Abstract
The strategy of combining genes from a regulatory protein and its antagonist within the same operon, but controlling their activities differentially, can lead to diverse regulatory functions. This protein-antagonist motif is ubiquitous and present in evolutionarily unrelated regulatory pathways. Using the sin operon from the Bacillus subtilis sporulation pathway as a model system, we built a theoretical model, parameterized it using data from the literature, and used bifurcation analyses to determine the circuit functions it could encode. The model demonstrated that this motif can generate a bistable switch with tunable control over the switching threshold and the degree of population heterogeneity. Further, the model predicted that a small perturbation of a single critical parameter can bias this architecture into functioning like a graded response, a bistable switch, an oscillator, or a pulse generator. By mapping the parameters of the model to specific DNA regions and comparing the genomic sequences of Bacillus species, we showed that phylogenetic variation tends to occur in those regions that tune the switch threshold without disturbing the circuit function. The dynamical plasticity of the protein-antagonist operon motif suggests that it is an evolutionarily convergent design selected not only for particular immediate function but also for its evolvability.
BACTERIAL survival is dependent on the ability to respond rapidly to shifting environmental conditions. The capacity for phenotypic exploration—or evolvability—can be manifested in multiple hierarchies in biology (Kirschner and Gerhart 1998). Evolutionary search is accelerated when systems are organized to reduce the number of simultaneous mutations required before beneficial variants are discovered (Kirschner and Gerhart 1998; Voigt et al. 2001). There are several organizational principles by which this can be achieved. For example, the combination of modularity and robustness minimizes the effect of mutations on other functions and accelerates the exploration of novel regions of parameter space by neutral drift (Huynen et al. 1996).
Cellular systems have evolved to deal with short-term adaptation to environmental fluctuation by implementation of the appropriate stress responses. On longer timescales, evolution modifies circuitry both by changing the kinetic parameters of the network and through the addition and subtraction of network components and reactions. A robust, yet evolvable, network architecture is designed such that a large fraction of possible mutations leads only to small or quantitative changes in circuit behavior, and a much smaller fraction can change the qualitative dynamics to new, dynamically important functions. In addition, the evolvable parameters should be able to sample diversity without traversing large, nonfunctional regions of parameter space.
Savageau's demand theory is one of the first formalisms for relating the architecture of a network to its evolvability (Savageau 1998). This theory asserts that frequently needed genes are controlled by repressors whereas rarely used genes are controlled by activators. This arrangement ensures that mutations that disrupt regulation maintain gene expression for high-demand genes, whereas low-demand genes are turned off, thus reducing metabolic load. Circuit designs that conform to the demand theory principle are relatively robust to mutation, thereby permitting a greater exploration of parameter space. The demand theory was first used to investigate differences in the regulatory mechanisms employed by Escherichia coli for metabolizing lactose, which is ubiquitous, vs. maltose, which is only present in a small region of the gut. The evolutionary capacity for changes in the circuit dynamics of the lac operon has also been studied. van Oudenaarden and co-workers experimentally demonstrated that a switch-like response to lactose induction could be changed to a graded response by perturbing a single parameter (Ozbudak et al. 2004). Considering multiple inputs, Alon and co-workers used a mathematical model to propose that the lac operon could be tuned from a fuzzy AND gate to other logic blocks, such as an OR and pure AND (Setty et al. 2003).
In addition to the lac operon, a number of other synthetic and natural genetic circuits have been analyzed for robustness and evolvability. A synthetic circuit that was designed to be an oscillator was shown to convert into a bistable switch when one of the interactions was removed (Atkinson et al. 2003). By recombining various promoters and repressors, Leibler and co-workers constructed random network topologies and tested their ability to integrate two inputs (Guet et al. 2002). In a relatively small library, they were able to identify circuits that behaved like different types of logic blocks (NOR, NOT IF, and NAND) and found that simple genetic changes could turn one logic block into another. Little and co-workers demonstrated that the bacteriophage λ lysis/lysogeny control circuit is mutationally robust and that mutations can easily tune the sensitivity and cooperativity of the switch (Little et al. 1999). The robustness of this circuit improves the evolvability of the virus by enabling it to tune the lysis/lysogeny distributions in bacteriophage populations so that the optimal distribution can be found for particular environmental niches (Mittler 1996; Arkin et al. 1998). In analyzing a model of the network that determines segment patterns in Drosophila embryos, O'Dell and co-workers demonstrated that different robust patterns could by achieved by tuning the underlying parameters (von Dassow et al. 2000; Meir et al. 2002). Finally, Hsp90 has been proposed to play a role in Drosophila phenotypic diversity by up- and downregulating specific signaling cascades (Rutherford and Lindquist 1998).
In this article, we study a portion of the Bacillus subtilis stress response network as an evolvable system. As a whole, this network is responsible for integrating input signals from the environment, such as cell density, ionic strength, and the presence of nutrients, and deciding whether to form a spore, initiate DNA uptake (competence), swim in a particular direction (chemotaxis and motility), scrounge for food (enzyme production), or kill competitors (antibiotic production; Msadek 1999). The optimal relationship between the input signals and the distribution of responses taken by a bacterial population will depend on the particular environmental niche that is occupied. In addition, it has been proposed that there is a role for stochastic effects in the governing network such that the population will diversify its responses to a particular environmental insult (Maughan and Nicholson 2004).
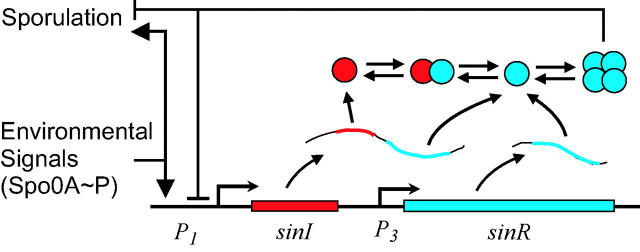
Sporulation is a dramatic response to stress and is a particularly expensive endeavor for the cell, in terms of both time and materials (Grossman 1995). The exact conditions and timing for sporulation are likely to be under strong selective pressure as both premature and belated spore production can have disastrous effects on cell growth and survival. Thus, an intricate phosphorelay, pheromone sensing, and a transcriptional regulatory network carefully control the onset of sporulation (Ireton et al. 1993; Grossman 1995). The sin (sporulation inhibition) operon is central to the timing and early dynamics of this network (Figure 1; Gaur et al. 1986, 1988, 1991; Mandic-Mulec et al. 1992; Bai et al. 1993; Shafikhani et al. 2002). When the environment provides sufficient nutrients and space for growth and maintenance of cells, the repressor SinR is constitutively expressed from an internal promoter in the operon. SinR represses the first committed (stage II) genes in the sporulation pathway. Resource depletion and high population densities lead to the phosphorylation of the transcription factor Spo0A. The accumulation of Spo0A∼P induces the expression of SinI, which binds to and inactivates SinR (Bai et al. 1993; Lewis et al. 1998; Shafikhani et al. 2002). The combined effect of positive regulation by Spo0A∼P and the inactivation of the negative regulator SinR activates the sporulation pathway.
Figure 1.—
A simplified schematic of the sin operon is shown. Mechanistic details are provided in the text.
We have built a model of the sin operon, parameterized it using available data from the literature, and investigated its dynamics using analytical and computational techniques. On the basis of these simulations, we propose that the sin operon improves the evolvability of the stress response network by several mechanisms. First, sin encodes an evolvable switch, which can provide a sink for phylogenetic variation, where the progression into sporulation can be controlled without disturbing the regulation of spore construction. Spore formation requires the coordination of protein machinery and scaffolds that slowly build the spore and time this process with cellular events, such as the partitioning of the chromosome (Stragier and Losick 1996). After initiation, the construction of the spore occurs in stages and is under the control of a cascade of alternative RNA polymerase σ-factors. Once the process of forming a spore has been optimized by evolution, it is advantageous to be able to alter the probability of progressing down that pathway without perturbing the pathway itself. We used a deterministic mathematical model and bifurcation analysis to study the switching behavior of the sin operon and to characterize those parameters that control the threshold of the switch. In addition, stochastic simulations were used to characterize how sin can implement population heterogeneity control.
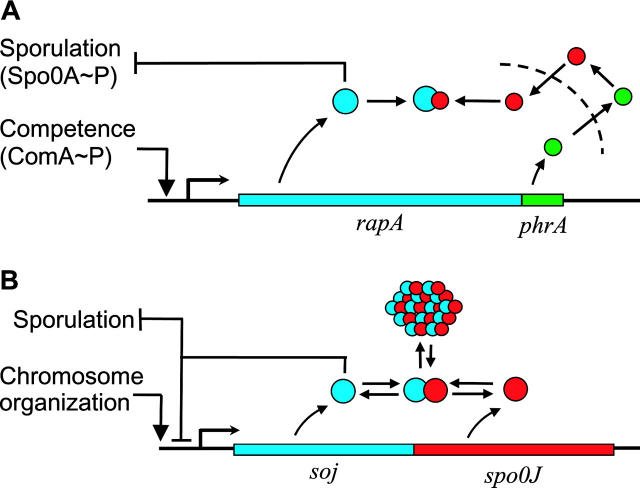
Second, we propose that the greater class of protein-antagonist operons (Hughes and Mathee 1998; Engelberg et al. 1999), of which sin is a member, have the ability to adopt a wide range of dynamical functions with a minimum amount of underlying genetic change. Instances of this ubiquitous network motif include operons encoding σ- and anti-σ-factors (Hughes and Mathee 1998), phosphatases and their inhibitory pheromone precursors (Perego 1997), toxins and their immunity factors (Baba and Schneewind 1998; Engelberg-Kulka and Glaser 1999), and all manner of transcriptional and post-translational regulators and their protein antagonists (Figure 2). These operons could emerge as the result of gene duplication events (as is likely for sin) or by other evolutionary pressures for operon formation (Lawrence 1997). Using sin as a model system, we demonstrate that protein-antagonist operons can behave like a graded or binary switch, oscillator, or pulse generator and can have multiple steady states, depending on the values of the kinetic parameters (Figure 3). We show that subtle changes in the kinetic parameters and the wiring of the network can bias the behavior toward different functions. Due to their dynamical plasticity, evolution may frequently converge on the protein-antagonist operon as a flexible solution to different regulatory needs.
Figure 2.—
Antagonist control motifs analogous to the sin operon are shown from the B. subtilis response pathway. (A) The rapA/phrA operon: PhrA is exported from the cell (dashed line), proteolytically modified (red), and reimported. The modified PhrA can bind to and inactivate the sporulation-inhibiting phosphatase RapA (Perego 1997). The function of this circuit may be to act like a timing device to create a pulse of RapA activity and delay sporulation (Perego 1997). (B) The soj/spo0J operon: Soj, a transcriptional inhibitor of the sporulation commitment signal spo0A and of stage II sporulation promoters, is rendered inactive through sequestration in a spatially oscillating multimeric complex with Spo0J (Quisel and Grossman 2000). The sequestration state of Soj is believed to be a signal relating the orientation and partition state of the chromosome to the sporulation initiation circuitry, presumably to indicate whether the cell is capable of forming a spore (Quisel and Grossman 2000).
Figure 3.—
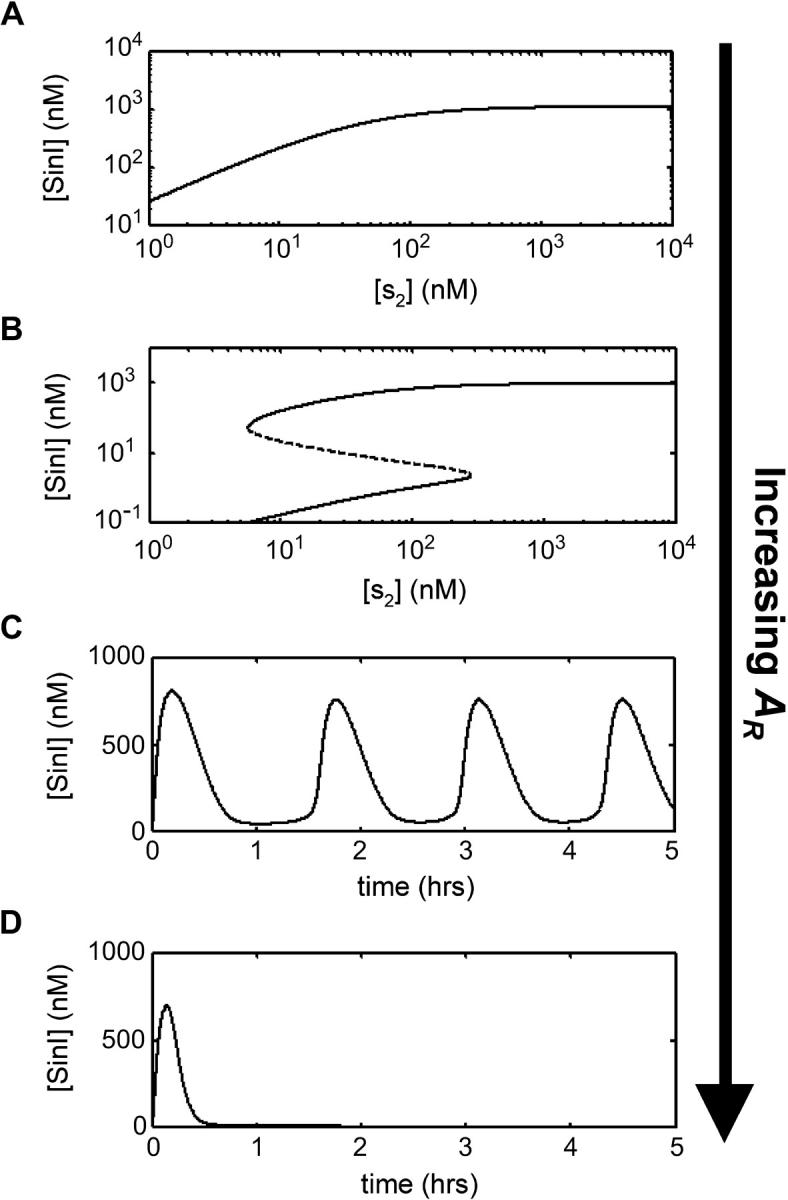
Depending on the kinetic parameters, sin can generate a graded response (A), a bistable switch (B), an oscillator (C), or a pulse generator (D). When situated in an appropriate region of parameter space, the expression rate of SinR (AR) has the unique ability to sample all of the functions. From top to bottom, the values of AR are 0.005 ([SinI] is a graded function of [S2]), 0.035 (a bistable switch controlled by [S2], where the dotted portion of the curve is the unstable solution branch and the solid portions of the curve are the two stable solution branches), 0.052 (oscillating [SinI] with time), and 0.08 (a single pulse of [SinI]) proteins/mRNA/sec. The slice of parameter space corresponding to this sequence of values is shown as a dashed line in Figure 7. Only the value of AR is varied in this series of graphs; the values of the other parameters are provided in Tables 1 and 2.
MATERIALS AND METHODS
Deterministic solutions, bifurcation analysis, and stochastic simulations:
The differential equation software package XPP was used to obtain deterministic numerical solutions for the differential equations, as well as an interface to AUTO (Ermentrout 2002). The stiff integrator, based on the Rosenbrock adaptive algorithm, was used with a tolerance of 0.0001 and minimum step size of 10−12. The bifurcation software package AUTO (Doedel 1981) was accessed through the XPP GUI interface. The perturbation studies were performed using MATLAB and the ode23s (Rosenbrock) stiff differential equation solver. The stochastic simulator, written in C, is based on the Gillespie algorithm (Gillespie 1977). The Shea-Ackers formulism was used to determine the Boltzman probability that the promoter forms the open complex. To convert between numbers of molecules and concentration, it was assumed that one molecule per cell equals 1.0 nm (Shea and Ackers 1985; Arkin et al. 1998). Note that the Shea-Ackers formulism requires the conversion of the equilibrium constant Ki of each state by the factor 10−9 for each bound molecule. This ensures that the equilibrium constants have nanomolar units. All codes and models can be retrieved from the web site www.lbl.gov/~aparkin.
Sequence comparisons:
Protein BLAST comparisons were performed using the NCBI web site (http://www.ncbi.nlm.nih.gov/BLAST/) (Tatiana et al. 1999). The BLOSUM62 amino acid similarity matrix was used with an open gap penalty of 10 and extension gap penalty of 1. ClustalW was used for nucleotide alignments using the IUB nucleotide weight matrix with a gap-opening penalty of 8.0, extension penalty of 0.2, and no end gap penalty (http://bioweb.pasteur.fr/cgi-bin/seqanal/clustalw.pl) (Thompson et al. 1994).
RESULTS
The sin operon—background and model:
The sin operon controls the production and activity of the repressor SinR, which in its active tetrameric form inhibits sporulation by repressing stage II and spo0A promoters (Gaur et al. 1986, 1988; Mandic-Mulec et al. 1992). sin also contains the gene for the SinR antagonist SinI and three promoters, two of which (P1 and P2) produce mRNA containing both sinR and sinI and an internal promoter (P3) from which only sinR is transcribed (Figure 1). During vegetative growth, a constant concentration of SinR is maintained through constitutive expression from P3 (Gaur et al. 1988; Mandic-Mulec et al. 1992; Bai et al. 1993). Transcription from P3 is strong and a high concentration of mRNA is maintained through early sporulation. However, SinR expression is weak because the ribosome-binding site overlaps with the transcription start site (Gaur et al. 1986; Smith 1993).
Phosphorylation induces Spo0A to form active dimers (Lewis et al. 2002), which activate transcription from the P1 promoter (Gaur et al. 1988). Transcripts initiated from P1 encode both sinI and sinR (Gaur et al. 1988). Despite the transcriptional linkage, induced SinI expression is 10-fold greater than that of SinR from P1 mRNA (Gaur et al. 1988). The sole phenotypic function of SinI is to inactivate SinR (Bai et al. 1993). SinI is a single-domain protein that disrupts the active tetramer by forming a tight 1:1 complex with SinR monomers (Bai et al. 1993; Lewis et al. 1998; Scott et al. 1999). Complete inhibition is achieved in vitro with a ratio of 1:2 monomers of SinR:SinI (Bai et al. 1993). Transcription occurs from an additional P2 promoter (immediately following the P1 promoter) after sporulation has been initiated. RNA generated from P2 is undetectable during vegetative growth and early sporulation and is not included in our model (Gaur et al. 1988).
In preliminary experiments, it has been found that SinR transcriptionally represses its antagonist SinI by binding to the P1 promoter (Smith et al. 1991). The SinR operator in P1 is downstream of the RNA polymerase (RNAP) binding site (Smith 1993). Due to the separation between their binding sites, it is unlikely that the binding of SinR displaces either RNAP or Spo0A∼P. A cross-repression motif is formed by the inhibition of SinR by SinI and the transcriptional repression of sinI by SinR. The cross-repression motif is ubiquitous in biological networks and can lead to complex dynamical behaviors, most famously implementing a bistable switch (Gardner et al. 2001; Wolf and Arkin 2003). In addition, the inactivation of SinR by SinI leads to the upregulation of both proteins from P1. This forms a positive feedback loop in the production of SinI, which can enhance bistability (Becskei et al. 2000). Because mRNA from P1 contains both the sinI and sinR genes, this positive feedback can also amplify SinR. This dual effect of amplification and inhibition can lead to oscillations. An additional feedback loop is formed by the repression of spo0A by SinR (Mandic-Mulec et al. 1995). This interaction is mechanistically redundant to the repression of P1 by SinR and will lead to similar circuit dynamics.
On the basis of these mechanistic details, a detailed model was built for the continuation analysis and stochastic simulations. Simpler analytical models are presented in appendixes a and b to demonstrate the underlying interactions that lead to bistability and oscillations. Both the computational and analytical models make mechanistic assumptions and do not explicitly deal with processes such as translational elongation. These assumptions are unlikely to change the basic conclusions, but their inclusion might add the possibility for other dynamical states not analyzed here.
The deterministic model consists of a set of differential equations to track the production and degradation of each protein and complex in the system. The following equations track the concentrations of SinI [I], SinR [R], mRNA from P1 [m1], mRNA from P3 [m3], SinR tetramers [R4], and SinI:SinR complexes [IR]:
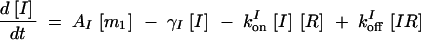 |
1 |
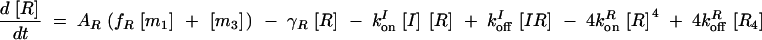 |
2 |
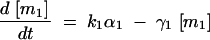 |
3 |
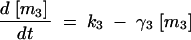 |
4 |
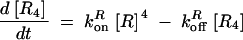 |
5 |
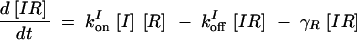 |
6 |
The production of m1 is dependent on the probability α1 that P1 is in the open complex as well as transcript production rate k1. In addition, it has been demonstrated that leaky transcription from P1 and PspoIIG in the absence of Spo0A∼P is minimal; in the model, it was assumed to be perfectly tight (Rowe-Magnus and Spiegelman 1998; Shafikhani et al. 2002). The probability of forming an open complex at P3 was assumed to be constant (Smith 1993) and the parameter k3 is the number of transcripts produced per unit time. SinI is expressed at rate AI from m1. SinR is expressed at rate AR from the combination of m1 and m3. The potential for differential expression of SinR from P1 and P3 is captured by the parameter fR, which was always set to one in our simulations.
The parameter values are provided in Table 1. The on rate for the formation of dimers kIon was fixed at the desolvation-mediated diffusion-limited rate 0.083 nm−1 sec−1 (Camacho et al. 2000) and only the off rates were varied in the simulations. The formation of SinR tetramers is via the dimerization of dimers (Scott et al. 1999) so we assumed the diffusion-limited rate for tetramer formation kRon is 0.00125 nm−3 sec−1 (Camacho et al. 2000).
TABLE 1.
Parameter values and ranges
| Nominala
|
Range
|
Notesb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Description | Bi | Osc | Units | Min | Max | Nominal | Range |
| k1 | Transcription rate from P1 | 0.15 | Trans sec−1 | 10−4 | 10 | c | d | |
| ΔGS | Binding energy of S2 to P1 | −9 | kcal mol−1 | −14 | −8 | e | f | |
| ΔGRNAP | Binding of RNAP to P1 | −10.5 | kcal mol−1 | −14 | −8 | e | f | |
| ΔGR | Binding energy of R4 to P1 | −12.5 | kcal mol−1 | −14 | −8 | e | f | |
| AI | Expression rate of SinI | 0.8 | Prot trans−1 sec−1 | 10−4 | 10 | g | h | |
| γI | Degradation rate of SinI | 0.02 | 1 sec−1 | 10−5 | 0.1 | i | j | |
| kIoff | Off rate for SinI:SinR | 0.5 | 1 sec−1 | 0.1 | 1000 | k | l | |
| k3 | Transcription rate from P1 | 0.28 | 10−4 | Trans sec−1 | 10−4 | 10 | m | d |
| AR | Expression rate of SinR | 0.014 | 0.07 | Prot trans−1 sec−1 | 10−4 | 10 | m,g | h |
| γR | Degradation rate of SinR | 0.002 | 1 sec−1 | 10−5 | 0.1 | n | j | |
| kRoff | Off rate for SinR tetramers | 0.5 | 1 sec−1 | 0.1 | 1000 | k | l | |
The nominal set of parameters around which the bistability (bi) and oscillatory (osc) bifurcation analyses are performed as in Figure 4. The two sets of parameters are identical except for k3 and AR.
References and notes are provided for the nominal value and the estimated parameter range.
Spo0A∼P-activated expression from P1 is very strong (Gaur et al. 1988).
The transcription rates (trans/sec) range from a minimum leakiness of a promoter to the maximum observed transcription rate (Vo et al. 2003).
See Table 2.
The range of transcription factor binding is estimated on the basis of data from (Shea and Ackers 1985; Fedoriw et al. 1998; Kunne et al. 1998).
Expression of SinI is 10-fold greater than that of SinR from P1 mRNA (Gaur et al. 1988).
The minimum expression rate accounts for one protein (prot) expressed per mRNA for a 2-hr half-life. The maximum rate is estimated from Farewell and Neidhardt (1998).
SinI is a small peptide, the free form of which is assumed to degrade rapidly (Lewis et al. 1998).
The range of protein degradation rates was picked to match half-lives of seconds to tens of hours.
It requires 2:1 molar quantities of SinI to disrupt SinR tetramers completely (Bai et al. 1993). It is assumed that the SinI:SinR equilibrium constant is 6 nm whereas the SinR:SinR4 equilibrium constant is 400 nm. The on rates are held fixed at their diffusion-mediated value (see text).
The range of off rates produces the range of equilibrium dissociation constants of 1 nm to 10 μm on the basis of the diffusion-limited on rates from Camacho et al. (2000).
The transcription rate from k3 is high, but the expression rate of SinR AR is low (Gaur et al. 1986; Smith 1993).
Chosen to reflect an ∼20-min half-life, considering the tetramerization reaction (Lewis et al. 1998).
The degradation rates of each of the species are represented by γ. SinI is a small protein that interacts with SinR through a domain-swapping mechanism. It does not drastically disturb the structure of SinR, so the degradation rates of SinR and the SinI:SinR complexes were assumed to be identical (Lewis et al. 1998). It was also assumed that the off rate for SinR tetramers is much faster than the tetramer degradation rate and therefore did not explicitly include a term for tetramer degradation. The degradation rates of m1 and m3 were held constant at γ1 = γ3 = 0.005 sec−1 (2-min half-life; Vilar et al. 2002).
The probability α1 that P1 is in the open complex was modeled using the Shea-Ackers formalism,
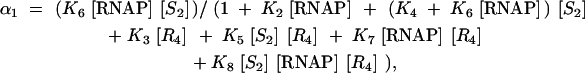 |
7 |
where [S2] is the concentration of Spo0A∼P dimers, [R4] is the concentration of SinR tetramers, and [RNAP] = 30 nm is the concentration of free RNA polymerase available for transcription (McClure 1980, 1983; Shea and Ackers 1985). The transcription factors were assumed to be at thermodynamic equilibrium between the DNA-bound and unbound states. Following this assumption, Equation 7 was derived on the basis of an enumeration of all possible bound states i of the transcription factors to the promoter and their equilibrium association constants Ki = exp(−βΔGi), where ΔGi is the free energy of the bound state (Table 2) and β = 1/RT = 1.62 mol/kcal, where R is the gas constant and T = 36.85°. The subscript i corresponds with the state in Table 2. The repressors AbrB and ScoC (previously named Hpr) also bind to the P1 promoter, but are not included in this model (see discussion; Shafikhani et al. 2002).
TABLE 2.
Binding states of promoterP1
| Binding region
|
|||||
|---|---|---|---|---|---|
| No. | O1 | O2 | O3 | Contributions | ΔG (kcal/mol) |
| 1 | — | — | — | — | 0.0a |
| 2 | — | RNAP | — | ΔGRNAP | −10.5b |
| 3 | — | — | (SinR)4 | ΔGR | −12.5 |
| 4 | (Spo0A)2 | — | — | ΔGS | −9.0c |
| 5 | (Spo0A)2 | — | (SinR)4 | ΔGR + ΔGS | −21.5d |
| 6 | (Spo0A)2 | RNAP | — | ΔGRNAP + ΔGS + ΔGRS | −21.5e |
| 7 | RNAP | (SinR)4 | ΔGRNAP + ΔGR | −22.5f | |
| 8 | (Spo0A)2 | RNAP | (SinR)4 | ΔGS + ΔGRNAP + ΔGR + ΔGRS | −33.5 |
Unbound DNA is defined as the reference state (Shea and Ackers 1985).
B. subtilis σA binds to a consensus −10 sequence with an affinity between 50 and 100 nm, as determined by in vitro footprint analysis (Camacho and Salas 1999). RNAP can bind without Spo0A (Bird et al. 1996).
The free energy of Spo0A binding was estimated from data from Rowe-Magnus and Spiegelman (1998).
Unlike the case for the stage II gene promoters, SinR tetramers and Spo0A dimers bind to different sites in the P1 promoter region (Cervin et al. 1998).
Spo0A dimers initiate transcription by binding to RNA polymerase and accelerating the formation of the open complex (Bird et al. 1996). The protein-protein contact between Spo0A and σA comprises ∼14 residues and is dominated by electrostatic forces (Cervin et al. 1998). In our simulations, we assume that this interaction energy is ΔGRS = −2.0 kcal/mol (Shea and Ackers 1985).
Preliminary evidence indicates that SinR binds to a 47-bp region between the P1 and P2 promoters (Smith et al. 1991; Smith 1993). This region occurs immediately downstream of the P2-binding site without overlapping it. This implies that SinR4 and RNAP bind independently, and the repressor activity is achieved by blocking RNAP initiation rather than RNAP binding.
sin as a switch:
Placing sporulation initiation under the control of a threshold-activated switch ensures that sufficient Spo0A∼P must accumulate before the cell commits to spore formation. A number of mechanisms can result in a switch, including multicomponent phosphorelay cascades operating near saturation (Huang and Ferrell 1996), cooperative mutually exclusive repressor/activator binding (Rossi et al. 2000; Biggar and Crabtree 2001), and positive feedback loops (Wolf and Arkin 2003). All of these mechanisms are present in the sporulation pathway and together they likely act to magnify and tune the switch-like behavior (Grossman 1995; De Jong et al. 2004). Here, we focus on the ability of the sin operon alone to produce a switch-like response.
Because SinR transcriptionally represses sinI and SinI represses SinR via a protein-protein interaction, a cross-repressive feedback network is completed (Figure 1). This architecture can introduce bistability into the switch (Figure 3B; McAdams and Arkin 1999; Gardner et al. 2000; Wolf and Arkin 2003). A bistable switch has two characteristic stable steady states (“on” and “off”) connected in phase space by an unstable steady state. A bistable switch has several advantages over a monostable switch in a biological network. First, the presence of bistability often results in a rapid, almost discontinuous induction from the off state to the on state (Gardner et al. 2000). This ultrasensitivity ensures that an adequate input concentration is reached before induction and creates the “all-or-none” response essential to developmental processes like sporulation. The second advantage of bistability is that it introduces hysteresis into the switch, which enables it to “latch” onto the on state, making the transition more resistant to fluctuations. We examined these properties of the sin architecture using the software package AUTO, which is able to computationally track steady states and bifurcations for a system of differential equations (Doedel 1981). These simulations were used to study the robustness of bistability to variations in the model parameters and to characterize those parameters that can tune the [S2]-activation threshold and hysteretic properties of the switch.
We found that expression of SinR from the internal promoter P3 is critical for bistability and that expression of SinR from the promoter P1 leads to complex dynamics. The role of the parameter k3 (transcription from the internal promoter P3) on bistability is shown in Figure 4A. The switch is monostable when k3 → 0. As k3 increases, bistability is introduced into the switch and the [S2]-activation threshold (right limit point) increases. In addition, the limit points separate, thus increasing hysteretic effects. Once k3 crosses a threshold, the high [S2] limit point goes to infinity. When one limit point goes to infinity, this is referred to as “type 2 bistability” and the system cannot transition from the off state to the on state in the absence of a significant perturbation (Guidi and Goldbeter 1997). When a switch can tunably exhibit both hysteretic and type 2 bistability, this provides exquisite control over the activation threshold, allowing it to be tuned essentially from zero to infinity.
Figure 4.—
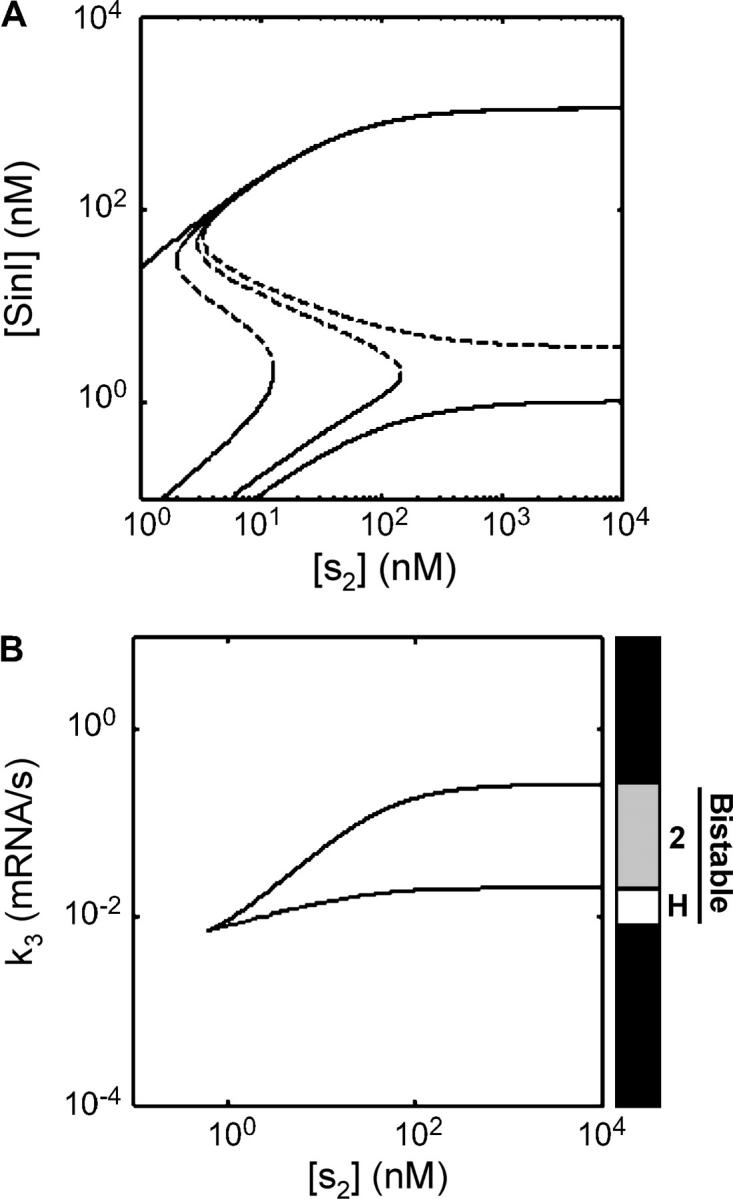
The sin operon can exhibit bistability when P3 constitutively generates SinR and SinR represses P1. The stable and unstable steady states are shown as solid and dashed lines, respectively. (A) Four curves are shown for (left to right) k3 = 0.05, 0.015, 0.02, and 0.022 mRNA/sec. When k3 = 0.05, sin produces a graded response. For k3 = 0.015 and 0.02, the switch exhibits hysteretic bistability with two limit points. When k3 > 0.021 the switch is bistable, but one limit point goes to infinity (type 2) and the higher steady state cannot be reached by increasing the [S2]. (B) The data from A can be viewed as a bifurcation plot with respect to k3. The top curve shows the left limit point from A as a function of [S2] and k3. The bottom curve is the right limit as a function of [S2] and k3. As k3 increases, the bottom curve goes to infinity faster than the top curve and it is at this point where type 2 bistability occurs. Our operational definition of hysteretic bistability is when two stable states exist at [S2] = 1 nm, but not 10,000 nm. The bar indicates the region of parameter space consistent with hysteretic (open) and type 2 (shaded) bistability and demonstrates how the bars are constructed in Figure 5. Values for the parameters not varied in these bifurcation analyses are provided in Tables 1 (“Bi” column) and 2.
While it is the cross-repression motif that introduces bistability into the switch, type 2 dynamics arise due to the independent binding of Spo0A and SinR to the P1 promoter (appendix A and Table 2). When the activator and repressor bind to the same operator and their binding is mutually exclusive (as is the case for the stage II promoters), then high concentrations of activator will eventually override the repressor. However, when the repressor binds independently and blocks RNAP by binding upstream of the transcription start site, the system cannot turn on until the repressor has been removed by some mechanism. This arrangement of binding sites in the P1 promoter enables the sin circuit to adopt type 2 bistability.
To identify regions of parameter space that exhibit hysteretic as well as type 2 bistability, we performed a bifurcation analysis for each parameter at [S2] = 1 nm and 10,000 nm, which covers the approximate range of concentrations for active transcription factors in bacteria. We operationally defined hysteretic bistability to be in effect if there were two stable states at [S2] = 1 nm and a single steady state at 10,000 nm; type 2 bistability was assumed if there were two steady states at [S2] = 1 and 10,000 nm (Figure 4B). A more rigorous definition of type 2 bistability is presented in appendix A. For each parameter, two bifurcation curves were calculated and the regions corresponding to each type of bistability were identified.
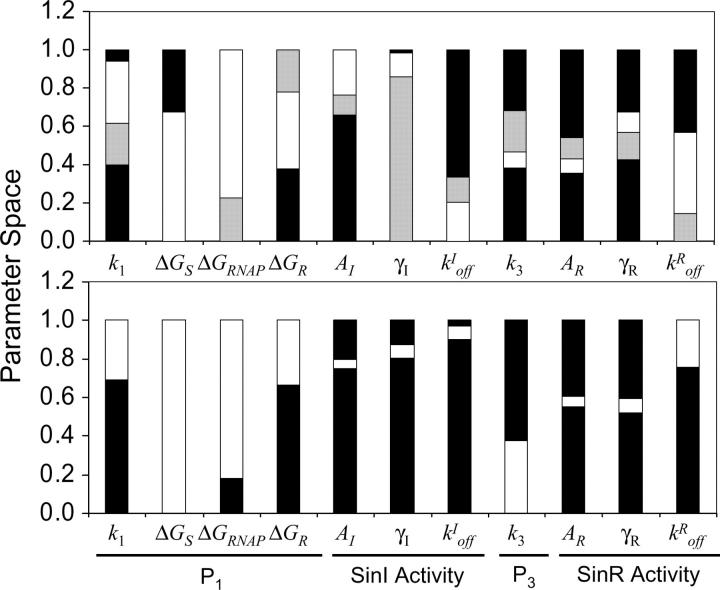
To compare the robustness of the various kinetic parameters, the bistable region is normalized by an estimation of the physically attainable parameter space (Figure 5; ranges provided in Table 1). The robustness of each parameter can be read as approximately the fraction of the corresponding bar that is either open or shaded. Those parameters associated with the expression and activity of SinR are the most fragile (kIoff, k3, AR, γR). Other parameters are more robust, such as those that control the activity of the P1 promoter and the SinI:SinR binding strength, and these parameters provide a means to tune the [S2]-activation threshold of the switch. For example, tuning the most robust parameter ΔGRNAP can tune the [S2]-activation threshold by two orders of magnitude (100–10,000 nm; not shown). Mutations that affect this parameter are more likely to sample a range of thresholds, without disturbing the underlying function of the switch. There are additional mechanisms in the sporulation pathway—other than the sin operon—by which the activation threshold can be tuned, such as the negative feedback from SinR to spo0A and other redundant positive feedback loops (not shown; Grossman 1995). The relative importance of each of these mechanisms in determining the overall threshold control for sporulation remains an open question (see discussion).
Figure 5.—
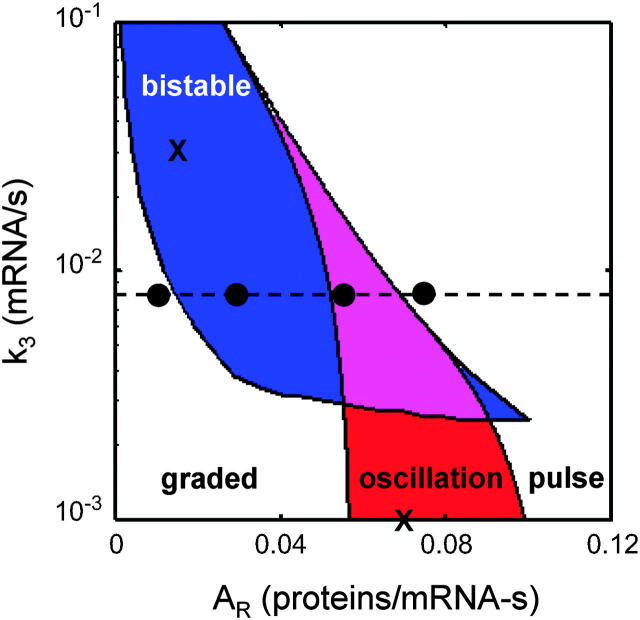
One-dimensional bifurcation analysis for the regions in parameter space consistent with bistable (top) and oscillatory (bottom) behaviors. For each parameter either the limit points or Hopf bifurcation points were identified using AUTO and then normalized by the biologically relevant region of parameter space (materials and methods). (Top) The region of parameter space consistent with hysteretic (open bars) and type 2 (shaded bars) bistability and monostability (solid bars) is shown on the basis of calculations like that performed in Figure 4B. (Bottom) The oscillatory regions of parameter space are shown for each parameter. Calculations were performed to identify the Hopf bifurcation points as in Figure 6. Note that the regions consistent with bistability and oscillatory behaviors overlap except for the parameters k3 and AR. Each one-dimensional bifurcation analysis was performed from a nominal point in parameter space where either bistable or oscillatory behavior dominates (marked by X's in Figure 7). The nominal values of the parameters and the biologically relevant ranges are provided in Tables 1 and 2.
Functional plasticity:
Because there is no terminator after sinI, the sinR gene is present on P1 mRNA. The expression of SinR from P1 can lead to complex dynamical responses to a Spo0A∼P input, including pulses and oscillations. Different dynamics are achieved by the relative expression levels of SinI and SinR from P1 mRNA. If SinI expression is large enough to continuously inactivate SinR, then a linear response to [S2] is observed. On the other hand, if SinR expression is strong, then it will be able to inactivate P1 and render the switch unable to respond to an input. It is between these extremes that complex dynamics arise, including pulse and oscillatory responses (Figures 3 and 6). We use AUTO to identify the parameter regimes where oscillatory and pulse behaviors occur (Doedel 1981). An analytical solution of a simplified set of equations that oscillate is presented in appendix B. Expression of SinR from the internal promoter damps the complex dynamics, so we initially consider the limit where k3 is small (k3 = 0.0001 mRNA/sec; Table 1).
Figure 6.—
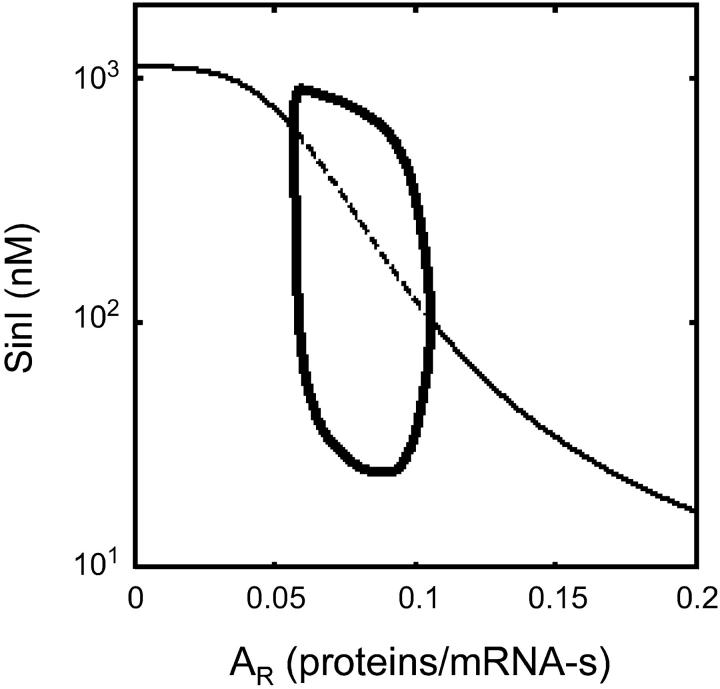
When mRNA transcripts from P1 contain the sinI and sinR genes and transcription from P3 is small (k3 = 0.0001 mRNA/sec), the system generates limit cycle oscillations or functions as a pulse generator (Figure 3, C and D). Using AUTO, a bifurcation curve was generated to identify oscillatory regions in parameter space as a function of the SinR expression rate AR. When AR is small, the system produces a linear response to a single stable steady state (solid line). Once AR crosses the left supercritical Hopf point, the steady state becomes unstable (dashed line) and limit cycles emerge (the minimums/maximums of the oscillations are shown as thick lines). When AR is larger than the right supercritical Hopf point, the steady state becomes stable again and the oscillations cease. For AR larger than this threshold, the system produces a large initial pulse and then returns to the lower steady state, creating a temporal pulse. The parameter values used in this analysis are provided in Tables 1 (“Osc” column) and 2.
Limit cycle oscillations occur when a stable steady state becomes unstable and a stable periodic orbit emerges. This transition is referred to as a supercritical Hopf bifurcation and AUTO can be used to identify and track Hopf points as a function of a model parameter. In Figure 6, a bifurcation plot is shown for the parameter AR (expression rate of SinR). Three regimes of behavior are associated with the bifurcation diagram. When AR is less than the left Hopf point, the system produces a linear response of [SinI] to increasing [S2]. Between the Hopf points, limit cycle oscillations occur (Figure 3C). When AR is greater than the right Hopf point, the system produces a temporal pulse of [SinI] in response to an input stimulus of increased [S2] (Figure 3D). As AR → ∞, the amplitude of the pulse decreases until the system no longer responds to input signals.
To investigate how the parameter space partitions into switching and oscillatory behaviors, we normalized the oscillatory regimes and compared them with the bistable regimes (Figure 5; ranges provided in Table 1). Oscillations and bistability both arise from an instability that is introduced as each parameter induces a transition from a high [SinI] state to a low [SinI] state and as a result these behaviors overlap strongly in parameter space. The parameters k3 and AR are exceptions as expression of SinR from P1 and P3 promotes and damps oscillations, respectively. This leads to the hypothesis that these parameters can tune the system to different dynamical behaviors as long as the remaining parameters position the system in a robust region of instability.
When the system is in an appropriate region of parameter space, the tuning of AR is adequate to sample all of the possible behaviors (Figures 3 and 7). When AR is low, the system behaves like a monostable switch (Figure 3A). When AR increases, bistability is introduced and the system becomes a bistable switch (Figure 3B) and then a bistable oscillator (Figure 3C). Finally, at large AR values, monostability is restored and sin behaves like a pulse generator (Figure 3D). The ability to sample all of these functions is unique to the parameter AR. The coexistence of bistability and oscillatory dynamics has been observed for biological (Guidi and Goldbeter 2000) and chemical (Olsen and Epstein 1993) systems, but these behaviors strongly overlap in these models and it is unclear whether a single model parameter could transition between them.
Figure 7.—
The bistable (blue) and oscillatory (red) regions of parameter space are shown for the critical parameters AR and k3. A region of parameter space is both bistable and oscillatory (purple). The parameter AR has the unique ability to sample all of the functions when varied. The dashed line represents a slice of parameter space (k3 = 0.008 transcriptions/sec), where as AR is tuned, sin is able to generate a graded response, bistability, oscillations, and pulse. The black circles on this line represent the values of AR used in Figure 3. The X's mark the nominal values used to produce the one-dimensional bifurcation analyses in Figure 5. The curves were generated using AUTO. The limit point loci are calculated for [S2] = 1 nm (left) and 10,000 nm (right) and the Hopf loci are for [S2] = 10,000 nm. The parameters not varied in the bifurcation analyses are fixed at the values presented in Tables 1 and 2.
Constitutive expression of SinR from P3 damps the oscillations, leading to a stable steady state (Figure 7). Indeed, the maintenance of a low, constant expression of repressor may ensure that oscillations do not occur when they are deleterious to the desirable network dynamics. This may be the case with the sin operon, as additional redundant feedback loops in the network (Grossman 1995) may bias the dynamics toward a bistable switch. Some protein-antagonist operons do not contain an internal promoter (Perego 1997; Quisel and Grossman 2000; Stephenson et al. 2003) and this may be important to achieve complex circuit dynamics, such as a pulse generator or an oscillator.
Noise rejection and exploitation:
Spo0A∼P is a regulator of multiple cellular processes and its concentration can fluctuate as a result of noisy environmental inputs as well as from cell-to-cell variation (Grossman 1995). The sin architecture can filter brief fluctuations of input so that sporulation is initiated only in response to sustained high [S2] (not shown). sin is able to filter short pulses because the input signal (Spo0A∼P) both activates stage II promoters and represses a repressor of the stage II promoters. Dynamically, this results in a time delay as SinR has to be depleted before the input can activate the downstream promoters. If the input signal is shorter than the time required to deplete SinR, then the sporulation pathway will not be activated. This aspect of the sin architecture resembles the feedforward topology, which was proposed to result in a filtering function, and our results agree with these predictions (Shen-Orr et al. 2002).
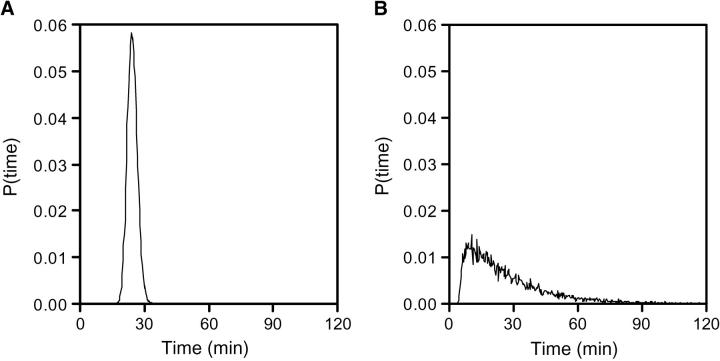
While sin is able to filter noisy inputs of Spo0A∼P, it also has the ability to be sensitive to cell-to-cell variations in the prestimulus steady-state SinR concentrations. This sensitivity enables individual cells within a population to differentiate their responses to an environmental signal. Noise sensitivity can arise in a network due to interactions between small numbers of competing regulators and by combining high levels of protein translation with low levels of gene transcription (Arkin et al. 1998; Rao et al. 2003). To examine the noise sensitivity of the sin operon, we ran stochastic simulations using the Gillespie algorithm (materials and methods; Gillespie 1977; Arkin et al. 1998). The noise sensitivity of sin can be tuned by altering the steady-state concentration of SinR expressed from P3 and the binding affinity of SinR to the P1 promoter. When the binding of SinR to P1 is weak and the number of SinR molecules is large, the population of cells tends to respond similarly (Figure 8A). However, when SinR binding is strong and there are fewer SinR molecules, then the switch is more sensitive to fluctuations and the distribution of response times broadens (Figure 8B).
Figure 8.—
The sin operon controls the stochastic characteristics of the sporulation pathway. The time required for SinI to reach its half-maximal value is shown for two cases: (left) when SinR binds weakly at high steady-state concentration and (right) binds tightly at low steady-state concentration. While the average of the two distributions is equal (∼25 min), tight SinR binding leads to a greater distribution of on times. The parameters are identical to the deterministic model (Tables 1 and 2) except ΔGR = 7.5 kcal/mol, AR = 0.02 proteins/mRNA/sec (left), and ΔGR = 14.0 kcal/mol, AR = 0.0045 proteins/mRNA/sec (right). The initial condition for each run is set at the steady-state P3 mRNA (4 nm) and [R4] concentrations (left, 6400 nm; right, 20 nm). The data represent 10,000 runs of the Gillespie algorithm and are binned in 20-sec intervals. When the control of SinR is stochastic (right graphs), cells switch randomly into the sporulation pathway. Those cells that do not sporulate may be able to choose alternative responses.
This broad distribution of response times translates into a heterogeneity of responses in the population (Maughan and Nicholson 2004), potentially a game theoretic form of bet hedging (Mittler 1996; Wolf and Arkin 2003). By staggering the sporulation initiation times of individual cells over a long time window, the population gradually commits a larger and larger fraction of its members to costly stress-resistant spores, all the while maintaining a shrinking subpopulation capable of rapid vegetative growth should environmental conditions improve. If the sin network is configured to exhibit hysteretic stability and environmental conditions push [S2] beyond the commitment threshold, all sporulation switches will eventually turn on. Such a noise-sensitive switch can lead the stochastic control of the progression into alternate pathways (Arkin et al. 1998). In the context of the complete stress response network, this might allow unaffected cells to progress down alternative response pathways.
Genomic comparisons:
Using the kinetic model, we have predicted two ways in which the sin operon can be diversified. First, to alter the sporulation probability given a set of conditions (and time of exposure), it is desirable to accumulate mutations that affect the P1 promoter and the SinI:SinR interaction strength. Second, variations in the transcription rate from P3 (k3) and the expression rate of SinR (AR) can fundamentally alter the dynamical behavior of the operon. It is expected that there will be strong selective pressure against dynamical instabilities, whereas there is more selective pressure to diversify the threshold required for sporulation in slightly different environments. To observe how these parameters have diversified in evolution, we aligned and compared the genomic sequences of several Bacillus species that occupy very different environmental niches (subtilis, halodurans, and anthracis) and between two more closely related species (anthracis and cereus; Takami et al. 2000; Read et al. 2002; Ivanova et al. 2003; materials and methods).
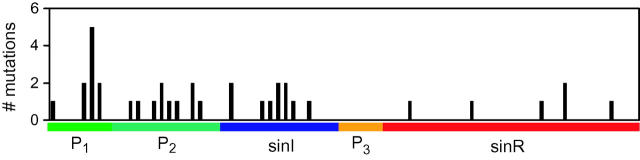
The model predicts that the most robust portions of the P1 promoter are the Spo0A- and RNAP-binding sites, and the transcription and translation rates are more fragile (Figure 5). The P1 promoter has diverged significantly among all the species. When subtilis, halodurans, and anthracis are compared, the 0A-boxes in the −35 regions differ both in orientation and nucleotide composition and the −10 σA-binding motifs are also divergent. It is noteworthy that the −35 and −10 regions of the stage II promoters are nearly identical between these species. When anthracis and cereus sequences are compared, the largest concentration of mutations occurs in these regions of sin, notably affecting the RNAP-binding region (Figure 9).
Figure 9.—
Evolutionary divergence of the sin operon. The nucleotide sequences of sin are aligned from the sequenced genomes of B. anthracis and B. cereus (materials and methods). Distance in base pairs is shown on the x-axis and the mutations are binned in 10-bp intervals. The P1 promoter is the most variable region between these species and the P3 promoter and SinR sequence are the most conserved. This trend is also present when the more divergent sequences of B. subtilis, B. anthracis, and B. halodurans are compared (see text). Only a single anthracis sequence is used for these analyses as there are no polymorphisms between anthracis species in this region (Read et al. 2002).
The model also predicts that the SinI:SinR-binding strength is a robust parameter that can tune the threshold of the switch. The SinI sequences have diverged in both length and amino acid content (25% average pairwise amino acid identity; materials and methods). In addition, the C-terminal domain of SinR to which SinI binds is also relatively variable (52% identity) with respect to the N-terminal DNA-binding domain (72% identity) and other regulatory proteins in the sporulation pathway (see below).
In contrast to P1, all of the P3 promoters are nearly identical and all contain perfect TG_TATAAT extended −10 σA-binding sites (Camacho and Salas 1999). The maintenance of strong transcription from the internal promoter improves the robustness of the switch by reducing the likelihood that instabilities, such as those that lead to oscillations, will be introduced into the system.
The expression rate of SinI is probably conserved as evidenced by the perfect Shine-Dalgarno ribosome-binding sites (RBS) present in all the species. There is more variability in the expression rate of SinR. In B. subtilis, the RBS and transcription start site overlap, which can reduce expression from P3 mRNA, implying that the pre-stimulus concentration of SinR is reduced. In B. halodurans and B. anthracis, the RBS and transcription start site do not overlap. The RBS in subtilis and halodurans differ from the Shine-Dalgarno sequence by a single nucleotide, whereas in anthracis it differs by two nucleotides. Finally, the start codon of SinR is ATG in halodurans and anthracis, as opposed to the unusual TTG start codon present in subtilis. By changing the relative expression and activity of SinI and SinR, different species may be able to tune their threshold for sporulation to be more or less stringent under different environmental conditions.
In the sporulation network, those components that act upstream and downstream of sin are strongly conserved among the species. The amino acid sequences of the phosphorelay proteins Spo0A and Spo0F are closely related (72 and 77% identity; Stephenson and Hoch 2002). Downstream from sin, the stage II spoIIE, spoIIA, and spoIIG promoters are conserved when compared to P1 and their 0A-box- and RNAP-binding motifs are nearly identical. In addition, the σ-factors they encode that regulate the next round of sporulation are also conserved (∼70% identity). The conservation of the phosphorelay and the stage II regulatory proteins ensures that the signal-processing properties of the response network and the program for spore formation are conserved. In this context, sin provides a mechanism by which the probability of sporulation can be tuned without affecting these processes.
DISCUSSION
Adaptive plasticity reflects the capacity of a bacterium to nongenetically alter gene expression or protein activities to suit environmental fluctuations during its life cycle, whereas evolvability reflects the ease by which genetic mutations enable the bacterium to invade a new environmental niche (Meyers and Bull 2002). The sin operon is part of the adaptive machinery that helps B. subtilis respond to stress by integrating environmental and intracellular signals into a probabilistic, all-or-none decision to sporulate (Schaeffer et al. 1965; Dawes and Mandelstam 1970; Chung et al. 1994; Maughan and Nicholson 2004). Bacteria evolved for one environment often fare poorly in other environments and a number of genetic changes are required to transition between the two. It is known, for example, that distinct Bacillus species have evolved to colonize the microenvironments of insects (thuringiensis) and mammals (anthracis), and that each subspecies does poorly in the other's host environment (Maagd et al. 2001; Read et al. 2003). Thus, when a new environmental niche is invaded where sporulation is an appropriate response to different conditions, or if a change in environmental conditions leads to a change in the optimal timing or distribution of spore formation, then genetic changes are required to reprogram the wiring between the phosphorelay and the downstream “actuation” circuitry in the sporulation pathway. An evolvable design minimizes the number of simultaneous mutations required to make such a shift.
The adaptive plasticity and evolvability of the sin operon in particular, and the protein-antagonist operon pair motif in general, stems from its diversity of behaviors under simple (evolvable) parametric control. Using a mathematical model, we have demonstrated that the sin operon can implement a bistable switch with exquisite control over both the activation threshold and the noise sensitivity of the switch. Together, these properties could change both the probability of spore formation and the population heterogeneity in sporulation initiation times. Our comparative genomics analysis supports the view that the sin operon—particularly the operators of the P1 promoter and SinI:SinR interaction—provide a means for evolution to rapidly alter the probability and timing of sporulation. It is at this point in the network where high relative phylogenetic variation can occur to optimize sporulation as a species invades a new environmental niche.
As a whole, the architecture of the sporulation pathway contains multiple circuit motifs that can act collectively to generate threshold control (De Jong et al. 2004). At least 13 positive feedback loops and additional redundant mechanisms occur between the phosphorelay and the activation of stage II genes (Grossman 1995). Notably, SinR represses the Spo0A promoter, which completes a cross-repression motif redundant to the proposed repression of P1 by SinR (Smith et al. 1991; Mandic-Mulec et al. 1995). In addition, Spo0A∼P represses abrB, which represses P1. Finally, AbrB activates scoC (hpr), which also represses P1 (Strauch et al. 1989; Shafikhani et al. 2002). This motif, where a gene is activated both directly and by repression-of-repression, has been proposed to lead to a threshold mechanism (Biggar and Crabtree 2001; Shen-Orr et al. 2002). There are three possibilities regarding the role of sin in this pathway. First, it could act as the final checkpoint upon which multiple pathways converge and subsequently has a critical role in setting the threshold required for sporulation. Second, the redundancy of the interactions ensures that no single system dominates, thus producing robust dynamics. Third, the redundancies are individually dominant under different environmental conditions. Further simulations of the pathway as a whole and experiments will be required to distinguish between these scenarios.
Depending on parameter values, protein-antagonist operon motifs like sin can also generate a graded or monostable switch-like response or function like an oscillator or pulse generator. This functional diversity is achieved with minimal changes in the architecture (such as the inclusion or removal of the internal promoter and the negative feedback of SinR on P1) and in several critical parameters. These subtle differences can bias the operon toward different functional dynamics. For example, some members of the Rap/Phr class of operons contain an internal promoter whereas others do not (Stephenson et al. 2003). It would be interesting to characterize the impact that this internal promoter may have on the pulse dynamics.
A number of underlying genetic mechanisms have been proposed to lead to operon formation and modification and many of these could lead to the transcriptional linkage of a protein with its antagonist (Lawrence 1997). In the case of sin, sinI most likely appeared via a gene duplication of sinR (Lewis et al. 1998). Using sin as a model system, we have demonstrated how such a simple evolutionary event can lead to complex controllers in genetic networks. As the need for different types of controllers emerges, the simplicity and flexibility of the protein-antagonist operon make it a solution on which evolution will frequently converge. Since protein-antagonist operons appear frequently in genetic networks, this implies that the bicistronic solution to control problems has been found by evolution independently many times.
The protein-antagonist control motif occurs frequently in the B. subtilis stress response pathway, as well as other heterologous networks. On the basis of experimental evidence, it has been postulated that different protein-antagonist motifs involved in B. subtilis stress responses can adopt switch-like (SigX-RsiX), pulse (RapA-PhrA), or spatial oscillatory (Spo0J-Soj) functions (Figure 2; Brutsche and Braun 1997; Perego 1997; Quisel and Grossman 2000). Wherever they occur, the functions of this motif can be biased through additional, redundant wiring on the level of the global network. There are also more complicated variations on the protein-antagonist operon theme, such as the control of the stress-induced σ-factor σB, which contains seven regulators that interact in a complex partner-swapping mechanism (Price 2002). This operon is responsible for integrating energy and environmental stresses and can produce both switch-like and pulse responses.
Similar complex protein-antagonist behaviors have been identified in heterologous organisms. σ-factors frequently occur with anti-σ-factors that regulate their activity via protein-protein interactions (Hughes and Mathee 1998). In addition, toxins and their immunity factors often exist in the same operon as well and are used either to control cell death (addiction modules) or in bacterial warfare (Baba and Schneewind 1998; Engelberg-Kulka and Glaser 1999). In bacteriophage λ and P1, genes involved in cell lysis are encoded along with their inhibiters, thus assuring a rapid pulse of activity once the proper signals have been received (Bläsi et al. 1990).
Directed evolution, where a library of mutant genes is constructed by mutagenesis and recombination, has emerged as a useful technique to build and refine de novo regulation (Yokobayashi et al. 2002). Using nonlinear dynamics to quantify the evolvability of a genetic circuit will be useful in targeting random mutagenesis toward regions most likely to produce the desired functional change. For example, to alter the switch threshold for the sin operon, our simulations predict that mutagenesis should be targeted toward the RNAP-binding region of the P1 promoter. To achieve a change in the global dynamics, for instance to create a pulse generator, mutagenesis should be targeted toward the P3 promoter. In the context of the sporulation pathway in B. subtilis, these changes will alter the timing and dynamics of sporulation. In addition, it may be possible to remove a protein-antagonist operon from its wild-type pathway and use it to drive novel processes in heterologous organisms.
Acknowledgments
We thank rotation student Sophie Dumont for her early contribution to this project. C.A.V. was supported by a DoE/Sloan postdoctoral fellowship for computational biology. A.P.A. and D.M.W. acknowledge the Defense Advanced Research Project Agency, the Department of Energy, and the Howard Hughes Medical Institute for support during the period of this project.
APPENDIX A:
MINIMAL SIN OPERON MODEL TO ACHIEVE TYPE 2 BISTABILITY
First, Equations 1–6 were simplified by making the following assumptions: (i) mRNA production and degradation is at steady state, (ii) the SinI:SinR and SinR:SinR4 multimerization reactions are at thermodynamic (quasi)equilibrium, (iii) SinI inhibits SinR irreversibly, (iv) the number of states associated with the P1 promoter was reduced, (v) there is no expression of SinR from P1 mRNA, and (vi) the pool of SinI is not depleted by binding to SinR. These changes result in the following set of simplified equations,
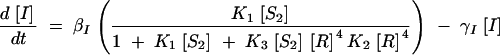 |
A1 |
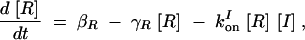 |
A2 |
where βI and βR are the expression rates of SinI and SinR, kIon is the rate at which SinI inactivates SinR, the equilibrium constants for promoter binding are given by K, and the degradation rates are given by γ. Note that the equilibrium constant for the formation of SinR tetramers KR is implicit in K2. Introducing the dimensionless parameters 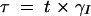 ,
, 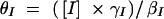 ,
, 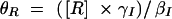 ,
, 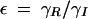 ,
, 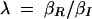 ,
, 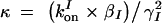 , the equations can be rewritten as
, the equations can be rewritten as
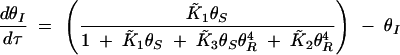 |
A3 |
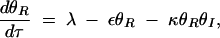 |
A4 |
where the overbar indicates dimensionless equilibrium constants. At steady state, the null clines are given by
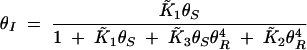 |
A5 |
and
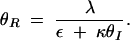 |
A6 |
Substituting (A6) into (A5) and solving for θS yield the polynomial
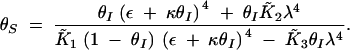 |
A7 |
The nonzero roots of the numerator are always imaginary. In other words, type 1 bistability, where the curve crosses the θS-axis (resulting in an irreversible switch due to unattainable negative θI concentrations), cannot occur.
Depending on the parameter values, Equation A7 can have three forms: (i) a single asymptote and no maxima, leading to a graded response, (ii) a single asymptote and a maximum, leading to a bistable switch, and (iii) three asymptotes, leading to type 2 bistability. Asymptotes arise when real solutions exist to the denominator polynomial of (A7). Using a Routh test, the condition for obtaining three real positive roots can be determined to be
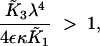 |
A8 |
when ε ≪ κ. The numerator of (A8) captures the strength of SinR expression and activity and the denominator represents the relative stability of SinI and its ability to inactivate SinR. When SinR is strong relative to SinI, then (A8) is large and type 2 bistability is present in the system.
APPENDIX B:
MINIMAL SIN MODEL TO ACHIEVE LIMIT CYCLE OSCILLATIONS
To determine the minimal system to achieve oscillations, we considered the simplified system of equations,
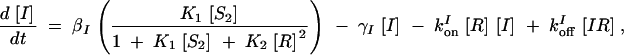 |
B1 |
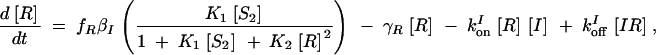 |
B2 |
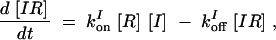 |
B3 |
where the number of promoter states has been reduced, the multimeric form of SinR was assumed to be dimers, and SinR multimerization is at equilibrium. Having SinR at least form dimers is critical in obtaining oscillations. Introducing the dimensionless parameters, 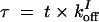 ,
, 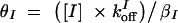 ,
, 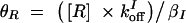 ,
, 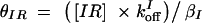 ,
, 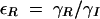 ,
, 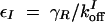 ,
, 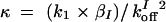 , and assuming that θI is a fast variable, the set of equations can be simplified to the following two-dimensional system:
, and assuming that θI is a fast variable, the set of equations can be simplified to the following two-dimensional system:
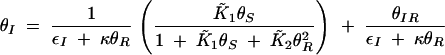 |
B4 |
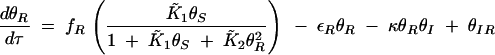 |
B5 |
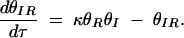 |
B6 |
Solving for null clines of (B5) and (B6) yields a single positive, real intersection that occurs at the fixed point
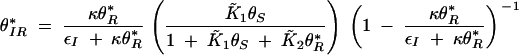 |
B7 |
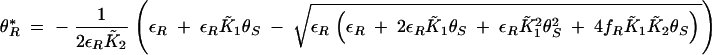 |
B8 |
Since the system is constrained to a closed bounded region and there are no other fixed points, the Poincaré-Bendixon theorem can be applied to identify oscillatory regimes in parameter space. When θ*I, θ*R is stable, the system will progress to this steady state. When θ*I, θ*R is unstable, oscillations will occur. The stability of a fixed point can be determined by analyzing the trace of the Jacobian matrix that is negative for stable fixed points and positive for unstable fixed points. The trace of the Jacobian associated with the differential equations (B5) and (B6) is
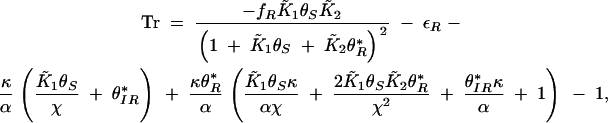 |
B9 |
where
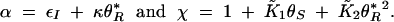 |
B10 |
Typical parameters that achieved oscillations in the numerical continuation analysis are εI = 0.07, εR = 0.007, κ = 4.0, fR = 0.125, K1θS = 8, K2 = 300, and, indeed, this results in a positive trace Tr = 0.03. Equation B8 can also be used to estimate the ranges of parameters that are consistent with oscillations. The trace is positive when κ → ∞, K2 → ∞, or εI → 0, although these limits result in the nonphysical situations of θIR → ∞, θIR → 0, and θIR → ∞, respectively. The parameters εI, K1θS, and fR have optimal values that maximize Tr and these parameters can be varied on average ±10-fold around this point. It is noteworthy that the simplified set of Equations B4–B6 oscillates more readily than the full set used in the continuation analysis.
References
- Arkin, A. P., J. Ross and H. H. McAdams, 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149: 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, M. R., M. A. Savageau, J. T. Myers and A. J. Ninfa, 2003. Development of a genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 13: 597–607. [DOI] [PubMed] [Google Scholar]
- Baba, T., and O. Schneewind, 1998. Instruments of microbial warefare: bacteriocin synthesis, toxicity, and immunity. Trends Microbiol. 6: 66–71. [DOI] [PubMed] [Google Scholar]
- Bai, U., I. Mandic-Mulec and I. Smith, 1993. SinI modulates the activity of SinR, a developmental switch protein by protein-protein interaction. Genes Dev. 7: 139–148. [DOI] [PubMed] [Google Scholar]
- Becskei, A., B. Seraphin and L. Serrano, 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20: 2528–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar, S. R., and G. R. Crabtree, 2001. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20: 3167–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, T. H., J. K. Grimsley, J. A. Hoch and G. B. Spiegelman, 1996. The Bacillus subtilis response regulator Spo0A stimulates transcription of the spoIIG operon through modification of the RNA polymerase promoter complexes. J. Mol. Biol. 256: 436–448. [DOI] [PubMed] [Google Scholar]
- Bläsi, U., C.-Y. Chang, M. T. Zagotta, K. Nam and R. Young, 1990. The lethal λS gene encodes its own inhibitor. EMBO J. 3: 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutsche, S., and V. Braun, 1997. SigX of Bacillus subtilis replaces the ECF sigma factor FecI of Escherichia coli and is inhibited by RsiX. Mol. Gen. Genet. 256: 416–425. [DOI] [PubMed] [Google Scholar]
- Camacho, A., and M. Salas, 1999. Effect of mutations in the “extended -10” motif of three Bacillus subtilis sigmA-RNA polymerase-dependent promoters. J. Mol. Biol. 286: 683–693. [DOI] [PubMed] [Google Scholar]
- Camacho, C. J., S. R. Kimura, C. DeLisi and S. Vajda, 2000. Kinetics of desolvation- mediated protein-protein binding. Biophys. J. 78: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervin, M. A., R. J. Lewis, J. A. Brannigan and G. B. Spiegelman, 1998. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 26: 3806–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J. D., G. Stephanopoulos, K. Ireton and A. D. Grossman, 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls initiation of sporulation. J. Bacteriol. 176: 1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes, I. W., and J. Mandelstam, 1970. Sporulation of Bacillus subtilis in continuous culture. J. Bacteriol. 103: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, H., J. Geiselmann, G. Batt, C. Hernandez and M. Page, 2004. Qualitative simulation of initiation of sporulation in Bacillus subtilis. Bull. Math. Biol. 66: 261–299. [DOI] [PubMed] [Google Scholar]
- Doedel, E. J., 1981. AUTO, a program for the automatic bifurcation analysis of autonomous systems. Congr. Numer. 30: 265–384. [Google Scholar]
- Engelberg-Kulka, H., and G. Glaser, 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53: 43–70. [DOI] [PubMed] [Google Scholar]
- Ermentrout, B., 2002 Simulating, Analyzing, and Animating Dynamical Systems. SIAM Press, Philadelphia.
- Farewell, A., and F. C. Neidhardt, 1998. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J. Bacteriol. 180: 4704–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw, A. M., H. Y. Liu, V. E. Anderson and P. L. deHaseth, 1998. Equilibrium and kinetic parameters of the sequence-specific interaction of Escherichia coli RNA polymerase with nontemplate strand oligodeoxyribonucleotides. Biochemistry 37: 11971–11979. [DOI] [PubMed] [Google Scholar]
- Gardner, T. S., C. R. Cantor and J. J. Collins, 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342. [DOI] [PubMed] [Google Scholar]
- Gaur, N. K., E. Dubnau and I. Smith, 1986. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J. Bacteriol. 168: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur, N. K., K. Cabane and I. Smith, 1988. Structure and expression of the Bacillus subtilis sin operon. J. Bacteriol. 170: 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur, N. K., J. Oppenheim and I. Smith, 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173: 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, D. T., 1977. Exact stochastic simulations of coupled chemical reactions. J. Phys. Chem. 81: 2340–2361. [Google Scholar]
- Grossman, A. D., 1995. Genetic networks controlling the initiation of sporulation and the development of competence in Bacillus subtilis. Annu. Rev. Genet. 29: 477–508. [DOI] [PubMed] [Google Scholar]
- Guet, C. C., M. B. Elowitz, W. H. Hsing and S. Leibler, 2002. Combinatorial synthesis of genetic networks. Science 296: 1466–1470. [DOI] [PubMed] [Google Scholar]
- Guidi, G. M., and A. Goldbeter, 1997. Bistability without hysteresis in chemical reaction systems: a theoretical analysis of irreversible transitions between multiple steady states. J. Phys. Chem. A 101: 9367–9376. [Google Scholar]
- Guidi, G. M., and A. Goldbeter, 2000. Oscillations and bistability predicted by a model for a cyclical bienzymatic system involving the regulated isocitrate dehydrogenase reaction. Biophys. Chem. 83: 153–170. [DOI] [PubMed] [Google Scholar]
- Huang, C.-Y., and J. E. Ferrell, Jr., 1996. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA 93: 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, K. T., and K. Mathee, 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52: 231–286. [DOI] [PubMed] [Google Scholar]
- Huynen, M. A., P. F. Stadler and W. Fontana, 1996. Smoothness within ruggedness: the role of neutrality in adaptation. Proc. Natl. Acad. Sci. USA 93: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton, K., D. Z. Rudner, K. J. Siranosian and A. J. Grossman, 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7: 283–294. [DOI] [PubMed] [Google Scholar]
- Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon et al., 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423: 87–91. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and J. Gerhart, 1998. Evolvability. Proc. Natl. Acad. Sci. USA 95: 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne, A. G. E., M. Sieber, D. Meierhans and R. K. Allemann, 1998. Thermodynamics of the DNA binding reaction of transcription factor MASH-1. Biochemistry 37: 4217–4223. [DOI] [PubMed] [Google Scholar]
- Lawrence, J. G., 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5: 355–359. [DOI] [PubMed] [Google Scholar]
- Lewis, R. J., J. A. Brannigan, W. A. Offen, I. Smith and A. J. Wilkinson, 1998. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J. Mol. Biol. 283: 907–912. [DOI] [PubMed] [Google Scholar]
- Lewis, R. J., D. J. Scott, J. A. Brannigan, J. C. Ladds, M. A. Cervin et al., 2002. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J. Mol. Biol. 316: 235–245. [DOI] [PubMed] [Google Scholar]
- Little, J. W., D. P. Shepley and D. W. Wert, 1999. Robustness of a gene regulatory circuit. EMBO J. 18: 4299–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maagd, R. A., A. Bravo and N. Crickmore, 2001. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 17: 193–199. [DOI] [PubMed] [Google Scholar]
- Maughan, H., and W. L. Nicholson, 2004. Stochastic processes influence stationary-phase decisions in Bacillus subtilis. J. Bacteriol. 186: 2212–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandic-Mulec, I., N. Gaur, U. Bai and I. Smith, 1992. sin, a stage-specific repressor of cellular differentiation. J. Bacteriol. 174: 3561–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandic-Mulec, I., L. Doukhan and I. Smith, 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spoOA. J. Bacteriol. 177: 4619–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H. H., and A. P. Arkin, 1999. It's a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 15: 65–69. [DOI] [PubMed] [Google Scholar]
- McClure, W. R., 1980. Rate-limiting steps in RNA chain elongation. Proc. Natl. Acad. Sci. USA 77: 5634–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, W. R., 1983 A biochemical analysis of the effect of RNA polymerase concentration on the in vivo control of RNA chain initiation frequency, pp. 207–217 in Biochemistry of Metabolic Processes: Proceedings of the 12th Steenbock Symposium, edited by D. Lenon, F. Stratman and R. Zahlman. Elsevier Scientific, New York.
- Meir, E., G. von Dassow, E. Munro and G. M. O'dell, 2002. Robustness, flexibility, and the role of lateral inhibition in the neurogenic network. Curr. Biol. 12: 778–785. [DOI] [PubMed] [Google Scholar]
- Meyers, L. A., and J. J. Bull, 2002. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 17: 551–557. [Google Scholar]
- Mittler, J., 1996. Evolution of the genetic switch in temperate bacteriophage. I. Basic theory. J. Theor. Biol. 179: 161–172. [DOI] [PubMed] [Google Scholar]
- Msadek, T., 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7: 201–207. [DOI] [PubMed] [Google Scholar]
- Olsen, R. J., and I. R. Epstein, 1993. Bifurcation analysis of chemical reaction mechanisms. II. Hopf bifurcation analysis. J. Chem. Phys. 98: 2805–2822. [Google Scholar]
- Ozbudak, E. M., M. Thattai, H. N. Lim, B. I. Shraiman and A. van Oudenaarden, 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740. [DOI] [PubMed] [Google Scholar]
- Perego, M., 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc. Natl. Acad. Sci. USA 94: 8612–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, C. W., 2002 General stress response, pp. 369–384 in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, edited by A. L. Sonenshein, J. A. Hoch and R. Losick. ASM Press, Washington, DC.
- Quisel, J. D., and A. D. Grossman, 2000. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J. Bacteriol. 182: 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, C. V., D. M. Wolf and A. P. Arkin, 2003. Control, exploitation and tolerance of intracellular noise. Nature 420: 231–237. [DOI] [PubMed] [Google Scholar]
- Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam et al., 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296: 2028–2033. [DOI] [PubMed] [Google Scholar]
- Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen et al., 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423: 81–86. [DOI] [PubMed] [Google Scholar]
- Rossi, F. M. V., A. M. Kringstein, A. Spicher, O. M. Guicherit and H. M. Blau, 2000. Transcriptional control: rheostat converted to on/off switch. Mol. Cell 6: 723–728. [DOI] [PubMed] [Google Scholar]
- Rowe-Magnus, D. A., and G. B. Spiegelman, 1998. DNA strand separation during activation of a developmental promoter by Bacillus subtilis response regulator Spo0A. Proc. Natl. Acad. Sci. USA 95: 5305–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S. L., and S. Lindquist, 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 338–342. [DOI] [PubMed] [Google Scholar]
- Savageau, M. A., 1998. Demand theory of gene regulation I: quantitative development of the theory. Genetics 149: 1665–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, P., J. Millet and J.-P. Aubert, 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D. J., S. Leejeerajumnean, J. A. Brannigan, R. J. Lewis, A. J. Wilkinson et al., 1999. Quaternary re-arrangement analyzed by spectral enhancement: the interaction of a sporulation repressor with its antagonist. J. Mol. Biol. 293: 997–1004. [DOI] [PubMed] [Google Scholar]
- Setty, Y., E. A. Mayo, M. G. Surette and U. Alon, 2003. Detailed map of a cis-regulatory input function. Proc. Natl. Acad. Sci. USA 100: 7702–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafikhani, S. H., I. Mandic-Mulec, M. A. Strauch, I. Smith and T. Leighton, 2002. Postexponential regulation of sin operon expression in Bacillus subtilis. J. Bacteriol. 184: 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea, M. A., and G. K. Ackers, 1985. The OR control system of bacteriophage lambda: a physical-chemical model for gene regulation. J. Mol. Biol. 181: 211–230. [DOI] [PubMed] [Google Scholar]
- Shen-Orr, S. S., R. Milo, S. Mangan and U. Alon, 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31: 64–68. [DOI] [PubMed] [Google Scholar]
- Smith, I., 1993 Regulatory proteins that control late-growth development, pp. 785–800 in Bacillus subtilis and Other Gram-Positive Bacteria, edited by A. L. Sonenshein, J. A. Hoch and R. Losick. American Society for Microbiology, Washington, DC.
- Smith, I., I. Mandic-Mulec and N. Gaur, 1991. The role of negative control in sporulation. Res. Microbiol. 142: 831–839. [DOI] [PubMed] [Google Scholar]
- Stephenson, K., and J. A. Hoch, 2002. Evolution of signaling in the sporulation pathway. Mol. Microbiol. 2: 297–304. [DOI] [PubMed] [Google Scholar]
- Stephenson, S., C. Mueller, M. Jiang and M. Perego, 2003. Molecular analysis of Phr peptide processing in Bacillus subtilis. J. Bacteriol. 185: 4861–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier, P., and R. Losick, 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30: 297–341. [DOI] [PubMed] [Google Scholar]
- Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys et al., 1989. The transition state transcription regulation abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8: 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki et al., 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28: 4317–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatiana, A., T. A. Tatusova and T. L. Madden, 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar, J. G. M., H. Y. Kueh, N. Barkai and S. Leibler, 2002. Mechanisms of noise-resistance in genetic oscillators. Proc. Natl. Acad. Sci. USA 99: 5988–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N. V., L. M. Hsu, C. M. Kane and M. J. Chamberlin, 2003. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry 42: 3798–3811. [DOI] [PubMed] [Google Scholar]
- Voigt, C. A., S. Kauffman and Z.-G. Wang, 2001. Rational evolutionary design: the theory of in vitro protein evolution. Adv. Protein Chem. 55: 79–160. [DOI] [PubMed] [Google Scholar]
- von Dassow, G., E. Meir, E. M. Munro and G. M. Odell, 2000. The segment polarity network is a robust developmental module. Nature 406: 188–192. [DOI] [PubMed] [Google Scholar]
- Wolf, D. M., and A. P. Arkin, 2003. Motifs, modules, and games in bacteria. Curr. Opin. Microbiol. 6: 125–134. [DOI] [PubMed] [Google Scholar]
- Yokobayashi, Y., R. Weiss and F. H. Arnold, 2002. Directed evolution of a genetic circuit. Proc. Natl. Acad. Sci. USA 99: 16587–16591. [DOI] [PMC free article] [PubMed] [Google Scholar]