Abstract
The majority of Candida albicans strains in nature are a/α and must undergo homozygosis to a/a or α/α to mate. Here we have used a mouse model for systemic infection to test the hypothesis that a/α strains predominate in nature because they have a competitive advantage over a/a and α/α offspring in colonizing hosts. Single-strain injection experiments revealed that a/α strains were far more virulent than either their a/a or α/α offspring. When equal numbers of parent a/α and offspring a/a or α/α cells were co-injected, a/α always exhibited a competitive advantage at the time of extreme host morbidity or death. When equal numbers of an engineered a/a/α2 strain and its isogenic a/a parent strain were co-injected, the a/a/α2 strain exhibited a competitive advantage at the time of host morbidity or death, suggesting that the genotype of the mating-type (MTL) locus, not associated genes on chromosome 5, provides a competitive advantage. We therefore propose that heterozygosity at the MTL locus not only represses white-opaque switching and genes involved in the mating process, but also affects virulence, providing a competitive advantage to the a/α genotype that conserves the mating system of C. albicans in nature.
CANDIDA albicans, which is diploid, contains a single mating-type locus, MTL (Hull and Johnson 1999). While 97% of natural strains are MTL heterozygous (a/α), 3% are MTL homozygous (a/a or α/α) (Lockhart et al. 2002). Only MTL-homozygous a/a and α/α strains can mate (Hull et al. 2000; Magee and Magee 2000; Miller and Johnson 2002; Lockhart et al. 2003a; Soll 2004). To become MTL homozygous, an a/α strain undergoes spontaneous MTL homozygosis either through loss of one homolog of chromosome 5, which harbors the MTL locus, followed by duplication of the retained homolog, or, less frequently, through mitotic crossing over (Wu et al. 2005). Expressing a mating type in C. albicans is, therefore, markedly different from expressing a mating type in Saccharomyces cerevisiae. S. cerevisiae possesses three different loci containing mating-type genes, two that are silent (HML, HMR) and one that is expressed (MAT). HML contains copies of a genes and HMR copies of α genes. MAT contains either a or α genes. Mating-type switching in S. cerevisiae occurs by recombination at the MAT locus with a copy of the silent locus harboring the alternative mating-type genes (Rine et al. 1979; Haber 1998). S. cerevisiae, therefore, conserves the alternative genetic information of a and α in silent cassettes when expressing either mating type. In contrast, C. albicans loses alternative MTL information when expressing a mating type.
Genetic studies of strain relatedness have revealed that the population structure of C. albicans is primarily clonal, with only hints of recombination (Graser et al. 1996; Xu et al. 1999; Pujol et al. 1993, 2004). Hence, mating most likely is a rare event and as such is an unlikely mechanism for returning MTL-homozygous strains to MTL heterozygosity. Since natural MTL-heterozygous strains can and do spontaneously generate MTL homozygotes (Lockhart et al. 2002; Pujol et al. 2003; Wu et al. 2005), but MTL homozygotes rarely replenish MTL heterozygotes, why do MTL homozygotes not accumulate and predominate in nature? Why are only 3% of natural strains MTL homozygous? One possible explanation is that MTL heterozygotes are more competitive than MTL homozygotes in natural settings. Here, we have tested this hypothesis, first by comparing virulence between a/α strains and their a/a and α/α offspring in the mouse model for systemic infection, and second by testing for competition by co-injecting mice with mixtures of a/α cells and either their a/a or α/α offspring cells. Our results demonstrate that three unrelated parental a/α strains are far more virulent than their a/a or α/α offspring, as measured by the time of extreme host morbidity or death in the mouse model for systemic infection, and that for four unrelated strains, a/α predominates at the time of extreme host morbidity or death in animals co-injected with equal volumes of a/α and either their a/a or α/α offspring. Furthermore, an engineered a/a/α2 strain had a competitive advantage at the time of extreme host morbidity or death when co-injected with its isogenic a/a parent strain in the mouse model, suggesting that the MTL genotype, not associated genes on chromosome 5, is responsible for the competitive advantage. We propose that the MTL genotype regulates C. albicans virulence and that the competitive advantage of the a/α genotype maintains a/α as the predominant genotype in nature, hence contributing to the conservation of the mating system.
MATERIALS AND METHODS
Strain maintenance and culture conditions:
The origins of the strains used in this study are presented in Table 1. Since cells can switch from white to opaque only when they are MTL homozygous, spontaneous MTL-homozygous offspring of each parent a/α strain were isolated by screening for opaque colonies on nutrient agar plates as previously described (Lockhart et al. 2002). The MTL genotypes of a/a or α/α offspring were immediately verified by PCR with primers for MTLa1 and MTLα2, as described below. All strains were maintained as glycerol stocks at −70°. For experimental purposes, cells from glycerol stocks were grown at 25° on agar plates containing the nutrient composition of Lee's medium (Lee et al. 1975) modified according to Bedell and Soll (1979). The agar was further supplemented with phloxine B to identify white-phase colonies (Anderson and Soll 1987) in MTL-homozygous populations.
TABLE 1.
Strains used in this study
| Strain | Reference |
|---|---|
| P37037a/α | Pujol et al. (2003) |
| P37037α/α-1 | Lockhart et al. (2002) |
| P37037α/α-2 | Wu et al. (2004) |
| P37037α/α-3 | Wu et al. (2004) |
| P37037a/a-1 | Wu et al. (2004) |
| P37039a/α | Pujol et al. (2003) |
| P37039α/α-1 | Lockhart et al. (2002) |
| P75063a/α | Pujol et al. (2002) |
| P75063a/a-1 | Lockhart et al. (2002) |
| P34048a/α | Wu et al. (2004) |
| P34048α/α-1 | Wu et al. (2004) |
| P34048a/a-1 | Wu et al. (2004) |
| P37005a/a | Lockhart et al. (2002) |
| P37005a/a/α2 | This study |
Virulence in a systemic mouse model:
Injections were performed as described by Kvaal et al. (1997). Five-day-old white colonies were selected from each strain for analysis. Cells from a colony were grown to late log phase in modified Lee's medium, washed twice in sterile phosphate buffered saline (PBS; 3 mm KCl, 137 mm NaCl, 2 mm KH2PO4, 7 mm NaH2PO4, pH 7.4) and resuspended in PBS at a concentration of 4 × 106 cells/ml. If two strains were mixed 1:1, the concentration of each was 2 × 106 cells/ml. Cell densities were estimated using a hemocytometer. Cell phenotype was checked microscopically to be sure >99% of cells were in the white phenotype (Anderson and Soll 1987; Slutsky et al. 1987). Six- to 8-week-old female ND4 mice (Harlan Sprague, Madison, WI) weighing 21–26 g were injected through their tail vein with 1 × 106 cells. Mice were examined every day. When a mouse showed the first signs of illness (i.e., tremors, hunched back), which we will refer to as “extreme morbidity,” it was euthanized using CO2. A previous study showed that death followed the selected moribund symptoms by 1 day or less (Kvaal et al. 1997). In the majority of cases, mice died. Either one or both kidneys were removed and ground in 2 ml of sterile PBS in a sterile mortar and pestle. Aliquots of each kidney macerate were plated on modified Lee's medium containing phloxine B (Anderson and Soll 1987) and incubated at 25° for 5 days. In control experiments in which mixtures of a/α and MTL-homozygous offspring were injected, we obtained similar results for alternative kidneys of the same animal and alternative organs (kidney vs. liver) (see Table 4). Therefore, using either one kidney or two kidneys ground together provided similar results.
TABLE 4.
a/α strains are more competitive than a/a or α/α offspring in the mouse model for systemic infection
| % genotypec
|
|||||
|---|---|---|---|---|---|
| Strain combinationa | Mouse | No. clones analyzedb | a/α | a/a | α/α |
| P37037a/α + P37037a/a-1 | 1 | 49 | 96 | 4 | 0 |
| 2 | 48 | 100 | 0 | 0 | |
| 3 | 50 | 98 | 2 | 0 | |
| Mean ± SD | 98 ± 2 | 2 ± 2 | 0 | ||
| P37037a/α + P37037α/α-1 | 1 | 50 | 64 | 0 | 36 |
| 2-1d | 50 | 82 | 0 | 18 | |
| 2-2d | 50 | 84 | 0 | 16 | |
| 3 | 50 | 88 | 0 | 12 | |
| Mean ± SD | 80 ± 11 | 0 | 20 ± 11 | ||
| P37037a/α + P37037α/α-2 | 1 | 50 | 100 | 0 | 0 |
| 2 | 50 | 96 | 0 | 4 | |
| Mean ± SD | 98 | 0 | 2 | ||
| P37037a/α + P37037α/α-3 | 1 | 49 | 100 | 0 | 0 |
| 2 | 50 | 100 | 0 | 0 | |
| 3 | 37 | 92 | 0 | 8 | |
| 4e | 45 | 91 | 0 | 9 | |
| Mean ± SD | 94 ± 5 | 0 | 6 ± 5 | ||
| P37039a/α + P37039α/α-1 | 1 | 50 | 98 | 0 | 2 |
| 2 | 50 | 100 | 0 | 0 | |
| 3 | 50 | 100 | 0 | 0 | |
| Mean ± SD | 99 ± 1 | 0 | 1 ± 1 | ||
| P34048a/α + P34048a/a-1 | 1 | 47 | 100 | 0 | 0 |
| 2 | 47 | 98 | 2 | 0 | |
| 3 | 50 | 94 | 6 | 0 | |
| Mean ± SD | 97 ± 3 | 3 ± 3 | 0 | ||
| P34048a/α + P34048αα-1 | 1 | 47 | 100 | 0 | 0 |
| 2 | 50 | 100 | 0 | 0 | |
| 3 | 48 | 88 | 0 | 12 | |
| Mean ± SD | 96 ± 7 | 0 | 4 ± 7 | ||
| P75063a/α + P75063a/a-1 | 1 | 49 | 98 | 2 | 0 |
| 2 | 46 | 87 | 13 | 0 | |
| Mean ± SD | 93 | 8 | 0 | ||
P37037a/a-1, P37037α/α-2, P37037α/α-3, P37039α/α-1, P34048a/a-1, and P34048α/α-1 were spontaneously generated by loss of one chromosome 5 homolog followed by duplication of the retained homolog, while P37037α/α-1 was spontaneously generated by mitotic recombination (Wu et al. 2005).
Clones were taken from the kidney of the injected mouse at the time of extreme host morbidity or death and analyzed for MTL genotype by PCR.
Mean ± SD was computed for each combination.
Mouse 2-1 and 2-2 represent different kidneys of the same mouse.
Mouse 4 is actually the liver from mouse 3 macerated and analyzed.
PCR analysis:
Following growth, 50 individual colonies were picked with sterile toothpicks and individually streaked on YPD (2% dextrose, 2% peptone, 1% yeast extract) agar plates. After 2 days, individual round colonies were picked, streaked on fresh nutrient agar plates to assure clonality, and grown for 2 days. Cells were then resuspended in 20 μl of sterile water in a 250-μl microfuge tube. The tube was heated to 94° for 6 min and then placed in a −70° freezer. To initiate a polymerase chain reaction (PCR) assay, the preparation was pelleted and 4 μl of cell supernatant, 2.5 μl of 10 × PCR buffer provided by the manufacturer (Invitrogen, Carlsbad, CA), 0.75 μl 50 mm MgCl2, 0.25 μl 10 mm dNTPs, 0.5 μl each of 5 μm forward and reverse primers, 0.15 μl Taq DNA polymerase (Invitrogen), and water were added to a fresh tube to a final volume of 25 μl. After an initial denaturation step at 94° for 5 min, the following reaction conditions were used for 40 cycles: 94° for 1 min, 47° for 1 min, and 68° for 1 min. A final elongation step at 72° for 7 min completed the reaction. If a preparation gave no PCR result, it was reanalyzed by PCR using fresh colonies.
Generation of an a/a/α2 strain:
To generate an a/a/α2 strain from the natural a/a strain P37005, which did not contain any auxotrophic markers, a plasmid that contained the mycophenolic acid resistance allele was first constructed. The mycophenolic acid resistance allele of the IMH3 gene was released by XhoI digestion from plasmid p3408 (Beckerman et al. 2001), a generous gift from P. T. Magee at the University of Minnesota. The sticky ends of the 2.7-kb fragment were blunt ended and then subcloned into the plasmid pCaExp at the Bg1II site to generate the plasmid pCaM. pCaExp (Care et al. 1999), a generous gift from P. Sudbery at Sheffield University, is an integrative plasmid designed to allow expression of a selected gene under the control of the MET3 promoter. It contains the C. albicans RP10 gene for site-directed integration via homologous recombination. MTLα2 was cloned by PCR using the primers 5′-ATT GGA TCC ATG AAT TCA CAT CTG GAG GCA-3′ and 5′-ATT CTG CAG TTA ACC TGT TAA TAG CAA AGC-3′, which contain engineered BamHI and PstI sites, respectively. Following digestion with BamHI and PstI, the MTLα2 fragment was ligated to BamHI- and PstI-digested pCaM, downstream of the MET3 promoter. The MET3 promoter was employed because it is leaky and allowed expression of MTLα2, as demonstrated by RT-PCR (see results). Sequence and orientation of the resulting plasmid pCaMα2 were confirmed by sequencing (data not shown).
To generate an a/a/α2 strain, 25 μg of pCaMα2 were linearized at the NcoI site within the RP10 gene, which encodes a ribosomal protein (Care et al. 1999). The RP10 gene has been identified as a neutral site for integration (Swoboda et al. 1995). The linearized DNA preparation was transformed into strain P37005, a natural a/a strain (Lockhart et al. 2002), using the lithium acetate method (Schiestl and Gietz 1989). Transformants were selected for mycophenolic acid resistance by growing cells on minimal medium supplemented with 5 μg/ml of mycophenolic acid (Sigma, St. Louis). Selected transformants were confirmed to be single-copy integrations at the RP10 locus by Southern analysis. Reverse transcription-polymerase chain reaction (RT-PCR) analysis was used to confirm MTLα2 gene expression.
RT-PCR:
To analyze expression of MTLa1, MTLα2, MTLα1 and the constitutively expressed gene HIS3, RT-PCR was performed according to the protocol of the Access RT-PCR System provided by Promega (Madison, WI). The primers are described in Table 2. RNA was extracted according to methods previously described (Lockhart et al. 2003a,b). Prior to RT-PCR, RNA samples were treated with RNase-free DNase (RQ1, Promega) at 37° for 1 hr to remove DNA contamination. One-tenth of a microgram of RNA was used as template for each reaction. Reverse transcription was performed at 48° for 45 min, immediately followed by denaturation at 94° for 5 min. The denatured template was then subjected to the following reaction regimen: 40 cycles at 94° for 30 sec, 45° for 1 min, and 68° for 2 min. The final elongation reaction was performed at 68° for 7 min. RT-PCR of the constitutively expressed C. albicans HIS3 gene served as a control. All PCR and RT-PCR reactions were performed in a Programmable Thermal Block II thermal cycler (Lab-line Instruments, Melrose Park, IL).
TABLE 2.
Primers used for RT-PCR
| Gene | Name | Sequence |
|---|---|---|
| MTLa1 | a1SmaIFT | 5′-ATC CCC CGG GAA TGA ACT CAG AAA TAG A-3′ |
| a1SmaIR | 5′-TCC CCC GGG CTA GGT TGA ATT TGA ACT-3′ | |
| MTLα2 | α2BamHIF | 5′-ATT GGA TCC ATG AAT TCA CAT CTG GAG GCA-3′ |
| α2PstIR | 5′-ATT CTG CAG TTA ACC TGT TAA TAG CAA AGC-3′ | |
| MTLα1 | α1BamH1F | 5′-CAG GGA TCC TGG CTT CAA CAG ATA TGG GAA-3′ |
| α1PstIR | 5′-TAA CTG CAG TTA CTT CAT TAT GTA AAC ATC-3′ | |
| HIS3 | HIS3F | 5′-ATG TCA CGA GAA GCT TTA-3′ |
| HIS3R | 5′-TCT ACT CAA TGC TTC ATC-3′ |
RESULTS
a/α strains are more virulent than their a/a or α/α offspring:
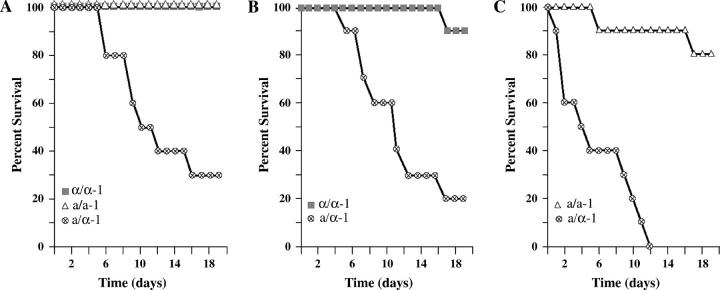
Kill curves were generated for mice injected with a/α strains alone or with their a/a or α/α offspring alone. The three tested parent strains and their offspring were as follows: P37037a/α, P37037a/a-1, and P37037α/α-1; P37039a/α and P37039α/α-1; and P75063a/α and P75063a/a-1. In each of the three tested combinations, the parent a/α strain killed mice much faster than the MTL-homozygous offspring, whether a/a or α/α (Figure 1, A–C). While the a/α parent strains P37037a/α, P37039a/α, and P75063a/α caused 50% host death after 10, 12, and 5 days, respectively, their MTL-homozygous offspring caused only 0%, 10%, and 20% host death, respectively, after 17 days (Figure 1, A–C, respectively). Natural a/α strains were, therefore, consistently far more virulent in this model than their MTL-homozygous offspring.
Figure 1.—
MTL-heterozygous (a/α) strains of Candida albicans are more virulent than their a/a and α/α offspring in the mouse model for systemic infection. For each strain, 10 mice were each injected with 1 × 106 cells. The percentage of surviving animals is plotted as a function of time for each tested strain. (A) P37037, (B) P37039, and (C) P75063 are independent a/α strains. The a/a and α/α offspring appeared spontaneously in vitro.
Stability of MTL heterozygotes in the mouse model:
To conclude that a/α strains were more virulent than a/a or α/α strains in single-strain infections, and to justify competition experiments between a/α and either a/a or α/α strains, we first had to demonstrate that the a/α genotype was stable in vivo—i.e., that a/α strains did not undergo high levels of homozygosis after injection into a mouse. To assess stability, cells of strains P37037a/α, P37039a/α, and P75063a/α were individually injected into mice. At the time of host death, 50 clones from the kidney macerate of each mouse were analyzed for MTL zygosity by PCR. One-hundred percent of the clones derived from kidneys of mice injected individually with strains P37037a/α and P37039a/α were a/α (Table 3). In the case of strain P75063a/α, 100% and 86% were a/α in the two test mice (Table 3). P75063 was previously observed to undergo very high rates of spontaneous MTL homozygosis in vitro (Wu et al. 2005). These results demonstrate that the a/α genotype is relatively stable during the course of a mouse model experiment.
TABLE 3.
MTL heterozygosity (a/α) is relatively stable in the mouse model for systemic infection
| % clones
|
|||||
|---|---|---|---|---|---|
| Experiment | Strain | No. of clones analyzeda |
a/α | a/a | α/α |
| 1 | P37037 | 50 | 100 | 0 | 0 |
| 2 | P37037 | 50 | 100 | 0 | 0 |
| 3 | P37039 | 50 | 100 | 0 | 0 |
| 4 | P37039 | 50 | 100 | 0 | 0 |
| 5 | P75063 | 50 | 100 | 0 | 0 |
| 6 | P75063 | 50 | 86 | 14 | 0 |
Clones were taken from the kidney of the injected mouse at the time of extreme host morbidity or death and analyzed for MTL zygosity by PCR.
Co-injection of a/α and their a/a or α/α offspring:
The kill curves for mice injected with single strains (Figure 1) revealed that a/α strains were more virulent than their a/a or α/α offspring. To test for competitiveness, equal numbers of a/α and either an a/a or an α/α offspring were co-injected into mice, and the proportions of MTL genotypes were tested at the time of extreme host morbidity or death by PCR analysis of ∼50 random clones, which were isolated from the kidneys of test animals. Four unrelated a/α strains and their respective offspring were analyzed (Table 4). For the a/α strain P37037a/α, one a/a and three α/α offspring were analyzed in mixtures with the a/α parent strain, and for strain P34048a/α, one a/a and one α/α offspring were analyzed in mixtures with the a/α parent strain. For two additional a/α strains, P37039a/α and P75063a/α, one α/α and one a/a strain, respectively, were analyzed in mixture. Multiple mice were tested for each mixture. In all eight tested mixtures, the genotype of the majority of yeast at the time of extreme host morbidity or death was a/α (Table 4). As controls in one combination (P37037a/α + P37037α/α-1), the two kidneys of one animal (2-1 and 2-2) were macerated separately and a PCR analysis was performed on yeast clones from each. In another combination (P37037a/α + P37037 α/α-3), yeast clones from the liver were tested. In all of these controls, a/α again predominated (Table 4). These results indicate that when mixtures of a/α cells and either their a/a or α/α offspring are co-injected with equal cell numbers into a host, the a/α parent strain had a competitive advantage in every case at the time of host death. Growth experiments in liquid nutrient medium or on agar failed to reveal a growth advantage for a/α cells, at least in vitro (data not shown).
Co-injection of a/a and α/α offspring:
Experiments in which a/α cells and their MTL-homozygous offspring were co-injected into the mouse model revealed that cells of every tested a/α strain exhibited a competitive advantage in the infection at the time of host death. These results revealed no differences between a/a and α/α offspring. To test for such differences, a/a and α/α cells from the same a/α parent were co-injected and colonization of the kidneys analyzed either at the time of extreme host morbidity or death or after 17 days, when surviving animals were killed. Three to six animals were injected in each cross, and 50 clones were analyzed for MTL genotype at the time of extreme host morbidity or death. Crosses were performed between P34048a/a-1 and P34048α/α-1 and between P37037a/a-1 and P37037 α/α-1, P37037α/α-2, or P37037α/α-3. In the case of P34048, the proportions of a/a and α/α cells at the time of host death were 38 ± 14% and 62 ± 14%, respectively. For the mixture P37037a/a-1 and P37037 α/α-1, the proportions of a/a and α/α cells at the time of host death or killing were 1 ± 2% and 99 ± 2%, respectively. However, for the mixtures P37037a/a-1 and either P37037α/α-2 or P37037α/α-3, the proportions of a/a and α/α cells at the time of host death in both cases were 100 and 0%, respectively. While P37037 α/α-1 was spontaneously generated by mitotic recombination, P37037α/α-2 and P37037α/α-3 were spontaneously generated by the loss of one chromosome 5 homolog followed by duplication of the retained homolog (Wu et al. 2005). Although these results demonstrate that, within a strain, either an a/a or an α/α derivative can have a competitive advantage in a mixed infection, there was no universal advantage by either MTL genotype over the other, as there was for the MTLa/α genotype over MTL-homozygous offspring.
Isogenic a/a vs. a/a/α2 competition:
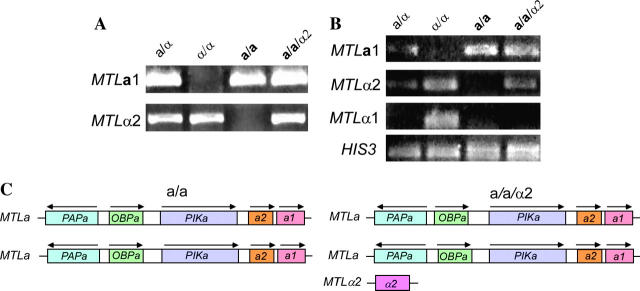
Our results demonstrated that a/α strains are more competitive than their a/a or α/α offspring in mixed systemic infections. Our results, however, did not distinguish between an advantage due to the heterozygosity of genes linked to the MTL locus on chromosome 5 (Wu et al. 2005) vs. an advantage due solely to heterozygosity at the MTL locus. To distinguish between these alternatives, we created an isogenic a/a/α2 strain from the a/a strain P37005 (Figure 2). This strain was generated by integrating a plasmid containing MTLα2 under control of the MET3 promoter and a dominant selection marker, the mycophenolic resistance allele of the IMH3 gene (Beckerman et al. 2001) at the neutral RP10 gene (Swoboda et al. 1995; Care et al. 1999). Hence, neither parent nor offspring harbored auxotrophic genes, a requirement for mouse model experiments. An RT-PCR analysis of the low level MTLa1 and MTLα2 transcripts from the parental and transformed strain revealed that both MTLa1 and MTLα2 were expressed in the a/a/α2 derivative (Figure 2B). When P37005a/a and P37005a/a/α2 cells were co-injected into the mouse model, a/a/α2 cells exhibited a competitive advantage at the time of host death (Table 5). These results suggest that it is the MTL genotype, not allelism of genes linked to MTL on the chromosome 5 homologs, that provides the competitive advantage exhibited by a/α strains over their a/a or α/α offspring.
Figure 2.—
Strain P37005 a/a/α2 was generated from the natural C. albicans strain P37005 a/a, using a dominant selection marker, the mycophenolic resistance allele of the IMH3 gene (Beckerman et al. 2001). (A) MTL genotyping by PCR demonstrated the addition of MTLα2 in the a/a/α2 strain. (B) RT-PCR demonstrated that while the a/a parent expressed only MTLa1, the a/a/α2 derivative expressed MTLa1 and MTLα2. The constitutively expressed gene HIS3 was assayed as a control. (C) The a/a/α2 derivative contains two copies of the entire MTLa locus and one copy of the MTLα2 gene.
TABLE 5.
The a/a/2 derivative of strain P37005a/a-1 has a competitive advantage in the mouse model for systemic infection
| % genotype reserved
|
|||||
|---|---|---|---|---|---|
| Strain combination | Mouse | No. clones analyzeda |
α/α | a/a | a/a/α2 |
| P37005a/a-1 + P37005a/a/α2 | 1 | 50 | 0 | 34 | 66 |
| 2 | 49 | 0 | 4 | 96 | |
| 3 | 47 | 0 | 6 | 94 | |
| Mean ± SD | 15 ± 17 | 85 ± 17 | |||
Clones were taken from the kidney of the injected mouse at the time of extreme host morbidity or death and analyzed for MTL genotype by PCR.
DISCUSSION
Our results first demonstrate that a/a and α/α offspring are far less virulent than their parental a/α strains when tested alone for the time of extreme host morbidity or death in the mouse model of systemic infection. This held true for three unrelated a/α strains and their MTL-homozygous offspring. It also held true for strains from different clades. P37037 is a member of clade I, while P75063 is a member of clade SA (Blignaut et al. 2002; Pujol et al. 2002, 2003; Soll and Pujol 2003). More importantly, in mixing experiments of four different a/α strains with their a/a or α/α offspring, the a/α parent strain in each case exhibited a strong competitive advantage at the time of extreme host morbidity or death. In the case of strain P37037, this held true for an α/α offspring (P37037α/α-1) that was generated spontaneously by mitotic recombination along chromosome 5, and for α/α offspring (P37037α/α-2 and P37037α/α-3) that were generated spontaneously by loss of one chromosome 5 homolog followed by duplication of the retained homolog (Wu et al. 2005). Mixing experiments of a/a and α/α offspring of the same parent strains revealed no universal advantage for either one, as we found for the a/α genotype when mixed with MTL-homozygous offspring.
Two alternative explanations could explain the increased virulence and competitive advantage of a/α strains over their a/a or α/α offspring in the mouse model. First, heterozygosity at the MTL locus alone may be the basis. MTL heterozygosity has been demonstrated to regulate genes associated with the mating process and to suppress phenotypic switching (Lockhart et al. 2002, 2003; Miller and Johnson 2002). Our combined results suggest that MTL heterozygosity is also involved in the regulation of genes that confer virulence and a competitive advantage in colonization. Alternatively, virulence and competitiveness could be conferred by other genes linked to the MTL locus along chromosome 5. A recent analysis revealed heterozygosity for a number of genes other than the MTL locus along chromosome 5, and association of particular alleles of these genes with either MTLa or MTLα (Wu et al. 2005). In this scenario, MTLa-linked and MTLα-linked alleles of different genes would combine to confer the a/α advantage. To test between these alternatives, we engineered an isogenic a/a/α2 strain from an a/a strain, co-injected the transformant and parent strain into mice, and analyzed the genetic composition of infecting yeast at the time of extreme host morbidity or death. The a/a/α2 strain exhibited a competitive advantage, supporting the hypothesis that the a/α genotype, not MTLa-linked or MTLα-linked genes, confers virulence and the competitive advantage of a/α strains over MTL-homozygous offspring in host colonization. Further experiments are now in progress to test this hypothesis.
Although we found that in mixed infections, a/α strains have a strong competitive advantage over either their a/a or α/α offspring, and that a/α strains are far more virulent than their MTL-homozygous offspring in the mouse model, we also know that ∼3% of natural strains are MTL homozygous and therefore are successful in nature. Some of these strains, such as strain WO-1, which is α/α (Lockhart et al. 2002), are quite virulent in the mouse model for systemic infection (Kvaal et al. 1997), suggesting that they have overcome the loss of MTL heterozygosity, presumably by compensatory changes in genes other than those at the MTL locus. However, the great majority of strains in nature are a/α (Lockhart et al. 2002), suggesting a general advantage to this genotype. We therefore propose that MTL heterozygosity in C. albicans plays a fundamental role not only in suppressing α-specific and “haploid”-specific genes (Johnson 2003) and phenotypic switching (Lockhart et al. 2002; Miller and Johnson 2002), but also in virulence. We further suggest that MTL heterozygosity, presumably through the regulation of genes involved in host colonization, provides a competitive advantage to a/α cells over MTL-homozygous offspring, which conserves a/α cells and hence the mating system in nature.
Acknowledgments
This research was supported by National Institutes of Health grant AI2392.
References
- Anderson, J. M., and D. R. Soll, 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169: 5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman, J., H. Chibana, J. Turner and P. T. Magee, 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, G., and D. R. Soll, 1979. The effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc resistant and zinc sensitive pathways for mycelium formation. Infect. Immun. 26: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blignaut, E., C. Pujol, S. Lockhart and D. R. Soll, 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J. Clin. Microbiol. 40: 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care, R. S., J. Trevethick, K. M. Binley and P. E. Sudbery, 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34: 792–798. [DOI] [PubMed] [Google Scholar]
- Graser, Y., M. Volovsek, J. Arrington, G. Schonian, W. Presber et al., 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93: 12473–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32: 561–599. [DOI] [PubMed] [Google Scholar]
- Hull, C. M., and A. D. Johnson, 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285: 1271–1275. [DOI] [PubMed] [Google Scholar]
- Hull, C. M., R. M. Raisner and A. D. Johnson, 2000. Evidence for mating of the “asexual” yeast Candida albicans in mammals. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Johnson, A., 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1: 106–116. [DOI] [PubMed] [Google Scholar]
- Kvaal, C. A., T. Srikantha and D. R. Soll, 1997. Misexpression of the white phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65: 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. L., H. R. Buckley and C. C. Campbell, 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13: 148–153. [DOI] [PubMed] [Google Scholar]
- Lockhart, S. R., C. Pujol, K. Daniels, M. Miller, A. Johnson et al., 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels and D. R. Soll, 2003. a Cell biology of mating in Candida albicans. Eukaryot. Cell 2: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., R. Zhao, K. J. Daniels and D. R. Soll, 2003. b α-Pheromone-induced shmooing and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2: 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, B. B., and P. T. Magee, 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Miller, M. G., and A. D. Johnson, 2002. White-opaque switching in Candida albicans is controlled by the mating type (MTL) locus and allows efficient mating. Cell 110: 293–302. [DOI] [PubMed] [Google Scholar]
- Pujol, C., J. Reynes, F. Renaud, M. Rayond, M. Tibayrenc et al., 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90: 9456–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, C., M. Pfaller and D. R. Soll, 2002. Ca3 fingerprinting of Candida albicans blood stream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 40: 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, C., S. A. Messer, M. Pfaller and D. R. Soll, 2003. Drug resistance is not directly affected by mating type locus zygosity in Candida albicans. Antimicrob. Agents Chemother. 47: 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, C., M. A. Pfaller and D. R. Soll, 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine, J., J. N. Strathern, J. B. Hicks and I. Herskowitz, 1979. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics 93: 877–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R. H., and R. S. Gietz, 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16: 339–346. [DOI] [PubMed] [Google Scholar]
- Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller et al., 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll, D. R., 2004. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. BioEssays 26: 10–20. [DOI] [PubMed] [Google Scholar]
- Soll, D. R., and C. Pujol, 2003. Candida albicans clades. FEMS Immunol. Med. Microbiol. 39: 1–7. [DOI] [PubMed] [Google Scholar]
- Swoboda, R. K., I. D. Broadbent, G. Bertram, S. Budge, G. W. Gooday et al., 1995. Structure and regulation of a Candida albicans RP10 gene which encodes an immunogenic protein homologous to Saccharomyces cerevisiae ribosomal protein 10. J. Bacteriol. 177: 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W., C. Pujol, S. R. Lockhart and D. R. Soll, 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 169: 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., T. G. Mitchell and R. Vilgalys, 1999. PCR-restriction fragment length polymorphism (RFLP) analyses reveal both extensive clonality and local genetic differences in Candida albicans. Mol. Ecol. 8: 59–73. [DOI] [PubMed] [Google Scholar]