Abstract
To understand long terminal repeat (LTR)-retrotransposon copy number dynamics, Ty1 elements were reintroduced into a “Ty-less” Saccharomyces strain where elements had been lost by LTR-LTR recombination. Repopulated strains exhibited alterations in chromosome size that were associated with Ty1 insertions, but did not become genetically isolated. The rates of element gain and loss under genetic and environmental conditions known to affect Ty1 retrotransposition were determined using genetically tagged reference elements. The results show that Ty1 retrotransposition varies with copy number, temperature, and cell type. In contrast to retrotransposition, Ty1 loss by LTR-LTR recombination was more constant and not markedly influenced by copy number. Endogenous Ty1 cDNA was poorly utilized for recombination when compared with LTR-LTR recombination or ectopic gene conversion. Ty1 elements also appear to be more susceptible to copy number fluctuation in haploid cells. Ty1 gain/loss ratios obtained under different conditions suggest that copy number oscillates over time by altering the rate of retrotransposition, resulting in the diverse copy numbers observed in Saccharomyces.
THE C-value paradox states that the haploid content of genomic DNA does not always follow phylogeny or developmental and behavioral complexity (Mirsky and Ris 1951; Hartl 2000). To a large extent the C-value paradox is due to the gain and loss of repetitive elements such as retrotransposons through processes that can occur over relatively short time periods and are not in equilibrium. How transposon-based genome expansion and contraction is modulated is not understood, despite the fact that >40% of mammalian genomes and the larger plant genomes are composed of mobile DNA (Kumar and Bennetzen 1999; Eichler and Sankoff 2003). A Saccharomyces version of the C-value paradox has been studied by reintroducing active Ty1 retrotransposons into Saccharomyces paradoxus strain 337, which is a close congener to S. cerevisiae (Wilke and Adams 1992; Garfinkel et al. 2003; Moore et al. 2004). Strain 337 lacks complete Ty1, Ty2, Ty4, and Ty5 elements and displays weak hybridization with a Ty3 element internal probe. Numerous degenerate solo long terminal repeats (LTRs) also are present in strain 337, many of which arose by intraelement LTR-LTR recombination at insertion sites that are occupied by full-length Ty elements in S. cerevisiae (Moore et al. 2004). Therefore, we use the term “Ty-less” to describe the strain 337 genome.
Ty elements are LTR-retrotransposons that replicate through an RNA intermediate (Sandmeyer et al. 2002; Voytas and Boeke 2002) and are present in essentially all eukaryotes (Craig et al. 2002). LTR-retrotransposons and retroviruses are similar in structure, encode functionally equivalent Gag and Pol proteins, including protease (PR), integrase (IN), and reverse transcriptase (RT), and undergo similar replication cycles. The Ty structural and enzymatic proteins form a virus-like particle within which protein maturation and reverse transcription occur. The resulting linear cDNA enters the genome through IN-mediated integration or, to a lesser degree, by homologous recombination with genomic elements. Ty retrotransposition is not infectious, which distinguishes Ty elements from retroviruses.
Since Ty element retrotransposition is additive and these elements compose ∼3% of the S. cerevisiae genome, element accumulation through retrotransposition must be modulated or offset by element loss to avoid uncontrolled genome expansion and rearrangement. Ty elements and yeast have coevolved several strategies to minimize genome “obesity” (Bennetzen and Kellogg 1997). Ty elements have pronounced target site preferences that avoid mutating genes (Boeke and Devine 1998); undergo transcriptional and post-transcriptional cosuppression, which limits retrotransposition in a copy-number-dependent manner (Jiang 2002; Garfinkel et al. 2003); respond to several host restriction systems; and require specific cofactors and environmental conditions for optimal levels of retrotransposition (Sandmeyer et al. 2002; Voytas and Boeke 2002). In particular, transcriptional cosuppression silences all Ty1 elements in a subset of cells by a mechanism that is independent of DNA methylation and polycomb-mediated repression (Jiang 2002). Post-transcriptional cosuppression blocks the utilization of Ty1 RNA for retrotransposition either prior to or during reverse transcription, possibly by titration of a positively acting factor (Garfinkel et al. 2003). Ty1 RNA levels also decrease ∼10-fold in diploid cells due to MATa/α repression (Elder et al. 1980; Herskowitz 1988). In addition, Ty1 retrotransposition peaks at 15°–20° and is severely inhibited at 37° (Paquin and Williamson 1984), apparently because Ty1 PR is heat sensitive (Lawler et al. 2002). Since Ty elements are bracketed by directly repeated LTRs, element loss can occur by intraelement homologous recombination (Farabaugh and Fink 1980). Solo-LTRs account for 85% of the Ty insertions in the S. cerevisiae genome (Kim et al. 1998) and may be the only element sequences remaining in strain 337 (Moore et al. 2004). However, spontaneous Ty-Ty recombination events are low when compared with other repeated sequences (Kupiec and Petes 1988a,b).
In contrast to many other host-mobile element interactions, the Saccharomyces-Ty relationship has the potential to be well understood at the molecular level. Here we have utilized a Ty-less strain to experimentally determine how Ty1 gain and loss fluctuates under genetic and environmental conditions known to modulate Ty1 retrotransposition. Our work strongly suggests that Ty1 copy number oscillates over time by altering the rate of retrotransposition, thereby contributing to the diverse copy numbers observed in different Saccharomyces species and strains (Wilke et al. 1992; Neuveglise et al. 2002) and, perhaps, other organisms as well.
MATERIALS AND METHODS
Genetic techniques, media, and strain construction:
Yeast genetic techniques and media were used as described previously (Sherman et al. 1986; Guthrie and Fink 1991). The strains used here (Table 1) were originally derived by crossing a mitotic derivative of strain 337 (Wilke and Adams 1992) called DG1768 with 155-5A (Garfinkel et al. 2003). Several strains resulted from extensive backcrosses with DG1768 and Gal+ Spo+ segregants were saved for further analysis. Strain DG2204 resulted from a cross between strains DG2196 and DG1768. Strain DG2503 was generated by introducing the URA3 gene into the Ty1 RT region of Ty1his3-AI(96) present in DG2196 by microhomologous recombination as described by Moore et al. (2004). Strains DG2602 and DG2547 resulted from a cross between strains DG2526 and DG2196 (Garfinkel et al. 2003; Moore et al. 2004). Diploid strains DG2567 and DG2646 were derived by mating strains DG2533 and DG2633 with DG2204, respectively. Diploid strains DG2566 and DG2601 were derived by mating strains DG2602 and DG2578 with DG1768, respectively. Ty1 elements were reintroduced into the genomes of strains DG2503, DG2533, and DG2602 by two serial galactose inductions of cells containing pGTy1-H3 (Boeke et al. 1985, 1991; Garfinkel et al. 2003), resulting in strains DG2525, DG2578, and DG2633, respectively. The repopulated strains were compared with their parent strains for cosuppression or Ty1 loss before reintroduction of the pGTy1-H3 plasmid. Candidate strains containing Ty1his3-AI and additional Ty1 elements had to exhibit cosuppression to be included in the second round of pGTy1-H3 induction. Strains containing Ty1::URA3 gave rise to 5-fluoroorotic acid (5-FOA)-resistant mutants at about the same frequency as determined by qualitative patch tests, regardless of the number of reintroduced Ty1 elements. Therefore, colonies were chosen at random for serial pGTy1-H3 induction. Mutations in Ty1 PR or IN were introduced into Ty1his3-AI(96) by cotransforming strain DG2547 with a mutant targeting fragment and a TRP1-2μ vector. The targeting fragment containing pr-1702(SacI) was generated by PCR using pGTy1-H3Neo/pr-1702 (Youngren et al. 1988) as template, Pfu Turbo high-fidelity polymerase (Stratagene, La Jolla, CA), and primers TyA700 (5′pr-AGTTGGAACGCCTCTGAGC-3′) and IN2700 (5′-AAATTCGATTGCAGAGAAGC-3′), according to the manufacturer's suggestions. The targeting fragment containing in-2600(MluI) was generated by PCR, using pGTy1-H3his3-AI/in-2600(MluI) (pDSM24) (Monokian et al. 1994; Moore et al. 1998) as template, and primers Ty1-1621 (5′-ACAGTAAATCATACTAATC-3′) and Ty1-3600 (5′-TTCCGGTGGAGAAGCATC-3′). The targeting fragment containing in-K596,597G was generated by PCR, using pGTy1-H3his3-AI/in-K596,597G (pDSM27) (Moore et al. 1998) as template, and primers IN-2600SEQ (5′pr-GGACCGTGGTTCTGAGTATACTAACAGAAC-3′) and Ty1-4851 (5′-GTTCATCCTGGTCTATATAAAGA-3′). Trp+ cotransformants containing the appropriate mutation in Ty1his3-AI(96) were identified by their inability to undergo Ty1his3-AI retrotransposition, as monitored by the formation of His+ colonies (Curcio and Garfinkel 1991). PCR products amplified from genomic DNA using primers TyA700 and HIS3-413-435 (5′-GCCGTACGCAGTTGTCGAACTTG-3′) were digested with SacI or MluI or sequenced to verify the presence of pr-1702(SacI), in-2600(MluI), or in-K596,597G mutations, respectively, in Ty1his3-AI(96). Similar PCR analyses using Ty1 (TyA700) and URA3 (URA3 5′out; 5′-ATTGTACTTGGCGGATAATGTC-3′) primers showed that the PR and IN mutations were not present in Ty1-4253::URA3. Trp− segregants obtained after nonselective growth were used in further experiments.
TABLE 1.
List of strains
| Strain name | Genotype | Source |
|---|---|---|
| DG1768 | MATα his3-Δ200hisG ura3 gal3 Ty-less Spo− | Garfinkel et al. (2003) |
| DG2196 | MATatrp1 ura3 his3-Δ200hisG Ty-less Ty1his3-AI(96) Gal+ Spo+ | Garfinkel et al. (2003) |
| DG2204 | MATahis3-Δ200hisG trp1 ura3 Ty-less Gal+ Spo+ | |
| DG2454 | MATα his3-Δ200hisG ura3 gal3 Ty1-4253 Ty-less Spo− | Garfinkel et al. (2003) |
| DG2568 | DG2454 + Ty1-4253 solo-LTR | |
| DG2503 | MATahis3-Δ200hisG ura3 trp1 Ty-less Ty1his3-AI(96)::URA3 Gal+ Spo+ | |
| DG2525 | DG2503 + 20 Ty1 elements | |
| DG2602 | MATahis3-Δ200hisG ura3 trp1 Ty-less Ty1-4253::URA3 Gal+ Spo+ | |
| DG2526 | MATα his3-Δ200hisG ura3 Ty-less Ty1-4253::URA3 Gal+ Spo+ | Moore et al. (2004) |
| DG2578 | DG2602 + 24 Ty1 elements | |
| DG2533 | MATα his3-Δ200hisG ura3 Ty-less Ty1-4253his3-AI gal3 Spo− | Moore et al. (2004) |
| DG2633 | DG2533 + 28 Ty1 elements | |
| DG2566 | DG2602 × DG1768 | |
| DG2601 | DG2578 × DG1768 | |
| DG2567 | DG2204 × DG2533 | |
| DG2547 | MATahis3-Δ200hisG ura3 trp1 Ty-less Ty1-4253::URA3 Ty1his3-AI(96) | |
| DG2575 | DG2547, Ty1his3-AI(96)pr-1702(SacI) | |
| DG2592 | DG2547, Ty1his3-AI(96)in-2600(MluI) | |
| DG2605 | DG2547, Ty1his3-AI(96)in-K596,597G | |
| DG2646 | DG2204 × DG2633 |
Ty1 location:
Vectorette PCR (Hui et al. 1998), as modified by C. Friddle and D. Botstein (http://www.genomics.princeton.edu/botstein/) was used to determine the chromosomal location of Ty1his3-AI(96). Both Ty1his3-AI(96) and our other reference element, Ty1-4253 (Moore et al. 2004), were located on chromosome X. Ty1his3-AI(96) was located between ASF1 and MDV1 and ∼90 bp from an Ala-tRNA, while Ty1-4253 was located between RAD7 and CDC8 and adjacent to a Gly-tRNA gene. Ty1his3-AI(96) and Ty1-4253 were inserted in inverted orientation relative to each other and bracketed by 5-bp target site duplications 5′-TAATA-3′ and 5′-CTTAT-3′, respectively. On the basis of S. cerevisiae genome sequence coordinates, the two Ty1 elements were separated by ∼350 kb. PCR analysis using primers ASFout118W (5′-GACAAGAAGAGGAGGAAAATAGAAG-3′) and MDVout5C (5′-GAGTTATTTGGTCGTTCACTGACAT-3′), which flank Ty1his3-AI(96), generated a 2.1-kb fragment from the preintegrated region.
Ty1 retrotransposition:
Ty1his3-AI retrotransposition events were detected as His+ colonies, as described by Curcio and Garfinkel (1991). Ty1 transposition rates and standard deviations were determined as described previously (Drake 1970; Rattray et al. 2000). For a given temperature (e.g., 30°), all liquid growth and plate incubations were performed at that temperature. The rate of Ty1 retrotransposition per element per generation was determined by multiplying the rate of His+ formation by a factor of 88 to correct for splicing efficiency and the inhibitory effects of the his3-AI indicator gene (Curcio and Garfinkel 1991). Splicing efficiency of the artificial intron varied less than twofold when cells were grown at 20° or 35°, as determined by reverse transcription-PCR (Garfinkel et al. 2003).
Ty1 loss and gene conversion:
Ty1 loss by LTR-LTR recombination, ectopic gene conversion, or loss of heterozygosity (LOH) events involving Ty1his3-AI(96)::URA3 or Ty1-4253::URA3 were detected using 5-FOA selection as described by Winston et al. (1984), except that all liquid growth and plate incubations were performed at 20°, 30°, or 35°. The rate of 5-FOAR per element per generation and standard deviations were estimated according to the method of Drake (1970). 5-FOAR (Ura−) colonies obtained in haploids could result from LTR-LTR recombination, ectopic gene conversion with the endogenous ura3 allele, or with other Ty1 elements present in the genome. PCR analyses of randomly selected 5-FOAR mutants, using primers specific for Ty1, URA3, his3-AI, and sequences flanking the reference elements, were used to determine how 5-FOAR occurred. Similar to strains containing Ty1-4253 or marked derivatives (Moore et al. 2004), the Ty1his3-AI(96) flanking primers ASFout118W and MDVout5C could not efficiently amplify the full Ty1his3-AI(96) or Ty1his3-AI(96)::URA3 elements because the PCR products were >9 kb, whereas LTR-LTR recombinants generated a 2.4-kb product. A 1.5-kb PCR product was amplified using primers XhoRTw (5′-CCGCCGCTCGAGGCTGTAAAAGCAGTA-3′) and His3-3′out (5′-GAGAAGCCACCTCGCCC-3′) if Ty1 gene conversion occurred in strain DG2525. A 3.9-kb PCR product was amplified using primers XhoRTw and CDC8out (5′-CTATCAGTATTAATTTGCCACGG-3′) if Ty1 gene conversion occurred in strain DG2578. PCR analysis with primers CDC8out and RAD7-286 (5′-TCCTTAGTTAACAACATCTC-3′) that flank Ty1-4253::URA3 (Moore et al. 2004) were used to determine how loss occurred in the single-element diploid DG2566. The presence of only the wild-type PCR product of 750 bp indicated that LOH had occurred, whereas an additional PCR product of 1100 bp indicated an LTR-LTR recombination event. Southern blot hybridization was also used to determine the types of Ty1-4253::URA3 loss events that occurred in diploid strains (see below). 5-FOAR His+ mutants from strains DG2547 and DG2592 were analyzed by PCR and Southern hybridization (see below) to determine whether Ty1HIS3 cDNA gene converted Ty1-4253::URA3. The primer pairs used to identify 5-FOAR and 5-FOAR His+ Ty1 gene conversion events were as follows: TyBout (5′-AAGAACATTGCTGATGTGATGAC-3′) + CDC8out (Moore et al. 2004), expected product 1.1 kb; Ura3600w (5′-CATTACGAATGCACACGGTG-3′) + CDC8out, expected product 2.6 kb; Ura35′out (5′-ATTGTACTTGGCGGATAATGTC-3′) + XhoRTw, expected product 0.95 kb; XhoRTw + CDC8out, expected product 3.9 kb; and His35′out (5′-CGCTAGGGGACCACCC-3′) + CDC8out, expected product 1.6 kb. Reconstruction experiments using a strain that had lost Ty1URA3 by LTR-LTR recombination indicated that cells grew equivalently on SC + 5-FOA plates at all temperatures. The rate of Ty1 loss was obtained by multiplying the rate of 5-FOAR-colony formation by the fraction of 5-FOAR events resulting from LTR-LTR recombination. PCR products containing solo-LTRs were gel-purified and cloned into a Topo TA cloning vector (Invitrogen, Carlsbad CA), according to the manufacturer's instructions. Solo-LTRs were sequenced with primer U3in (5′-TGTTGGAATAGAAATC-3′). Additional technical details will be provided upon request.
Ty1 gain/loss ratio:
The gain vs. loss of Ty1 elements was expressed as a ratio by dividing the retrotransposition rate by the loss rate. Ratios were not corrected for the slight difference in Ty1 copy number used to estimate Ty1 gain (DG2633; 28 additional Ty1 elements) or loss (DG2578; 24 additional Ty1 elements).
Chromosome separation:
Yeast chromosomes were prepared for CHEF analysis as described previously (Carle and Olson 1985; Chu et al. 1986; Moore et al. 2004). Chromosomes were separated using a CHEF-MAPPER gel electrophoresis apparatus (Bio-Rad, Hercules, CA) and the following program (Moore et al. 2004): block 1—20 hr, 60 sec switching time; and block 2—12 hr, 90 sec switching time. Both blocks were run at a voltage of 6 V/cm, angle 120° in 0.5× TBE at 13°.
Southern analysis:
Southern blot hybridization was used to determine the copy number of Ty1 elements added back to the Ty-less genome, how Ty1-mediated recombination occurred, and the fate of Ty1 cDNA in strains DG2547 and DG2592. Total genomic DNA was digested with PvuII, separated on a 0.7% agarose gel and transferred to Hybond N (Amersham, Piscataway, NJ). A 32P-labeled probe containing sequences from the Ty1 RT region (nucleotides 3944–5562) was made by randomly primed DNA synthesis (Amersham). The number of the “right-end” junction fragments was determined by phosphorimage analysis and ImageQuant 1.2 software using the reference element's PvuII (nucleotide 3944)-cellular junction PvuII fragment as a single-copy control. Southern analysis was used to assign Ty1 integration events to specific yeast chromosomes using the numbering system developed for the Ty-less strain by Moore et al. (2004). The CHEF gel, stained with ethidium bromide, was photographed and treated with ultraviolet light to introduce single-strand nicks. DNA transfer and hybridization were performed as described above, except that a 32P-labeled Ty1-specific probe was derived from TYA1 sequence. A chromosome X-specific probe was derived from plasmid pJLS1 (kindly provided by F. Lacroute) containing the URA2 gene. Southern analysis was used to determine how Ty1-4253::URA3 loss occurred in diploid strains DG2566 and DG2601 by comparison with the hybridization pattern obtained with haploid strains DG1768 (Ty-less), DG2454 (Ty1-4253), DG2526 (Ty1-4253::URA3), DG2578 (Ty1-4253::URA3 + 24 additional Ty1 elements), and DG2568 (Ty1-4253 solo-LTR). Total DNA digested with EcoRI was processed for Southern analysis and the resulting blot was hybridized with a 32P-labeled 500-bp HindIII fragment that mapped between CDC8 and the Ty1-4253 integration site and was present on plasmid pBDG1213 (Moore et al. 2004). The Ty1-4253::URA3 element present in the haploid strains DG2526 and DG2578 produced a 3.5-kb EcoRI fragment that hybridized with the pBDG1213 probe. When LOH occurred, a 6-kb EcoRI fragment hybridized with the flanking-sequence probe that was identical in size to the fragment present prior to Ty1-4253 integration. When gene conversion with an ectopic Ty1 element occurred, a 2.5-kb EcoRI fragment was detected that was identical in size to the EcoRI fragment from DG2454 (Ty1-4253). An intraelement LTR-LTR recombinant produced a 6.4-kb EcoRI fragment that was identical in size to the EcoRI fragment from DG2568 (Ty1-4253 solo-LTR).
RESULTS
Repopulating the Ty-less genome:
To determine the rates of Ty1 gain through retrotransposition vs. loss by LTR-LTR recombination or reassortment by gene conversion, we individually tagged a single transposition-competent element, Ty1-4253 (Garfinkel et al. 2003), located on chromosome X between CDC8 and RAD7 with either his3-AI to measure retrotransposition (Curcio and Garfinkel 1991) or the counter-selectable URA3 gene to measure loss via LTR-LTR recombination or gene conversion between elements (Roeder and Fink 1982; Winston et al. 1984) (Figure 1). Another competent element, Ty1his3-AI(96) (Garfinkel et al. 2003), which was located on chromosome X between ASF1 and MDV1 also was marked with URA3. We then repopulated the Ty-less genome with de novo Ty1 element insertions by serial pGTy1 induction. Ty1 copy number and chromosomal occupancy were determined by Southern analysis of genomic DNA digested with PvuII (data not shown) or separated chromosomes (Figure 2). The number of right-end Ty1-chromosomal junction fragments that hybridized with a 32P-labeled probe from Ty1 RT was used to determine the number of Ty1 transposition events in strains DG2633, DG2578, and DG2525, which were found to harbor 28, 24, and 20 additional Ty1 elements, respectively.
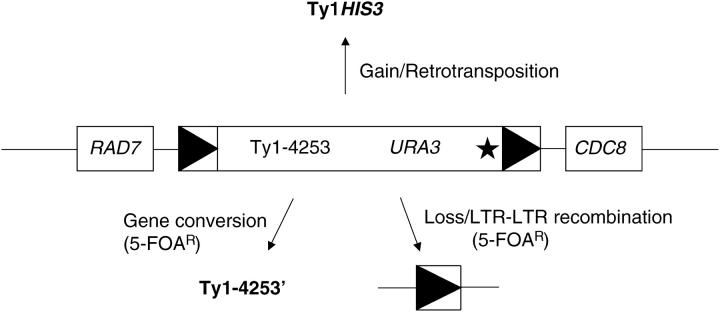
Figure 1.—
Ty1 copy number dynamics. Ty1-4253 is located between RAD7 and CDC8 on chromosome X (Moore et al. 2004). Ty1-4253 was tagged with his3-AI, represented by the star, to determine the rate of retrotransposition. Ty1-4253 was separately tagged with URA3 to determine the level of gene conversion or element loss by LTR-LTR recombination. LTRs are represented by solid triangles. Ty1HIS3 represents Ty1 gain through retrotransposition. Ty1-4253′ and a solo-LTR represent gene conversion and LTR-LTR recombination events that remove URA3 and confer 5-FOAR, respectively.
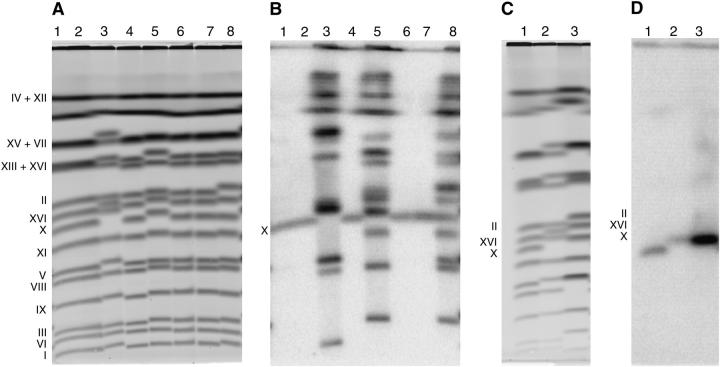
Figure 2.—
Electrophoretic karyotype of strains repopulated with Ty1. (A and B) Lane 1, DG2454 (Ty1-4253); lane 2, DG2533 (Ty1-4253his3-AI); lane 3, DG2633 (DG2533 + 28 Ty1's); lane 4, DG2602 (Ty1-4253::URA3); lane 5, DG2578 (DG2602 + 24 Ty1's); lane 6, DG2196 [Ty1his3-AI(96)]; lane 7, DG2503 [Ty1his3-AI(96)::URA3]; and lane 8, DG2525 (DG2503 + 20 Ty1's). (A) CHEF gel stained with ethidium bromide. (B) Southern blot hybridization of gel in A with a 32P-labeled probe derived from the TYA1 region. (C and D) Lane 1, DG2533 (Ty1-4253his3-AI); lane 2, DG2633 (DG2533 + 28 Ty1's); lane 3, S. cerevisiae S288c. (C) CHEF gel stained with ethidium bromide. (D) Southern blot hybridization of gel in C with a 32P-labeled chromosome X-specific probe derived from the S. cerevisiae URA2 gene. Chromosomal assignments are indicated at the left of A–D, respectively.
The electrophoretic karyotype (Figure 2A, lanes 1–8) and the hybridization pattern obtained with a 32P-labeled Ty1-specific probe derived from TYA1-gag (Figure 2B, lanes 1–8) were used to determine whether chromosomal alterations occurred during Ty1 repopulation. The hybridization pattern also allowed chromosomal assignment of new Ty1 insertions. As expected, modifying the reference element Ty1-4253 (lane 1; DG2454) with marker genes his3-AI (lane 2; DG2533) or URA3 (lane 4; DG2602) or modifying Ty1his3-AI(96) (lane 6; DG2196) with URA3 (lane 7; DG2503) did not noticeably alter chromosome X mobility or the Ty1 hybridization pattern. However, mobility of several yeast chromosomes was altered in the repopulated strains DG2633 (lane 3), DG2578 (lane 5), and DG2525 (lane 8) when compared with parental strains DG2533 (lane 2), DG2602 (lane 4), and D2503 (lane 7), respectively. In particular, chromosomes I, V, X, XIV, XIII/XVI, and XV/VII in DG2633 (lane 3); III, XI, X, XIV, II, and XIII/XVI in DG2578 (lane 5); and III, XI, XIV, and II in DG2525 (lane 8) contained noticeable size alterations that correlated with insertion of additional Ty1 elements. Larger chromosomes, such as IV, XII, XV, and VII, also contained new Ty1 insertions but their eletrophoretic mobility remained the same.
The increase in size of chromosome X in strain DG2633 was analyzed further since the reference element Ty1-4253his3-AI also was present on chromosome X and this strain displayed strong cosuppression (Table 2; compare DG2533 with DG2633). Therefore, the additional Ty1 insertions on chromosome X might influence cosuppression by an intrachromosomal interaction with Ty1-4253 that is stronger than that conferred when the elements are dispersed (Jiang 2002; Garfinkel et al. 2003). To verify that chromosome X was altered, we performed CHEF gel (Figure 2C, lanes 1–3) and Southern analyses using a 32P-labeled chromosome X-specific probe derived from the S. cerevisiae URA2 gene (Figure 2D, lanes 1–3). The results show that chromosome X increased in size in DG2633 (Figure 2D, lane 2), when compared with chromosome X in the single-element parental strain DG2533 (Figure 2D, lane 1), and that the URA2 probe hybridized reasonably well with chromosome X in the Ty-less strains (lanes 1 and 2), which are closely related to S. paradoxus (Moore et al. 2004). As expected, the URA2 probe from S. cerevisiae hybridized strongly with chromosome X from the wild-type S. cerevisiae strain S288c (lane 3), which was included as a positive control.
TABLE 2.
Retrotransposition of Ty1-4253his3-AI elements
| Transposition rate,a × 10−6 (SD)
|
||||||
|---|---|---|---|---|---|---|
| Strainb | Ty1 no. | 20° | 23° | 26° | 30° | 35° |
| Haploid | ||||||
| DG2533 | 1 | 240 (64) | 270 (119) | 126 (27) | 10.6 (0.7) | <0.62 |
| DG2633 | 29 | 0.7 (0.4) | 0.18 (0.07) | <0.13 | <0.05 | <0.11 |
| Diploid | ||||||
| DG2567 | 1 | 15.3 (4.4) | ND | ND | 0.53 (0.4) | <0.44 |
| DG2646 | 29 | 0.18 (0.13) | ND | ND | <0.01 | ND |
SD, standard deviation. ND, not determined.
Transposition rates were averaged from at least two trials.
Strain DG2633 was derived from DG2533 by the addition of unselected Ty1 insertions. Strains DG2533 and DG2633 were haploid. Strains DG2567 and DG2646 were diploid and heterozygous for Ty1-4253his3-AI. DG2646 also harbored the 28 Ty1 insertions present in DG2633 in a heterozygous configuration.
We also performed tetrad analysis after mating the single-element parental strain DG2533 or the repopulated strain DG2633 with the Ty-less strain DG2204 to determine whether the chromosomal alterations present in DG2633 affected spore viability. When the DG2533 cross was analyzed, 29 tetrads gave rise to four ascosporal colonies and all markers segregated normally, 2 tetrads produced three colonies, and 1 tetrad produced one colony. When the DG2633 cross was analyzed, 31 tetrads produced four ascosporal colonies that displayed normal segregation, 3 tetrads produced three colonies, and 2 tetrads produced two colonies. Therefore, the Ty1-mediated chromosomal alterations in DG2633 do not affect meiosis or spore viability.
In addition, strains DG2602, DG2196, and DG2503 (Figure 2A, lanes 4, 6, and 7) displayed a chromosome-length polymorphism involving the XIII/XVI doublet that distinguished these strains from the mitotic derivatives of 337, strains DG2454 (lane 1) and DG2533 (lane 2) (Garfinkel et al. 2003; Moore et al. 2004). We have not investigated this polymorphism further, but it may be correlated with the improved sporulation and galactose utilization in strains DG2602, DG2196, and DG2503.
Ty1 gain by retrotransposition:
We determined the rate of Ty1-4253his3-AI-mediated His+ formation in haploid strains DG2533 and DG2633 or heterozygous diploid strains DG2567 and DG2646 that were closely related but differed in the number of Ty1 elements present in the genome (Table 2). The rate of Ty1 retrotransposition was determined by correcting the rate of His+ formation by the splicing efficiency and inhibitory effects of the his3-AI indicator gene (Curcio and Garfinkel 1991). Since Ty1 transposition is progressively inhibited at temperatures above 20° (Paquin and Williamson 1984) and temperature is a critical variable for growth, transposition rates were determined at temperatures ranging from 20°, which is permissive for retrotransposition, to 35°, which severely inhibits retrotransposition. Splicing efficiencies varied less than twofold when cells were grown at 20° or 35° (data not shown), as determined by reverse transcription-PCR (Garfinkel et al. 2003). In addition, we assumed that each His+ prototroph resulted from the insertion of a single Ty1HIS3 element (Figure 1). This probably resulted in a modest underestimate of transposition rates obtained in single-element strains since 28% of the His+ colonies contain two Ty1 transposition events when cosuppression is not active (Garfinkel et al. 2003).
The single competent Ty1-4253his3-AI element transposed at a rate of 240 × 10−6 per element per generation when the haploid strain DG2533 was grown at 20° (Moore et al. 2004); however, the retrotransposition rate decreased 2-fold (240/126 × 10−6) and 23-fold (240/10.6 × 10−6) when incubation temperature was increased to 26° and 30°, respectively. Ty1-4253his3-AI retrotransposition was undetectable at 35°, decreasing >387-fold (240/<0.62 × 10−6). Repopulating DG2533 with 28 chromosomal Ty1 insertions decreased the Ty1-4253his3-AI retrotransposition rate 342-fold (compare DG2533 with DG2633 at 20°; 240/0.7 × 10−6) in strain DG2633 at 20°, which supports recent studies on Ty1 cosuppression (Jiang 2002; Garfinkel et al. 2003). Increasing the growth temperature also inhibited Ty1-4253his3-AI retrotransposition in the repopulated haploid strain DG2633. In addition, Ty1-4253his3-AI retrotransposition occurred at the same rate in MATa or MATα haploids (data not shown), which is in agreement with previous work (Curcio and Garfinkel 1991; Conte et al. 1998; Lee et al. 1998).
Ty1-4253his3-AI retrotransposition was determined in diploid strains containing only the tagged element (DG2567) or with 28 additional elements (DG2646) in a heterozygous configuration (Table 2). MATa/α repression decreased Ty1-4253his3-AI transposition ∼20-fold (compare DG2533 with DG2567 at 20° and 30°; 240/15.3 × 10−6 and 10.6/0.53 × 10−6, respectively) in the single-element heterozygote DG2567, which is comparable to the decrease in Ty1 RNA observed in diploid laboratory strains (Elder et al. 1980). When reintroduced Ty1 elements were present as heterozygotes, retrotransposition remained under MATa/α and copy number control with about a 4-fold (compare DG2633 with DG2646 at 20°; 0.7/0.18 × 10−6) decrease due to MATa/α repression and an 85-fold decrease due to cosuppression (compare DG2567 with DG2646 at 20°; 15.3/0.18 × 10−6). We could not detect Ty1-4253his3-AI retrotransposition events in repopulated haploid (DG2633) or diploid (DG2646) strains at 30° or 35°.
Ty1 LTR-LTR recombination, ectopic gene conversion, and LOH:
Ty elements undergo a variety of recombinational events that result in loss or sequence reassortment (Figure 1). In diploid strains, most LOH events involving Ty1 result from mitotic gene conversion with the unoccupied homolog (Kupiec and Petes 1988a). We have shown that Ty1-4253::URA3 undergoes intraelement LTR-LTR recombination at a rate of ∼1 × 10−6 per element per generation at 20° in single-element MATa or MATα haploids (Moore et al. 2004), which is comparable to results obtained with a Ty1-induced mutation at HIS4 called his4-912(URA3a) (Winston et al. 1984) where URA3 is in the same position and orientation as in Ty1-4253. A similar rate of 5-FOAR (Table 3) due to LTR-LTR recombination (Table 4) also was obtained with a different insertion, Ty1his3-AI(96)::URA3, in strain DG2503 at 30°. Recombination increased slightly in the single-element strain DG2602 at 35°, as monitored by the rate of 5-FOAR colony formation (Table 3), and 10 independent 5-FOAR mutants recovered at each temperature contained a solo Ty1-H3 LTR present at the Ty1-4253 insertion site, as indicated by PCR analysis and DNA sequencing (Table 4). Furthermore, a temperature-dependent increase in mitotic recombination using an ade2-based inverted-repeat substrate has been reported (Rattray and Symington 1995).
TABLE 3.
Rate of 5-FOAR when Ty1 copy number increases
| 5-FOAR rate, × 10−6 (SD)a
|
||||
|---|---|---|---|---|
| Strainb | Ty1 no. | 20° | 30° | 35° |
| Haploid | ||||
| DG2503 | 1 | ND | 4.5 (1.6) | ND |
| DG2525 | 21 | ND | 3.7 (0.8) | ND |
| DG2602 | 1 | 0.7 (0.1) | 1.8 (1.3) | 2.3 (1.0) |
| DG2578 | 25 | 1.0 (0.26) | 1.25 (0.4) | 4.3 (2.0) |
| Diploid | ||||
| DG2566 | 1 | 3.5 (1.5) | 12.0 (0.8) | 28.5 (5.0) |
| DG2601 | 25 | 3.8 (1.3) | ND | ND |
Rate of 5-FOAR per cell per generation as determined as described previously (Drake 1970; Rattray et al. 2000).
Strain DG2525 was derived from DG2503, and DG2578 was derived from DG2602 by the addition of unselected Ty1 insertions. All strains were haploid except for DG2566 and DG2601, which were diploid and heterozygous for Ty1-4253::URA3. DG 2601 also harbored the 25 Ty1 insertions present in DG2578 in a heterozygous configuration.
TABLE 4.
Ty1 recombination in haploid strains
| 20°
|
30°
|
35°
|
|||||
|---|---|---|---|---|---|---|---|
| Strain | Ty1 no. | LTR-LTR | Gene conversion |
LTR-LTR | Gene conversion |
LTR-LTR | Gene conversion |
| DG2503 | 1 | ND | ND | 32/32 | 0/32 | ND | ND |
| DG2525 | 21 | ND | ND | 16/20 | 4/20 | ND | ND |
| DG2602 | 1 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 |
| DG2578 | 25 | 8/10 | 2/10 | 7/10 | 3/10 | 8/10 | 2/10 |
5-FOAR colonies were analyzed by PCR. Also refer to footnotes in Table 3.
To determine whether additional Ty1 insertions influenced the loss of Ty1::URA3, we generated repopulated haploid strain DG2525 from DG2503 [Ty1his3-AI(96)::URA3] and strain DG2578 from DG2601(Ty1-4253::URA3) that now contained 20 and 24 additional Ty1 element insertions, respectively. Repopulating the haploid strains with additional Ty1 elements by pGTy1 induction did not markedly alter the rate of 5-FOAR (Table 3). We determined the spectrum of recombination events leading to 5-FOAR in these strains by PCR analysis using primers derived from sequences flanking the insertion site, Ty1-H3, and the marker genes (Table 4; materials and methods). Ty1 gene conversion events retained the his3-AI marker in strain DG2525, since there was 1 kb of DNA homology between URA3 and his3-AI and only 338 bp between his3-AI and the end of Ty1his3-AI(96). The fraction of LTR-LTR recombinants (70–80%) vs. ectopic gene conversions (20–30%) remained constant in strains DG2525 and DG2578, regardless of the growth temperature or the chromosomal location of the reference element.
We determined the rate of LOH in diploid strains containing Ty1-4253::URA3 alone (DG2566) or in the presence of additional elements (DG2601) (Table 3). There was an eightfold increase in the rate of 5-FOAR with increasing growth temperature in the single-element heterozygote DG2566. Almost all LOH events (57/60) at 20° resulted from a complete loss of Ty1-4253::URA3, as determined by PCR and Southern analyses, while three 5-FOAR mutants resulted from LTR-LTR recombination (Table 5). Even though the diploid strain DG2601 contained 24 additional Ty1 elements dispersed in the genome in a heterozygous configuration, the rate of 5-FOAR was similar to that obtained in the single-element diploid DG2566 at 20° (Table 3).
TABLE 5.
Ty1 recombination in diploid strains
| Strain | Ty1 no. | LTR-LTR | LOH | Gene conversion |
|---|---|---|---|---|
| DG2566 | 1 | 3/60 | 57/60 | 0/60 |
| DG2601 | 25 | 1/45 | 38/45 | 6/45 |
5-FOAR colonies obtained at 20° were analyzed by PCR and Southern hybridization. Also refer to footnotes in Table 3.
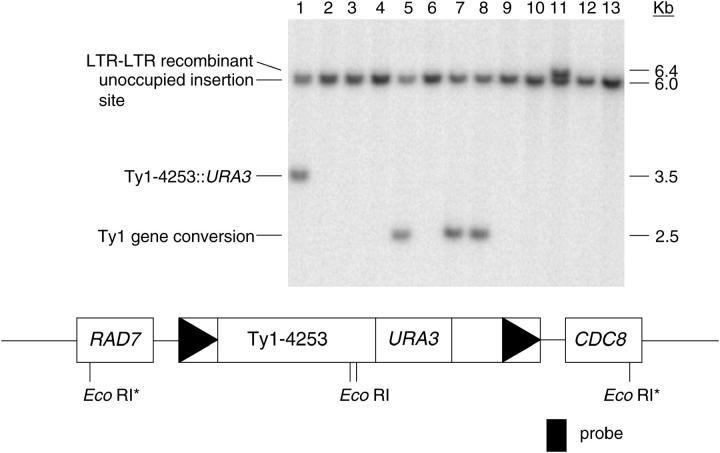
To determine whether the 5-FOAR mutants with strain DG2601 were generated by LOH, ectopic gene conversion with chromosomal Ty1 elements, or LTR-LTR recombination, 45 mutants isolated at 20° were analyzed by Southern hybridization. For example (Figure 3; materials and methods), genomic DNA from the parental strain DG2601 (lane 1) and 12 of the 5-FOAR mutants (lanes 2–13) were digested by EcoRI and analyzed by Southern hybridization using a 32P-labeled probe derived from a 500-bp region of chromosome X adjacent to CDC8. One LTR-LTR recombinant was recovered (lane 11), as indicated by loss of the Ty1-4253::URA3 junction fragment and the presence of a novel EcoRI fragment ∼350 bp larger (6.4 kb) than the unoccupied 6-kb fragment on chromosome X. LOH events resulted in 8 5-FOAR mutants (lanes 2, 3, 4, 6, 9, 10, 12, and 13), as indicated by loss of the Ty1-4253::URA3 junction fragment (3.5 kb) and the presence of only the unoccupied EcoRI fragment (6 kb). Ty1 gene conversions resulted in 3 5-FOAR mutants (lanes 5, 7, and 8), as indicated by loss of the Ty1-4253::URA3 junction fragment and the presence of a Ty1-4253 junction fragment that lacked URA3. Overall, the hybridization patterns showed that 1 5-FOAR event (2.2%) resulted from LTR-LTR recombination, 38 (84.5%) resulted from LOH, and 6 (13.3%) resulted from gene conversion with the chromosomal Ty1 elements present in DG2601 (Table 5). We were unable to determine the level of Ty1 loss by LTR-LTR recombination at 30° and 35° in DG2601 because the rate of 5-FOAR caused by LOH increased dramatically with temperature, making LTR-LTR recombination events very difficult to detect (data not shown).
Figure 3.—
Ty1 recombination events recovered in strain DG2601. Total DNA from the parental strain DG2601 (lane 1) and 12 5-FOAR mutants was digested with EcoRI and processed for Southern analysis. The resulting filter was hybridized with a 32P-labeled probe that was derived from sequences adjacent to CDC8. At the bottom is a map of the RAD7-Ty1-4253-CDC8 interval on chromosome X. LTRs are represented by solid triangles and the hybridization probe is denoted by the solid rectangle. Ty1-4253/chromosome X junction fragments and sizes are noted on either side of the figure. The location of the EcoRI sites flanking Ty1-4253 has not been determined and, therefore, is denoted by asterisks.
Participation of Ty1 cDNA in chromosomal gene conversion events:
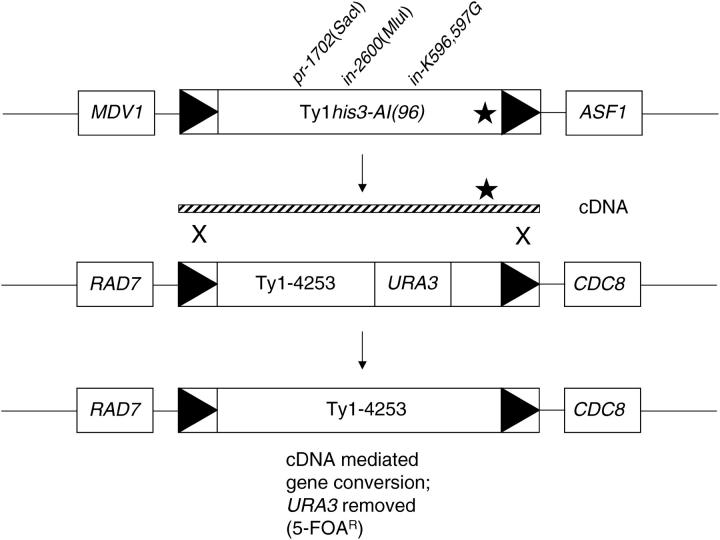
Ty1 cDNA can undergo recombination with chromosomal or plasmid-borne elements (Melamed et al. 1992; Sharon et al. 1994). Since these experiments have been performed with Ty1 elements that are highly expressed from the GAL1 promoter on a multicopy plasmid, utilization of the cDNA recombination pathway may be artificially elevated. To determine whether cDNA recombination contributes to Ty1 sequence redistribution under more natural conditions, a Ty-less strain (DG2547) was constructed that contained Ty1his3-AI(96) and Ty1-4253::URA3 (Figure 4). Note that both Ty1 insertions were present in their normal chromosomal locations and were expressed from their native promoters. Ty1his3-AI(96) is competent for retrotransposition (Garfinkel et al. 2003) while Ty1-4253::URA3 contained a complete URA3 gene inserted into the RT coding sequence, which should result in a truncated Ty1-4253 transcript defective for reverse transcription and production of RT. Ty1 mutations in PR [pr-1702(SacI)] and IN [in-2600 (MluI) and in-K596,597G] were introduced into the chromosomal Ty1his3-AI(96) element. PR mutants produce very little cDNA and are blocked for both transposition and cDNA recombination (Youngren et al. 1988; Sharon et al. 1994). IN mutants DG2592 [in-2600(MluI)] and DG2605 (in-K596,597G) are defective in transpositional integration and IN nuclear localization, respectively, but undergo cDNA recombination and wild-type Ty1 cDNA production when the IN mutations are present in a pGTy1 plasmid (Sharon et al. 1994; Moore et al. 1998). The Ty1IN mutants also produced wild-type levels of cDNA when present in their native context in the Ty-less background, whereas the PR mutant produced very little cDNA, as determined by quantitative Southern analysis (Lee et al. 1998; data not shown). Since the URA3 and his3-AI markers were well separated, homologous recombination between Ty1his3-AI(96) cDNA and the chromosomal Ty1-4253::URA3 element should not be inhibited by spacing of the markers. In addition, strain DG2547 displayed little if any cosuppression (data not shown).
Figure 4.—
Detection of cDNA-mediated gene conversion using native Ty1 elements. Ty1his3-AI(96) and Ty1-4253 are both located on chromosome X, in inverted orientation relative to each other and separated by ∼350 kb, based on coordinates from the S. cerevisiae genome. Ty1his3-AI(96) and Ty1-4253 inserted between MDV1 and ASF1, and RAD7 and CDC8, respectively. The Ty1 mutations pr-1702(SacI), in-2600(MluI), or in-K596,597G were introduced into Ty1his3-AI(96) by homologous recombination. The pr-1702(SacI) mutant accumulates little if any Ty1 cDNA and does not undergo cDNA recombination, while the IN mutants accumulate wild-type levels of cDNA and can undergo cDNA recombination (Youngren et al. 1988; Sharon et al. 1994; Moore et al. 1998). The star represents the retrotransposition indicator gene, his3-AI, which was not utilized here. Ty1his3-AI(96) or Ty1HIS3(96) cDNA is depicted by the striped bar. Loss of the Ty1-4253::URA3 marker was detected by plating on SC + 5-FOA medium. Also refer to Table 6.
If Ty1 cDNA played a major role in maintaining Ty1 element sequence integrity, then cDNA-mediated gene conversion events should compose a significant fraction of 5-FOAR mutants, especially when cDNA accumulated in the IN-defective mutants (Figure 4). However, both the overall rate of 5-FOAR colony formation at 20° and the spectrum of recombination events remained the same regardless of whether Ty1his3-AI(96) was transposition competent or an element was transpositionally blocked prior to or after cDNA production (Table 6).
TABLE 6.
Involvement of Ty1 cDNA in ectopic gene conversion
| Strain | Relevant genotype | Rate of 5-FOAR, × 10−6 (SD) | LTR-LTR | Gene conversion |
|---|---|---|---|---|
| DG2547 | Wild-type | 1.6 (0.23) | 20/21 | 1/21 |
| DG2575 | pr-1702 (SacI) | 0.9 (0.14) | 17/21 | 4/21 |
| DG2547 | Wild-type | 0.8 (0.1) | 9/10 | 1/10 |
| DG2592 | in-2600 (MluI) | 0.9 (0.13) | 10/10 | 0/10 |
| DG2547 | Wild-type | 0.8 (0.12) | 8/10 | 2/10 |
| DG2605 | in-K596,597G | 0.7 (0.1) | 10/10 | 0/10 |
5-FOAR mutants were obtained at 20°. Also refer to Figure 4.
To determine whether endogenous Ty1 cDNA recombination occurred at all, 5-FOAR His+ colonies were selected from strains DG2547 (wild-type) and DG2592 [in-2600(MluI)] at 20°. Although 5-FOAR His+ colonies were extremely rare, we analyzed six colonies from the wild-type strain and 2 from the in-2600(MluI) mutant by PCR and Southern analyses (data not shown). Three of six His+ 5-FOAR events from the wild-type strain DG2547 were caused by Ty1HIS3 cDNA-mediated gene conversion of Ty1-4253::URA3, while the rest were the product of two events, LTR-LTR recombination and Ty1HIS3 retrotransposition. In the two 5-FOAR His+ colonies recovered from the in-2600(MluI) mutant DG2592, one resulted from a Ty1HIS3 cDNA gene conversion, while the other underwent two events, conversion of URA3 by Ty1 DNA or cDNA and a separate Ty1HIS3 insertion elsewhere in the genome.
DISCUSSION
Here we have used an experimental system based on a Ty-less strain to study Ty1 copy number dynamics and genome restructuring in Saccharomyces. Our work suggests that the rate of Ty1 retrotransposition is the key variable required to establish copy number, because the rate of Ty1 loss by LTR-LTR recombination remains fairly constant even in the presence of additional Ty1 elements. Ty1 elements also appear to be more susceptible to copy number fluctuation in haploid than in diploid cells. Taken together, our results strongly suggest that Ty1 element copy number can oscillate in nature, allowing for the considerable variability observed in Saccharomyces (Wilke et al. 1992; Neuveglise et al. 2002), including complete loss in S. paradoxus strain 337.
Ty1 repopulation:
We have identified yeast chromosomes containing “bursts” of new Ty1 insertions that result from serial induction of a pGTy1 plasmid. Although new Ty1 element insertions are present in many yeast chromosomes in the repopulated strains, a number of these insertions are associated with chromosome size alterations. However, the ability to detect chromosomal alterations by CHEF gel analysis is biased by the size distribution of the 16 Saccharomyces chromosomes and the distribution of tRNA target genes (Hani and Feldmann 1998). Smaller chromosomes display Ty1-mediated alterations more readily because of their size, but are poorer targets since these chromosomes contain fewer tRNA genes. Conversely, larger chromosomes have more Ty1 targets, but more insertions would have to occur before a size alteration is evident.
Nonetheless, chromosomal alterations can result from several Ty1-mediated events, such as DNA rearrangement associated with Ty integration (Sutton and Liebman 1992) or between resident elements (Liebman et al. 1981; Downs et al. 1985; Fischer et al. 2000), multimeric Ty insertions (Weinstock et al. 1990), as well as independent insertion events in the same chromosome. Of particular interest are the additional chromosome X insertions present in DG2633. Cosuppression of Ty1 occurs at both the transcriptional and the post-transcriptional levels (Jiang 2002; Garfinkel et al. 2003); therefore, the additional elements present on chromosome X may enhance these forms of copy number control. Although chromosome X is also altered in strains chosen at random for analysis of Ty1 loss, the role of neighboring elements in cosuppression should be investigated further using more specific assays. About five additional elements are present in chromosome X in DG2633, yet results from Southern analysis suggest that large multimeric elements, such as those observed at the silent mating locus HMLα (Weinstock et al. 1990), are not present (data not shown). Chromosome X may also contain preferred Ty1 integration regions, since two independent transpositional insertions, Ty1his3-AI(96) and Ty1-4253, are both located on chromosome X adjacent to tRNA genes but hundreds of kilobases apart. In support of this idea, certain tRNA targets are preferred by Ty1 over others (Bachman et al. 2004) and, surprisingly, the mouse Steel gene is a hotspot for Ty1 integration when present on an artificial chromosome in yeast (Dalgaard et al. 1996). The several Ty1-mediated chromosomal alterations in DG2633 also do not affect meiosis or spore viability, suggesting that genetic isolation has not occurred, as is observed for other Ty-mediated chromosome alterations (Garfinkel 2005).
Ty1 gain and loss:
Ty1 retrotransposition remains under copy number control at different temperatures in both haploid and MATa/α diploid cells. In contrast to the multiple ways Ty1 retrotransposition is modulated, Ty1 loss remains relatively constant in haploid cells even in the presence of additional Ty1 elements. All Ty1 loss events in single-element haploids result from intraelement LTR-LTR recombination. However, 20–30% of the 5-FOAR recombinants result from mitotic gene conversion between ectopically placed Ty1 elements in repopulated haploids, a process that maintains copy number and probably homogenizes Ty sequence. Although our results are in general agreement with previous work in S. cerevisiae (Chaleff and Fink 1980; Roeder and Fink 1980, 1982; Ciriacy and Williamson 1981; Liebman et al. 1981; Winston et al. 1984; Kupiec and Petes 1988a,b; Vincent and Petes 1989), additional features of the Ty-less strain are worth noting. Ty elements and their solo-LTR derivatives can undergo recombination (Downs et al. 1985); however, we have not observed this class of recombinants in the Ty-less background. A likely explanation is that the remnant solo-LTRs present in the Ty-less strain have diverged beyond the level of sequence complementarity required for efficient homologous recombination with Ty1 (Moore et al. 2004). We might also expect LTR-Ty1 recombinants to return as the repopulated strains evolve. Similarly, gene conversion with the endogenous URA3 locus in the Ty-less strain has not been observed in our study. In addition, it is unlikely that the repopulated strains recapitulate the pattern of Ty1 insertions present in the laboratory strains. Therefore, the repopulated and laboratory strains may have different levels of gene conversion between particular Ty1 elements.
Another important consideration is that the Ty1 loss and conversion events described here occur between elements in their preferred chromosomal regions upstream of tRNA genes (Devine and Boeke 1996; Boeke and Devine 1998). Recent results suggest that Ty1 elements at their preferred insertion sites are selectively neutral (Blanc and Adams 2004) and do not markedly influence adjacent tRNA transcription (Bolton and Boeke 2003). However, earlier work on Ty recombination has predominantly utilized elements that have mutated genes, which introduces new variables such as selective pressure, different chromatin configuration, and proximity to recombinational hotspots (Ben-Aroya et al. 2004).
We have shown that Ty1 loss by LTR-LTR recombination occurs in MATa/α diploids at about the same level in the presence or absence of additional Ty1 insertions; however, these events are overshadowed by LOH. LOH events involving Ty1 probably result from mitotic gene conversion with the unoccupied homolog although more complex rearrangements, chromosome loss, or nondisjunction also can occur (Kupiec and Petes 1988a). Mitotic and meiotic gene conversion events usually show parity, which should result in the gain or loss of a Ty element with equal efficiency. Since 5-FOAR selection detects only loss of an element carrying URA3, we have assumed that the frequency of LOH events where a complete Ty1 is lost by gene conversion or some other chromosomal event will be the same as the frequency where both chromosomes gain an element. Therefore, Ty1 copy number should remain unchanged if Ty1 gene conversion shows parity. However, Ty-induced insertion mutations are removed by mitotic or meiotic gene conversion less frequently than expected for ectopically placed repeats (Vincent and Petes 1989), a feature Ty may confer by adopting a chromatin structure that suppresses meiotic recombination (Ben-Aroya et al. 2004). Since most previous work on Ty recombination has used Ty-induced mutations at rare insertional targets rather than at Ty1's preferred locations upstream of genes transcribed by RNA polymerase III, it will be interesting to determine whether gene conversion bias also exists for Ty1 elements at their preferred location in single-element and repopulated genomes. Furthermore, Ty retrotransposition events at ADH2 and ADH4 are reported to increase during meiosis (Ribeiro-dos-Santos et al. 1997). Taken together, the recombinational properties of the MATa/α diploid cell type minimize Ty1 loss by LTR-LTR recombination, even though MATa/α repression reduces Ty1 transcription and, hence, retrotransposition from occurring.
Our results suggest that Ty1 cDNA recombination plays a minor role in converting chromosomal elements when the elements are in their native context. In contrast, overexpression of Ty1, Ty5, and Schizosaccharomyces pombe Tf1 elements results in high levels of cDNA recombination especially when IN-mediated integration is blocked (Melamed et al. 1992; Sharon et al. 1994; Hoff et al. 1998; Ke and Voytas 1999). We suggest that the large increase in reverse transcription intermediates and complete cDNA present in pGTy1-induced cells and the presence of an episomal pGTy1 element (Boeke et al. 1985; Garfinkel et al. 1985), which is an excellent recombination partner (Sharon et al. 1994), lead to the high level of Ty1 cDNA recombination reported in previous studies.
Ty1 oscillation:
Like many other mobile elements, Ty1 retrotransposition depends on an interplay between element, host, and environmental factors (Bushman 2001). In this work, three major determinants of Ty1 copy number control have been analyzed: the copy number of the element itself, cell type, and growth temperature. We have attempted to determine both gain and loss rates using appropriately tagged reference elements to derive a gain/loss ratio (Table 7), correcting for the effects of the Ty1 marker his3-AI (Curcio and Garfinkel 1991). In haploid cells containing a single element grown at 20°, conditions that are very favorable for retrotransposition, the gain/loss ratio is well over 100. But even in the absence of Ty1 cosuppression, loss will predominate if single-element haploid cells are grown at 35° (gain/loss <0.21), where Ty1 retrotransposition is inhibited. When additional Ty1 elements are present in haploids, the gain/loss ratio is ∼0.9 at 20°, which should result in element homeostasis. Correspondingly, Ty1 loss is greatly favored as growth temperature increases, with gain/loss ratios of <0.03 at 30° and <0.02 at 35°. Given the effects that Ty1 cosuppression and growth temperature have on retrotransposition in haploid cells, additional environmental and genetic modulators (Scholes et al. 2001; Staleva Staleva and Venkov 2001; Griffith et al. 2003) are likely to cause Ty1 copy number to oscillate with time. Our results also suggest that Ty1 elements are more vulnerable to copy number fluctuations in haploid cells, and repopulation by Ty1 probably occurs frequently, making Ty-less genomes transitory.
TABLE 7.
Ty1 gain and loss
| Ty1 gain/loss
|
|||
|---|---|---|---|
| Ty1 copy number | 20° | 30° | 35° |
| Single element (haploid) | Gain (342) | Gain (8.4) | Loss (<0.21) |
| Multiple elements (haploid) | Gain ≅ loss (0.9) | Loss (<0.03) | Loss (<0.02) |
| Single element (diploid heterozygote) | Gain (87) | ND | ND |
| Multiple elements (diploid heterozygote) | Gain (2) | ND | ND |
Even though the Ty1 copy number dynamics outlined for haploid cells seem clear, many natural Saccharomyces isolates are homothallic diploids due to mating type interconversion (Herskowitz 1988). Our analyses of MATa/α diploid cells suggest that Ty1 gain is favored at 20° with gain/loss ratios of 87 for a single element and 2 when additional elements are present. We have not been able to estimate Ty1 gain and loss at 30° and 35° in repopulated diploids because Ty1 retrotransposition is greatly inhibited and LOH increases significantly with temperature, making LTR-LTR recombinants difficult to detect. Ty1 copy number may remain constant in diploids, depending on whether native Ty1 insertions show parity of LOH events. Interestingly, a Ty gain/loss ratio of 1.2 has been obtained from Ty sequence analysis using a demographic computational model where solo-LTRs are viewed as “element death” (Promislow et al. 1999). The gain/loss ratio derived computationally is comparable to our data obtained experimentally for either haploid or diploid cells containing multiple elements grown at 20°. These results suggest that the evolutionary history of the sequenced S. cerevisiae laboratory strain has included periods of growth under conditions favoring Ty1 retrotransposition. In support of this idea, the 32 Ty1 elements present in S. cerevisiae laboratory strains are on the upper end of the distribution of Ty1 copy number (Goffeau et al. 1996; Kim et al. 1998) when compared with 88 natural isolates (Wilke et al. 1992). This is striking when one considers that 85% of the Ty elements in the laboratory strain have been lost by recombination, leaving solo-LTRs behind. Therefore, the laboratory strain may have had a much higher Ty1 copy number and is now in a “loss phase” of an oscillation cycle.
Larger genomes:
Genome analyses have shown that closely related eukaryotes maintain retrotransposons with different activities and copy numbers. For example, retrotransposon gain is considerably slower among great-ape species than in old world monkeys (Liu et al. 2003), yet both types of genomes maintain a large transposable element load. There is evidence for both rapid gain and loss of LTR-retrotransposons in different plant lineages. LTR-retrotransposons in all plants analyzed are typically young, originating ∼10 million years ago, despite evidence that older elements have been present in plants for hundreds of millions of years (Kumar and Bennetzen 1999). Recently, Ma and Bennetzen (2004) have shown that at least some of the rapid element gain in rice is counterbalanced by rapid loss via LTR-LTR or illegitimate recombination, suggesting that other genomes may oscillate in size and not continue to expand indefinitely (Bennetzen and Kellogg 1997). Post-transcriptional cosuppression mechanisms based on RNA interference (RNAi) are also likely to minimize element gain in plants and animals (Tijsterman et al. 2002). However, a novel form of post-transcriptional cosuppression probably limits Ty1 retrotransposition (Jiang 2002; Garfinkel et al. 2003), since conserved RNAi genes are not present in S. cerevisiae. In addition, Saccharomyces may have countered Ty invasion in part by coevolving extremely active homologous recombination systems, which may be less active in larger eukaryotes (Garfinkel 2005).
Acknowledgments
We thank F. Lacroute for plasmid pJLS1; N. Copeland and N. Jenkins for use of their CHEF apparatus; G. Liti and N. O'Sullivan for technical advice; and S. Martin, K. Turner, and two anonymous reviewers for comments on the manuscript. We also thank J. Bennetzen, S. Wessler, J. Westpheling, and members of the Gene Regulation and Chromosome Biology Laboratory for stimulating discussions. This work was sponsored by the Center for Cancer Research of the National Cancer Institute, Department of Health and Human Services. The contents of this publication do not necessarily reflect views or policies of the Department of Health and Human Services nor does the mention of trade names, commercial products, or organizations imply endorsement from the U.S. Government.
References
- Bachman, N., Y. Eby and J. D. Boeke, 2004. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 14: 1232–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya, S., P. A. Mieczkowski, T. D. Petes and M. Kupiec, 2004. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae. Mol. Cell 15: 221–231. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, V. M., and J. Adams, 2004. Ty1 insertions in intergenic regions of the genome of Saccharomyces cerevisiae transcribed by RNA polymerase III have no detectable selective effect. FEMS Yeast Res. 4: 487–491. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., and S. E. Devine, 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93: 1087–1089. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., D. J. Eichinger and G. Natsoulis, 1991. Doubling Ty1 element copy number in Saccharomyces cerevisiae: host genome stability and phenotypic effects. Genetics 129: 1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., D. J. Garfinkel, C. A. Styles and G. R. Fink, 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500. [DOI] [PubMed] [Google Scholar]
- Bolton, E. C., and J. D. Boeke, 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 13: 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman, F., 2001 Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor Laboratory Press, Woodbury, NY.
- Carle, G. F., and M. V. Olson, 1985. An electrophoretic karyotype for yeast. Proc. Natl. Acad. Sci. USA 82: 3756–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleff, D. T., and G. R. Fink, 1980. Genetic events associated with an insertion mutation in yeast. Cell 21: 227–237. [DOI] [PubMed] [Google Scholar]
- Chu, G., D. Vollrath and R. W. Davis, 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234: 1582–1585. [DOI] [PubMed] [Google Scholar]
- Ciriacy, M., and V. M. Williamson, 1981. Analysis of mutations affecting Ty-mediated gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 182: 159–163. [DOI] [PubMed] [Google Scholar]
- Conte, D., Jr., E. Barber, M. Banerjee, D. J. Garfinkel and M. J. Curcio, 1998. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 18: 2502–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, N. L., R. Craigie, M. Gellert and A. M. Lambowitz (Editors), 2002 Mobile DNA II. ASM Press, Washington, DC.
- Curcio, M. J., and D. J. Garfinkel, 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, J. Z., M. Banerjee and M. J. Curcio, 1996. A novel Ty1-mediated fragmentation method for native and artificial yeast chromosomes reveals that the mouse Steel gene is a hotspot for Ty1 integration. Genetics 143: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine, S. E., and J. D. Boeke, 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Downs, K. M., G. Brennan and S. W. Liebman, 1985. Deletions extending from a single Ty1 element in Saccharomyces cerevisiae. Mol. Cell. Biol. 5: 3451–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., 1970 The Molecular Basis of Mutation. Holden-Day, Oakland, CA.
- Eichler, E. E., and D. Sankoff, 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797. [DOI] [PubMed] [Google Scholar]
- Elder, R. T., T. P. St. John, D. T. Stinchcomb and R. W. Davis, 1980. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. Cold Spring Harbor Symp. Quant. Biol. 45: 581–584. [DOI] [PubMed] [Google Scholar]
- Farabaugh, P. J., and G. R. Fink, 1980. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature 286: 352–356. [DOI] [PubMed] [Google Scholar]
- Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver and E. J. Louis, 2000. Chromosomal evolution in Saccharomyces. Nature 405: 451–454. [DOI] [PubMed] [Google Scholar]
- Garfinkel, D. J., 2005 Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet. Genome Res. (in press). [DOI] [PubMed]
- Garfinkel, D. J., J. D. Boeke and G. R. Fink, 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42: 507–517. [DOI] [PubMed] [Google Scholar]
- Garfinkel, D. J., K. Nyswaner, J. Wang and J. Y. Cho, 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165: 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon et al., 1996. Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed] [Google Scholar]
- Griffith, J. L., L. E. Coleman, A. S. Raymond, S. G. Goodson, W. S. Pittard et al., 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991 Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Hani, J., and H. Feldmann, 1998. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 26: 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. L., 2000. Molecular melodies in high and low C. Nat. Rev. Genet. 1: 145–149. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I., 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52: 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff, E. F., H. L. Levin and J. D. Boeke, 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination. Mol. Cell. Biol. 18: 6839–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, E. K., P. C. Wang and S. J. Lo, 1998. Strategies for cloning unknown cellular flanking DNA sequences from foreign integrants. Cell. Mol. Life Sci. 54: 1403–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. W., 2002. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev. 16: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, N., and D. F. Voytas, 1999. cDNA of the yeast retrotransposon Ty5 preferentially recombines with substrates in silent chromatin. Mol. Cell. Biol. 19: 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel and D. F. Voytas, 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., and T. D. Petes, 1988. a Allelic and ectopic recombination between Ty elements in yeast. Genetics 119: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec, M., and T. D. Petes, 1988. b Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler, J. F., Jr., D. P. Haeusser, A. Dull, J. D. Boeke and J. B. Keeney, 2002. Ty1 defect in proteolysis at high temperature. J. Virol. 76: 4233–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. S., C. P. Lichtenstein, B. Faiola, L. A. Rinckel, W. Wysock et al., 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148: 1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman, S., P. Shalit and S. Picologlou, 1981. Ty elements are involved in the formation of deletions in DEL1 strains of Saccharomyces cerevisiae. Cell 26: 401–409. [DOI] [PubMed] [Google Scholar]
- Liu, G., S. Zhao, J. A. Bailey, S. C. Sahinalp, C. Alkan et al., 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed, C., Y. Nevo and M. Kupiec, 1992. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky, A. E., and H. Ris, 1951. The desoxyribonucleic acid content of animal cells and its evolutionary significance. J. Gen. Physiol. 34: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monokian, G. M., L. T. Braiterman and J. D. Boeke, 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene 139: 9–18. [DOI] [PubMed] [Google Scholar]
- Moore, S. P., L. A. Rinckel and D. J. Garfinkel, 1998. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, S. P., G. Liti, K. M. Stefanisko, K. M. Nyswaner, C. Chang et al., 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21: 649–660. [DOI] [PubMed] [Google Scholar]
- Neuveglise, C., H. Feldmann, E. Bon, C. Gaillardin and S. Casaregola, 2002. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 12: 930–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin, C., and V. M. Williamson, 1984. Temperature effects on the rate of Ty transposition. Science 226: 53–55. [DOI] [PubMed] [Google Scholar]
- Promislow, D. E., I. K. Jordan and J. F. McDonald, 1999. Genomic demography: a life-history analysis of transposable element evolution. Proc. R. Soc. Lond. B Biol. Sci. 266: 1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., and L. S. Symington, 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., B. K. Shafer and D. J. Garfinkel, 2000. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-dos-Santos, G., A. C. Schenberg, D. C. Gardner and S. G. Oliver, 1997. Enhancement of Ty transposition at the ADH4 and ADH2 loci in meiotic yeast cells. Mol. Gen. Genet. 254: 555–561. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., and G. R. Fink, 1980. DNA rearrangements associated with a transposable element in yeast. Cell 21: 239–249. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., and G. R. Fink, 1982. Movement of yeast transposable elements by gene conversion. Proc. Natl. Acad. Sci. USA 79: 5621–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer, S. B., M. Aye and T. Menees, 2002 Ty3, a position specific, gypsy-like element in Saccharomyces cerevisiae, pp. 663–683 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Scholes, D. T., M. Banerjee, B. Bowen and M. J. Curcio, 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, G., T. J. Burkett and D. J. Garfinkel, 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14: 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Woodbury, NY.
- Staleva Staleva, L., and P. Venkov, 2001. Activation of Ty transposition by mutagens. Mutat. Res. 474: 93–103. [DOI] [PubMed] [Google Scholar]
- Sutton, P. R., and S. W. Liebman, 1992. Rearrangements occurring adjacent to a single Ty1 yeast retrotransposon in the presence and absence of full-length Ty1 transcription. Genetics 131: 833–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman, M., R. F. Ketting and R. H. A. Plasterk, 2002. The genetics of RNA silencing. Annu. Rev. Genet. 36: 489–519. [DOI] [PubMed] [Google Scholar]
- Vincent, A., and T. D. Petes, 1989. Mitotic and meiotic gene conversion of Ty elements and other insertions in Saccharomyces cerevisiae. Genetics 122: 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D. F., and J. D. Boeke, 2002 Ty1 and Ty5 of Saccharomyces cerevisiae, pp. 631–662 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Weinstock, K. G., M. F. Mastrangelo, T. J. Burkett, D. J. Garfinkel and J. N. Strathern, 1990. Multimeric arrays of the yeast retrotransposon Ty. Mol. Cell. Biol. 10: 2882–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke, C. M., and J. Adams, 1992. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics 131: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke, C. M., E. Maimer and J. Adams, 1992. The population biology and evolutionary significance of Ty elements in Saccharomyces cerevisiae. Genetica 86: 155–173. [DOI] [PubMed] [Google Scholar]
- Winston, F., D. T. Chaleff, B. Valent and G. R. Fink, 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren, S. D., J. D. Boeke, N. J. Sanders and D. J. Garfinkel, 1988. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 8: 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]