Abstract
We report molecular genetic studies of three genes involved in early germ-line proliferation in Caenorhabditis elegans that lend unexpected insight into a germ-line/soma functional separation of autosomal/X-linked duplicated gene pairs. In a genetic screen for germ-line proliferation-defective mutants, we identified mutations in rpl-11.1 (L11 protein of the large ribosomal subunit), pab-1 [a poly(A)-binding protein], and glp-3/eft-3 (an elongation factor 1-α homolog). All three are members of autosome/X gene pairs. Consistent with a germ-line-restricted function of rpl-11.1 and pab-1, mutations in these genes extend life span and cause gigantism. We further examined the RNAi phenotypes of the three sets of rpl genes (rpl-11, rpl-24, and rpl-25) and found that for the two rpl genes with autosomal/X-linked pairs (rpl-11 and rpl-25), zygotic germ-line function is carried by the autosomal copy. Available RNAi results for highly conserved autosomal/X-linked gene pairs suggest that other duplicated genes may follow a similar trend. The three rpl and the pab-1/2 duplications predate the divergence between C. elegans and C. briggsae, while the eft-3/4 duplication appears to have occurred in the lineage to C. elegans after it diverged from C. briggsae. The duplicated C. briggsae orthologs of the three C. elegans autosomal/X-linked gene pairs also display functional differences between paralogs. We present hypotheses for evolutionary mechanisms that may underlie germ-line/soma subfunctionalization of duplicated genes, taking into account the role of X chromosome silencing in the germ line and analogous mammalian phenomena.
GENOMES are shaped over evolution by a variety of forces, including gene duplication. The functions of duplicated genes can change over evolutionary time as well. One copy of a duplicated gene pair may acquire deleterious mutations, ultimately rendering it useless to the organism. Alternatively, both genes may remain functional. In the latter case, the duplicates may remain fully redundant or they may take on nonredundant roles. If the ancestral gene fulfilled multiple roles (e.g., was expressed in multiple regions of an organism), subfunctionalization can follow gene duplication such that the duplicated genes acquire different functional roles (Thomas 1993; Hughes 1994; Lynch and Conery 2000).
One potential factor postulated to influence the fate of duplicated genes is germ-line-specific X chromosome inactivation (McKee and Handel 1993). This phenomenon occurs in the genomes of many organisms, including the nematodes Caenorhabditis elegans and C. briggsae and male mammals. In mammals, the XY body forms during spermatogenesis (see Handel 2004 and references therein). In the nematodes, repressing histone modifications have been observed on the X throughout much of hermaphrodite and all of male germ-line development (Fong et al. 2002; Kelly et al. 2002).
Silencing of the X chromosome during germ-line development could conceivably place constraints on genome organization. Taking cell-essential or housekeeping genes into account, this phenomenon suggests that (1) cell-essential genes are not located on the X; (2) if located on the X, these genes are not subject to inactivation; or (3) germ-line-active copies of these genes must be located in at least one additional place in the genome (i.e., on an autosome). The first possibility is suggested by functional-genomic and microarray studies that have demonstrated a dearth of essential genes (Piano et al. 2002; Kamath et al. 2003) and germ-line-enriched genes (Reinke et al. 2000) on the X of C. elegans and of late spermatogenesis-acting genes in mammals (Khil et al. 2004). The second possibility is still unresolved. The last possibility was suggested by McKee and Handel (1993) and may be a determining factor for the existence of several autosomal intronless genes that function late in mouse spermatogenesis and that are closely related to intron-containing genes on the X (Handel 2004).
Here, we provide genetic evidence suggesting that the activity of certain X-linked housekeeping genes in C. elegans is restricted to the soma and that the presence of an autosomal duplicate in the genome ensures that this activity is maintained in the germ line. In a forward genetic screen for genes required for early germ-line proliferation, we identified three alleles representing three different germ-line-active genes (rpl-11.1, pab-1, and glp-3/eft-3), all of which have a paralog on the X chromosome. Further functional analysis identified a fourth gene with these properties, rpl-25.2, and suggests that all four of these gene pairs are subfunctionalized with respect to the germ line and soma. Additional analysis of the C. elegans genome and results of large-scale RNAi screens suggests that members of other autosome/X gene pairs may be similarly subfunctionalized. An additional rpl gene, rpl-24, is an autosome/autosome duplicate pair and does not display the same degree of soma/germ-line subfunctionalization. Of the five duplications we examined in the rpl, pab, and eft genes, eft-3/eft-4 occurred recently over the course of C. elegans evolution, while the others preceded the divergence of C. elegans and C. briggsae lineages. Functional analysis of the orthologous pairs of the four duplicated C. briggsae genes suggests that the members of the gene pairs are functionally distinct, but may exhibit a lesser degree of germ-line/soma distinction. We present several hypotheses for evolutionary mechanisms that may underlie germ-line/soma subfunctionalization of duplicated genes, X chromosome silencing, and genome organization.
MATERIALS AND METHODS
Worm handling and strains:
Wild types were the C. elegans var. Bristol N2 strain and Hawaiian CB4856 strain. The following mutations were used [from Brenner (1974) unless indicated]: LGI—lin-11(n566) (Trent et al. 1983), unc-75(e950) (Anderson and Brenner 1984), and rrf-1(pk1417) (Sijen et al. 2001); LGIII—unc-32(e189), glp-1(q175) (Austin and Kimble 1987), glp-3(q145) (Kadyk et al. 1997), sma-3(e491), and unc-36(e251); and LGV—dpy-11(e224) and unc-34(e566). Deficiencies used were qDf7 (Ellis and Kimble 1995), sDf70 (Stewart et al. 1991), sDf50 (Johnsen and Baillie 1988), and Df(e2173) (Johnsen 1990). The following rearrangements were used: eT1 (III;V) (Rosenbluth and Baillie 1981); hT2[let-? qIs48] (I;III), hereafter referred to as hT2[let GFP]; nT1[unc-?(n754) let-? qIs50](IV;V) (Mathies et al. 2003); and the latter nT1 without qIs50 (Ferguson and Horvitz 1985). The C. briggsae strain was PB800.
Phenotypic analyses:
For time course analysis of ar228 and ar232 germ-line proliferation, worms (homozygous mutants and sibling heterozygotes expressing a dominant GFP marker) were synchronized by hatch-off at 25° and cultured at either 15° and 25° (ar228) or 25° (ar232) before being examined live under Nomarski optics and/or individually fixed and stained with DAPI [synchronization and staining as described in Pepper et al. (2003a)] at three time points corresponding to early L1 (within 2 hr of hatching at 25°), L3 (48 hr at 15° or 24–26 hr at 25°), and early adult (96 hr at 15° or 48 hr at 25°).
All C. elegans RNAi experiments were conducted by feeding at 25° as described (Timmons et al. 2001). L4 animals were fed RNAi-inducing or control [L4440 vector in HT115(DE3)] bacteria and removed as adults after 24 hr. Progeny from these animals were maintained on the same bacteria until being scored as adults, 48 hr later. All animals were scored first under low magnification and sterile animals were inspected under higher magnification—both live and after fixation and staining with DAPI. The RNAi feeding plasmid was pGC10 for rpl-11.1: a PCR product was amplified from N2 worms (using sjj_T22F3.4 primers; Kamath et al. 2003), digested with SacI, and cloned into SacI/HincII of L4440 (Timmons et al. 2001). Feeding constructs were used for all additional experiments (MRC gene service; Fraser et al. 2000; Kamath et al. 2003).
Genomic DNA templates for C. briggsae RNAi were PCR amplified from the following primers (each forward primer sequence was preceded by gggaaggtacc and each reverse primer was preceded by caaaacggccg, where the underlined sequence is the KpnI or EagI recognition site in the case of each forward and reverse primer, respectively, that was subsequently used to clone each fragment into the same sites in the L4440 vector):
CBG01314—F:CAAGATTCAAAAGCTCTGCC, R:CACCTGAAAAAAGTCCGAAC;
CBG14053—F:GAGATAGACTTACCCGTGCC, R:TCATACTTCTGTTGGAACCAC;
CBG22273—F:ATCAGATGGACCGTCCTTTACAG, R:TTTATCGCTTTCCTCCGACGC;
CBG03702—F:CTGCTCTTCTCCGATCTACCC, R:TCTTCTTCTCCTTTGTTGCTG;
CBG14529—F:AGAAGGTCGCCAAGGGAC, R:GTTGGAGTGGGGTGATGAG;
CBG04080—F:CCTCGGTTCATTTCAGACG, R:TAGTCGGAAGCCAATCGCAC;
CBG02207—F:CAAGATGGTCTGCTCGAAGC, R:GTCCACCGATGACGATTCC;
CBG07431—F:GGATTCGGATTCGTTGCC, R:GGATAGATTGGAGCCATTCC.
The identity of each construct was verified by sequencing with M13F(-21) primer. Prior to in vitro transcription (Ribomax; Promega, Madison, WI), each construct or the “empty” L4440 (negative control) was linearized with appropriate (blunt or 5′-overhang) enzymes. Transcribed RNA products were checked by gel electrophoresis prior to injection. dsRNA was injected into both arms of the germ line as described by Guo and Kemphues (1995) and Fire et al. (1998). Injected animals were raised at 20°, transferred to a new plate after overnight recovery from the injection, and subsequently transferred as necessary to score for maternal sterility (injected worm) and progeny survivorship. The negative control was the L4440 plasmid carried through the entire protocol in parallel. A positive control dsRNA (Cb PAR-1) was also injected and a highly penetrant maternal embryonic lethality phenotype was observed.
The glp-1; rpl-11.1 double-mutant analysis was carried out at 15° and 25° by picking Unc-32 Dpy progeny of unc-32 glp-1/eT1; dpy-11 rpl-11.1(ar228)/eT1 mutant animals as L4 larvae and examining the germ line after fixation and DAPI staining 24 hr later. Control animals were Unc-32 Dpy progeny from the unc-32/eT1; dpy-11 rpl-11.1(ar228)/eT1.
Molecular identification of rpl-11.1(ar228), pab-1(ar232), and glp-3/eft-3(ar229):
rpl-11.1(ar228):
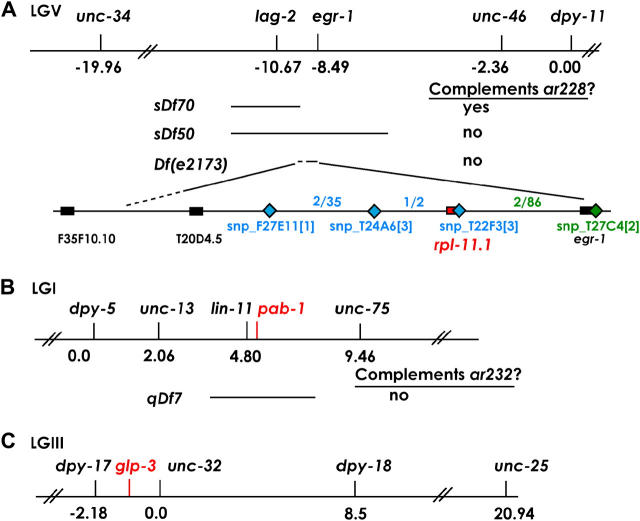
Two- and three-factor mapping and deficiency complementation tests narrowed the ar228 mutation to the area deleted by the deficiency e2173 (Johnsen 1990), previously identified as an allele of lin-40 (WormBase). In the course of mapping ar228, we positioned the left breakpoint of the e2173 deletion between ORFs F35F10.10 and T20D4.5, while the right breakpoint was located near egr-1, between T27C4.4 and T27C4.1 (Figure 1A). This deletion is homozygous viable yet sterile, suggesting that no genes causing zygotic embryonic or larval lethality are located in this region. We determined the left endpoint of e2173, by amplifying PCR fragments from homozygous sterile e2173 animals (with a positive control amplification outside the region). On the basis of these data, we determined that the left breakpoint of Df(e2173) is within the ∼100-kb interval between T20D4.5 and F35F10.10 (Figure 1).
Figure 1.—
Mapping of (A) rpl-11.1(ar228), (B) pab-1(ar232), and (C) glp-3/eft-3(ar229). Relevant sections of linkage groups V, I, and III are depicted with genetic map positions indicated for markers used in the analysis. Complementation information is given for deficiencies tested in each region. For A, SNPs used to map ar228 from the left (blue) and right (green) are given with the relevant intervals and recombinants observed in each interval [see materials and methods for details and further information regarding Df(e2173)].
SNP mapping using the Hawaiian strain CB4856 further reduced the interval containing ar228 by taking advantage of marked recombinants that lost ar228 and thereby produced progeny. From the right side, 86 Dpy nonsterile self-progeny of ar228 dpy-11/Hawaiian mothers were selected, animals homozygous for the recombinant chromosome were identified in the next generation, and the strain was tested for the N2 or Hawaiian sequence of snp_T27C4[2]. Two recombinants that had undergone a crossover event to the left of snp_T27C4[2] (N2 DNA) both contained Hawaiian sequence at snp_T22F3[3]. From the left side, 35 Unc nonsterile self-progeny were picked from unc-34 ar228/Hawaiian mothers. Two of the 35 recombinant chromosomes had crossed over to the right of snp_F27E11[1] and were further tested: in one of two recombinants, the crossover occurred between snp_T24A6[3] and snp_T22F3[3].
DNA was amplified by PCR from the rpl-11.1 locus (ATG to stop, including introns) from animals homozygous for ar228 and our laboratory stock of the wild-type N2 strain. Comparison of the sequence indicated a single base pair change (verified on both strands) in the coding region of the rpl-11.1 gene as indicated in results.
pab-1(ar232):
After linkage testing placed ar232 on LGI, three-factor analysis was performed: from the self-progeny of lin-11 unc-75/ar232, 1/10 Lin non-Unc recombinants segregated sterile worms and 6/9 Unc non-Lin segregated sterile worms (Figure 1B). The deficiency qDf7 failed to complement ar232: all (∼50) non-GFP progeny of qDf7/hT2[let GFP] × ar232/hT2[let GFP] males were sterile. RNAi feeding was performed on 8 of 16 candidate genes among the ∼270 genes in the region (WormBase version 110) that were previously identified as sterile (Ste or Stp) by RNAi (Fraser et al. 2000; Maeda et al. 2001). Among this set, only pab-1(RNAi) (clone Y106G6H.2) conferred a highly penetrant low-proliferation sterile phenotype. DNA was amplified by PCR from the pab-1 locus and analyzed as noted above for rpl-11.1(ar228).
glp-3/eft-3(ar229):
After linkage testing placed ar229 on chromosome III, three-factor crosses narrowed ar229 to the left of dpy-18 (16/16 Unc-25 non-Dpy-18 and 0/13 Dpy non-Unc segregated ar229) and between dpy-17 and unc-32 (22/42 Unc non-Dpy and 25/53 Dpy non-Unc segregated ar229) (Figure 1C). Complementation between glp-3(q145) and ar229 was tested as follows: (1) of 60 F1 non-Unc cross-progeny of glp-3/sma-3 unc-36 hermaphrodites crossed to ar229/eT1 males, 48 were fertile and segregated Unc or Sma Unc and 12 were sterile (presumed genotype glp-3/ar229) and (2) from the reciprocal cross, of 36 F1 non-Unc cross-progeny, 27 were fertile and segregated Unc or Sma Unc and 12 were sterile (presumed genotype glp-3/ar229).
Life-span and size assays:
Life-span and size assays were conducted at 20°. Individual L4 animals were transferred every other day until the end of the reproductive period, after which they were maintained on the same plate (sterile animals were maintained on the same plates throughout). Missing animals were not included in life-span calculations. Worms were considered dead when they no longer responded to prodding. Body length and width at midbody were measured at ×50 magnification using a KR-851 1 mm/100 divisions stage micrometer, at ages indicated, after immobilization with 0.1 m NaN3. For volume calculations, worms were treated as cylinders: V = π(1/2D)2L, where D is the width and L is the length (McCulloch and Gems 2003).
Analysis of RNAi results for X autosome gene pairs:
Reciprocal whole proteome BLASTP analysis (Altschul et al. 1997) and comparative analysis of RNAi results were performed as described (Fernandez et al. 2005). Homologous gene pairs were defined on the basis of BLASTP matches between C. elegans and human or between C. elegans protein pairs that met a reciprocal best hit criterion and had an expectation value of ≤10−50 and ≥75% identity over the length of both proteins.
Phylogenetic analysis:
BLASTP was used with WormBase (release WS128, http://www.WormBase.org) to identify predicted proteins in C. briggsae that are homologous to the pab-1/2, rpl-11.1/11.2, rpl-24.1/24.2, rpl-25.1/25.2, and eft-3/4 genes of C. elegans. Corresponding gene sequences (minus inferred introns) were aligned using Clustal X version 1.81 (Thompson et al. 1997) using a gap-open penalty of 90.0, a gap-extension penalty of 0.1, and default parameters otherwise. These alignments were imported into MacClade version 4.05 (Maddison and Maddison 2002) and refined by hand relative to the encoded amino acids. These final alignments were phylogenetically analyzed with maximum likelihood (GTR + SS, general time-reversible model, allowing site-specific rates for the three different codon positions, with all parameters estimated by likelihood from the data), implemented in PAUP* 4.0b10 (Swofford 2002). For each set of homologs, jackknife analyses (using 50% deletion) and log-likelihood tree comparisons were performed using maximum likelihood with the GTR + I + G model (Swofford 2002), with parameters estimated from the data. In all cases, the root was assumed to occur in the longest branch, which was always the internal branch of each set of homologs. Orthologs were inferred to be the closest gene relatives occurring in different species.
RESULTS
To investigate zygotic requirements for the reestablishment and maintenance of early germ-line proliferation, we performed an EMS mutagenesis screen and looked for adult worms that, as judged by Nomarski optics, had a normal somatic gonad but displayed little or no germ line (Pepper et al. 2003a; E. J. A. Hubbard, unpublished data). To identify mutants with an early germ-line proliferation defect (as opposed to germ-line specification defects), we concentrated on mutants in which Z2 and Z3 were present in the L1 gonad primordium but subsequently underwent few or no additional rounds of division and did not differentiate. This phenotype is distinct from the glp-1(loss-of-function) mutant phenotype, in which Z2 and Z3 undergo several rounds of division, complete meiosis, and mature as sperm (Kimble and Hirsh 1979), and is more similar to that of glp-3 (Kadyk et al. 1997) and glp-4 (Beanan and Strome 1992). A severe germ-line proliferation defect without subsequent maturation is also observed when RNAi is directed against the chromodomain-containing protein mrg-1, although the requirement for this gene in early germ-line proliferation is maternal (Fujita et al. 2002). Other genes required for germ-line proliferation have been identified using RNAi (Hanazawa et al. 2001; Maeda et al. 2001; Colaiacovo et al. 2002), and a large-scale analysis of RNAi-mediated germ-line proliferation defects is in progress (E. J. A. Hubbard, unpublished data).
Phenotypic analysis of ar228:
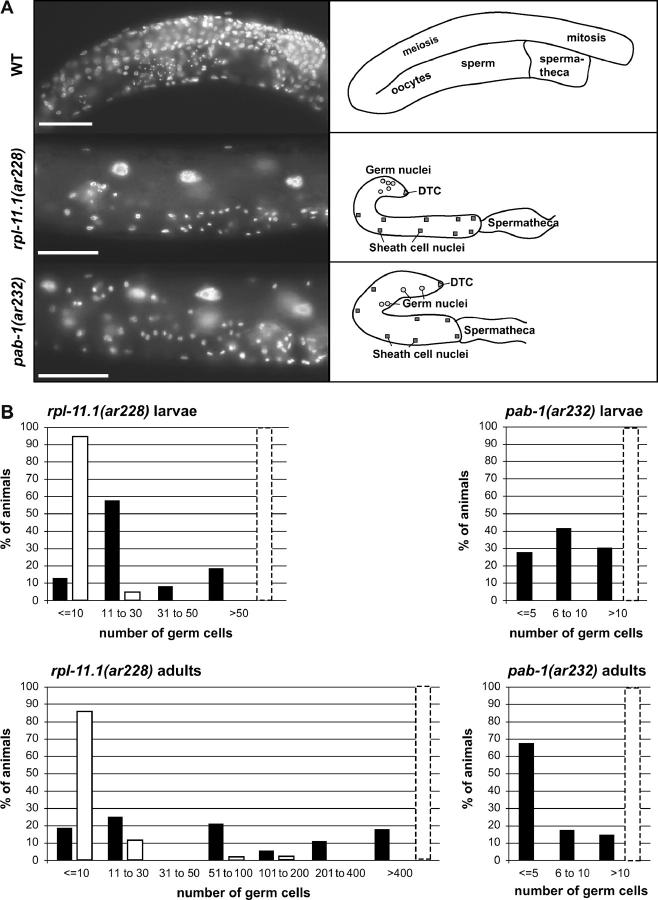
ar228 is recessive and causes sterility at all temperatures, although the most severe and most penetrant proliferation defect (>80% of adults with 10 or fewer germ cells per animal) is seen at a lower temperature (Figure 2). Further phenotypic analysis of the ar228 mutant revealed that Z2 and Z3 are present in the early L1 gonad (n > 40), but they do not appear to proliferate well in subsequent larval stages. On the basis of the distribution of germ cell counts at two time points (L2/L3 and adult; Figure 2), germ cells in animals with very few germ cells in the L2/L3 did not appear to proliferate further, while germ cells that underwent some early proliferation did appear to proliferate further over time (Figure 2). In some cases, germ cells appeared to be in various stages of disintegration. Because Z2 and Z3 were visible in the L1 primordium of all animals examined, we assume that Z2 and Z3 or their early descendents disintegrated completely in those animals with no visible germ cells in the L3 or adult.
Figure 2.—
Germ-line proliferation in rpl-11.1(ar228) and pab-1(ar232). (A) Images of DAPI-stained early adult animals: wild type (WT), rpl-11.1(ar228), and pab-1(ar232) and corresponding cartoons. Bars, 50 μm. (B) Histograms depicting distributions of germ cell counts in rpl-11.1(ar228) and pab-1(ar232). Solid bars represent total counts taken per animal at 25° and open bars represent counts taken from animals at 15°. Larval counts were obtained 24 hr post-hatch at 25° (early L3) or 48 hr post-hatch at 15° (late L2). Adult counts were taken 48 hr post-hatch at 25° and 96 hr post-hatch at 15° (both early adult). Dashed bar indicates the cell count category for the wild type at the equivalent time point (Hirsh et al. 1976; Kimble and Hirsh 1979; Pepper et al. 2003b).
To assess whether the few germ cells produced in ar228 animals were capable of entering meiosis, we examined the phenotype of glp-1(q175); ar228 double-mutant animals under restrictive (15°) and less restrictive (25°) conditions for the ar228 proliferation defect (see materials and methods). In glp-1 loss-of-function mutants, all germ cells enter meiosis early and differentiate as sperm. At 15°, the majority of the double-mutant animals (26/28), like the ar228 single mutants (n = 28), did not produce any sperm and were morphologically similar to ar228 single mutants. At 25°, the germ lines of 4/19 animals contained a total of 0–2 germ cells that did not differentiate (similar to ar228 single mutants), while in 5/19 animals, the germ line contained a total of 5–30 sperm [similar to glp-1(q175) single mutants]. Genetically matched control animals produced 0–9 (n = 28 animals) and 0–17 (n = 19) total undifferentiated germ cells per animal at 15° and 25°, respectively. Surprisingly, the germ lines of the remaining 10 double-mutant animals at 25° produced 46–200 sperm, far more than are normally seen in glp-1(q175) mutants alone. One possibility is that the germ cells produced by ar228 mutants are resistant to differentiation and proliferated abnormally in the glp-1 mutant background prior to differentiating en masse. Nevertheless, the results at the restrictive temperature suggest that the ar228 proliferation defect precludes differentiation.
Molecular identification of rpl-11.1(ar228):
ar228 was mapped to a region of the left arm of LGV containing ∼37 ORFs (materials and methods; Figure 1). One gene in the region, rpl-11.1, was reported to confer sterile (Ste), sterile progeny (Stp), or embryonic-lethal (Emb) and growth defective (Gro) phenotypes in previous RNAi experiments (Piano et al. 2000, 2002; Simmer et al. 2003). This gene also displayed a strong germ-line expression pattern (NEXTDB http://nematode.lab.nig.ac.jp; Y. Kohara, personal communication). Sequence analysis of DNA from the rpl-11.1 gene amplified from ar228 homozygotes revealed a single C-to-T change that causes a premature termination at position Q175 (materials and methods). Therefore, we refer to this allele as rpl-11.1(ar228). RPL-11.1 is a large ribosomal subunit L11 protein. These results suggest that robust translation is an absolute requirement for early germ-line proliferation.
Because the mutant phenotype of rpl-11.1(ar228) is zygotic sterility rather than embryonic lethality, L11 may be provided to the embryo maternally. To investigate this possibility we turned to RNA interference (RNAi) that degrades both the maternal and the zygotic mRNAs and found that the rpl-11.1(RNAi) phenotype is more severe than the rpl-11.1(ar228) phenotype. Early progeny of animals consuming rpl-11.1(RNAi) bacteria (see materials and methods) exhibit a proliferation-defective sterility phenotype identical to that seen in rpl-11.1(ar228) homozygous animals. However, consistent with previous RNAi observations (Piano et al. 2000, 2002; Simmer et al. 2003), animals that continue to feed on RNAi-inducing bacteria produce dead embryos and, eventually, no embryos at all. These results suggest that the homozygous rpl-11.1(ar228) self-progeny of heterozygous mothers are rescued from embryonic lethality by the maternal contribution of rpl-11.1. The similarity of the RNAi-induced sterility defect to that of the mutant phenotype lends further credence to the supposition that the mutation at 15° confers an essentially null phenotype in the germ line despite the fact that the predicted protein product from rpl-11.1(ar228) is truncated by only 22 of 196 amino acids.
Germline-specific zygotic role of rpl-11.1:
Among the genes encoding proteins highly similar to RPL-11.1 from other phyla is a second C. elegans L11-encoding gene, rpl-11.2, located on the X chromosome (WormBase). Given previous reports of a dearth of germ-line-enriched genes on the X chromosome (Reinke et al. 2000, 2004), underrepresentation of expression of essential X-linked genes (Piano et al. 2002; Reinke et al. 2004), and reports of X chromosome silencing in the germ line (Fong et al. 2002; Kelly et al. 2002), we wondered if there might be a clear functional separation of the L11-encoding genes in the germ line and soma, with the autosome-linked rpl-11.1 functioning in the germ line and the X-linked rpl-11.2 functioning in the soma. We used several assays to assess the germ-line vs. soma activities of these two genes.
First we examined carefully the RNAi phenotypes of both rpl-11 genes. Consistent with previous reports, we found that rpl-11.2 confers a penetrant growth defect (Gro) that is not observed in parallel rpl-11.1 RNAi experiments (Table 1). We also investigated the RNAi phenotypes of the two genes in a genetic background that reduces the efficacy of RNAi in the soma while retaining it in the germ line, rrf-1(pk1417) (Sijen et al. 2001). We found that in an rrf-1 mutant background, rpl-11.1 still conferred a penetrant germ-line proliferation defect, while the somatic growth defect of rpl-11.2 was largely suppressed. Other adult defects were observed in association with rpl-11.2(RNAi) in the rrf-1 mutant background, including sterility (Table 1).
TABLE 1.
RNAi analysis of duplicatedrpl genes in wild-type (N2) andrrf-1(pk1417) mutant animals
| Phenotypea
|
|||||||
|---|---|---|---|---|---|---|---|
| Germ-line defect
|
Somatic defect
|
||||||
| RNAi target | LG | Strain | WTb (%) | Sterilec (%) | Unhealthyd (%) | Gro/Lvae (%) | n |
| (None) | N2 | 76 | 0 | 21 | 0.7 | 275 | |
| rrf-1 | 24 | 25f | 40 | 0 | 114 | ||
| rpl-11.1 | V | N2 | 0 | 77f | 5 | 0.7 | 298 |
| rrf-1 | 0 | 75f,g | 5 | 0 | 59 | ||
| rpl-11.2 | X | N2 | 3 | 12 | 3 | 78 | 295 |
| rrf-1 | 0 | 15g | 45 | 10 | 80 | ||
| rpl-25.1 | X | N2 | 73 | 0 | 25 | 0 | 149 |
| rrf-1 | 44 | 43f,g | 7 | 4 | 99 | ||
| rpl-25.2 | I | N2 | 8 | 86f | 6 | 0 | 152 |
| rrf-1 | 10 | 68f,g | 5 | 0 | 19 | ||
| rpl-24.1 | I | N2 | 1 | 22f,g | 15 | 62 | 112 |
| rrf-1 | 0 | 76f | 17 | 0.2 | 58 | ||
| rpl-24.2 | I | N2 | 24 | 10g | 18 | 47 | 180 |
| rrf-1 | 0 | 89f | 11 | 5 | 121 | ||
Data are reported here for one parallel experiment; several experiments gave similar results (see materials and methods). Percentages for phenotypes that were most penetrant for each treatment are in italic type. Remaining animals fell into “other” phenotypic categories that were not scoreable, such as lethal (Let) or ruptured (Rup).
Normal-sized fertile adults.
Sterile adults (both normal sized and reduced size).
Fertile adults but small or sick looking.
Larval arrest (Lva) or larval-sized animals.
Majority of sterility due to strong Glp (germ-line proliferation defective) phenotype; this phenotype is observed variably in the rrf-1 background and may be temperature sensitive. In the particular experiment reported here, the penetrance is on the high side of the range of variability compared to other experiments.
Sterility largely due to mild germ-line proliferation defect often accompanied by signs of gametogenesis (f,g indicates that a large number of worms exhibited both types of sterility defect).
Second, we compared the available expression patterns of rpl-11.1 and rpl-11.2 images from the NEXTDB expression pattern database (http://nematode.lab.nig.ac.jp; Y. Kohara, personal communication). Weakly detectable levels of rpl-11.1 mRNA are observed in a limited number of cells in the embryo. Strong levels are detected, however, in a small number of cells located centrally in the larvae (likely Z2 and Z3 and their descendants) and thereafter in the germ line, primarily in the distal proliferating regions. In contrast, rpl-11.2 mRNA is detectable in a subset of cells in the embryo, with high levels in what appear to be intestinal cells in later embryonic stages. During larval development, rpl-11.2 mRNA is weakly detectable, with somewhat higher levels in the adult. These observations suggest that vastly differential levels of mRNAs encoding ribosome subunits could be synthesized in different tissues, a phenomenon observed in other organisms as well (Agarwal et al. 1999). Given the presumably cell-essential function of the L11 protein, we found the dynamic and nonubiquitous levels of expression surprising. These results likely reflect tissue-specific requirements for high translational capacity. Previously reported microarray analysis comparing wild type with the germ-line-deficient glp-4(bn2) mutant also firmly places rpl-11.1 in the germ-line-enriched class (log2 ratio of +4.1; Reinke et al. 2004) while rpl-11.2 is not enriched in the germ line (log2 ratio of −1.3; Reinke et al. 2004).
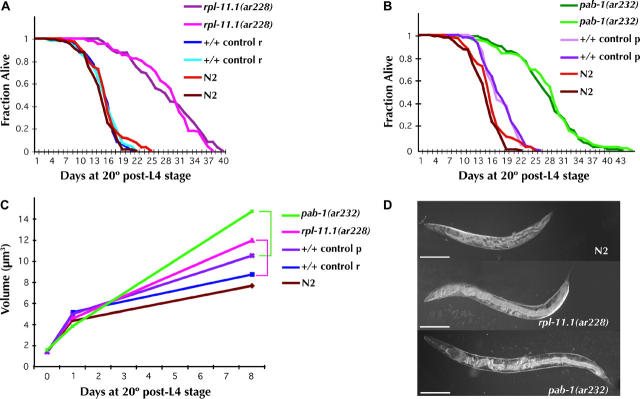
Third, previous studies documented two dramatic germ-line-associated phenotypes after removal of Z2 and Z3 by laser microsurgery: life-span extension (Hsin and Kenyon 1999) and gigantism (Patel et al. 2002). We reasoned that if only the germ line is affected by rpl-11.1(ar228), we should see the same phenotypes previously reported for germ-line-ablated animals. If, on the other hand, rpl-11.1 also plays a role in somatic development that would compromise the somatic contribution to life-span or size determination, these phenotypes may be less severe in our mutant than those observed in the cell-ablation studies. We found that in rpl-11.1(ar228) animals, both life-span extension and overall increase in the volume of the worms is similar to that observed in germ-line-ablation experiments (Figure 3). Life span is extended in ar228 sterile homozygotes to 27 days (starting from the L4 stage) from a mean life span of 14 days in control animals, an increase of 1.9-fold. These results are comparable to the increase in mean life span observed after ablation of Z2 and Z3 (an extension from 19 to 32 days or 1.7-fold starting from day of birth; Hsin and Kenyon 1999). We also observed a positive correlation between rpl-11.1(ar228) mutant body length, width, and volume. Homozygous mutant and wild-type L4 larvae and early adults were similar in size, but adult size increased more in the mutant than in the wild type as the animals aged (Figure 3). The overall volume increase in adult sterile rpl-11.1(ar228) animals (as compared to genetically matched controls) is from 8.7 μm3 to 11.9 μm3 8 days post-L4 (or 7 days after reaching adulthood), an increase of 1.4-fold. These results are similar to the previously reported increase in volume from 5.8 to 8.4 μm3, also 1.4-fold, 7 days postadulthood after Z2/Z3 cell ablations (Patel et al. 2002).
Figure 3.—
Life span and gigantism in rpl-11.1(ar228) and pab-1(ar232). Survival curves (A) for rpl-11.1(ar228) and (B) for pab-1(ar232) and (C) worm volumes, with respective control strains and the wild-type strain N2, are shown. All non-N2 strains (mutant and respective controls) are homozygous progeny of heterozygous mothers balanced by nT1[unc-?(n754) let-?] for rpl-11.1(ar228) (control r) and by hT2[let-? qIs48] for pab-1(ar232) (control p). Survival curves are given for two independent experiments for each non-N2 strain. Taken together, for N2 wild type (brown and orange), mean life span (m) = 14.2 ± 3.5 days and number of animals observed (n) = 163; for +/+ control r, m = 14.2 ± 3.3 days and n = 127; for rpl-11.1(ar228), m = 27.4 ± 6.9 days and n = 126; for +/+ control p, m = 16.9 ± 3.4 days, n = 126; and for pab-1(ar232), m = 26.4 ± 6.9 days, n = 185. (C) Volume calculations are based on length and width measurements (see materials and methods); n = 20 worms of each genotype per time point. Brackets link mutants with relevant genetically matched controls. Measurements were taken at the mid-L4 (day 0), as early adults (day 1), and a week later (day 8) as older adults. Actual volumes (in cubic micrometers) calculated for the three indicated time points (±1 SD) are as follows: for pab-1(ar232), 1.55 ± 0.27, 3.82 ± 0.43, and 14.7 ± 1.12; for rpl-11(ar228), 1.37 ± 0.31, 4.55 ± 0.90, and 11.94 ± 5.84; for control p, 1.37 ± 0.19, 4.98 ± 0.77, and 10.50 ± 1.67; for control r, 1.37 ± 0.23, 5.14 ± 1.00, and 8.72 ± 2.12; and for N2, 1.37 ± 0.27, 4.43 ± 1.27, and 7.64 ± 1.21. (D) Digital images of wild-type (N2), rpl-11.1(ar228), and pab-1(ar232) animals 10 days after the L4 stage at 20°. Bars, 200 μm.
Taken together, the results of RNAi experiments, expression data, and phenotypic analysis suggest that the autosomal copy of the gene encoding the L11 protein of the large ribosome subunit is dedicated to germ-line function and affects early embryogenesis as a consequence of its germline role.
rpl-25 genes are duplicated and are organized similarly to rpl-11:
We next asked if other rpl genes were duplicated in the C. elegans genome and if their genome distribution correlated with a separation of germ-line/soma function. Of the 43 rpl genes in the database (WormBase Release WS130), we found 2 additional duplicated genes: rpl-24 (likely encoding the L24 protein of the large ribosome subunit), with both duplicates (rpl-24.1 and rpl-24.2) on LGI, and rpl-25 (likely encoding the L23a protein of the large ribosome subunit) with an X-linked paralog, rpl-25.1, and an autosomal paralog, rpl-25.2, on LGI. Previous RNAi studies indicate that reduction of rpl-24.1 and rpl.25.2 function can cause embryonic lethality and sterility (Fraser et al. 2000; Simmer et al. 2003) and that rpl-25.2 transcripts are germ-line enriched (log2 ratio for +/glp-4(bn2) of +1.6 vs. −1.2 for rpl-25.1; Reinke et al. 2004) while rpl-24.1 and rpl-24.2 are not (log2 ratios of 0.174 and 0.5, respectively; Reinke et al. 2004).
To assess the relative germ-line and somatic roles of these gene pairs, we examined the effects of RNAi targeted against these genes individually in the wild-type (N2) and the rrf-1 mutant backgrounds (Sijen et al. 2001; Table 1; see materials and methods). In this experiment, rpl-11 and rpl-25, the two gene pairs with an autosome/X chromosome duplication, behave more similarly to each other than does the autosome/autosome duplicated rpl-24 gene pair. For the rpl-11 and rpl-25 pairs in the N2 background, the primary defect after RNAi of the autosome-linked genes of each pair (rpl-11.1 and rpl-25.2) is a germ-line proliferation defect (77 and 86%). However, RNAi directed against the X-linked genes of each pair confers either growth/larval arrest (rpl-11.2, 78%) or a weak overall health defect (rpl-25.1, 25%) but not a significant germ-line proliferation defect (12 and 0%, respectively). As expected for genes acting primarily in the germ line, RNAi directed against the autosomal copies of these genes confers similar phenotypes in the rrf-1 mutant background and in the wild type, while as expected for genes acting in the soma, the phenotypic profile changed significantly between N2 and rrf-1 when the X-linked paralogs were targeted. Specifically, the percentage of growth defective/larval arrest animals in rpl-11.2(RNAi) is reduced from 78 to 10% in N2 vs. rrf-1 and the unhealthy phenotype observed in rpl-25.1(RNAi) is reduced from 25 to 7%. In addition, rpl-25.1(RNAi) displayed a significant increase in sterility in the rrf-1 background, but the germ-line proliferation defect was not as severe as that observed with rpl-25.2 (Table 1).
Finally, the effect of RNAi-mediated depletion of rpl-24.1 and rpl-24.2 resulted in quite similar phenotypic profiles, but differed from that of rpl-11 and rpl-25 (Table 1). After RNAi depletion of each gene in the rpl-24 pair in wild type and rrf-1, each displayed a similar decrease in severe growth/larval arrest phenotypes and concomitant increase in sterility in the rrf-1 background as compared to the wild-type background. Thus, in contrast to the germ-line-specific roles of the autosomal copies of rpl-11 and rpl-25, it is likely that both rpl-24 genes (both autosomal) are redundant in the soma and the germ line.
In summary (Table 2), our functional analysis of the three rpl genes (and the pab genes, see below) is consistent with the hypothesis that the two X-linked members of the X/A gene pairs act in the soma while the autosome-linked members of each pair act either in the germ line alone (e.g., rpl-11.1) or in the germ line and soma (e.g., rpl-25.2). In contrast, the pair of rpl-24 genes (both autosomal) appears to act in both the soma and the germ line.
TABLE 2.
Functional comparison ofC. elegans andC. briggsae orthologs
| C. elegans ortholog | A/Xa | Phenotypeb | C. briggsae ortholog | Phenotypec (n) |
|---|---|---|---|---|
| rpl-11.1 | A | Sterile | CBG01314 | Sterile (7/7) |
| rpl-11.2 | X | Fertile (Gro) | CBG14053 | Sterile (1/8), sub-Fertiled (7/8) |
| Progeny: Lva/Gro | ||||
| rpl-24.1 | A | Fertile (Gro) | CBG22273 | Sterile (2/8), sub-Fertiled (6/8) |
| Progeny: Lva/Gro | ||||
| rpl-24.2 | A | Fertile (Gro) | CBG03702 | sub-Fertiled (7/7) |
| Progeny: Lva/Gro | ||||
| rpl-25.1 | X | Fertile (WT) | CBG14529 | Fertilee (5/5) |
| Progeny: WT | ||||
| rpl-25.2 | A | Sterile | CBG04080 | Sterile (3/3) |
| pab-1 | A | Sterile | CBG02207 | Sterile (8/8) |
| pab-2 | X | Fertile (WT) | CBG07431 | sub-Fertiled (7/7) |
| Progeny: WT/Gro | ||||
| L4440 (−)ctrl | Fertile (5/5)f | |||
| Progeny: WT |
Lva, larval arrest.
Autosome or X-linked locus in C. elegans; Gro, growth defect.
Summarized from Table 1 and from the text. Phenotype refers to progeny phenotype from RNAi feeding experiments (see materials and methods).
Phenotype of injected animals (n, number of animals displaying phenotype per number of surviving injected animals) as assessed after overnight transfer to deplete animals of embryos produced prior to the injection (see materials and methods). In addition, few of the adult progeny produced by these fertile injected animals exhibited sterility compared to a high penetrance of sterility observed in the progeny (first 12 hr) from injected animals that quickly became sterile.
These animals were subfertile, producing few embryos, some embryonic lethality, and few live progeny: CBG14053, average <4 surviving progeny per injected animal; CBG22273 and CBG03702, average <2 surviving progeny per injected animal; CBG07431, average <5 surviving progeny per injected animal.
These animals were fertile, producing many embryos but few live progeny due to embryonic lethality: CBG14529, average <2 surviving progeny per injected animal.
Negative control (see materials and methods), ≥20 progeny per injected animal over 2–3 days scored.
Identification and characterization of pab-1(ar232):
We positionally cloned pab-1(ar232), another mutation identified in our screen (Figure 1; materials and methods). DNA sequence revealed a C-to-T transition, resulting in a conceptual translation of Q408 in Y106G6H2.a and Q428 in Y106G6H2.b and Y106G6H2.c to an amber termination. This mutation causes a recessive fully penetrant severe germ-line proliferation defect (n = 120) and an incompletely penetrant protruding vulva (Pvl) phenotype. pab-1 encodes a cytoplasmic poly(A)-binding protein (PABP) and is the autosomal copy of an autosome/X-linked gene pair: pab-1 and pab-2.
Further analysis suggested that pab-1 has an essential germ-line function whereas pab-2 does not [consistent with recent findings of Ciosk et al. (2004)]. RNAi directed against pab-1 resulted in several phenotypes, including a high penetrance of sterility (43%, n = 84) and Pvl (32%, n = 84) and, at lower penetrance, a ruptured vulva (Rup) and flaccid-looking body morphology. In the rrf-1 mutant background all pab-1(RNAi) animals were sterile (n = 54) and 25% also displayed the Pvl phenotype. These results suggest that the sterility is a result of germ-line function of pab-1. In contrast, RNAi directed against the X-linked pab-2 in N2 resulted in 94% wild-type animals with low-penetrance sterility, Rup, and Pvl phenotypes (2%, 4%, and 0.3%, respectively; n = 325). Very similar observations were made in the rrf-1 mutant background where pab-2(RNAi) produced 90% wild-type, 7% sterile, and 4% Rup animals (n = 119). These results indicate that pab-1 and pab-2 act redundantly in the soma but that pab-1 is required in the germ line.
Consistent with a role for pab-1 in the germ line and similar to rpl-11.1(ar228), pab-1(ar232) extends life span (from an average of 17 days post-L4 in the control strain to an average of 26 days in the mutant, an increase of 1.5-fold; Figure 3) and causes gigantism, with worms averaging 1.4-fold greater volume (Figure 3). Previous microarray analysis does not indicate strong germ-line enrichment for pab-1 or pab-2 transcripts (log2 ratio for +/glp-4 of +0.1 for pab-1 and −1.25 for pab-2; Reinke et al. 2004).
Identification and characterization of glp-3(ar229); glp-3 is eft-3:
A third allele from our screen mapped to a small interval on LGIII (see materials and methods). We noted that mutations in glp-3, a gene that maps genetically to the region, had been analyzed previously at the phenotypic level and conferred a similar zygotic germ-line proliferation defect (Kadyk et al. 1997). We performed complementation analysis (see materials and methods) with glp-3(q145) and ar229, and they failed to complement, indicating that ar229 is an allele of glp-3.
We next examined genes in the region, looking for predicted essential genes with an X-linked paralog. eft-3, an EF-1-α ortholog, satisfied these criteria. One of several closely related genes, eft-4, resides on the X chromosome. Interestingly, one alternative splice form of eft-4 apparently encodes a protein identical to eft-3 [WormBase WS130]. In addition to identical protein sequences, the nucleotide sequences of the coding regions of the two genes are very similar (94% identical over 1392 bases), precluding the possibility of separate RNAi-based functional analysis of these two genes. Sequence analysis of glp-3(q145) in the eft-3 region revealed a single base pair change (G to A) that would result in an the amino acid change A301T. Our mutant, ar229, contains a C-to-T transition in eft-3 that encodes a S414F amino acid change. This analysis thus identified glp-3 as eft-3.
Another gene, glp-4, confers a severe zygotic germ-line proliferation defect (Beanan and Strome 1992). We examined the predicted genes in the region to which glp-4 maps (LGI, 21.4 ± 2 cM) for candidate genes for which previously reported RNAi experiments conferred a sterility or lethality phenotype (Stp, Ste, Lvl, Lva, or Emb) and for which a closely related paralog exists on the X chromosome. For genes with no available RNAi information, we checked for X-linked paralogs. No obvious candidates were identified. rpl-31 maps very close to glp-4, but no change in the rpl-31 coding sequence was detected in the DNA of glp-4(bn2) animals (data not shown).
X/autosome duplications of highly conserved genes in the C. elegans genome:
We examined the C. elegans genome for other X/autosome gene pairs and found that 395 X-linked genes have a paralog on an autosome. Of these, 168 have a highly conserved homolog in human (E ≤ 10−50 and ≥75% identity over the length of the protein), an indication of likely “cell-essential” function. Of these 168 X-linked genes, 156 have been assayed by RNAi, and 128 have no reported phenotype in any large-scale RNAi study (i.e., were scored as “wild-type” in every reported assay) (Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000; Maeda et al. 2001; Kamath et al. 2003; Simmer et al. 2003). Of these 128 genes, 23 have an autosomal paralog that conferred an Emb, Ste, or Stp phenotype in one or more of the above studies, indicating likely germ-line function. Large-scale studies thus provide evidence suggestive of germ-line/soma subfunctionalization for ∼16% of these highly conserved X/A gene pairs. No definitive conclusion can be reached regarding the remainder of these X/A pairs for various technical reasons, including likely false negatives in these studies (Piano and Gunsalus 2002; Fernandez et al. 2005), lack of an RNAi assay for the autosomal paralog (19 cases), or possible redundancy among members of multigene families (which may prevent the detection of phenotypes for single-gene knockouts). Thus, a more directed combined bioinformatic and functional analysis of highly conserved paralogous gene pairs will be required to assess on a genomic scale the prevalence of the X/A trend we observed. Nonetheless, our finding that three alleles from a nonbiased screen for germ-line proliferation defects led to the identification of three X/A duplicated genes is striking.
Unlike many autosome/X-linked gene pairs in mammals and Drosophila, retrotransposition is not the mechanism for the rpl, pab, and eft duplications:
The implications of gene duplication, genome organization, and large-scale gene silencing mechanisms in the germ line are relevant to other organisms. Indeed, in an analogous process to C. elegans X chromosome germ-line silencing (Fong et al. 2002; Kelly et al. 2002), the X chromosome is inactivated in mouse during meiosis, a general phenomenon referred to as meiotic sex chromosome inactivation (MSCI) (McKee and Handel 1993). The possibility of an autosome/X-linked distribution of gene duplicates and correlated germ-line/soma functional separation has been noted and proposed as one possible “coping” mechanism to allow important genes to function despite MSCI in mammals (McKee and Handel 1993 and references therein; Handel 2004). There are six documented cases of mammalian genes with autosome/X duplications in which only the autosomal copy is expressed during spermatogenesis. For five of these genes, the X-linked copies contain introns while the autosomal genes are intronless, a hallmark of duplicative retrotransposition to the autosome from an ancestral X-linked gene (see Handel 2004 and references therein). Moreover, in humans there are five active duplicates of the cytoplasmic PABP, at least two of which, including the X-linked PABPC5, are intronless, again suggesting retrotransposase-mediated gene duplication (see Mangus et al. 2003 and references therein). Studies in Drosophila also suggest a high rate of X-to-autosome gene duplication by way of retrotransposons with a significant enrichment of testes-specific expression in the intronless autosomal members of the gene pairs (Betran et al. 2002). We therefore examined the predicted intron structure (WormBase WS130) of the duplicated C. elegans rpl, pab, and eft genes identified in this study and found no evidence for retrotransposition: all autosomal and X-linked copies contain introns and, in the cases of the rpl-11, rpl-25, and eft duplicates, some predicted intron/exon boundaries are conserved to the base. These results suggest that mechanisms of gene duplication other than retrotransposition may be important for duplications that evolve separate germ-line and somatic functions in C. elegans.
Duplicated genes in C. briggsae:
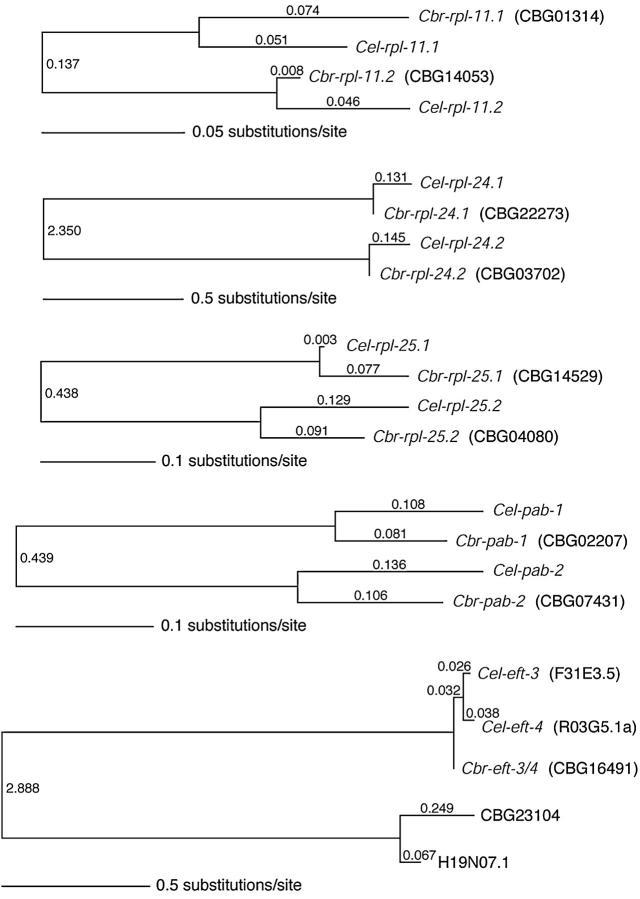
To determine if the duplication of the three rpl genes, the pab genes, and the eft genes arose prior to the evolutionary separation of lineages that gave rise to C. elegans and C. briggsae or if these duplications are derived within the C. elegans branch, we searched for closely related homologs of these genes in C. briggsae. The presence of closely related paralogs within the C. elegans genome required some care in assignment of the corresponding orthologs in C. briggsae, in part because similarity alone may not reflect the degree of phylogenetic relationship since rates of evolution can be very different in different gene lineages. Therefore, rather than relying solely on the best BLASTP match, we examined syntenic regions between the two species and performed a maximum likelihood phylogenetic analysis (see materials and methods; Figure 4). The results indicate that each of the rpl-11, rpl-24, rpl-25, and pab genes has a single corresponding ortholog in C. briggsae. Our BLASTP, synteny, and phylogenetic analyses agree for the three rpl genes and the pab genes, identifying the same pairs of genes as putative orthologs. Our analysis is consistent with the hypothesis that four duplications (three rpl genes and the pab genes) preceded the divergence between C. elegans and C. briggsae. Unlike the rpl and pab genes, the duplication event that produced eft-3 and eft-4 appears to be more recent, having occurred within the C. elegans lineage after diverging from the lineage leading to C. briggsae (Figure 4). It is likely that the C. briggsae chromosome that shares synteny with the C. elegans X chromosome is the C. briggsae sex chromosome, but genetic analyses in C. briggsae will be required to verify this prediction.
Figure 4.—
Phylogenetic relationships of pab, rpl, and eft gene pairs in C. elegans (Cel) and C. briggsae (Cbr). Branch lengths (substitutions per site, estimated by likelihood) are indicated on the branches and with a scale for each set of genes. For each set of homologs, all possible unrooted phylogenies were compared using a log-likelihood ratio test; in all cases, the maximum likelihood (ML) tree was significantly supported (P < 0.05) over all other trees. Jackknife analyses demonstrated 100% support for the internal branch in 2000 Monte-Carlo replicates for each set of four or five homologous genes. Note that H19N07.1 is the C. elegans ortholog of the CBG23104 gene in C. briggsae; these genes are paralogs of the eft-3/4 genes in these two species. After diverging from C. briggsae, the eft-3/4 ortholog in a C. elegans ancestor duplicated to produce eft-3 and eft-4, which are therefore both orthologs of the single C. briggsae eft-3/4 gene.
To determine if the related duplicated C. briggsae genes (Figure 4) also show functional separation in the germ line and soma, we performed RNAi for each gene of the four gene pairs (Table 2). Because C. briggsae is not amenable to RNAi analysis by feeding, double-stranded RNA was delivered by injection into the germ line. We found that injection of dsRNA corresponding to the C. briggsae orthologs of the autosome-linked member of each A/X pair caused complete sterility (no embryos) in the injected animal within 12 hr of injection (Table 2). In contrast, all or most of the worms injected with the orthologs of the X-linked C. elegans genes produced embryos and/or live progeny. The latter injected animals, however, displayed various degrees of embryonic lethality and/or were subfertile, suggesting the possibility of germ-line function for these genes. Larval arrest and growth defects were also observed among the progeny of these animals (Table 2), however, suggesting that these genes act in the soma as well as the germ line. As was observed from RNAi in C. elegans, injection of dsRNA corresponding to each gene of the C. briggsae rpl-24 pair (an A/A pair in C. elegans) produced almost identical results. In addition, for the C. briggsae orthologs of the C. elegans autosome members of X/A pairs, many progeny produced within the first 12 hr after injection became sterile adults, whereas adult sterility was not a prevalent phenotype among early progeny from the other experiments (Table 2).
Therefore, the paralogs in C. briggsae that correspond to C. elegans autosome/X paralogs display consistent functional differences between paralogs, including a potential somatic role for the orthologs of C. elegans X-linked members of X/A pairs. Because both genes in each pair caused phenotypes that are associated with germ-line function, however, there may be a lesser degree of germ-line/soma subfunctionalization between these genes in C. briggsae. The extent to which this difference between C. elegans and C. briggsae is a consequence of differences in experimental methods, species-specific response to RNAi, or other developmental differences is unclear. In summary, our analysis in C. briggsae suggests that RNAi directed against the C. briggsae orthologs of C. elegans autosomal members of the three autosome/X gene pairs (rpl-11, rpl-25, pab) caused a more profound germ-line defect than did RNAi directed against the C. briggsae orthologs of C. elegans X-linked members of these pairs or against the autosome/autosome pair (rpl-24).
DISCUSSION
RPL, PAB, and EFT proteins and early germ-line proliferation:
Using forward genetics, we identified rpl-11.1, pab-1, and glp-3/eft-3 as genes required zygotically for early germ-line proliferation. These genes encode the L11 protein of the large ribosome subunit, a cytoplasmic PABP, and an elongation factor (EF-1-α), respectively. All are important for translation and are likely essential for cell proliferation. Their critical roles in early germ-line proliferation suggest that the processes of reinitiation and maintenance of early proliferation from the quiescent state of the germ-line precursor cells Z2 and Z3 is dependent on proper ribosome biogenesis and/or active translation. That the primordial germ cells are born and locate properly to the somatic gonad in the mutants suggests that the maternal copy is sufficient for these events and/or that the X-linked paralogs of each gene are functionally redundant in the germ line until Z2 and Z3 resume cell division. We also identified a critical germ-line proliferation function for rpl-25.2 that is not shared by its X-linked paralog, rpl-25.1.
Gene duplication, genome organization, and the germ line:
Our phenotypic analysis of the three duplicated rpl genes and pab genes and previous analysis of glp-3 (Kadyk et al. 1997) in C. elegans suggest that the functional role of autosomal members of each autosome/X-linked pair is critical in the germ line and that the X-linked copy is largely if not exclusively active in the soma. We also identified orthologous genes in the C. briggsae genome, and in four of the five cases (all but eft-3/eft-4), the gene duplication preceded the evolutionary divergence of these two species. Among the paralogous pairs within C. briggsae that correspond to X/A pairs in C. elegans, some degree of subfunctionalization of these gene duplicates occurs, whereas the pair corresponding to the autosome/autosome duplication does not display the same degree of subfunctionalization. Given that the germ-line phenotypes were much more severe in the C. briggsae orthologs of the autosome-linked members of X/A pairs in C. elegans, if the chromosome that carries the paralogs is, indeed, the C. briggsae X chromosome, we speculate that germ-line-expressed genes may be similarly underrepresented on the X chromosome of C. briggsae, as are germ-line-expressed genes in C. elegans (Reinke et al. 2000, 2004). Interestingly, the same mode of X chromosome silencing that operates in the C. elegans germ line appears to operate in C. briggsae (Kelly et al. 2002).
That a similar pattern of germ-line function of the autosomal copy of autosome/X-linked paralogs is abundant among many cell proliferation-essential genes is evidenced by our genetic identification of three independent loci (from three alleles) that are required for robust germ-line proliferation. Given that these three mutants (of seven obtained from a forward genetic screen covering 11,586 genomes that were screened in a way that could identify putative severe proliferation defective mutants; Pepper et al. 2003a; E. J. A. Hubbard, unpublished data) uncovered autosomal copies of duplicated genes with the same kind of autosome/X chromosome distribution, we speculate that other genes essential for early germ-line proliferation may be encoded by the autosome duplicate of autosome/X-linked pairs of genes encoding proteins essential for cell proliferation.
Recent studies of X-linked genes in mammals suggest that only genes expressed after the onset of male germ-line X chromosome silencing (MSCI) are underrepresented on the X chromosome (Khil et al. 2004). In C. elegans the X chromosome lacks histone modifications that correlate with transcriptional activation throughout the adult male germ line and in the zone of mitosis and early meiosis in adult hermaphrodites (Fong et al. 2002; Kelly et al. 2002). Therefore, it is conceivable that in addition to genes required in spermatogenesis, genes required in earlier stages of germ-line development in C. elegans may be particularly sensitive to selective pressure keeping them off the X chromosome.
Identification of correlations between the genomic distribution of gene duplicates and germ-line function may be mutually informative in other species with X chromosome silencing. That is, the presence of X-linked “cell-essential” genes with autosomal duplicates could identify genes required for germ-line development during X chromosome silencing, while those that are not duplicated could point to genes that are required at a time in development when the X is not silenced. This approach may be useful in identifying autosomal genes required for fertility.
Evolutionary hypotheses:
We propose three possible evolutionary hypotheses regarding functional gene duplicates of cell-essential genes, X chromosome germ-line silencing, and genome organization. From our study and from other published reports, we found data supporting each of the hypotheses. Therefore, we suggest that each of these hypothetical mechanisms is likely at work.
One hypothesis is that gene duplication (regardless of whether it occurred before or after the evolution of germ-line X chromosome silencing) generally results in “subfunctionalization” (Lynch and Conery 2000) into germ-line/soma roles. That is, when there is positive selection to subfunctionalize into different expression compartments, it is generally the case that these compartments correspond to germ-line and soma compartments. Such a general trend might result, for example, if many regulatory regions in the genome had elements controlling germ-line vs. soma expression and if, by chance alone, transposed duplicates “land” near such regulatory elements or if germ-line- or soma-specific elements are lost during duplication/transposition. This hypothesis predicts that autosome/autosome and autosome/X duplicates should show similar degrees of germ-line/soma subfunctionalization. An example of an autosome/autosome duplicate pair that exhibits germ-line/soma subfunctionalization is the iff-1 and iff-2 gene pair that encodes eIF5A homologs. These duplicated genes fulfill separate germ-line/soma functions (Hanazawa et al. 2004). An alternative hypothesis to account for the behavior of this pair is that the member of this pair that is functioning in the soma, although it is not located on the X, may be silenced by the same mechanism that silences genes on the X chromosome.
A second hypothesis is that gene duplication (again, regardless of whether it occurred before or after the evolution of X chromosome silencing) results in subfunctionalization due to X chromosome silencing. That is, X chromosome silencing provides the conditions for positive selection for subfunctionalization. In this case, we predict that relative to autosome/autosome duplicates, the autosome/X duplicates should show a bias toward germ-line/soma subfunctionalization. The rpl-11 duplicate genes are examples of this kind of subfunctionalization: RNAi directed against rpl-11.1 disrupts germ-line development, while rpl-11.2(RNAi) has a more profound effect on somatic development. While autosome/autosome duplicates may show subfunctionalization, under this mechanism they would not necessarily be biased toward germ-line/soma compartmentalization. This hypothesis would also predict that strict germline/soma subfunctionalization would not occur unless one duplicate is on the X chromosome. In this regard, the rpl-24 genes provide an example of autosomal duplicates that do not show germ-line/soma subfunctionalization and remain active in both the germ line and the soma.
A third hypothesis is that subfunctionalization results from genetic drift, not from positive selection. Here, the only role of selection would be to maintain the new function that resulted from random drift (i.e., purifying selection). In this case, gene copies that end up on the X chromosome lose their germ-line expression capability by drift, since another autosomal gene is redundant for this function. This hypothesis predicts that X-linked copies are expressed only somatically, but autosomal copies are expressed in both the germ line and the soma where there might be strong purifying selection to maintain somatic expression and identical protein sequences. Examples of this phenomenon are the rpl-25 and pab genes. For both of these pairs, removal of the X-linked pair had little phenotypic consequence (suggesting that the autosomal gene acts redundantly with the X copy in the soma) while removal of the autosome-linked duplicate resulted in sterility due to germ-line proliferation defects.
To understand the relationships between gene duplication, X chromosome silencing, and germ-line function and the relative roles of the proposed evolutionary mechanisms, it will be important to correlate specific gene function with silencing and duplication over the entire genome. Germ-line-acting genes required specifically for early germ-line proliferation will likely appear among the sterile and embryonic-lethal phenotypic classes that have been identified by large-scale RNAi screens (as are the autosomal rpl and pab duplicates) (Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000; Hanazawa et al. 2001; Maeda et al. 2001; Colaiacovo et al. 2002; Kamath et al. 2003; Simmer et al. 2003). Although available data from large-scale RNAi screens identify genes required for fertility, these data do not delineate the precise germ-line defect that underlies the sterility phenotype. A large-scale screen to pinpoint more precisely the defects that cause sterility in C. elegans (e.g., proliferation defects vs. gametogenesis defects) is underway (E. J. A. Hubbard, unpublished data). Together with genome-wide informatic analysis of gene duplicates, these data should help elucidate further the correlation between gene duplication, X chromosome silencing, genome organization, and germ-line development.
Acknowledgments
We thank Dan Culliford and Tamer Hadi for technical assistance; Yuji Kohara for permission to include information from NEXTDB; Iva Greenwald in whose laboratory ar228, ar229, and ar232 were originally isolated; and the Hubbard and Fitch laboratory members (especially Karin Kiontke), Fabio Piano, Scott Baird, and Bill Kelly for helpful discussions. We also thank the C. elegans Genetics Center for strains. This work was supported in part by National Institutes of Health (NIH) grant GM-61706 to E.J.A.H., National Science Foundation (NSF) grant DEB-0228692 to D.H.A.F., the National Sciences and Engineering Research Council of Canada (R.C.J. and D.L.B.), NIH grant HD046236 to Fabio Piano, and NSF grant DBI-0137617 to K.C.G.
References
- Agarwal, A. K., S. N. Parrish and D. D. Blumberg, 1999. Ribosomal protein gene expression is cell type specific during development in Dictyostelium discoideum. Differentiation 65: 73–88. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P., and S. Brenner, 1984. A selection for myosin heavy chain mutants in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 81: 4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, J., and J. Kimble, 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. [DOI] [PubMed] [Google Scholar]
- Beanan, M., and S. Strome, 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Betran, E., K. Thornton and M. Long, 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk, R., M. DePalma and J. R. Priess, 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131: 4831–4841. [DOI] [PubMed] [Google Scholar]
- Colaiacovo, M. P., G. M. Stanfield, K. C. Reddy, V. Reinke, S. K. Kim et al., 2002. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics 162: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R. E., and J. Kimble, 1995. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139: 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, A. G., K. C. Gunsalus, J. Huang, L.-S. Chuang, N. Ying et al., 2005. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 15: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fong, Y., L. Bender, W. Wang and S. Strome, 2002. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296: 2235–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A., R. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Fujita, M., T. Takasaki, N. Nakajima, T. Kawano, Y. Shimura et al., 2002. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech. Dev. 114: 61–69. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., C. Echeverri, K. Oegema, A. Coulson, S. J. Jones et al., 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Guo, S., and K. J. Kemphues, 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. [DOI] [PubMed] [Google Scholar]
- Hanazawa, M., M. Mochii, N. Ueno, Y. Kohara and Y. Iino, 2001. Use of cDNA subtraction and RNA interference screens in combination reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98: 8686–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa, M., I. Kawasaki, H. Kunitomo, K. Gengyo-Ando, K. L. Bennett et al., 2004. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Dev. 121: 213–224. [DOI] [PubMed] [Google Scholar]
- Handel, M. A., 2004. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 296: 57–63. [DOI] [PubMed] [Google Scholar]
- Hirsh, D., D. Oppenheim and M. Klass, 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. [DOI] [PubMed] [Google Scholar]
- Hsin, H., and C. Kenyon, 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., 1994. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B Biol. Sci. 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Johnsen, R. C., 1990 Genetic analysis of the left half of linkage group V in Caenorhabditis elegans. Ph.D. Thesis, Simon Fraser University, Burnaby, BC, Canada.
- Johnsen, R. C., and D. L. Baillie, 1988. Formaldehyde mutagenesis of the eT1 balanced region in Caenorhabditis elegans: dose-response curve and the analysis of mutational events. Mutat. Res. 201: 137–147. [DOI] [PubMed] [Google Scholar]
- Kadyk, L., E. Lambie and J. Kimble, 1997. glp-3 is required for mitosis and meiosis in the Caenorhabditis elegans germ line. Genetics 145: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim et al., 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil, P. P., N. A. Smirnova, P. J. Romanienko and R. D. Camerini-Otero, 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36: 642–646. [DOI] [PubMed] [Google Scholar]
- Kimble, J., and D. Hirsh, 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Maddison, D. R., and W. P. Maddison, 2002 MacClade 4: Analysis of Phylogeny and Character Evolution, Version 4.05. Sinauer Associates, Sunderland, MA.
- Maeda, I., Y. Kohara, M. Yamamoto and A. Sugimoto, 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11: 171–176. [DOI] [PubMed] [Google Scholar]
- Mangus, D. A., M. C. Evans and A. Jacobson, 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies, L. D., S. T. Henderson and J. Kimble, 2003. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development 130: 2881–2892. [DOI] [PubMed] [Google Scholar]
- McCulloch, D., and D. Gems, 2003. Body size, insulin/IGF signaling and aging in the nematode Caenorhabditis elegans. Exp. Gerontol. 38: 129–136. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., and M. A. Handel, 1993. Sex chromosomes, recombination, and chromatin conformation. Chromosoma 102: 71–80. [DOI] [PubMed] [Google Scholar]
- Patel, M. N., C. G. Knight, C. Karageorgi and A. M. Leroi, 2002. Evolution of germ-line signals that regulate growth and aging in nematodes. Proc. Natl. Acad. Sci. USA 99: 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A. S., D. J. Killian and E. J. Hubbard, 2003. a Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A. S., T. W. Lo, D. J. Killian, D. H. Hall and E. J. Hubbard, 2003. b The establishment of Caenorhabditis elegans germline pattern is controlled by overlapping proximal and distal somatic gonad signals. Dev. Biol. 259: 336–350. [DOI] [PubMed] [Google Scholar]
- Piano, F., and K. C. Gunsalus, 2002. RNAi-based functional genomics in C. elegans. Curr. Genomics 3: 69–81. [Google Scholar]
- Piano, F., A. J. Schetter, M. Mangone, L. Stein and K. J. Kemphues, 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619–1622. [DOI] [PubMed] [Google Scholar]
- Piano, F., A. J. Schetter, D. G. Morton, K. C. Gunsalus, V. Reinke et al., 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren et al., 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605–616. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Rosenbluth, R. E., and D. L. Baillie, 1981. The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish et al., 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. Van Der Linden, E. Kuijk, P. V. Van Den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, H. I., R. E. Rosenbluth and D. L. Baillie, 1991. Most ultraviolet irradiation induced mutations in the nematode Caenorhabditis elegans are chromosomal rearrangements. Mutat. Res. 249: 37–54. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2002 PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0b10. Sinauer Associates, Sunderland, MA.
- Thomas, J. H., 1993. Thinking about genetic redundancy. Trends Genet. 9: 395–399. [DOI] [PubMed] [Google Scholar]
- Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin and D. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Trent, C., N. Tsuing and H. R. Horvitz, 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104: 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]