the genome during germline development.
MATERIALS AND METHODS
Yeast strains, plasmids, and genetic techniques:
Supplementary Table 1 (part A) at http://www.genetics.org/supplemental/ lists the S. pombe strains, their genotypes, and the crosses or sporulations in which they were used. For meiotic crosses, single colonies were transferred to 5 ml of supplemented yeast extract liquid medium (YEL + 5S; Davis and Smith 2003) and grown at 30° until saturated. An equal volume of each parental strain was mixed, washed in 0.85% NaCl, and spotted on supplemented sporulation agar plates (SPA; Gutz et al. 1974). The plates were incubated for 3 days at 25°, after which the cell-ascus mixture was suspended in 1 ml of H2O and treated with glusulase and ethanol to kill vegetative cells, essentially as described by DeVeaux et al. (1992).
For chromosome-by-chromosome ade6 intragenic recombinant frequency measurements, spores were plated on supplemented yeast extract agar plates (YEA + 4S; Davis and Smith 2003) for total spore count and on YEA + 4S + guanine (100 μg/ml; Cummins and Mitchison 1967) for Ade+ recombinant count. YEA + 4S contains a limited amount of adenine, which allows Ade6− strains to form red or pink colonies, depending on the ade6 allele. For plasmid-by-chromosome ade6 intragenic recombinant frequency measurements, plasmid-containing cells were grown in supplemented liquid EMM2 minimal medium (Davis and Smith 2003) devoid of uracil to select for the plasmid and crossed with strain GP3336 grown in YEL + 5S. Spores from such crosses were plated on supplemented EMM2 agar plates devoid of uracil for total plasmid-containing spore count and on YEA + 4S + guanine for total Ade+ recombinant count. Plates were incubated at 32° for 3–4 days before counting the colonies. Except as noted, statistical analyses used Student's paired t-test.
For intergenic recombination measurements (Table 6B), individual spore colonies were gridded on YEA + guanine (selected Ade+ colonies) or on YEA + adenine (total colonies) and incubated for 3 days at 25°. The plates were replica plated onto properly supplemented nitrogen-base minimal agar plates (NBA; Ponticelli and Smith 1989) to score ura4+-aim and onto supplemented YEA + phloxin B plates (20 μg/ml; Moreno et al. 1991) at 37° to score tps16. Ura4+ Tps16+ Arg1+ diploids, detected as large cells by microscopic analysis (for the rec12Δ mus81+ crosses) or by segregation of Ura− colonies (for the rec12Δ mus81Δ crosses), were omitted from the analysis.
TABLE 6.
Mus81 is essential for generating crossovers associated with M-pal-dependent Ade+ convertants
| Parental genotypea |
Ade+ recombinant/106 viable sporesb |
||||||
|---|---|---|---|---|---|---|---|
| Cross series | mus81 | rec12 | SI × 52 | M-pal × 52 | |||
| A. Gene conversion at ade6 | |||||||
| 19 | + | + | 185 ± 20 | 1260 ± 100 | |||
| 20 | Δc | + | NAd | NAd | |||
| 21 | + | Δ152 | 8 ± 3 | 410 ± 65 | |||
| 22 | Δ | Δ152 | 2 | 140 ± 45 | |||
| Parental genotypef |
% among Ade+ spores
|
||||||
| NCOg
|
COg
|
||||||
| Cross | rec12 | mus81 | P1 | P2 | R1 | R2 | Total Ade+ tested |
| B. Crossovers among the M-pal-dependent Ade+ convertantse | |||||||
| 23 | Δ152 | + | 58.6 | 3.3 | 36.9 | 1.2 | 336 |
| 24 | Δ152 | Δ | 88.4 | 4.4 | 5.8 | 1.3 | 292 |
Strains with either ade6-3034 (SI) or ade6-3036 (M-pal) were crossed with strain GP3813 (relevant genotype: h− ade6-52 rec12Δ mus81Δ). Crosses were homozygous for the indicated mutations or heterozygous for the indicated wild-type (+) alleles.
Values are the means of four independent experiments ±SEM. For cross series 22 (SI × 52), the total Ade+ recombinants obtained was divided by the combined total viable spores (from the four experiments). Crosses were performed at 25°.
mus81::kanMX6 (Δ) is a deletion of the mus81 ORF (Boddy et al. 2000).
Not available; the very low spore yield precluded reliable measurement of the Ade+ recombinant frequency.
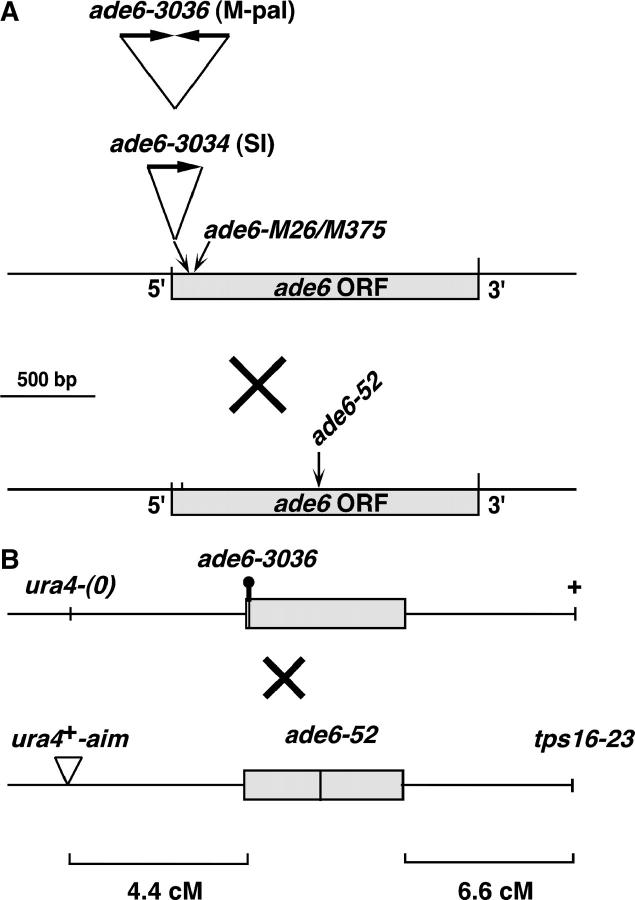
Selected Ade+ convertant spore colonies were classified according to configuration of the flanking markers ura4+-aim and tps16 (Figure 1B). The parental (NCO) configurations were P1 (ade6-3036 tps16+), corresponding to parents GP4182 and GP4183, and P2 (ura4+-aim ade6-52 tps16-23), corresponding to parents GP4198 and GP4199. The recombinant (CO) configurations were R1 (ura4+-aim tps16+) and R2 (tps16-23).
Crosses were homozygous for the rec12 and mus81 alleles.
NCO, noncrossover; CO, crossover.
Diploid strains were constructed by mixing h+ leu1-32 strains with h− his7-366 strains on supplemented SPA plates; leu1 and his7 are closely linked on chromosome 2. After 10–12 hr at 25° cells from the edge of each spot were streaked and maintained on NBA + adenine + uracil plates. For the experiments shown in Table 4, a diploid colony was grown in YEL + 5S at 30° until saturation, and 1 ml was washed in H2O and spotted onto supplemented EMM2 plates. After 3 days, when most of the cells had sporulated, spores were harvested and analyzed as above.
TABLE 4.
M-pal on a plasmid is a meiotic recombination hotspot in the absence of Rec12
| Parental genotypea |
Ade+ recombinants/106 viable sporesb
|
|||
|---|---|---|---|---|
| Cross series | rad50 | rec12 | SI × 52 | M-pal × 52 |
| 11 | + | + | 580 ± 52 | 1600 ± 170 |
| 12 | S | + | 480 ± 70 | 510 ± 190 |
| 13 | + | Δ152 | 9 | 310 ± 43 |
| 14 | S | Δ152 | 20 ± 6 | 40 ± 11 |
Strains harboring the ade6-D19 deletion and either plasmid pJF141 [containing the ade6-3034 (SI) allele] or plasmid pJF138 [containing the ade6-3036 (M-pal) allele] were crossed with strain GP3336 (ade6-52 rec12Δ rad50S). Crosses were homozygous for the indicated mutations or heterozygous for the indicated wild-type (+) alleles. Crosses were performed at 25°.
Values are the means of three independent experiments ±SEM. For cross series 13 (SI × 52), the total Ade+ recombinants obtained were divided by the combined total viable spores (from the three experiments).
Plasmids pJF138 and pJF141 (Farah et al. 2002) were introduced into cells by electroporation (Prentice 1992).
Meiotic inductions and DNA break analysis:
Supplementary Table 1 (part B) at http://www.genetics.org/supplemental/ lists the S. pombe strains used for meiotic induction and physical analysis of DNA. Meiotic inductions and preparation of DNA in agarose plugs were performed as described (Young et al. 2002). The agarose-embedded DNA was digested with PacI, separated by pulsed-field gel electrophoresis, blotted, and hybridized as described previously (Steiner et al. 2002). Strains GP4207 (SI rad50+ rec12+), GP4802 (M-pal rad50S rec12Δ), and GP4899 (SI rad50+ rec12Δ) were induced and analyzed by Southern blot hybridization only once (Figure 2, B, D, and G). All other strains were induced and analyzed at least twice and showed reproducible results (Figure 2, A, C, E, and F). Samples (1 ml) collected at the indicated time points from the meiotic induction cultures were processed and analyzed for DNA content by flow cytometry as described (Li and Smith 1997).
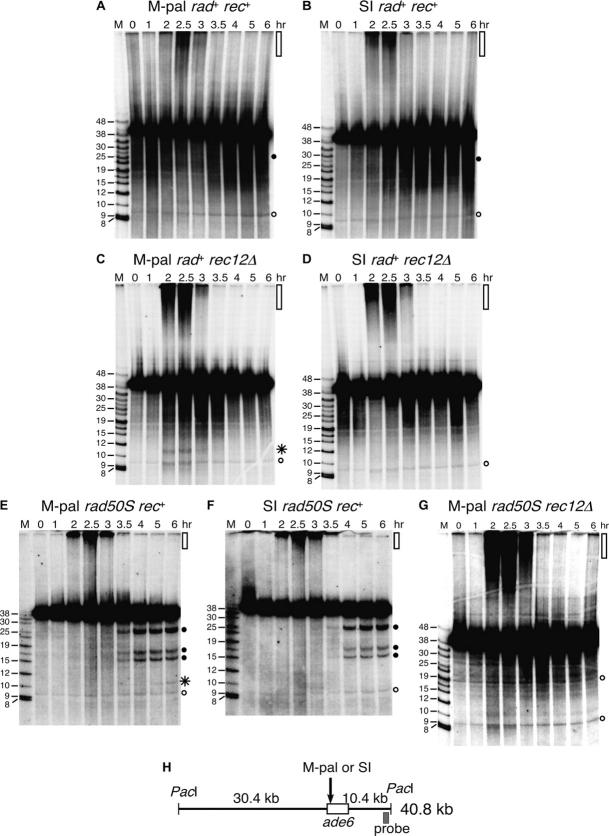
Figure 2.—
Early (Rad50-dependent) and late (Rec12-dependent) DSBs at or near the M-pal. DNA was prepared at the indicated times after thermal induction (34°) of pat1-114 meiosis, digested with PacI, separated by pulsed-field gel electrophoresis, blotted, and probed from the right as indicated in H. Size markers (in kilobases) are in lane M. (A) Strain GP4653 [ade6-3036 (M-pal) rad50+ rec12+]. (B) Strain GP4207 [ade6-3034 (SI) rad50+ rec12+]. (C) Strain GP4388 (ade6-3036 rad50+ rec12Δ). (D) Strain GP4899 (ade6-3034 rad50+ rec12Δ). (E) Strain GP3279 (ade6-3036 rad50S rec12+). (F) Strain GP3276 (ade6-3034 rad50S rec12+). (G) Strain GP4802 (ade6-3036 rad50S rec12Δ). (H) Diagram of the PacI fragment containing the ade6 gene. Shown are the positions of the M-pal or SI, ade6 ORF, and probe used. Rec12-dependent meiosis-specific DSBs are indicated by solid circles. M-pal-dependent breaks are indicated by an asterisk. Nonspecific hybridization, present at each time point, is indicated by open circles. High-molecular-weight DNA, between 2 and 3 hr likely representing replicating branched DNA, is indicated by open vertical bars at the top of the scans.
RESULTS
M-pal is a meiotic recombination hotspot dependent on both the Rec12 protein and the Rad32/Rad50/Nbs1 complex:
We measured meiotic recombination at the ade6 locus, which harbors the 160-bp palindrome allele ade6-3036 (or M-pal) near the beginning of the ade6 ORF (Figure 1A). This allele is a mitotic recombination hotspot relative to a control allele consisting of a single insertion of one arm of the palindrome (ade6-3034 single insertion allele or SI; Farah et al. 2002). We determined whether the M-pal was also a meiotic recombination hotspot by measuring the recombination frequency in crosses involving the M-pal or SI allele and the ade6-52 test allele, as depicted in Figure 1. The hotspot activity of the M-pal was determined as the ratio of the recombinant frequency in crosses between the M-pal and the test allele divided by that between the SI and the same test allele. The M-pal-dependent recombination frequency was fivefold higher than that involving the SI control allele (Table 1, cross series 1 and 5), indicating that the M-pal is a meiotic recombination hotspot. Because the size of the M-pal (160 bp) rather than the potential secondary structure that it could adopt might be responsible for the observed hotspot effect, we also tested a direct-repeat insertion of the 80-bp fragment (ade6-3035 direct-repeat insertion allele or DR). The Ade+ recombination frequency in crosses involving the DR allele and the ade6-52 test allele was not significantly different from that involving the SI control allele (data not shown), indicating that the M-pal-dependent hotspot was not due to the size of the inserted fragment in ade6.
Figure 1.—
Genetic markers used in this study. (A) ade6 alleles. The ade6-3034 and ade6-3036 alleles, described by Farah et al. (2002), consist of one or two copies, respectively, of an 80-bp DNA segment inserted at the BamHI site (bp 24) of the ade6 ORF (GenBank accession no. M37264). The DNA corresponds to 76 bp from the S. cerevisiae mat-a-stk locus (bp 2044–2119 of GenBank accession no. V01313; Ray et al. 1991), plus a 5′ 4-bp BamHI compatible extension. The inserted DNA is not drawn to scale. Both alleles confer an Ade− phenotype. The ade6-M26 allele is a G-to-T transversion at bp 136 of the ade6 ORF and the ade6-M375 allele is a G-to-T transversion at bp 133 of the ade6 ORF (Szankasi et al. 1988). The common test allele ade6-52 is a G-to-A transition at bp 796 of the ade6 ORF (M. E. Fox, unpublished results) and is separated by 773 and 661 bp from ade6-3036 and ade6-M26, respectively. (B) Markers flanking ade6. The ura4+-aim insertion and tps16-23 (temperature-sensitive) markers were described previously (Grimm et al. 1994); tps16 is identical to ags1, 56.1 kb centromere distal to ade6 (Hochstenbach et al. 1998). ura4+-aim is an insertion of a 1.8-kb ura4+ cassette ∼15 kb from ade6. (0) indicates the absence of an insertion. The diagram is not drawn to scale.
TABLE 1.
M-pal is a meiotic recombination hotspot dependent on Rec12, Rad50, and Rad32
| Parental genotypea
|
Ade+ recombinants/106 viable sporesb |
||||
|---|---|---|---|---|---|
| Cross series | rad50 | rad32 | rec12c | SI × 52 | M-pal × 52 |
| 1 | + | + | + | 265 ± 15 | 1455 ± 50 |
| 2 | S | + | + | 220 ± 22 | 420 ± 40 |
| 3 | + | + | Δ152 | 4 | 470 ± 45 |
| 4 | S | + | Δ152 | 11 | 27 ± 5 |
| 5 | + | + | + | 210 ± 30 | 1900 ± 500 |
| 6 | + | D65N | + | NAd | NAd |
| 7 | + | + | Δ171 | 1 | 410 ± 20 |
| 8 | + | D65N | Δ171 | 14 ± 5 | 11 ± 2 |
Strains with either the ade6-3034 (SI) allele or the ade6-3036 (M-pal) allele were crossed with either strain GP3335 (ade6-52 rec12Δ rad50S; cross series 1–4) or strain GP4077 (ade6-52 rec12Δ rad32-D65N; cross series 5–8). Crosses were homozygous for the indicated mutations or heterozygous for the indicated wild-type (+) alleles. Crosses were performed at 25°.
Values are the means of four (cross series 1–4) or three (cross series 5–8) independent experiments ±SEM. For cross series 3, 4, and 7 (SI × 52), the total Ade+ recombinants obtained were divided by the combined total viable spores from the four (cross series 3 and 4) or three (cross series 7) experiments.
The rec12-152::LEU2 allele (Δ152) partially deletes, and the rec12-171::ura4+ allele (Δ171) completely deletes, the rec12 ORF (Lin and Smith 1994; Davis and Smith 2003).
Not available; the very low viable spore yield precluded reliable measurement of the Ade+ recombinant frequency.
Previous ade6 recombination hotspots determined by mutations creating the M26 heptamer and closely related sequences are completely dependent on Rec12, the protein that introduces meiosis-specific DSBs (DeVeaux et al. 1992; Fox et al. 2000). To determine if the M-pal hotspot was also dependent on Rec12, we performed crosses homozygous for a rec12 disruption and measured recombination involving either the M-pal or the SI control allele (Table 1, cross series 3 and 7). As expected, the rec12Δ mutation nearly abolished the SI-dependent recombination frequency. In contrast, the M-pal-dependent recombination frequency was reduced by a factor of only 3, to a level higher than that of the wild-type (rec12+) control involving the SI allele. Therefore, recombination involving the M-pal was only partially dependent on Rec12. In its absence, the M-pal was still a hotspot, perhaps because another protein can generate recombinogenic lesions at or near the M-pal.
Since the putative nuclease activity of the MRN complex is essential for the mitotic hotspot activity of the M-pal (Farah et al. 2002), we tested whether this complex was also involved in the meiotic hotspot activity of the M-pal. In particular, we tested the rad50S-K81I allele (hereafter denoted rad50S), a separation-of-function allele of rad50 that completely abolishes the mitotic hotspot activity of the M-pal (Farah et al. 2002). In crosses involving the M-pal, the Ade+ recombinant frequency was reduced three- to fourfold in the rad50S background relative to that in the rad50+ background (Table 1, cross series 2). Recombination involving the SI control allele was not significantly altered by rad50S (P > 0.2), indicating that the rad50S mutation affected specifically the M-pal. This conclusion was substantiated by crosses involving the ade6-M26 hotspot: the Ade+ recombinant frequencies in these crosses were also not significantly affected by the rad50S allele (P > 0.3, Table 2). The M-pal-dependent hotspot activity was not abolished in rad50S crosses: a hotspot activity of ∼2 was still observed in crosses involving the M-pal when compared to the SI-dependent crosses (Table 1, cross series 2; P < 0.015). Because the rad50S mutation completely abolishes the mitotic hotspot activity of the M-pal (Farah et al. 2002), the residual meiotic hotspot activity observed in rad50S crosses suggested that the M-pal rendered the DNA in its vicinity more accessible to Rec12. As expected, the M-pal-dependent recombination was strongly reduced in the rad50S rec12Δ double mutant (Table 1, cross series 4). These results show that the M-pal-dependent meiotic recombination was dependent on both Rec12 and the MRN complex, whereas M26 recombination was dependent on only Rec12 (Table 2; DeVeaux et al. 1992).
TABLE 2.
Therad50S allele has no effect on theM26 hotspot
| Ade+ recombinants/106 viable sporesb
|
|||
|---|---|---|---|
| Cross series | Parental genotypearad50 | M375 × 52 | M26 × 52 |
| 9 | + | 510 ± 70 | 9000 ± 1750 |
| 10 | S | 680 ± 100 | 7100 ± 170 |
Strains with either the ade6-M375 allele or the ade6-M26 allele were crossed with strain GP3337 (relevant genotype: h− ade6-52 rad50S). Cross series 9 was heterozygous, and cross series 10 was homozygous for rad50S. Crosses were performed at 25°.
Values are the means of three independent experiments ±SEM.
We postulated previously that the MRN complex was the nuclease responsible for generating the mitotic recombinogenic lesions at or near the M-pal (Farah et al. 2002). To test whether the nuclease domain of the Rad32 protein was involved in generating the meiotic M-pal hotspot, we used the rad32-D65N allele, which substitutes Asn for Asp at codon 65, a highly conserved residue in esterase motif II among the Mre11 family members (Moreau et al. 1999). The rad32-D65N allele had a strong deleterious effect on viable spore yield, which precluded determination of the recombinant frequency at ade6 (Table 1, cross series 6; see also Table 3, sporulation series V, discussed below). The very low viable spore yield in rad32-D65N crosses was suppressed in the absence of Rec12 (Table 3, sporulation series VI), suggesting that this mutant makes but does not repair meiotic DSBs, as shown previously in S. cerevisiae and for S. pombe rad32Δ mutants (Moreau et al. 1999; Young et al. 2004). The M-pal-dependent recombination observed in the absence of Rec12 (Table 1, cross series 7) was essentially eliminated by the rad32-D65N allele (Table 1, cross series 8), suggesting that the nuclease activity of the MRN complex was the likely source of the Rec12-independent recombinogenic lesions. In turn, these results suggest that at 25° the rad50S allele abolishes the ability of the Rad32 nuclease domain to cleave secondary structures in the DNA, such as extruded palindromes, but not its ability to promote other aspects of meiotic recombination (see below).
TABLE 3.
Therad50S allele is temperature sensitive for viable spore formation and mimics a putativerad32nuclease mutation at the restrictive temperature
| Genotypea
|
Relative viable spore yieldb (% of wild type) |
||||
|---|---|---|---|---|---|
| Sporulation series |
rad50 | rad32 | rec12c | 25° | 34° |
| I | + | + | + | 100 | 100 |
| II | + | + | 117 | 16 ± 5 | 10 ± 3.1 |
| III | S | + | + | 23 ± 9.5 | 0.035 ± 0.013 |
| IV | S | + | 117 | 15 ± 3.2 | 5.6 ± 0.3 |
| V | + | D65N | + | 0.12 ± 0.095 | 0.011 ± 0.008 |
| VI | + | D65N | 117 | 70 ± 27 | 6.2 ± 0.83 |
Diploid strains were homozygous for the alleles shown.
Cultures were sporulated on supplemented EMM2 plates. The results are from three independent experiments. The number of viable spores from each experiment was divided by the corresponding number of viable diploid cells deposited on the sporulation plate; the mean (±SEM) for each series was determined and expressed relative to the value of the wild type at each temperature. Wild-type values were 1.3 ± 0.55 (25°) and 6.8 ± 2.5 (34°).
The rec12-117 allele results in the G202E substitution (data not shown) and appears to be null (Lin and Smith 1994).
Because the M-pal is a mitotic recombination hotspot when present on a plasmid (Farah et al. 2002), we determined whether the M-pal on a plasmid was a hotspot also during meiosis. The M26 heptamer and related hotspots are clearly not active when present on a plasmid (Ponticelli and Smith 1992; Virgin et al. 1995; Fox et al. 2000). We transformed S. pombe strains having a chromosomal ade6 deletion (ade6-D19 allele) with plasmids harboring either the M-pal or the SI control allele and crossed these strains with an ade6-52 tester strain. M-pal on a plasmid was clearly a meiotic recombination hotspot, relative to the SI control, that was dependent on the MRN complex (Table 4, cross series 12 compared to cross series 11) but not on Rec12 (cross series 13 compared to cross series 11). This result suggested that the plasmid-borne M-pal extruded into a hairpin susceptible to the nuclease activity of the MRN complex. In the presence of the rad50S mutation the M-pal hotspot activity was completely eliminated, suggesting that the M-pal on the plasmid did not stimulate Rec12 action, contrary to what we inferred previously from the chromosome-by-chromosome recombination experiments (Table 1, cross series 2).
Taken together, the above results reveal a novel recombination pathway dependent on the MRN complex that may specifically eliminate deleterious DNA elements such as palindromes during germline development.
rad50S is a ts allele that eliminates the MRN pathway but allows processing of meiotic DSBs at 25°:
All rad50S crosses gave high viable spore yields (Tables 1, 2, and 4), despite the fact that meiotic DSBs accumulate and remain unrepaired in rad50S strains (Young et al. 2002). If DSBs accumulate in rad50S crosses, we would expect the viable spore yield to be dramatically lower than that observed. Because the genetic crosses were done at 25° but meiotic DSBs were assessed at 34° (Young et al. 2002), the rad50S allele might have affected meiosis differently at the two temperatures. Hence, we tested the effect of temperature on the viable spore yield of a rad50S strain, and we used a rad32 nuclease (rad32-D65N) mutant strain as a control for low viable spore yield at all temperatures (Table 3, sporulation series V).
rad50S was temperature sensitive with respect to viable spore yield (Table 3). At 25° the viable spore yield of a rad50S strain was only moderately reduced (∼4-fold) compared to wild type (compare sporulation series I and III). In contrast, the viable spore yield of the rad32-D65N strain was reduced by ∼800-fold relative to that of wild type (compare sporulation series I and V). At 34°, however, both the rad50S and the rad32-D65N strains gave very low viable spore yields, ∼3000 and 8000-fold lower than that of wild type, respectively (sporulation series III and V). Importantly, a rec12 null allele when combined with either rad50S or rad32-D65N strongly suppressed the viable spore yield defects (sporulation series IV and VI), suggesting that the defect was dependent on DSB formation. This conclusion is supported by the suppression of the low viable spore yield of rad50Δ and rad32Δ mutations by rec12Δ and rec6Δ mutations (Young et al. 2004).
The rad50S allele was neither completely null nor temperature sensitive during mitotic growth: at both 25° and 34°, a rad50S haploid strain was less resistant to UV, MMS, and bleomycin than a rad50+ strain but clearly more resistant to these agents than a rad50Δ strain (data not shown). The M-pal-dependent mitotic recombination hotspot was reduced to the same extent at both 23° and 30° by the rad50S mutation (Farah et al. 2002; data not shown). Taken together, the above results suggested that the MRSN (Rad32/Rad50S/Nbs1) complex was deficient in the nuclease activity essential for processing meiotic DSBs only at 34°, but deficient in a structure-specific nuclease activity that cleaves palindromes at both 25° and 34°. The Rad32-D65N/Rad50/Nbs1 complex, with the Asp-to-Asn change at the putative nuclease active site (Moreau et al. 1999), may completely lack both nuclease activities.
DSBs at or near the M-pal and dependent on MRN are temporally separable from those dependent on Rec12:
In our model of M-pal-dependent mitotic hotspot activation, we proposed that during lagging-strand DNA synthesis the M-pal opposite a single-strand gap extrudes into a hairpin and becomes susceptible to cleavage by the MRN complex, thereby forming a recombinogenic DSB (Farah et al. 2002). We wished to determine whether DSBs were made at or near the M-pal during meiosis of wild-type (rad+ rec+) or mutant strains. The ade6 locus was analyzed by Southern blot hybridization of DNA isolated from synchronously induced cultures. We analyzed first a wild-type (rad+ rec+) strain but failed to detect any M-pal-dependent DSBs over a time course of 6 hr that spanned both meiotic divisions (compare Figure 2A with Figure 2B; Li and Smith 1997; see below); meiotic DNA breakage is detected at prominent chromosomal sites from ∼3 to 4 hr in wild-type cells (Cervantes et al. 2000).
In contrast, in both rad50S and rec12Δ mutant strains, M-pal-dependent DSBs were visible but at different times after induction. In the rec12Δ strain, where only the MRN-dependent pathway was active, DSBs appeared ∼2 hr after induction of meiosis, coinciding with premeiotic DNA replication, as determined both by bulk DNA synthesis assayed by flow cytometry (supplementary Figure 1 at http://www.genetics.org/supplemental/) and by DNA accumulating in the wells of the gels and likely representing replicating, branched DNA (Figure 2C). These “early” Rec12-independent DSBs were M-pal specific because they were not detectable in the rec12Δ strain harboring the SI control allele at ade6 (compare Figure 2C with Figure 2D). These early breaks were repaired by 3.5 hr after meiotic induction, when replication was complete. In the rad50S strain, presumably with a nuclease-deficient MRN complex at 34°, the temperature used for meiotic induction, meiosis-specific DSBs appeared at ∼4 hr after meiotic induction, slightly later than Rec12-dependent DSBs elsewhere on chromosome 3, and remained unrepaired at least until 6 hr (Figure 2E). These “late” DSBs were M-pal specific because they were not detectable in the rad50S strain harboring the SI control allele at ade6 (compare Figure 2E with Figure 2F). Neither “early” nor “late” DSBs were detected in a rad50S rec12Δ strain (Figure 2G).
It is unclear why the early and late DSBs seen in the rec12Δ and the rad50S mutant backgrounds, respectively, were not more readily visible in the wild-type (rad+ rec+) M-pal-containing strain. Perhaps, in the rec12Δ background, the absence of other nearby meiotic DSBs reduced the amount of background hybridization, making the faint early M-pal-dependent breaks more readily visible. In the rad50S background, the absence of break processing at 34° (see Table 3) allows the accumulation and therefore the detection of faint breaks, such as those arising late at the M-pal (Steiner et al. 2002; Young et al. 2002).
The MRN-dependent meiotic recombination pathway requires both Rad16 and Mus81 for full activity:
The ade6-3036 (M-pal) allele was constructed by inserting 160 bp of heterologous DNA into the ade6 ORF (Figure 1). If the MRN complex cleaves within the M-pal, the DSBs would have heterologous 3′-ends that might need to be removed for productive strand invasion and subsequent recombination. The human ERCC1/XPF complex and the orthologous S. cerevisiae Rad1/Rad10 complex are involved in nucleotide excision repair but also remove 3′-end heterologous tails from recombining DSB ends during mitotic growth (Fishman-Lobell and Haber 1992; Adair et al. 2000). The S. pombe Rad16/Swi10 putative complex has properties similar to those of the Rad1/Rad10 complex (Carr et al. 1994).
We found that Rad16 was indeed required for full activity of the M-pal-dependent meiotic hotspot. In crosses involving the M-pal, recombination was reduced ∼2.5-fold in the absence of Rad16 (Table 5; compare cross series 15 and 16; P < 0.01). This reduction was specific to crosses involving the M-pal, since crosses involving the SI control allele were not significantly affected by the rad16Δ mutation (cross series 15 and 16; P > 0.1). This result suggested that meiosis-specific lesions with 3′ heterologies were made at the M-pal. In the absence of Rec12, when only the MRN-dependent recombination pathway was available, we found that recombination was reduced ∼4-fold in the absence of Rad16 (Table 5, cross series 17 and 18; P < 0.05). This suggested that the majority (∼75%) of the MRN-dependent recombinants originated from lesions inside or near the M-pal and hence required Rad16 for removing 3′ heterology. The remainder of the Rad16-independent recombinants generated in the MRN-dependent pathway presumably originated from lesions outside or at the 5′ edge of the M-pal and thus provided homologous recombinogenic 3′-ends. Alternatively, these recombinants could have originated from a minor pathway, perhaps involving a 3′–5′ exonuclease (such as that of DNA polymerase δ or the MRN complex itself), that removed the 3′ heterologies from the DSB ends generated inside the M-pal by the MRN complex.
TABLE 5.
Rad16 stimulates specifically M-pal-dependent recombination
| Parental genotypea |
Ade+ recombinants/106 viable sporesb |
|||
|---|---|---|---|---|
| Cross series | rad16 | rec12 | SI × 52 | M-pal × 52 |
| 15 | + | + | 150 ± 7 | 1150 ± 57 |
| 16 | Δc | + | 140 ± 11 | 440 ± 32 |
| 17 | + | Δ152 | 2 ± 0.3 | 405 ± 89 |
| 18 | Δ | Δ152 | 6 ± 1 | 100 ± 19 |
Strains with either ade6-3034 (single insertion; SI) or ade6-3036 (M-pal) were crossed with strain GP4655 [relevant genotype: h−(smt-0) ade6-52 rec12Δ rad16Δ]. Crosses were homozygous for the indicated mutations and heterozygous for the indicated wild-type (+) alleles. Crosses were performed at 25°.
Values are the means of three independent experiments ±SEM.
The rad16::ura4+ (Δ) is a deletion of the rad16 ORF (Carr et al. 1994).
The Rec12-dependent pathway shows a high association of crossovers among gene convertants at ade6 and other loci, and crossovers depend upon Mus81 (Grimm et al. 1994; Osman et al. 2003; Smith et al. 2003; Cromie et al. 2005). We wanted to know whether this property was also shared by the MRN-dependent pathway. Hence, we determined MRN-dependent recombination frequencies in the presence or absence of Mus81/Eme1, the complex likely responsible for resolving crossover products in S. pombe. As a meiotic Holliday junction (HJ) resolvase, the Mus81/Eme1 complex is essential for the successful completion of meiosis in S. pombe, but not when DSB formation is blocked, such as in the absence of Rec12 (Boddy et al. 2001). Crosses homozygous for the mus81Δ mutation gave a very low viable spore yield (0.1–0.01% of that of a mus81+ cross), which precluded reliable measurement of the Ade+ recombinant frequency (Table 6A, cross series 20).
The occurrence of M-pal-dependent recombination in the absence of Rec12 allowed us to determine the effect of a mus81Δ mutation on the ade6 intragenic recombination in a situation where the viable spore yield was comparable to that of a rec12Δ cross (10–20% of wild type, Table 3). In the absence of Rec12, mus81Δ reduced the Ade+ recombinant frequency with the M-pal by a factor of ∼3 (Table 6A, cross series 21 and 22). This reduction was statistically significant (P < 0.004) and indicates that ∼30% of the Ade+ convertants originated through a pathway that did not involve resolution of a HJ by the Mus81/Eme1 complex. Hence, these results suggest that events that generate conversion associated with crossovers by the MRN-dependent pathway are lost and not channeled into a conversion-only pathway in the absence of Mus81.
The Rec12-dependent recombinants likely arise during meiosis, since rec12 is a meiosis-specific gene (Lin and Smith 1994; Mata et al. 2002). The data in Table 6A indicate that most of the Rec12-independent recombinants also arise during meiosis. The mus81Δ mutation had no significant effect on mitotic recombination [recombination rate of 670/106 division compared to 700/106 division for a mus81+ strain harboring ade6-3036 (M-pal) on the chromosome and ade6-469 on a plasmid (Farah et al. 2002; data not shown)]. Thus, the reduction in the M-pal-dependent, Rec12-independent meiotic recombinants by mus81Δ indicates that approximately two-thirds of these recombinants arose from postmitotic events, i.e., after entry into meiosis.
We next looked directly at the effect of mus81Δ on the frequency of flanking marker crossovers among the M-pal-dependent Ade+ convertants in the absence of Rec12. Crossovers show a high association with meiotic conversion events at the ade6 locus (Grimm et al. 1994; Cromie et al. 2005), reflecting the common origin of gene conversions and crossovers via DSB repair. Thirty-eight percent of the Ade+ convertants generated by the MRN-dependent pathway showed crossing over between the flanking markers diagrammed in Figure 1B (Table 6B, cross series 23), while among unselected spores <2% had crossovers (no crossovers among 165 colonies tested). If, as expected, the M-pal generates the recombinogenic lesions and converts to wild type, the configuration of flanking markers corresponding to the parent with the M-pal should be more frequent than that of the other parent, as observed (Table 6B, cross series 23). Thus, the M-pal is preferentially eliminated by conversion to wild type. The frequency of crossing over among convertants generated by the MRN-dependent pathway dropped to ∼7% in the absence of Mus81 (Table 6B, cross series 23 and 24). Therefore, as in rec12+ crosses (Osman et al. 2003; Smith et al. 2003), Mus81 is specifically required to generate crossover convertants, but not noncrossover convertants.
DISCUSSION
DNA palindromes have the propensity to extrude into hairpins or cruciforms and hence form discontinuities in the genome, especially during active processes such as transcription and replication. The consequences could be stalling of the replication machinery and eventually fork breakage (Leach 1994). Cells may remove palindromes by the action of structure-specific nucleases that cleave and eliminate such DNA elements. This process is often detected as an increase in homologous recombination when appropriate genetic markers are used (Nag and Kurst 1997; Lobachev et al. 1998, 2002; Waldman et al. 1999; Farah et al. 2002). But cleaving the DNA at a palindrome could also be harmful, because this could lead to translocation of chromosome arms with consequent mutation or aneuploidy, such as the palindrome-associated human translocations responsible for severe congenital abnormalities (Edelmann et al. 2001; Kurahashi and Emanuel 2001a; Kurahashi et al. 2003). Hence, palindrome elimination must be carefully regulated to avoid debilitating outcomes. We have begun characterizing the palindrome elimination pathways and their regulation during meiosis in S. pombe to reveal how palindromes promote recombination that may lead to translocations.
A palindrome-associated meiotic recombination hotspot dependent on two pathways of DNA breakage by Rec12 or by the MRN complex:
The M-pal inserted at ade6 behaved as a meiotic recombination hotspot (Table 1). The requirement of both Rec12 and the MRN complex for full hotspot activity of the M-pal strongly suggested that each of these components could initiate recombinogenic lesions at or near the M-pal by two genetically separable pathways. This feature seems not to be shared by S. cerevisiae, since in that yeast a 140-bp palindrome is fully (>98%) dependent on Spo11 for stimulating meiotic recombination; hence, any MRN-dependent, Spo11-independent recombination is below the level of detection (Nasar et al. 2000; for simplicity, the S. cerevisiae MRX complex will be referred to as the MRN complex in this discussion). Unlike the situation in S. pombe where the MRN complex is not required for generating Rec12-dependent meiotic DSBs, the MRN complex is essential for generating Spo11-dependent DSBs in S. cerevisiae (Cao et al. 1990; Young et al. 2004). This requirement for MRN explains the full dependence of the palindrome recombination on the MRN complex in S. cerevisiae (Nasar et al. 2000). In S. cerevisiae a longer palindrome is, however, an MRN-dependent mitotic recombination hotspot, but DNA break repair, not cleavage, is MRN dependent (Lobachev et al. 2002).
The Rec12-dependent pathway was accompanied by formation of DSBs after premeiotic DNA replication at a time when meiosis-specific DSBs accumulate at other locations on chomosome 3 (late breaks; Figure 2E). Since in a rad50S strain the M-pal was not active on a plasmid (Table 4), local chromatin structure may be an important determinant for M-pal activity by the Rec12-dependent pathway. In that view, extrusion of the M-pal on the chromosome would perturb neighboring chromatin, which in turn would stimulate Rec12 binding or DNA breakage. Chromatin of a plasmid, on the other hand, may not be susceptible to meiosis-specific perturbation. This may explain why, in the few tested cases, hotspots not dependent on secondary structure formation, such as M26 and related sequences, are not active when present on a plasmid (Ponticelli and Smith 1992; Virgin et al. 1995; Fox et al. 2000). Alternatively, chromatin configuration on a plasmid may not be appropriate for Rec12 action under any circumstance.
The MRN-dependent pathway was completely dependent on the putative nuclease activity of the MRN complex (Table 1) and formed DSBs during premeiotic DNA replication (early breaks; Figure 2C). Interestingly, this pathway was active when the M-pal was present on a plasmid (Table 4), consistent with the proposal that MRN-dependent breaks are dependent on extrusion of the M-pal. The M-pal-dependent, MRN-dependent, and Rec12-independent recombinant frequency measured in Tables 1, 5, and 6 (∼420 Ade+ recombinants/106 viable spores) was higher than the M-pal-dependent Ade+ mitotic recombinant rate of a rad+ diploid strain (280 Ade+ recombinants/106 divisions; Farah et al. 2002). Furthermore, most of the meiotic MRN-dependent Ade+ recombinants were Mus81 dependent (Table 6), unlike most of the mitotic recombinants (as noted in the results). Therefore, we conclude that many of the MRN-dependent Ade+ recombinants arose during meiosis.
Since MRN-dependent recombination was strongly reduced by a rad16Δ mutation (Table 5), we infer that the breaks were located inside the extruded palindrome, perhaps at the tip of the hairpin. This view is substantiated by the ability of purified S. cerevisiae MRN complex to cleave hairpin loops in an ATP-stimulated reaction (Trujillo and Sung 2001; Trujillo et al. 2003). If the extruded M-pal is cleaved by the MRN complex during replication on the lagging strand, then a DSB could readily ensue, since the nicked M-pal would be opposite a gap (Figure 3A).
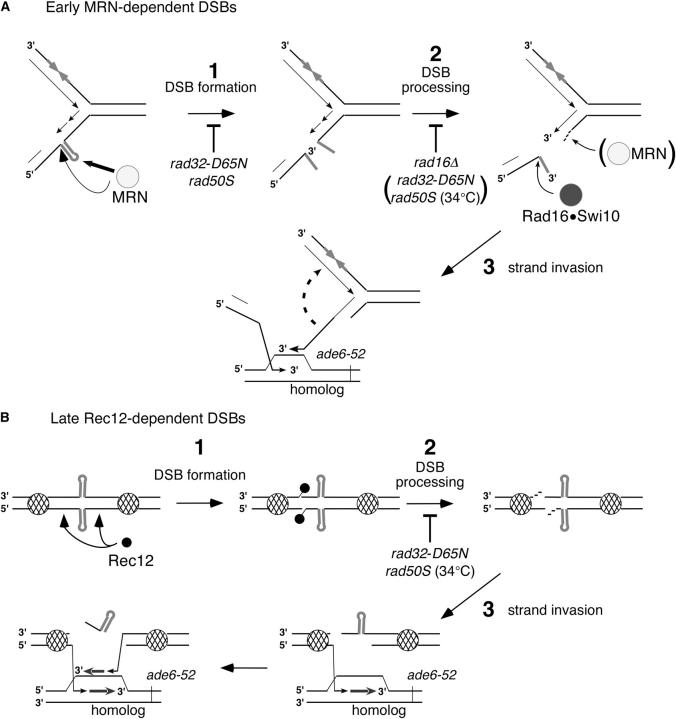
Figure 3.—
Model for the meiotic elimination of the M-pal by the MRN and Rec12-dependent pathways. (A) MRN-dependent pathway. During premeiotic S phase, the M-pal extrudes on the lagging DNA strand, generating a hairpin (Leach 1994). The MRN complex recognizes and cleaves the hairpin. Cleaveage by the MRN complex could happen at the base of the stem, thus generating a homologous 3′-end ready for strand invasion. Alternatively, the MRN complex could cleave inside the hairpin, generating a heterologous 3′-end that needs to be removed by Rad16/Swi10 to allow the subsequent steps of recombination. In the presence of the rad50S allele at 25°, cleavage of the hairpin is severely impaired but subsequent processing of the DSB is active. In the presence of the rad32-D65N allele, cleavage of the hairpin is eliminated, and it is unknown whether DSB end processing is inhibited. These MRN-dependent DSBs are likely the lesions responsible for generating the Rec12-independent Ade+ recombinants. The MRN-dependent DSBs may also recombine with the newly synthesized sister chromatid, as indicated by the dashed arrow, and therefore may not generate an observable recombinant. (B) Rec12-dependent pathway. DNA in the vicinity of the M-pal is devoid of nucleosomes (cross-hatched circles) perhaps upon extrusion of the M-pal. This allows Rec12 to introduce DSBs in the vicinity of the hairpin after premeiotic DNA synthesis. The rad50S allele at 25° has negligible effects on this pathway as observed in crosses involving either the SI control allele or the M26 hotspot (Tables 1, 2, and 4). In the presence of the rad32-D65N allele (or the rad50S allele at 34°), processing of Rec12-generated DSBs in ade6 and elsewhere in the genome is blocked and spore formation is severely defective.
The S. pombe rad50S allele separates two critical nuclease activities of the MRN complex:
The MRN complex is a critical component of the genome-safeguarding machinery of cells from bacteria (where it is denoted SbcCD) to humans. Among its numerous activities, the MRN complex is important for the intra-S-phase checkpoint, telomere maintenance, and genome stability (Symington 2002). A crucial role of the MRN complex occurs during meiosis when it is essential for processing Spo11 (Rec12)-dependent DSBs (Cao et al. 1990; Young et al. 2004). Processing of meiotic DSBs is dependent on the nuclease activity of the MRN complex, as nuclease-deficient MRN complexes fail to process meiotic DSBs (Moreau et al. 1999). Because hypomorphic mutations in other components of the MRN complex, such as rad50S mutations, display a phenotype very similar to that of MRN nuclease mutations, it is tempting to suggest that rad50S mutations inhibit the nuclease activity of the MRN complex.
The finding that Rad50S/S mutant mice complete meiosis efficiently and are fertile suggests that the rad50S mutation does not affect the end-processing nuclease activity of the MRN complex in all organisms (Bender et al. 2002). In S. pombe, the accumulation of unrepaired DSBs during the thermal induction of meiosis in a rad50S strain at 34° indicates that the rad50S mutation blocks a step essential for meiotic DSB processing (Young et al. 2002). However, this result was difficult to reconcile with the mild effect of the mutation on the viable spore yield at 25° (Table 3). We found, however, that the S. pombe rad50S mutation was temperature sensitive for viable spore formation and that this phenotype was dependent on the presence of Rec12 (Table 3). Hence, the S. pombe MRN complex has at least two activities, both of which need a functional Rad32 nuclease subunit. One “structure-specific” activity involves nicking DNA secondary structures (such as the M-pal). This activity is strongly inhibited by the rad50S allele at 25°. The other “end-processing” activity processes meiotic DSBs and is not affected by the rad50S allele at 25°. At higher temperature (34°), the rad50S mutation inactivates both activities and hence displays a rad32 nuclease-deficient phenocopy. In mice Rad50S/S mutant cells, the “structure-specific” nuclease activity of the MRSN complex may be absent and this deficiency may be responsible for the observed chromosome instability phenotype, underlining the importance of this activity for genome maintenance (Bender et al. 2002). The fertility of female Rad50S/S mice implies, however, that the MRSN complex is functional in processing meiotic DSBs at 37° (Bender et al. 2002).
A model for M-pal-dependent meiotic recombination in S. pombe:
The M-pal is a small foreign element that introduces a discontinuity in the genome when extruded into a hairpin or cruciform. Hence, cells may eliminate such an element to avoid its interfering with DNA metabolism. Elimination of the M-pal is reflected by the preference with which it is converted to wild type in an MRN-dependent manner during both mitotic growth and meiosis (Table 6B; Farah et al. 2002). We propose that the main pathway for eliminating the M-pal or other similar DNA elements operates during DNA replication, when the M-pal is most likely to extrude and become susceptible to the MRN nuclease (Figure 3A). The MRN complex cleaves either inside the M-pal, an event that requires further processing by the Rad16/Swi10 endonuclease, or near the base of the M-pal. In both cases breaks are made early in meiosis during the time of replication. It is unclear whether, in the absence of covalently bound Rec12, the MRN complex is involved in further processing of the 5′-end to generate long 3′ single-strand tails required for strand invasion. Because this pathway of recombination is initiated before full juxtaposition of the homologous chromosomes, its fidelity may not be as high as that involving Rec12 and “late” breaks.
The extruded M-pal may perturb the chromatin in its vicinity, facilitating Rec12 access or action. We propose that the late DSBs formed by Rec12 are at the M-pal or close by (as shown in Figure 3B) in a chromatin-accessible region. Processing of the DSB ends is fully dependent on the MRN complex: M-pal dependent DSBs accumulate unrepaired in the rad50S background at high temperature (Figure 2E). Hence, the Rec12-dependent hotspot observed at the M-pal also contributes to the elimination of this element, but indirectly through perturbation of the local chromatin structure.
Palindrome menace:
We have shown that a small, artificially inserted DNA element with a propensity to disrupt the continuity in the DNA double helix actively recombines during meiosis in S. pombe. Most of the time recombination is homologous and involves either a sister chromatid or a homologous chromosome. In some rare cases, however, we envision that ectopic recombination occurs and leads to chromosomal translocations. This could happen because Mus81-dependent crossovers do occur in the MRN-dependent pathway (Table 6B) and could well involve ectopic sequences, such as two palindromes located on different chromosomes that become simultaneously cleaved by the safeguarding machinery of the cell. Frequent chromosomal translocations are observed when two DSBs, but not one, are introduced in the genome of mouse embryonic stem cells (Richardson and Jasin 2000). In humans chromosomal translocations arise in the germline between chromosome 22 and either chromosome 11 or chromosome 17. Interestingly, palindromes are present at each of these translocation breakpoints; the smallest palindrome involved, on chromosome 17, is only ∼195 bp (Edelmann et al. 2001; Kurahashi and Emanuel 2001a,b; Kurahashi et al. 2003). The translocation breakpoints occur at the center of symmetry of the palindromes, lending support to the notion that during meiosis two simultaneous DSBs at the tip of two extruded palindromes favor ectopic rejoining. We propose that such events happen during premeiotic DNA replication when the palindromes are extruded and the homologs are not yet juxtaposed or synapsed.
The deleterious effect of palindromes might explain why nearly identical, closely spaced Alu repeats in inverted orientation are underrepresented in the human genome (Lobachev et al. 2000). Large palindromes introduced into mice are rearranged at a high rate during both germline development and somatic growth by a mechanism presumably involving hairpin nicking of an extruded palindrome followed by imprecise rejoining (Akgun et al. 1997; Cunningham et al. 2003).
The MRN-dependent pathway is not specific for meiosis, as MRN-dependent palindrome elimination also occurs in mitotic cells with the potential formation of translocations and other genomic aberrations (Farah et al. 2002). However, unless associated with other tumor-promoting events, reciprocal translocations would be of limited risk for the organism. During meiosis, on the other hand, such translocations may be much more deleterious because they would be transmitted through the germline and compromise the next generation. It will be important to determine whether the two recombination pathways described here for palindrome elimination during S. pombe meiosis operate in multicellular organisms and, if so, how they are regulated.
Acknowledgments
We thank Sue Amundsen, Linda Breeden, Luther Davis, Harmit Malik, Michael McMurray, Andrew Taylor, and Julio Vazquez for critical reading of the manuscript. We also thank Nick Boddy, Paul Russell, Edgar Hartsuiker, Tony Carr, and Felicity Watts for strains. G.C. was supported by a long-term European Molecular Biology Organization fellowship (ALTF 123-2001) and W.W.S. by a Leukemia and Lymphoma Society Special Fellowship (3230-05). This work was supported by research grant GM-32194 from the National Institutes of Health to G.R.S.
References
- Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson et al., 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19: 5552–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgun, E., J. Zahn, S. Baumes, G. Brown, F. Liang et al., 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17: 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, C. F., M. L. Sikes, R. Sullivan, L. E. Huye, M. M. Le Beau et al., 2002. Cancer predisposition and hematopoietic failure in Rad50S/S mice. Genes Dev. 16: 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer et al., 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20: 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., P.-H. L. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates et al., 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Cao, L., E. Alani and N. Kleckner, 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carr, A. M., H. Schmidt, S. Kirchhoff, W. J. Muriel, K. S. Sheldrick et al., 1994. The rad16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell and N. Rhind, 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23: 6564–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, I., and C. S. Newlon, 1994. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell 76: 65–75. [DOI] [PubMed] [Google Scholar]
- Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco et al., 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8: 137–147. [DOI] [PubMed] [Google Scholar]
- Cromie, G. A., C. Rubio, R. W. Hyppa and G. R. Smith, 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins, J. E., and J. M. Mitchison, 1967. Adenine uptake and pool formation in the fission yeast Schizosaccharomyces pombe. Biochim. Biophys. Acta 136: 108–120. [DOI] [PubMed] [Google Scholar]
- Cunningham, L. A., A. G. Cote, C. Cam-Ozdemir and S. M. Lewis, 2003. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 23: 8740–8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and S. P. Jackson, 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15: 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163: 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann, L., E. Spiteri, K. Koren, V. Pulijaal, M. G. Bialer et al., 2001. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 68: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta and G. R. Smith, 2002. A 160-bp palindrome is a Rad50•Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell, J., and J. E. Haber, 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- Fox, M. F., T. Yamada, K. Ohta and G. R. Smith, 2000. A family of CRE-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics 156: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., J. Bähler and J. Kohli, 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 135: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974 Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Haber, J. E., 1998. The many interfaces of Mre11. Cell 95: 583–586. [DOI] [PubMed] [Google Scholar]
- Hochstenbach, F., F. M. Klis, H. van den Ende, E. van Donselaar, P. J. Peters et al., 1998. Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc. Natl. Acad. Sci. USA 95: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi, H., and B. S. Emanuel, 2001. a Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 10: 2605–2617. [DOI] [PubMed] [Google Scholar]
- Kurahashi, H., and B. S. Emanuel, 2001. b Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 29: 139–140. [DOI] [PubMed] [Google Scholar]
- Kurahashi, H., T. Shaikh, M. Takata, T. Toda and B. S. Emanuel, 2003. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 72: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, D. R., 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., R. A. Mitchell, A. Cheng and E. A. Hendrickson, 1997. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 17: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. F., and G. R. Smith, 1997. The Schizosaccharomyces pombe rec16 gene product regulates multiple meiotic events. Genetics 146: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and G. R. Smith, 1994. Transient meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev, K. S., B. M. Shor, H. T. Tran, W. Taylor, J. D. Keen et al., 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev, K. S., D. A. Gordenin and M. A. Resnick, 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Lobachev, K. S., J. E. Stenger, O. G. Kozyreva, J. Jurka, D. A. Gordenin et al., 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19: 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl et al., 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96: 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams et al., 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21: 6006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., R. Lyne, G. Burns and J. Bahler, 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32: 143–147. [DOI] [PubMed] [Google Scholar]
- Moreau, S., J. R. Ferguson and L. S. Symington, 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. J. S. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nag, D. K., and A. Kurst, 1997. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics 146: 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar, F., C. Jankowski and D. K. Nag, 2000. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol. 20: 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Paull, T. T., and M. Gellert, 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1992. Context dependence of a eukaryotic recombination hotspot. Proc. Natl. Acad. Sci. USA 89: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice, H. L., 1992. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 20: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, B. L., C. I. White and J. E. Haber, 1991. Heteroduplex formation and mismatch repair of the “stuck” mutation during mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5372–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C., and M. Jasin, 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405: 697–700. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., M. N. Boddy, P. Shanahan and P. Russell, 2003. Fission yeast Mus81•Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, W. W., R. W. Schreckhise and G. R. Smith, 2002. Meiotic DNA breaks at the S. pombe recombination hotspot M26. Mol. Cell 9: 847–855. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi, P., W. D. Heyer, P. Schuchert and J. Kohli, 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe: wild-type and mutant alleles including the recombination hotspot allele ade6–M26. J. Mol. Biol. 204: 917–925. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., and P. Sung, 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50•Mre11 complex. J. Biol. Chem. 276: 35458–35464. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., D. H. Roh, L. Chen, S. Van Komen, A. Tomkinson et al., 2003. Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J. Biol. Chem. 278: 48957–48964. [DOI] [PubMed] [Google Scholar]
- Virgin, J. B., J. Metzger and G. R. Smith, 1995. Active and inactive transplacement of the M26 recombination hotspot in Schizosaccharomyces pombe. Genetics 141: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman, A. S., H. Tran, E. C. Goldsmith and M. A. Resnick, 1999. Long inverted repeats are an at-risk motif for recombination in mammalian cells. Genetics 153: 1873–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. A., R. W. Schreckhise, W. W. Steiner and G. R. Smith, 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Young, J. A., R. W. Hyppa and G. R. Smith, 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. H., E. Akgun and M. Jasin, 2001. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl. Acad. Sci. USA 98: 8326–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., S. Petersen, L. Tessarollo and A. Nussenzweig, 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11: 105–109. [DOI] [PubMed] [Google Scholar]