Abstract
We developed a classification approach to multiple quantitative trait loci (QTL) mapping built upon a Bayesian framework that incorporates the important prior information that most genotypic markers are not cotransmitted with a QTL or their QTL effects are negligible. The genetic effect of each marker is modeled using a three-component mixture prior with a class for markers having negligible effects and separate classes for markers having positive or negative effects on the trait. The posterior probability of a marker's classification provides a natural statistic for evaluating credibility of identified QTL. This approach performs well, especially with a large number of markers but a relatively small sample size. A heat map to visualize the results is proposed so as to allow investigators to be more or less conservative when identifying QTL. We validated the method using a well-characterized data set for barley heading values from the North American Barley Genome Mapping Project. Application of the method to a new data set revealed sex-specific QTL underlying differences in glucose-6-phosphate dehydrogenase enzyme activity between two Drosophila species. A simulation study demonstrated the power of this approach across levels of trait heritability and when marker data were sparse.
THE fact that we can map variation in complex phenotypes to chromosomal regions by exploiting the linkage between random genetic markers and causal genetic variants in related individuals has long been understood. Since the formalization of statistical approaches to this type of inference by Lander and Botstein (1989) and the advent of high-throughput methodologies for constructing genetic maps with high marker density, quantitative trait locus (QTL) mapping in organisms from crops to mice has provided a rich knowledge of genes underlying important socioeconomic traits. It also has provided a better understanding of the genetic architecture of complex traits both within and between species. QTL mapping promises the improvement of crops of international importance, such as drought-resistant rice (for review see Price and Courtois 1999; Price et al. 2002), and the advancement of treatments for complex physiological diseases like high blood pressure (Sugiyama et al. 2001). QTL mapping has also been used to map traits that may be the target of intense selection both in natural populations, such as sexually dimorphic pigmentation patterns in Drosophila (Kopp et al. 2003), and in crop domestication (Doebley and Stec 1991). As such, QTL mapping is not simply a gene-finding tool. QTL mapping provides critical information regarding quantitative evolutionary genetic processes.
Traditional approaches to QTL mapping primarily involve multiple regression models and maximum-likelihood estimation and are powerful for detecting QTL of moderate to large effect. However, detecting multiple smaller genetic effects that may modify or interact with larger effects is necessary and remains a challenge. These smaller effects are important, as they can potentially enhance crop breeding and further our understanding of genetic background effects on complex disease. Quantifying the abundance of these types of effects for any given trait also fills a gap in our knowledge regarding the distribution of genetic effects.
The most popular approach for QTL mapping is interval mapping (IM). Proposed by Lander and Botstein (1989), IM conducts likelihood-ratio tests for each possible QTL by densely gridding chromosomes using linkage information in the available marker data. It tacitly assumes that the trait of interest is regulated by a single gene. Under this single-QTL model, IM may fail to separate closely linked QTL and instead report ghost QTL that have no true effect on the trait (Knott and Haley 1992; Martinez and Curnow 1992; Wright and Kong 1997). Furthermore, epistatic interactions between QTL are not identified by IM. Many approaches have therefore been developed on the basis of multiple-QTL models that generalize the single-QTL model. Conditioning on selected markers outside a region of interest to account for background effects, composite-interval mapping (CIM) and multiple-QTL mapping (MQM) search for QTL across a series of intervals covering chromosomes (Jansen 1993; Zeng 1993, 1994; Jansen and Stam 1994). Multiple-interval mapping (MIM) directly regresses the trait on a set of markers, which densely grid the chromosomes (Kao et al. 1999). Identification of multiple QTL is subject to the statistical issue of variable selection (Piepho and Gauch 2001; Broman and Speed 2002; Sillanpää and Corander 2002), and Bayesian methodology using Markov chain Monte Carlo algorithms has been developed for this problem (Satagopan et al. 1996; Sillanpää and Arjas 1998; Stephens and Fisch 1998; Ball 2001; Sen and Churchill 2001; Xu 2003; Yi et al. 2003).
The Bayesian approach provides a natural framework for modeling multiple QTL, as it can accommodate multiple imputation of missing values in phenotypes as well as genotypes and include all markers as random variables in a single model. The ability to incorporate available information into QTL mapping and update with newly observed data is an advantage provided uniquely by Bayesian analysis. Access to powerful computational resources and efficient algorithms makes it realistic to implement Bayesian analysis, and the direct interpretation of the results from a Bayesian analysis also makes it particularly applicable for the scientific community (Shoemaker et al. 1999; Beaumont and Rannala 2004).
Many Bayesian QTL-mapping methods capitalize on the complex reversible-jump Markov chain Monte Carlo algorithm (Green 1995) to estimate the number of QTL and their effects on the trait (Satagopan et al. 1996; Sillanpää and Arjas 1998; Stephens and Fisch 1998). To avoid the problematic issue of Markov chain mixing introduced by uncertain dimensionality of parameter space, Yi et al. (2003) developed an alternative Bayesian method for identifying multiple QTL in experimental designs based on stochastic search variable selection (George and McCulloch 1993). For those markers that have negligible effects on the trait, they assume the effects follow mean-zero Gaussian distributions with arbitrarily specified small standard deviations. In this way the dimension of the parameter space is fixed and a more tractable Gibbs sampler can be constructed. The posterior probability that a marker has a large effect is estimated and used to indicate significance of QTL. However, by using Gaussian distributions with small standard deviations to model negligible effects, Yi et al. (2003) reduce the efficiency in the mapping procedure, resulting in small posterior probabilities for the effects of QTL on the trait even if the corresponding effects are large.
We propose a new Bayesian framework to identify multiple QTL. We categorize all genetic markers into three classes, a positive-effect class (including all QTL that have detectable positive effects on the phenotypic values), a negative-effect class (including all QTL that have detectable negative effects on the phenotypic values), and a negligible-effect class (including all non-QTL markers and all nondetectable QTL). In modeling the population distribution for each class, we construct a three-component mixture prior distribution for the effect of each investigated marker. The proposed procedure is able to incorporate the a priori information that most of the markers under investigation have negligible effect on the trait and that the positive-effect class and negative-effect class may have different sizes. Two truncated Gaussian distributions are used to model the population distributions for the positive-effect class and negative-effect class. Using an a priori inverse gamma distribution for their variance parameters, the corresponding prior distributions are essentially truncated t-type distributions so as to be sufficiently flexible heavy-tailed prior distributions. This incorporates the empirical observation that the distribution of genetic effects is heavy tailed (Lopez and Lopez-Fanjul 1993; Keightley 1994; Keightley and Ohnishi 1998). These partially informative prior distributions not only shrink the estimates of the QTL effects toward zero to avoid the “curse of dimensionality,” but also allow for the estimation of the a posteriori probabilities that a marker belongs to the positive-effect class, the negative-effect class, or the negligible-effect class. Although point estimates of these a posteriori probabilities provide information to discover the corresponding effects' classes (as in Yi et al. 2003), the distributional departure from probability 0.5 delivers additional information to help investigators make informed decisions when determining QTL significance. As a graphical display, we propose a “heat map” to visually display the posterior probabilities of membership in the positive-, negative-, or negligible-effect class.
To validate our proposed approach we analyzed publicly available data from a study of agronomic traits in a doubled-haploid (DH) population of barley (North American Barley Genome Project). Data sets simulated across three trait heritabilities suggest that the proposed approach is powerful for detecting a broad range of QTL effects, even when genotype data are missing. As a further application, we used the method to detect sex-specific QTL underlying glucose-6-phosphate dehydrogenase activity in a set of recombinant inbred introgression lines between Drosophila simulans and D. sechellia.
THE MODEL AND BAYESIAN CLASSIFICATION
Multiple-linear-regression model:
We focus on mapping multiple QTL in a set of homozygous lines, such as doubled-haploid lines or recombinant inbred lines, generated from an initial cross between two isogenic parental lines. In practice this model could be extended to include inferences from crosses with resulting heterozygous individuals, such as backcrosses or intercrosses. Assume genotypic data for m markers and phenotypic data for one complex trait of interest are collected from n individuals. Further assume the m markers are densely located on the chromosomes of interest such that putative QTL will be cotransmitted with some of these m markers. Subject to additive main effects from putative QTL, the phenotypic value of individual i (yi) is modeled as
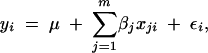 |
1 |
where μ is the overall mean, xji is the genotypic value of the jth marker of individual i, and εi is the disturbance error from environmental factors, which is assumed to be distributed as N(0, σ2ε). Therefore, βj describes the main effect of the jth putative QTL.
When the markers are widely spaced across the genome, we can tightly grid the genome by imputing genotypes between markers (Lander and Botstein 1989; Ball 2001; Sen and Churchill 2001; Kilpikari and Sillanpää 2003; Xu 2003). This is equivalent to assuming that the genotypic values of some markers are missing for all individuals. In practice, some marker genotypes are also partially missing. All of these missing genotypic values can be inferred using the known linkage information and the available marker genotype data (see Jiang and Zeng 1997). This model can incorporate both observed and imputed marker information.
Identifying QTL from the markers under investigation using the above multiple-linear-regression model is equivalent to selecting variables xji, which have nonzero coefficients βj. Although previous approaches for QTL mapping have considered classical model selection approaches in statistics (e.g., Kao et al. 1999; Zeng et al. 1999; Ball 2001; Broman and Speed 2002), effects of imputed missing values on model selection have been largely ignored due to the potential difficulty. Classical model selection approaches are severely challenged when there are numerous highly correlated markers and a small sample size. We therefore propose a Bayesian classification method that incorporates the important prior information that the QTL effects of most genotypic markers are negligible and naturally exploits the linkage information in the genetic linkage map to impute missing values.
Bayesian framework:
We first classify all markers under investigation into three classes, the positive-effect class 𝒫(β) = {j : βj > 0}, the negative-effect class 𝒩(β) = {j : βj < 0}, and the negligible-effect class 𝒵(β) = {j : βj = 0}. Therefore, for each j in 𝒩(β) or 𝒫(β), the corresponding marker has a negative or positive effect on the trait, respectively, and for each j in 𝒵(β) the corresponding marker has no detectable effect on the trait. Often, many markers may belong to the negligible-effect class 𝒵(β), and the sizes of the positive-effect class and the negative-effect class may be small and varied. Classifying effects into three classes provides the foundation for modeling and incorporating prior information as shown below.
Assume the population distribution for the positive-effect class and the negative-effect class to be Fβ+ and Fβ−, respectively. Let pβ+ be the probability for any marker to be included in 𝒫(β) and pβ− be the probability for any marker to be included in 𝒩(β). Then, each βj with j ∈ 𝒫(β) [or j ∈ 𝒩(β)] can be considered as independently sampled from an unknown distribution Fβ+ (or Fβ−). Hence, we have a three-component mixture prior distribution for the effect of each marker; that is,
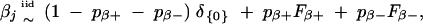 |
2 |
where δ{0} is a Dirac function with value one at zero and value zero otherwise. This three-component mixture prior distribution is able to incorporate the a priori information that most of the markers under investigation have negligible effects on the trait and that the sizes of the positive-effect class and negative-effect class may be different. Note that this prior does not use indicators to specify each marker's classification and avoids the unnecessary sampling of the indicator variables in the Gibbs sampler.
In practice, we can simply take 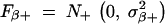 ,
, 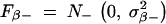 . The probability density functions of the two truncated Gaussian distributions N+(μ, σ2) and N−(μ, σ2) are, respectively,
. The probability density functions of the two truncated Gaussian distributions N+(μ, σ2) and N−(μ, σ2) are, respectively,
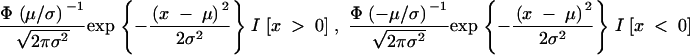 |
3 |
The generality of the above priors can be guaranteed by putting a further hierarchy of prior distributions on the hyperparameters σ2β+ and σ2β−; that is, assuming the prior distributions
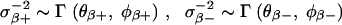 |
4 |
These priors (e.g., setting θβ+ = θβ− = 0.5 and φβ+ = φβ− = 2 for χ21-distributions) lead to truncated t-type distributions that are heavy tailed for the positive βj and negative βj, respectively. They will shrink the estimated effects toward zero but at the same time provide the flexibility to model the population distributions for 𝒫(β) and 𝒩(β). Furthermore, t-type prior distributions confer desirable decision-theoretic properties for the Bayes estimators (Fourdrinier et al. 1998).
Results from previous QTL mapping may provide information about the probability of a marker having a positive, negative, or negligible effect on the trait. This a priori information may be incorporated into the following conjugate prior distribution for pβ+ and pβ−,
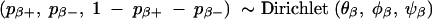 |
5 |
In the case that no prior information is available for pβ+ and pβ−, we can assume each is uniformly distributed on the interval [0, 1] [i.e., the joint Dirichlet(1, 1, 1) distribution, which describes the characteristics of no prior information]. Typically the number of markers m is large relative to the sample size n, and it is unrealistic to assume both pβ+ and pβ− are uniformly distributed on the interval [0, 1]. Instead, we can restrict both pβ+ and pβ− to be smaller than min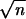 /m, 1. This restriction also accounts for the sample size. Accordingly, the prior distribution for pβ+ and pβ− should follow a truncated Dirichlet distribution. The intercept μ has a uniform prior while σ2ε has a prior proportional to 1/σ2ε, both of which are noninformative. These priors, together with priors defined by (2)–(5), provide a proper joint posterior distribution for the model (1), which is shown in the appendix.
/m, 1. This restriction also accounts for the sample size. Accordingly, the prior distribution for pβ+ and pβ− should follow a truncated Dirichlet distribution. The intercept μ has a uniform prior while σ2ε has a prior proportional to 1/σ2ε, both of which are noninformative. These priors, together with priors defined by (2)–(5), provide a proper joint posterior distribution for the model (1), which is shown in the appendix.
Single-site Gibbs sampler:
A single-site Gibbs sampler can be developed following the above formulation of the Bayesian model. Let yn collect all phenotypic values of the trait and xn collect all genotypic values of the m putative QTL. Let β = (β1, … , βm), β−j be β excluding βj, and x−j,i = (x1i, … , xj−1,i, xj+1,i, … , xmi). Each iteration of the Gibbs sampler proceeds by recursively drawing each missing genotypic value and each parameter value from its full conditional posterior distribution. Details for the implementation of the Gibbs sampler with the imputation of missing genotypic values are presented in the appendix.
This Gibbs sampler starts from initial values for missing genotypic values and all other parameters. Initial values for missing genotypes can be sampled on the basis of the nearest neighboring observed genotypic values and available genetic linkage information. Initial values for μ and σ2ε can simply take the sample mean and variance of yn. Regressing the phenotypic value of the trait only on the jth genotypic value provides suitable initial values for the βj. Then, the initial values for σ2β+ and σ2β− can be calculated by using min2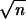 , m components of the initial values of β, which have the largest absolute values.
, m components of the initial values of β, which have the largest absolute values.
Starting from these initial values and running the Gibbs sampler for a sufficient burn-in period (5000 steps in our analysis), the Gibbs sampler reaches stationarity that can be confirmed by diagnostic tools (Cowles and Carlin 1996). Each subsequent iteration of the Gibbs sampler provides a random draw of the missing values and all other parameters from their posterior distributions. All the draws after the burn-in period form a multivariate Markov chain on which inferences can be based.
Marker classification and effect estimation:
After the sufficient burn-in period, we run the above Gibbs sampler for T additional iterations. Then, for each βj, we have two assumably stationary chains, i.e., 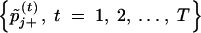 and
and 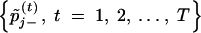 , from
, from
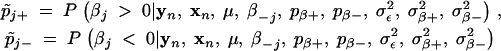 |
The chain 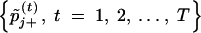 or
or 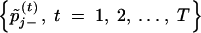 can be used to evaluate whether the jth marker has a positive or negative effect on the trait, respectively. Furthermore, the posterior probabilities pj+ = P(βj > 0|yn, xn) and pj− = P(βj < 0|yn, xn) can be estimated from these two chains, and it is these posterior probabilities that provide information on the classification of markers into the positive- and negative-effect classes. In other words, these posterior probabilities can be used as statistics for evaluation of whether or not a marker is linked to a QTL for the trait of interest. A value of the posterior probability pj+ > 0.5 indicates that the jth marker has a positive effect on the trait, while a value of pj− > 0.5 indicates a negative effect of the jth marker on the trait. Otherwise, we infer that the jth marker has a nondetectable effect on the trait.
can be used to evaluate whether the jth marker has a positive or negative effect on the trait, respectively. Furthermore, the posterior probabilities pj+ = P(βj > 0|yn, xn) and pj− = P(βj < 0|yn, xn) can be estimated from these two chains, and it is these posterior probabilities that provide information on the classification of markers into the positive- and negative-effect classes. In other words, these posterior probabilities can be used as statistics for evaluation of whether or not a marker is linked to a QTL for the trait of interest. A value of the posterior probability pj+ > 0.5 indicates that the jth marker has a positive effect on the trait, while a value of pj− > 0.5 indicates a negative effect of the jth marker on the trait. Otherwise, we infer that the jth marker has a nondetectable effect on the trait.
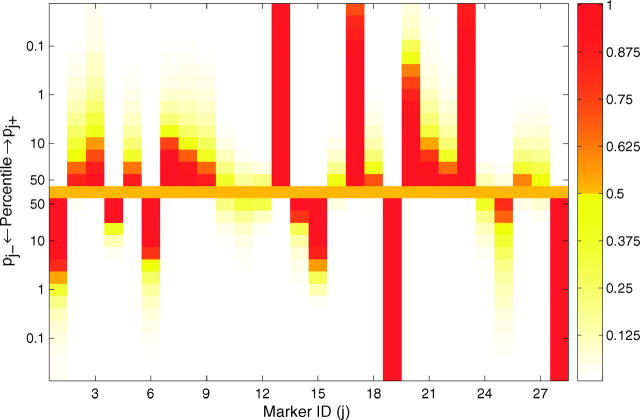
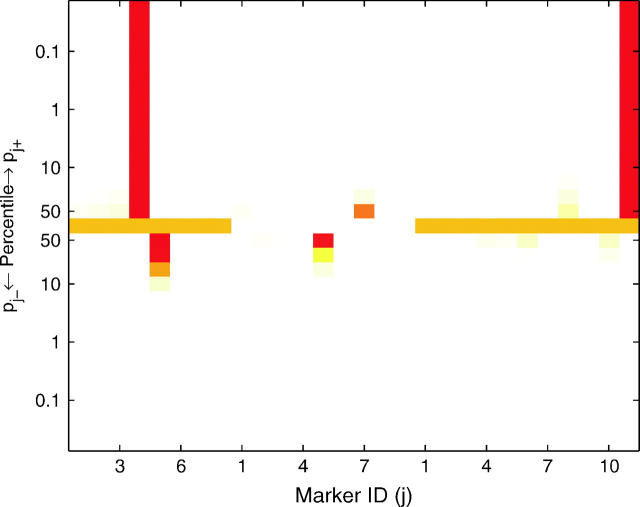
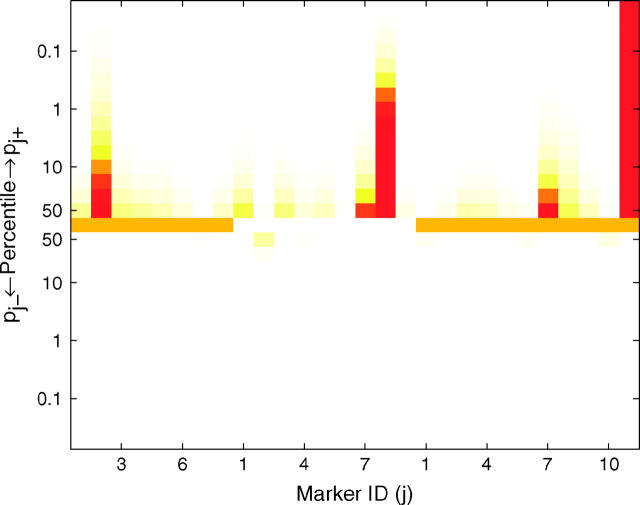
A heat map (Figure 1) can be used to graphically view the values of pj+ and pj− at different percentiles of their posterior distributions, allowing the investigator to visualize the posterior probabilities of a marker having a positive or negative effect with different levels of stringency. In this way, the heat map provides a visual device for determining the significance of QTL. The values of pj+ and pj− at different percentiles of their distributions are shown using a color scheme that maps a value of zero to white, 0.5 to orange, and 1 to red. A spot at the α × 100 percentile in the top (or bottom) half of the heat map with color ranging from orange to red implies that the probability of the corresponding marker belonging to the positive-effect (or negative-effect) class is >0.5 with a credibility of (1 − α) × 100%. For example, the first marker in Figure 1 can be inferred as a QTL with negative effect at the 90% credibility level but not at the 99% credibility level, as its tenth percentile spot in the bottom half is red (pj− > 0.5), but its first-percentile spot in the bottom half is less than that of yellow (pj− < 0.5). The heat map provides flexibility to investigators, allowing them to be more or less conservative when identifying QTL.
Figure 1.—
Heat map for posterior probabilities pj+ = P(βj > 0|yn, xn) and pj− = P(βj < 0|yn, xn). These are the probabilities of being in either the positive or the negative genetic-effect class. The values of pj+ and pj− at different percentiles of the posterior distribution are shown using different colors according to the color scheme on the right. If the color of pj+ (or pj−) at the α × 100 percentile ranges from orange to red, it implies that the probability of the jth marker belonging to the positive-effect (or negative-effect) class is >0.5 with a credibility of (1 − α) × 100%.
For each βj, we may use the chain 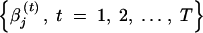 to estimate its value. However, we are more interested in estimating the size of βj given the class it belongs to. The corresponding chain may provide an unreliable estimate because of the limited number of βtj in some of the three classes. We propose to calculate the median values at each iteration of the Gibbs sampler,
to estimate its value. However, we are more interested in estimating the size of βj given the class it belongs to. The corresponding chain may provide an unreliable estimate because of the limited number of βtj in some of the three classes. We propose to calculate the median values at each iteration of the Gibbs sampler,
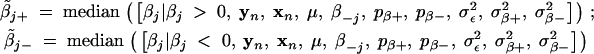 |
Then, if βj ∈ 𝒫(β) [or βj ∈ 𝒩(β)], the chain 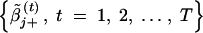 [or
[or 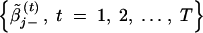 ] will provide an estimate of βj. With μ̃j+, μ̃j−, σ̃j+, and σ̃j− defined in the appendix, the two median values can be calculated as
] will provide an estimate of βj. With μ̃j+, μ̃j−, σ̃j+, and σ̃j− defined in the appendix, the two median values can be calculated as
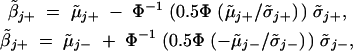 |
where Φ(·) is the cumulative distribution function of a standard normal distribution, and Φ−1(·) is its inverse function.
Extensions:
Our Bayesian framework can be easily adapted to include imputation of genotypes between markers, as well as epistatic interactions. The extensions of the genetic model to non-Gaussian phenotypic data may complicate the development of the corresponding Gibbs sampler. However, this type of data could be handled conceptually. In particular, drawing random samples of βj from its full conditional distribution may lose its easy computability. In this case, while p̃j+ and p̃j− may be calculated numerically, computation of β̃j+ and β̃j− may need to be approximated using a Metropolis-type algorithm.
The model (1) and its Bayesian framework can be further extended. Continuous and discrete nonmarker cofactors, can be incorporated into the multiple-linear-regression model. For example, let zi include, for individual i, all nonmarker cofactors that affect the corresponding phenotypic value. Then, subject to additive main effects from putative QTL and nonmarker factors, the phenotypic value of individual i (yi) can be modeled as
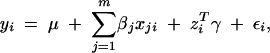 |
where γ describes the effects of the nonmarker factors. Usually, we incorporate nonmarker factors into the above model to control for their potential effects on the trait. In QTL mapping the selection of nonmarker cofactors is not our primary interest. We can simply partition all nonmarker factors into different groups such that the coefficients for all factors within the same group can be assigned independently and identically distributed prior distributions. The Bayesian framework and Gibbs sampler can therefore be developed adaptively.
Instead of collecting one observation, we may collect replicate observations for each inbred line. For this type of clustered data, efficiency consideration and nonmarker cofactors may prevent summarizing the observations from each line into one phenotypic value. The above genetic model and Bayesian framework are quite amenable to this type of data. Since individuals from the same line share marker genotypes, a common value should be imputed to each missing marker genotype for all individuals within the same line.
VALIDATION AND SIMULATION
Days to heading QTL in barley:
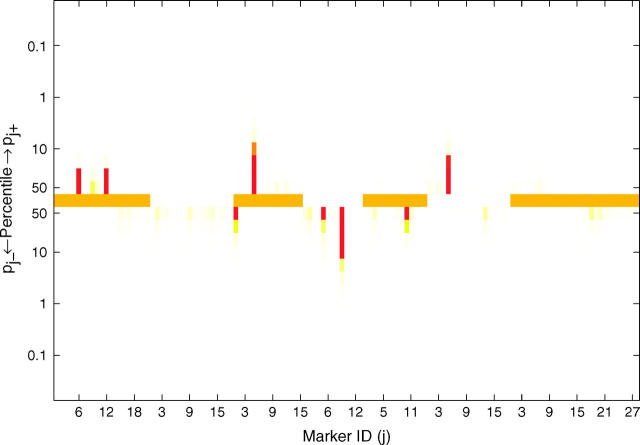
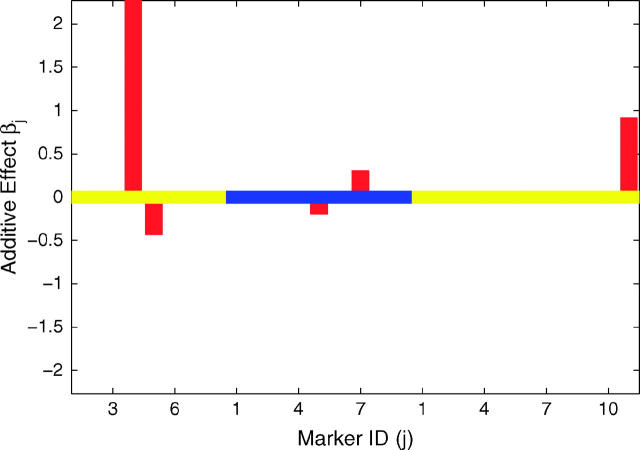
To validate the model, we analyzed line means for days from planting until emergence of 50% of heads on main tillers for 145 barley doubled-haploid lines that were genotyped for 127 markers across seven linkage groups (Tinker et al. 1996). Yi et al. (2003) analyzed this data set using stochastic search variable selection. Using a critical threshold value of 0.5 for the posterior probability of a marker being in the nonnegligible class, Yi et al. (2003) mapped QTL at markers I.12, III.5, IV.9, V.10, and VI.5 (the Roman number refers to the linkage group and the Arabic number refers to the marker index within the group). However, simply using the point estimates of these posterior probabilities to indicate significance of the corresponding markers ignores the variability of these statistics. Using the distributional departure of these posterior probabilities from probability 0.5 provides a more informative approach for QTL detection. With our three-component prior approach, QTL are mapped by using the distributional departure of the posterior probabilities pj+ and pj− from probability 0.5. Figure 2 shows the result of mapping QTL by our proposed approach. Markers III.5 and IV.9 are significant with credibility level at 90%, but the evidence for significance of markers I.12, VI.5, and V.10 is weak. In this example, if we simply threshold the medians of posteriors pj+ and pj− at 0.5, 8 markers, including those above, appear to have significant nonnegligible effects, demonstrating the drawback to using a point estimate as a critical threshold for QTL detection.
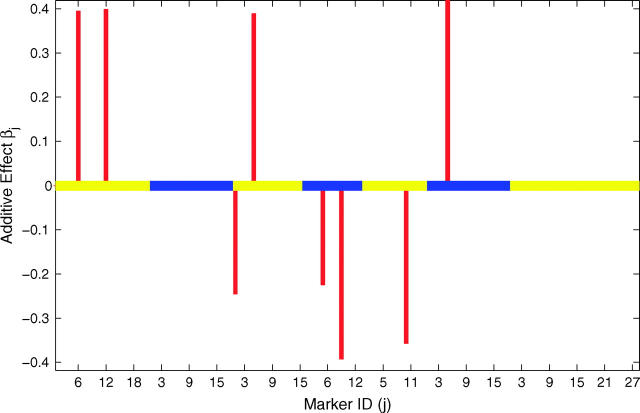
Figure 2.—
Results of Bayesian classification for heading trait in the North American Barley Genome Mapping Project. Shown are the heat map for posterior probabilities pj+ and pj− (top) and the estimated additive effects (bottom). In the top and bottom, the central lines represent different chromosomes by using colors alternating between orange and white and between yellow and blue, respectively. The marker identifications (IDs) along the x-axis are the IDs within the corresponding chromosomes.
We further analyzed the data set using IM, CIM, and MIM implemented in QTL Cartographer 2.0 (Wang et al. 2004). We identified significant QTL using a 5% experimentwise critical threshold value for the LOD score obtained from 1000 permutations of the phenotypic data. In concordance with results obtained from our method and by Yi et al. (2003), IM identified significant QTL around markers I.12, III.5, IV.9, and V.10 plus several additional QTL around markers IV.5, VI.3, and VII.18. Background markers for CIM were chosen by forward selection with background elimination regression using inclusion and exclusion probabilities of 0.1. CIM identifies QTL around markers I.6, I.12, III.5, III.9, III.12, IV.9, V.10, and VII.18 and better localizes the QTL to a more narrow region around marker IV.9. Implementation of MIM using the forward/backward selection method with a significance level of 0.01 identified 15 QTL. Using the standard Bayes information criterion model selection, we were able to detect three additional QTL.
While all methods detect QTL neighboring markers I.12, III.5, IV.9, and V.10, some methods detect unique QTL, with the results from CIM and MIM depending upon the model selection criterion employed. In particular, MIM detects many more significant QTL than the other methods. A comprehensive simulation study is necessary to fully assess the relative strengths and weaknesses of these different approaches. However, one advantage of the method we propose is better evaluation of the significance of a QTL.
Simulation study:
The ability to detect QTL is strongly influenced by the trait heritability, with most statistical methods being able to detect QTL for highly heritable traits. However, for many phenotypes of interest, the genetic component of the variance may be small relative to the environmental variance, making QTL detection challenging. In these cases, even QTL of relatively large effect may be difficult to detect when the random environmental effects on the trait are also large. To assess the performance of our approach we analyzed 10 randomly generated QTL models with phenotypes simulated under three levels of heritability and with either no or 10% missing data. The data sets simulated were for 225 recombinant inbred lines with three linkage groups containing a total of 27 markers. The number of recombination events per chromosome per generation was drawn from a Poisson distribution with mean equal to the length of the chromosome in morgans (Haldane 1919).
The 10 QTL models each contained four QTL with effects drawn from a Γ(2, 1) distribution. At the jth QTL of the ith line, the effect is defined as 2αj for marker genotype AA (i.e., αij = αj) and 0 for marker genotype aa (i.e., αij = 0). The genotypic value of a line is the sum of these effects across the four true QTL, and the genetic variance (σ2g) is the sample variance of the genotypic values across the lines. The phenotypic value for each line (Yi) is calculated as 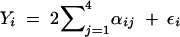 , where the random environmental effect (εi) is drawn from N(0, σ2ε). The environmental variance (σ2ε) is defined as 1 − h2/h2σ2g, where h2 is the heritability (0 < h2 < 1). We simulated phenotypic values for the 10 QTL models using h2 = 0.2, 0.4, and 0.6, which correspond to the environmental variance being 4 times, 1.5 times, and two-thirds of the genetic variance. Simulations were performed using QTL Cartographer version 1.13 (Basten et al. 1994, 1999), and simulated data sets with and without missing data were analyzed by our Bayesian classification method to infer the true- and false-positive rates.
, where the random environmental effect (εi) is drawn from N(0, σ2ε). The environmental variance (σ2ε) is defined as 1 − h2/h2σ2g, where h2 is the heritability (0 < h2 < 1). We simulated phenotypic values for the 10 QTL models using h2 = 0.2, 0.4, and 0.6, which correspond to the environmental variance being 4 times, 1.5 times, and two-thirds of the genetic variance. Simulations were performed using QTL Cartographer version 1.13 (Basten et al. 1994, 1999), and simulated data sets with and without missing data were analyzed by our Bayesian classification method to infer the true- and false-positive rates.
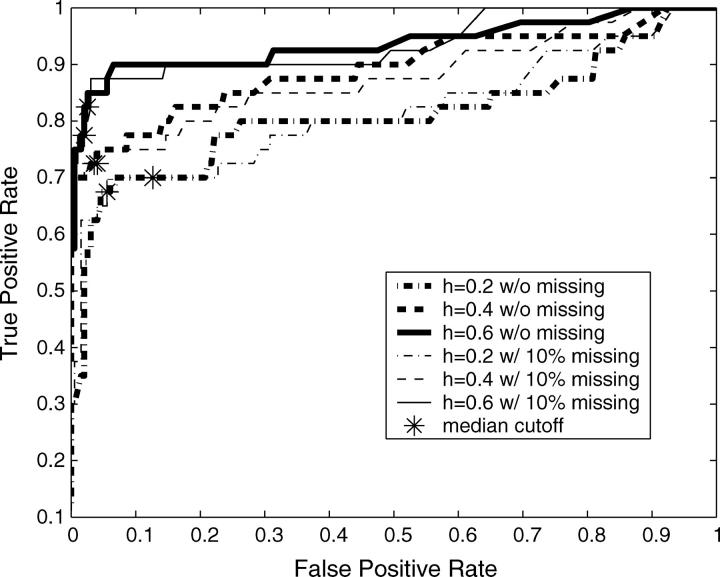
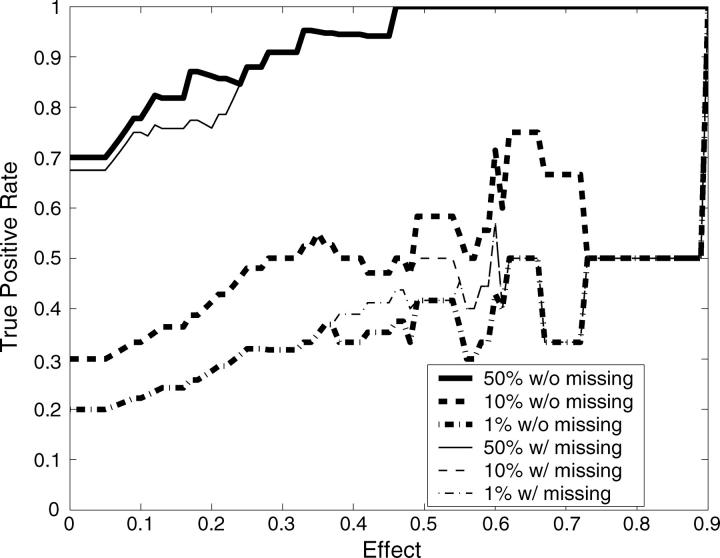
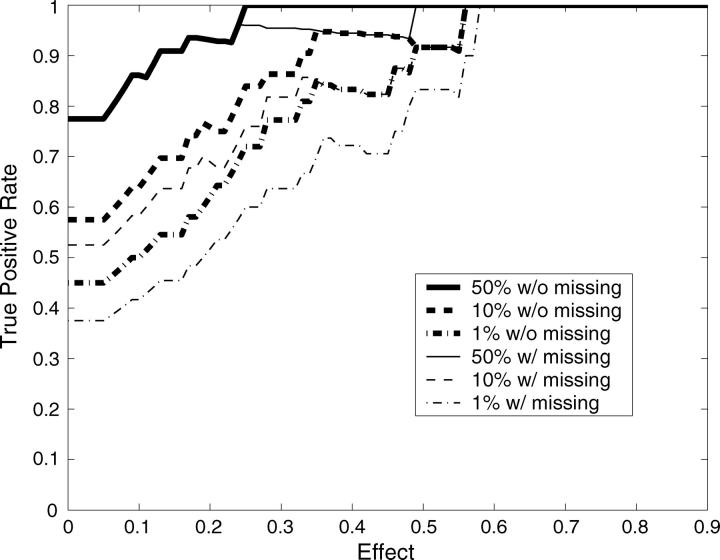
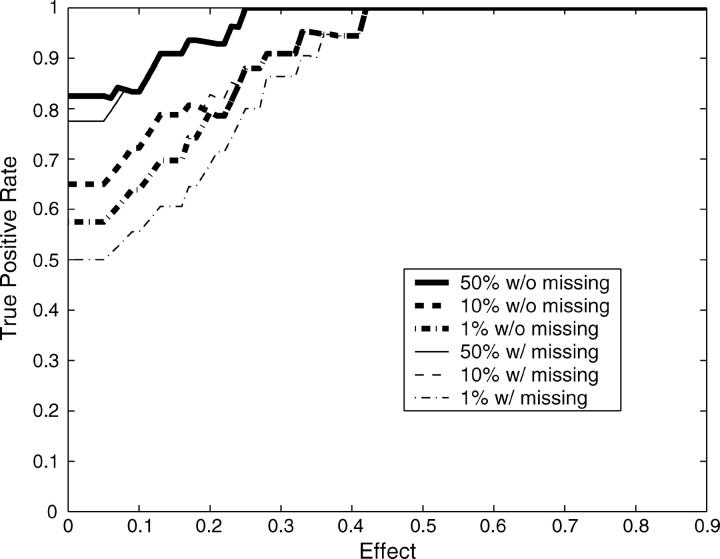
In total there were 10 mapping data sets with 40 true QTL simulated across the range of heritabilities, both with and without missing data. With sufficient recombination between markers, each QTL should be detected only by its neighboring markers. We therefore considered any significant markers not directly neighboring simulated QTL as false positives. This will inflate our false-positive rate when markers are tightly linked. Following this definition, 198 negatives are in each 10-data set group. The receiver operating characteristic (ROC) curves (Metz 1978) are drawn by using the Bayesian classification approach on each 10-data set group (Figure 3). ROC curves assess the trade-off between the true- and false-positive rates. Our ability to detect the 40 QTL effects drawn from a Γ(2, 1) distribution improved significantly with increasing heritability and was only slightly affected by missing values. Using the median values from the distributions of pj+ and pj− as critical threshold values for mapping QTL is equivalent to making decisions at the turning part of the ROC curve (i.e., Figure 3, asterisks). More liberal QTL mapping may favor some decision rules at the flat part of the ROC curve to improve the true-positive rate by allowing an increased false-positive rate. This liberal approach to QTL mapping may be particularly useful when the goal is to identify large numbers of QTL candidates, such as in marker-assisted selection programs (Spelman and Bovenhuis 1998; Beuzen et al. 2000; Dekkers and Hospital 2002). However, as is often the case, more conservative QTL mapping will require decision rules at the steep part of the ROC curve, decreasing the false-positive rate but potentially missing some true QTL. The heat map for posterior probabilities pj+ and pj− is designed to allow investigators to make these types of decisions when scanning genomes for QTL.
Figure 3.—
ROC curves plotting the true- vs. the false-positive rates from the simulation study. The asterisks correspond to mapping QTL by reading the median values from the distributions of pj+ and pj−. The flat part of the ROC curve corresponds to mapping QTL using more liberal decision rules to allow a higher false-positive rate to improve the true-positive rate. On the other hand, more conservative QTL mapping may prefer some decision rules at the steep part of the ROC curve, where the false-positive rate is decreased at a cost to the detection of true positives.
Given a trait's heritability, QTL detection will also depend upon the magnitude of the single-QTL effect. Figure 4 demonstrates the true-positive rates vs. effect sizes at different heritabilities when using the Bayesian classification approach. The true-positive rates here are calculated by counting only those QTL with effects higher than each given effect size. With heritability 0.2, conservative QTL mapping makes it difficult to identify QTL even if these QTL have large effects. Mapping QTL by reading the median values from the distributions of pj+ and pj− identified large-effect QTL, but this approach may lead to more false positives (Figure 3). With increasing heritability, more conservative decision rules could be adopted to lower false-positive rates without loss of power to detect large-effect QTL (Figure 4). Note that many markers that are one marker away from the markers neighboring QTL were significant and classified as false positives according to our stringent definition of true positives. A looser definition of true positives will significantly improve the results reported in Figures 3 and 4.
Figure 4.—
True-positive rate vs. effect size (α) at different heritabilities: h2 = 0.2 (top), h2 = 0.4 (middle), and h2 = 0.6 (bottom). The true-positive rates are calculated by counting only those QTL with effects higher than the given effect size (x-axis). The different lines refer to the different decision rules with and without missing data; 50%, 10%, and 1% refer to the percentiles of the posterior probabilities pj+ and pj− that were used as threshold values.
DATA ANALYSIS
Glucose-6-phosphate dehydrogenase (EC1.1.1.49, G6PD) catalyzes the conversion of glucose-6-phosphate (G6P) to 6-phospho-d-glucono-1,5-lactone, shunting G6P from the main backbone of glycolysis through the pentose-phosphate pathway and creating reducing power for the cell in the form of NADPH. In Drosophila, patterns of nucleotide variation at G6PD (Eanes et al. 1993, 1996), as well as covariance in enzyme activities of G6PD and its neighboring enzyme, 6-phosphogluconate dehydrogenase, across Drosophila species (Clark and Wang 1994), suggest that G6PD activity may come under selection in natural populations. Enzyme activities may evolve via mutations at the enzyme-encoding loci or rather through mutations at trans-acting loci that alter the quantity or function of the enzyme. QTL mapping provides a way to determine whether variation in enzyme activity (Mitchell-Olds and Pedersen 1998; Montooth et al. 2003) or protein quantity (Damerval et al. 1994) is the result of genetic variation cis or trans to the enzyme-encoding locus.
Introgression lines between closely related species allow us to map QTL underlying interspecific differences in quantitative traits. We quantified male and female G6PD activity in 221 inbred introgression lines between the sibling species D. simulans and D. sechellia that were genotyped at 28 markers across the X, second, and third chromosomes. Details for the construction and genotyping of these lines can be found in Dermitzakis et al. (2000) and Civetta et al. (2002). We measured G6PD activity as in vitro maximal activity from whole-fly homogenates using a standard spectrophotometric assay to monitor the NADPH that accumulates when G6P is converted to 6-phospho-d-glucono-1,5-lactone (Clark and Keith 1989). The data set for male G6PD activity (G6PDM) contained 864 trait measures across 210 lines, while that for females (G6PDF) contained 832 measures across 206 lines.
We applied our Bayesian classification approach to detect interspecific QTL for G6PD activity and to determine whether the same loci underlie G6PD activity in males and females. This is a particularly challenging data set for QTL detection, as the percentage of missing genotype data is high (18%) and, due to the nature of the introgression (see Dermitzakis et al. 2000), the frequency of the D. sechellia genotype at certain markers can range from 2 to 66%. There were also a number of covariates that we needed to incorporate into the model to control for both biological (fly weight and total protein content) and experimental effects.
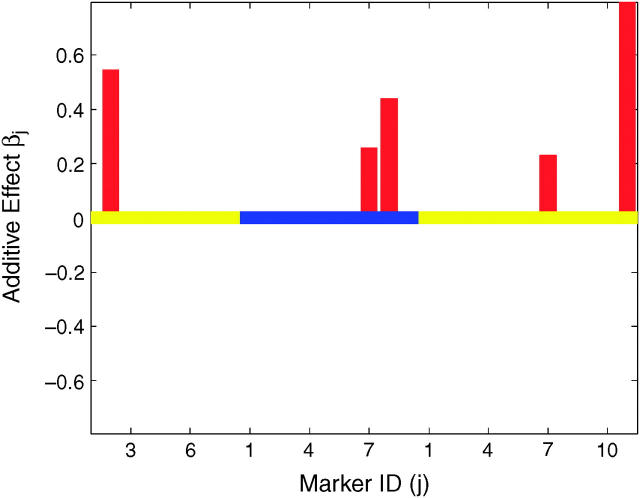
We identified a QTL on the tip of the right arm of chromosome 3 (marker III.11) that has strong effects on G6PD activity in both males and females (Figure 5). It is interesting to observe that while this QTL had the same magnitude of effect in both sexes, there was an additional X-linked QTL (marker I.4), distinct from the X-linked structural locus of G6PD, that had a rather outstanding effect on male G6PD activity only (Figure 5). The residual variances for G6PDM and G6PDF were estimated to be 0.5697 and 0.7089, respectively. Because the phenotypic values are standardized in our analysis, the markers and covariates in this model explained ∼43 and 29% of the phenotypic variation in G6PD activity for males and females, respectively.
Figure 5.—
Results of Bayesian classification for male G6PD (left) and female G6PD (right) enzyme activities. Shown are the heat map for posterior probabilities pj+ and pj− (top) and the estimated additive effects (bottom). In the top and bottom, the central lines represent different chromosomes by using colors alternating between orange and white and between yellow and blue, respectively. The marker IDs along the x-axis are the IDs within corresponding chromosomes.
To assess the performance of our method with this data set, we simulated five data sets using the observed marker genotypes and the parameter estimates from the above analysis for both G6PDM and G6PDF. Analyzing data simulated in this fashion can reveal the effects of imputing missing genotype data, as the missing data are imputed independently for each simulated dataset. Among the two most outstanding effects on G6PDM in Figure 5, marker I.4 was strongly significant in four of five simulated data sets and was mildly significant in the fifth data set, while marker III.11 was highly significant in all simulated data sets. The remaining three weak effects on G6PDM were occasionally detected in the simulated data sets. Although marker I.4 had a larger effect than marker III.11, more missing genotype data for marker I.4 than for marker III.11 slightly compromised its significance in mapping QTL.
The estimated effects on G6PDF were much smaller (Figure 5). The most outstanding effect on G6PDF at marker III.11 was strongly significant in three out of five simulated data sets and mildly significant in the other two data sets. Because of unbalanced genotypes at marker I.2 (∼1:50), marker I.2 is seldom significant in the five simulated data sets, although it is only slightly smaller in effect size than marker III.11. Nonnegligible effects in the initial data analysis were detected as weakly significant effects in one of the five simulated G6PDM data sets and in two of the five G6PDF data sets.
As illustrated in this simulation study, equal segregation of marker genotypes can improve the ability to accurately map QTL. The extent of missing genotype data may also affect QTL detection, particularly when the marker genotypes are unbalanced. False nonnegligible effects seldom appear in the results from our approach and, when observed, their significance as QTL was marginal.
DISCUSSION
The three-component mixture prior as a natural specification of genetic effects:
Model selection based on multiple-regression models of phenotypic data on multiple genetic markers is increasingly accepted as a general framework for mapping multiple QTL, with a large number of proposed methodologies being developed (e.g., see Hoeschele 2001; Piepho and Gauch 2001; Broman and Speed 2002; Sillanpää and Corander 2002; Yi 2004). QTL mapping is an inherently challenging problem. Large amounts of missing marker data, due to failure in genotyping or selective genotyping, are quite common in practice. When markers are sparse, the missing genotype information between markers must also be inferred (Kao et al. 1999; Zeng et al. 1999). In addition, the number of markers to test can be very large relative to the number of observed individuals (Meuwissen et al. 2001; Xu 2003), a problem that has been notoriously difficult in statistics.
The majority of genetic markers across a genome will not be linked to QTL for the trait of interest. From a statistical theory perspective, the parameter space in a QTL identification problem is quite sparse. Most classical methods for QTL mapping work well for a small number of QTL candidates. The challenge is then to develop an easy-to-implement framework for QTL mapping that efficiently detects sparse effects with a sufficiently low false-positive rate, precisely estimates their effects, and does so in the face of missing data and small numbers of observations. Two typical parameter spaces used to model sparseness are “nearly black” spaces, where the proportion of the nonzero parameter components is no more than a positive η, and Besov spaces, where the normalized lp-norm is bounded by η (Johnstone and Silverman 2004). We conjecture that the estimation methods proposed here achieve an optimal estimation rule as the sample size increases and as η goes to zero, in which sense it adapts automatically to the parameter space's sparseness. Johnstone and Silverman (2004) study a general class of estimation problems in sparse parameter spaces and show that a two-component mixture prior is adaptive and has some optimal estimation properties. The modeling strategy using a two-component mixture prior has been quite successful in attacking similar issues of false positives and false negatives in gene expression identification (Zhang et al. 2004) where one needs to identify a small number of differentially expressed genes from a large number of candidates.
The specification of the prior distribution for the genetic effects is critical and can influence the performance of the Bayesian approach to QTL mapping. Motivated by the above observations and the need to incorporate biologically relevant information into the prior specification of the genetic effects, we developed a three-component mixture prior on the basis of a natural classification of the marker effects (i.e., positive-, negative-, and negligible-effect classes) in a new Bayesian inference framework. The posterior probability of a marker belonging to one of the three categories is a natural statistic for assessing the significance of any marker being linked to a QTL for the trait of interest. This posterior probability of a marker's classification can be sharply inferred, and the marker effect on the phenotype can also be efficiently estimated using the proposed Gibbs sampler. Furthermore, the uncertainty associated with these estimates is naturally available from the corresponding posterior distributions, providing an advantage over classical approaches. Simulation experiments revealed that the approach is powerful for QTL detection and has relatively low false-positive rates, even when there are large amounts of missing data.
The three-component prior approach that we advocate for here has four significant advantages over existing methods for QTL inference. First, three-component priors incorporate the known information that most markers are not cotransmitted with QTL or their QTL effects are not detectable, which is important in controlling false-positive inference. In particular, if the number of available markers is on the same scale as the number of lines (or even if there are more markers than lines), it is necessary to incorporate this prior expectation of rarity of QTL to guarantee the model identifiability in multiple-linear regression. Second, the three-component prior approach is flexible and allows an imbalance between sizes/distribution of positive- and negative-effect classes. Third, unlike the two-component priors used by Yi et al. (2003), we classify all effects into three classes and describe the population distribution of each class. This avoids the disadvantage of stochastic search variable selection, which has difficulty in specifying many prior parameters and relies on assorting of each marker into either the small-effect or the large-effect class. Note that Xu (2003) models each putative QTL effect with a Gaussian distribution having its own variance parameter and further specifies noninformative priors for each variance parameter to avoid the above difficulty. However, the efficiency in extracting information from the data may be lowered by ignoring that most markers have negligible effects on the trait. Tuning parameters is a general problem with reversible-jump Markov chain Monte Carlo that we can avoid in our method. The fourth advantage of our approach is that the Gibbs sampler exports parameters β̃j+, β̃j−, p̃j+, and p̃j−, which can be used to make inference more efficiently than the β-chain only.
Identification of sex-specific QTL in D. simulans and D. sechellia:
Application of our Bayesian classification approach to a data set of metabolic enzyme activities from inbred introgression lines revealed QTL underlying G6PD activity differences between the closely related Drosophila species, D. simulans and D. sechellia. We identified a QTL on the tip of the right arm of chromosome 3 at cytological position 99E2 where the D. sechellia allele increased G6PD activity in both males and females. We also identified a male-specific QTL on the X chromosome at cytological position 7C1, which is distinct from the X-linked G6PD enzyme-encoding locus at cytological position 18D13. These results suggest that genetic differences in G6PD activity between D. simulans and D. sechellia are caused by trans-acting and sex-specific genetic effects.
In D. melanogaster sex-specific genetic architecture is common. Sex-specific QTL underlie neuro-sensory phenotypes (Long et al. 1995; Mackay and Fry 1996; Fanara et al. 2002), as well as life-history traits, such as longevity (Nuzhdin et al. 1997) and starvation resistance (Harbison et al. 2004). Sex-specific genetic effects also shape global expression variation within D. melanogaster (Anholt et al. 2003) and between Drosophila species (Ranz et al. 2003). Our results demonstrate that in Drosophila these sex-specific genetic effects also contribute to interspecific differences between species in metabolic processes.
Genome-wide analyses of gene expression, protein abundance, and function are shedding light on the relative contribution of cis- and trans-acting genetic variants to both inter- and intraspecific variation. QTL mapping results indicate that trans-acting effects predominate intraspecific variation in yeast (Schadt et al. 2003) and mouse (Brem et al. 2002) expression profiles, protein quantity in maize (Damerval et al. 1994), and enzyme activity in both D. melanogaster (Montooth et al. 2003) and Arabidopsis (Mitchell-Olds and Pedersen 1998). However, cis-acting effects are also detected, and in yeast these effects are of larger magnitude (Schadt et al. 2003). A recent analysis of differential allelic expression in D. melanogaster and D. simulans hybrids found that cis-acting effects could largely explain interspecific expression differences between the two closely related Drosophila species (Wittkopp et al. 2004). The interspecific G6PD QTL identified in our analysis have trans-acting effects in both males and females, suggesting that differences in G6PD activity have evolved between D. simulans and D. sechellia via genetic variants located away from the enzyme-encoding locus. QTL mapping is an important tool in our continued attempts to understand the role of cis- and trans-acting genetic effects in the evolution of gene expression, protein quantity, and enzymatic activity regulation.
Implementation and extension of the Bayesian classification approach:
The proposed Gibbs sampling algorithm for our Bayesian classification approach is implemented in MATLAB as software called QTLBayes (free for academic usage), which, due to its flexibility, can be readily applied to most QTL data. The framework is currently for the analysis of inbred lines derived from two inbred parental lines, and it can accommodate multiple covariates, as well as replicate measures for individuals from the same line. The heat map provides an informative visual tool for identifying significant QTL at varying levels of stringency.
One disadvantage of Bayesian analysis is the intensive computation involved (Nakamichi et al. 2001). If there are only a small number of missing values, the computation will not be an issue. Although imputation of missing data can be easily handled statistically within our framework, imputation of large amounts of missing genotype data may be computationally slow. An alternative strategy is to assume that there is at most one QTL between each pair of neighboring markers and adopt the composite space representation by Yi (2004). Prior specification can also impact algorithm performance in Bayesian analysis. The only informative priors in our analysis are the specification of inverse gamma distributions for σ2β+ and σ2β− to provide heavy-tailed priors for the distribution of genetic effects. When available, additional information can be readily incorporated into the prior specification, increasing the efficiency of estimation.
While the software currently analyzes data from isogenic lines, the model can be readily modified to accommodate a variety of experimental designs. The approach could also be extended to more complicated cases in QTL mapping, such as clustered data, multiple phenotypes, and pairwise epistasis. Detecting epistatic interactions between pairs of QTL is an important challenge, driven by the biological interest in detecting genetic interactions, but hampered by the extreme multiplicity of tests in performing an exhaustive search. The ability of our approach to select variables in the case of many tests with a small number of observations makes it possible to directly extend the approach to identify pairwise epistasis underlying complex traits.
Acknowledgments
We thank Carlos Bustamante for stimulating the collaboration between the authors, Hengli Liang for her contribution to the early stages of this project, and Lei Wang for collection of the primary enzyme kinetics data. We also thank Steven D. Tanksley, Gary Churchill, Patricia Wittkopp, and two anonymous reviewers for suggestions that improved this article. Research support by National Science Foundation (NSF) grant DMS-0204252 to M.T.W. as well as National Institutes of Health grant AI46409 and NSF grant DEB-0242987 to A.G.C. is gratefully acknowledged.
APPENDIX:
IMPLEMENTATION OF THE SINGLE-SITE GIBBS SAMPLER
Let the vector yn collect all phenotypic values of the trait and xn collect all genotypic values of the m putative QTL, and let β = (β1, … , βm) and β−j be β excluding βj, x−j,i = (x1i, … , xj−1,i, xj+1,i, … , xmi). Denote the conditional distribution of A given B as [A|B] and the marginal distribution of A as [A]. Each of the conditional distributions below are based on the fact that [A|B] ∝ [B|A][A].
Each iteration of the Gibbs sampler can proceed as follows:
Specify initial values as described in the Bayesian framework section.
- Sample each missing genotypic value xji from its full conditional posterior distribution,
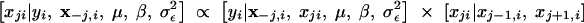
- Sample μ from its full conditional distribution,
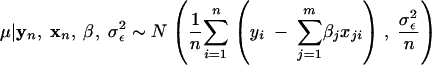
- For each j = 1, … , m, sample βj from its full conditional distribution,
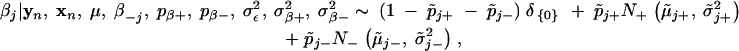
- where
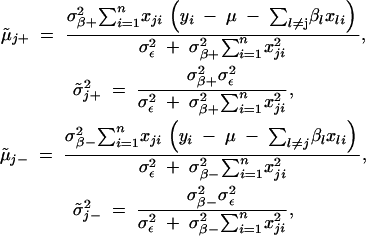
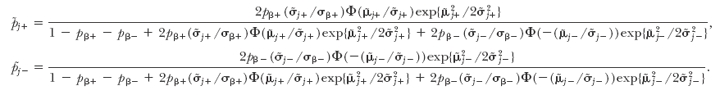
- Sample σ2ε from its full conditional distribution,
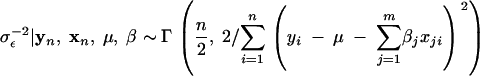
- Sample pβ+ and pβ− from the full conditional distribution,
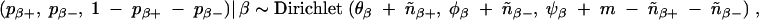
where ñβ+ = #{βj : βj > 0, 1 ≤ j ≤ m} and ñβ− = #{βj : βj < 0, 1 ≤ j ≤ m}. If the prior distribution of pβ+ and pβ− is restricted to be less than min
 /m, 1, the full conditional distribution should be a truncated Dirichlet distribution.
/m, 1, the full conditional distribution should be a truncated Dirichlet distribution.- Sample σ2β+ and σ2β− from the full conditional distributions,
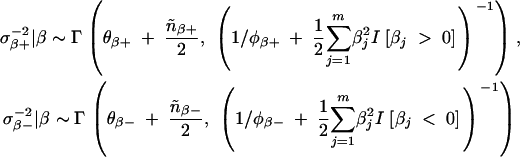
Repeat steps 1–7 until stationarity and the desired number of samples has been obtained.
References
- Anholt, R. R. H., C. L. Dilda, S. Chang, J.-J. Fanara, N. H. Kulkarni et al., 2003. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat. Genet. 35: 180–184. [DOI] [PubMed] [Google Scholar]
- Ball, R. D., 2001. Bayesian methods for quantitative trait loci mapping based on model selection: approximate analysis using the Bayesian information criterion. Genetics 159: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1994 Zmap—a QTL cartographer, pp. 65–66 in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, edited by C. Smith, J. S. Gavora, B. Benkel, J. Chesnais, W. Fairfull et al. Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production, Guelph, Ontario, Canada.
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1999 QTL Cartographer. Department of Statistics, North Carolina State University, Raleigh, NC.
- Beaumont, M. A., and B. Rannala, 2004. The Bayesian revolution in genetics. Nat. Rev. Genet. 5: 251–261. [DOI] [PubMed] [Google Scholar]
- Beuzen, N. D., M. J. Stear and K. C. Chang, 2000. Molecular markers and their use in animal breeding. Vet. J. 160: 42–52. [DOI] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., and T. P. Speed, 2002. A model selection approach for the identification of quantitative trait loci in experimental crosses. J. R. Stat. Soc. B 64: 641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., H. M. Waldrip-Dail and A. G. Clark, 2002. An introgression approach to mapping differences in mating success and sperm competitive ability in Drosophila simulans and D. sechellia. Genet. Res. 79: 65–74. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and L. E. Keith, 1989. Rapid enzyme kinetic assays of individual Drosophila and comparisons of field-caught D. melanogaster and D. simulans. Biochem. Genet. 27: 263–277. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and L. Wang, 1994. Comparative evolutionary analysis of metabolism in nine Drosophila species. Evolution 48: 1230–1243. [DOI] [PubMed] [Google Scholar]
- Cowles, M. K., and B. P. Carlin, 1996. Markov chain Monte Carlo convergence diagnostics: a comparative review. J. Am. Stat. Assoc. 91: 883–904. [Google Scholar]
- Damerval, C., A. Maurice, J. M. Josse and D. de Vienne, 1994. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics 137: 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers, J. C., and F. Hospital, 2002. The use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 3: 22–32. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E. T., J. P. Masly, H. M. Waldrip and A. G. Clark, 2000. Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes, W. F., M. Kirchner and J. Yoon, 1993. Evidence for adaptive evolution of the G6pd gene in the Drosophila melanogaster and Drosophila simulans lineages. Proc. Natl. Acad. Sci. USA 90: 7475–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes, W. F., M. Kirchner, J. Yoon, C. H. Biermann, I. N. Wang et al., 1996. Historical selection, amino acid polymorphism and lineage-specific divergence at the G6pd locus in Drosophila melanogaster and D. simulans. Genetics 144: 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanara, J. J., K. O. Robinson, S. M. Rollmann, R. R. H. Anholt and T. F. C. Mackay, 2002. Vanasco is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162: 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourdrinier, D., W. E. Strawderman and M. T. Wells, 1998. On the construction of Bayes minimax estimators. Ann. Stat. 26: 660–671. [Google Scholar]
- George, E. I., and R. E. McCulloch, 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88: 881–889. [Google Scholar]
- Green, P. J., 1995. Reversible jump Markov Chain Monte Carlo computation and Bayesian model determination. Biometrika 82: 711–732. [Google Scholar]
- Haldane, J. B. S., 1919. The combination of linkage values, and the calculation of distance between the loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Harbison, S. T., A. H. Yamamoto, J. J. Fanara, K. K. Norga and T. F. C. Mackay, 2004. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166: 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeschele, I., 2001 Mapping quantitative trait loci in outbred pedigrees, pp. 599–644 in Handbook of Statistical Genetics, edited by D. J. Balding, M. Bishop and C. Cannings. John Wiley & Sons, New York.
- Jansen, R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., and P. Stam, 1994. High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., and Z-B. Zeng, 1997. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica 101: 47–58. [DOI] [PubMed] [Google Scholar]
- Johnstone, I. M., and B. W. Silverman, 2004. Needles and straw in haystacks: empirical Bayes estimates of possibly sparse sequences. Ann. Stat. 32: 1594–1649. [Google Scholar]
- Kao, C.-H., Z-B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P. D., 1994. The distribution of mutation effects on viability in Drosophila melanogaster. Genetics 138: 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P. D., and O. Ohnishi, 1998. EMS-induced polygenic mutation rates for nine quantitative characters in Drosophila melanogaster. Genetics 148: 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpikari, R., and M. J. Sillanpää, 2003. Bayesian analysis of mulitlocus associations in quantitative and qualitative traits. Genet. Epidemiol. 25: 122–135. [DOI] [PubMed] [Google Scholar]
- Knott, S. A., and C. S. Haley, 1992. Aspects of maximum likelihood methods for mapping of quantitative trait loci in line crosses. Genet. Res. 60: 139–151. [Google Scholar]
- Kopp, A., R. M. Graze, S. Xu, S. B. Carroll and S. V. Nuzhdin, 2003. Quantitative trait loci responsible for variation in sexually dimorphic traits in Drosophila melanogaster. Genetics 163: 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [corrigendum: Genetics 136: 705 (1994)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., A. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melangogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, M. A., and C. Lopez-Fanjul, 1993. Spontaneous mutation for a quantitative trait in Drosophila melanogaster. II. Distribution of mutant effects on the trait and fitness. Genet. Res. 61: 117–126. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., and J. D. Fry, 1996. Polygenic mutations in Drosophila melanogaster: genetic interactions between selection lines and candidate quantitative trait loci. Genetics 144: 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, O., and R. N. Curnow, 1992. Estimating the locations and the size of the effects of quantitative trait loci using flanking markers. Theor. Appl. Genet. 85: 480–488. [DOI] [PubMed] [Google Scholar]
- Metz, C. E., 1978. Basic principles of ROC analysis. Semin. Nucl. Med. 8: 283–298. [DOI] [PubMed] [Google Scholar]
- Meuwissen, T. H., B. J. Hayes and M. E. Goddard, 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and D. Pedersen, 1998. The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth, K. L., J. H. Marden and A. G. Clark, 2003. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics 165: 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi, R., Y. Ukai and H. Kishino, 2001. Detection of closely linked multiple quantitative trait loci using a genetic algorithm. Genetics 158: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, C. L. Dilda, Z-B. Zeng and T. F. C. Mackay, 1997. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho, H.-P., and H. G. Gauch, Jr., 2001. Marker pair selection for mapping quantitative trait loci. Genetics 157: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. H., and B. Courtois, 1999. Mapping QTLs associated with drought resistance in rice: progress, problems and prospects. Plant Growth Reg. 29: 123–133. [Google Scholar]
- Price, A. H., J. E. Cairns, P. Horton, H. G. Jones and H. Griffiths, 2002. Linking drought-resistance mechanisms to drought avoidance in upland rice using a QTL approach: progress and new opportunities to integrate stomatal and mesophyll responses. J. Exp. Bot. 53: 989–1004. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Satagopan, J. M., B. S. Yandell, M. A. Newton and T. C. Osborn, 1996. A Bayesian approach to detect quantitative trait loci using Markov chain Monte Carlo. Genetics 144: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E. E., S. A. Monks, T. A. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Sen, S., and G. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, J. S., I. S. Painter and B. S. Weir, 1999. Bayesian statistics in genetics: a guide for the uninitiated. Trends Genet. 15: 354–358. [DOI] [PubMed] [Google Scholar]
- Sillanpää, M. J., and E. Arjas, 1998. Bayesian mapping of multiple quantitative trait loci from incomplete inbred line cross data. Genetics 148: 1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää, M. J., and J. Corander, 2002. Model choice in gene mapping: what and why. Trends Genet. 18: 301–307. [DOI] [PubMed] [Google Scholar]
- Spelman, R. J., and H. Bovenhuis, 1998. Moving from QTL experimental results to the utilization of QTL in breeding programmes. Anim. Genet. 29: 77–84. [DOI] [PubMed] [Google Scholar]
- Stephens, D. A., and R. D. Fisch, 1998. Bayesian analysis of quantitative trait locus data using reservible jump Markov chain Monte Carlo. Biometrics 54: 1334–1347. [Google Scholar]
- Sugiyama, F., G. A. Churchill, D. C. Higgins, C. Johns, K. P. Makaritsis et al., 2001. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics 71: 70–77. [DOI] [PubMed] [Google Scholar]
- Tinker, N. A., D. E. Mather, B. G. Rossnage, K. J. Kasha and A. Kleinhofs, 1996. Regions of the genome that affect agronomic performance in two-row barley. Crop Sci. 36: 1053–1062. [Google Scholar]
- Wang, S., C. J. Basten and Z-B. Zeng, 2004 Windows QTL Cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm).
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wright, F. A., and A. Kong, 1997. Linkage mapping in experimental crosses: the robustness of single-gene models. Genetics 146: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., 2003. Estimating polygenic effects using markers of the entire genome. Genetics 163: 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, N., 2004. A unified Markov chain Monte Carlo framework for mapping multiple quantitative trait loci. Genetics 167: 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, N., V. George and D. B. Allison, 2003. Stochastic search variable selection for identifying multiple quantitative trait loci. Genetics 164: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., C.-H. Kao and C. J. Basten, 1999. Estimating the genetic architecture of quantitative traits. Genet. Res. 74: 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang, D., M. T. Wells, C. D. Smart and W. E. Fry, 2004. Bayesian normalization and inference for differential gene expression data. J. Comp. Biol. 12: 391–406. [DOI] [PubMed] [Google Scholar]