Abstract
Estrogens stimulate proliferation and enhance survival of the prolactin (PRL)-producing lactotroph of the anterior pituitary gland and induce development of PRL-producing pituitary tumors in certain inbred rat strains but not others. The goal of this study was to elucidate the genetic bases of estrogen-induced pituitary tumorigenesis in reciprocal intercrosses between the genetically related ACI and Copenhagen (COP) rat strains. Following 12 weeks of treatment with the synthetic estrogen diethylstilbestrol (DES), pituitary mass, an accurate surrogate marker of absolute lactotroph number, was increased 10.6-fold in ACI rats and 4.5-fold in COP rats. Composite interval mapping analyses of the phenotypically defined F2 progeny from the reciprocal crosses identified six quantitative trait loci (QTL) that determine the pituitary growth response to DES. These loci reside on chromosome 6 [Estrogen-induced pituitary tumor (Ept)1], chromosome 3 (Ept2 and Ept6), chromosome 10 (Ept9), and chromosome 1 (Ept10 and Ept13). Together, these six Ept loci and one additional suggestive locus on chromosome 4 account for an estimated 40% of the phenotypic variance exhibited by the combined F2 population, while 34% of the phenotypic variance was estimated to result from environmental factors. These data indicate that DES-induced pituitary mass behaves as a quantitative trait and provide information that will facilitate identification of genes that determine the tumorigenic response of the pituitary gland to estrogens.
ESTROGENS play a central role in the regulation of cell proliferation and survival in numerous mammalian tissues and are implicated in the etiology of several types of cancer (Shull 2002). The prolactin (PRL)-producing lactotroph of the anterior pituitary gland provides a well-defined cell model for studying estrogen action. It is well established that estrogens enhance transcription of the PRL gene, stimulate lactotroph proliferation, and promote lactotroph survival (Spady et al. 1999b). Continuous treatment with either naturally occurring or synthetic estrogens induces rapid and sustained growth of the anterior pituitary gland in male or female rats of the Fischer 344 (F344) (Segaloff and Dunning 1945; Wiklund et al. 1981a,b; Wiklund and Gorski 1982), A × C Irish (ACI) (Segaloff and Dunning 1945; Holtzman et al. 1979; Shull et al. 1997; Spady et al. 1999c,d), Copenhagen (COP) (Spady et al. 1998a, 1999d), and several other inbred rat strains (Noble et al. 1940; Furth et al. 1973; Spady et al. 1999b). The grossly enlarged pituitary glands, commonly referred to as pituitary tumors, exhibit diffuse lactotroph hyperplasia histologically and result in marked hyperprolactinemia (Spady et al. 1998b, 1999b,c). In contrast, continuous estrogen treatment induces very little pituitary growth in other rat strains, such as the outbred Holtzman strain (Wiklund et al. 1981b; Wiklund and Gorski 1982) or the inbred Brown Norway (BN) strain (Wendell et al. 1996; Wendell and Gorski 1997; Spady et al. 1999d).
The genetic bases of sensitivity of the F344 rat strain to estrogen-induced pituitary growth have been investigated in crosses to the insensitive Holtzman and BN strains. Following 8 weeks of treatment with the synthetic estrogen diethylstilbestrol (DES), average pituitary mass was increased 7.8-fold in female F344 rats but was unaffected in female Holtzman rats (Wiklund et al. 1981a). Average pituitary mass was increased 1.8- and 2.2-fold in DES-treated (Holtzman × F344)F1 and (F344 × Holtzman)F1 rats, respectively, suggesting that sensitivity to DES-induced pituitary growth is recessive or incompletely dominant in these crosses. More recently, Wendell and colleagues (Wendell et al. 1996, 2000; Wendell and Gorski 1997) have evaluated DES-induced pituitary growth in crosses between the F344 and BN strains and have mapped six genetic loci, each of which harbors one or more genes that control the pituitary growth response to DES in these crosses. The effects of two of these loci have now been evaluated using congenic rat lines (Wendell et al. 2002; Pandey et al. 2004). Together, these studies indicate that estrogen-induced pituitary growth behaves as a quantitative trait in these crosses between the F344 and BN rat strains.
We are studying the genetically related ACI and COP rat strains to define the genetic bases of the differing sensitivities of these strains to estrogen-induced pituitary growth (Spady et al. 1999d; Harvell et al. 2003) as well as susceptibility to estrogen-induced mammary cancer (Shull et al. 1997; Spady et al. 1998a; Harvell et al. 2000; Shull et al. 2001; Gould et al. 2004). The goals of this study were to characterize DES-induced pituitary growth in F1, F2, and backcross (BC) progeny from reciprocal crosses between the ACI and COP rat strains and to map the genetic loci that determine the differing pituitary growth responses of the ACI and COP rat strains to DES. We demonstrate that sensitivity to DES-induced pituitary growth behaves as a complex trait determined by at least six quantitative trait loci (QTL). For the most part, these loci are distinct from those mapped previously by Wendell et al. in crosses between the F344 and BN rat strains (Wendell et al. 1996, 2000; Wendell and Gorski 1997). Moreover, these loci are distinct from loci that we mapped previously that determine susceptibility to estrogen-induced mammary cancer in reciprocal crosses between the ACI and COP rat strains (Gould et al. 2004). These data indicate that the genes that determine the tumorigenic potential of estrogens act in a rat strain- and tissue-specific manner.
MATERIALS AND METHODS
Analysis of phenotypes:
The Institutional Animal Care and Use Committee of the University of Nebraska Medical Center approved all procedures involving live animals. ACI rats were obtained from Harlan Sprague Dawley (Indianapolis). COP rats were obtained from the breeding program of the National Cancer Institute. Animals were housed in a barrier facility under controlled temperature, humidity, and 12-hr light/12-hr dark conditions. This facility was accredited by the American Association for Accreditation of Laboratory Animal Care and operated in accordance with the standards outlined in the Guide for the Care and Use of Laboratory Animals (DHHS Pub. 85-23). The animals were caged and fed as described previously (Spady et al. 1999d). Female COP rats were mated to male ACI rats to produce (COP × ACI)F1 progeny, F1 progeny were mated to generate (COP × ACI)F2 progeny, and ACI females were mated to F1 males to produce (COP × ACI)BC progeny. Pups were weaned at 20–24 days of age. Implants, either empty or containing 5 mg of DES (Sigma, St. Louis), were made from Silastic tubing and medical adhesive (Dow Corning, Midland, MI) and were inserted subcutaneously in the interscapular region as described previously (Spady et al. 1998a). Treatment with DES was initiated when the animals were 63 ± 4 days of age. Small populations of male rats of each genetic type received empty implants. The rats were killed by decapitation after 12 weeks of DES or sham treatment. The pituitary gland was immediately removed and weighed. Pituitary mass in estrogen-treated rats correlates highly with pituitary DNA content (Wiklund et al. 1981b; Wendell et al. 2000) and with circulating PRL (Spady et al. 1999d), indicating that pituitary mass serves as an accurate surrogate indicator of absolute lactotroph number. The spleen was collected as a source of DNA and stored at −80°.
Analysis of genotypes:
The phenotypically defined (COP × ACI)F2 population described herein and the (ACI × COP)F2 population described by us previously (Spady et al. 1999d) were subjected to genotypic analyses. DNA was isolated from the spleen of 162 of 163 (COP × ACI)F2 rats and 102 of 103 (ACI × COP)F2 rats using DNeasy columns (QIAGEN, Valencia, CA). One rat from each cross was excluded from analysis because tissue was not available for isolation of DNA. The marker panel for QTL mapping consisted of simple sequence length polymorphisms (SSLP) that were selected at ∼20-cM intervals from those distributed across the 20 autosomes (Rat Genome Database, http://rgd.mcw.edu). Oligonucleotide primers specific to each SSLP marker were obtained from Invitrogen (Carlsbad, CA). Genotypes were determined at each of the 178 SSLP markers for the F2 progeny from each cross that exhibited the highest and lowest pituitary mass values. For the remaining F2 progeny, genotypes were determined at the 58 SSLP markers that resided on the six chromosomes that yielded evidence of the presence of a QTL. Each DNA sample was amplified in a 10-μl reaction containing 30 ng of DNA; 1.25 units Taq polymerase (Invitrogen); 20 mm Tris (pH 8.4); 1.5 mm MgCl2; 50 mm KCl; 198 nm of each of the forward and reverse primers; 200 μm each of dATP, dGTP, dCTP, dTTP; and 1.0 μCi [α-32P]dATP (Amersham, Arlington Heights, IL). The reaction mixtures were incubated at 94° for 5 min and subjected to 30 cycles of PCR as follows: (1) 94° for 30 sec; (2) 55° for 30 sec; (3) 72° for 1.5 min, with the final cycle followed by incubation at 72° for 3 min. The DNA products were denatured, resolved on 5, 6, or 8% polyacrylamide gels, and visualized using a PhosphorImager and ImageQuant 5.0 for Windows NT software (Molecular Dynamics, Sunnyvale, CA).
Interval mapping:
Genetic maps were estimated and interval mapping (IM) analyses were performed using MapManager QTX version 0.29 (Manly et al. 2001). The IM function of MapManager QTX is most accurate when the phenotypic data are normally distributed (K. F. Manly, personal communication). Therefore, the pituitary mass data were log10 transformed, resulting in a normal distribution of values, prior to IM analyses. Two subpopulations consisting of the 45 (COP × ACI)F2 and the 44 (ACI × COP)F2 rats that exhibited extreme phenotypes were selected for the initial IM analyses (Lander and Botstein 1989). Experiment-wise threshold values were calculated by performing 1000 permutations of the phenotypic data (Churchill and Doerge 1994). IM was performed using data for 178 markers distributed across the 20 autosomes. Because similar regions of the genome were indicated to harbor QTL during the initial analyses of the two phenotypically extreme populations and there was no significant difference in the pituitary mass phenotype between the two F2 populations, these two populations were combined to provide additional power to detect QTL. Permutation-derived thresholds were again calculated and IM was performed on all autosomes for the combined selective population of 89 F2 animals. The remaining 117 (COP × ACI)F2 and 58 (ACI × COP)F2 animals were subsequently genotyped across those chromosomes that exhibited at least suggestive evidence of a QTL upon analysis of the phenotypically extreme F2 animals. Permutation-derived thresholds were then calculated and IM was performed using the data from the 264 F2 animals.
Composite interval mapping:
Composite interval mapping (CIM) was performed using MapManager QTX to reduce the effect of unlinked and linked QTL on each locus mapped (Zeng 1993, 1994; Teuscher et al. 1999; Doerge 2002). The markers included as cofactors in the CIM analyses were identified using a stepwise selection process and the marker regression function of MapManager QTX. For each step, marker regression was performed across the six chromosomes of interest using the log10-transformed pituitary mass as the phenotype, and the marker exhibiting the most statistically significant likelihood-ratio statistic (LRS) was added to the background. This process was repeated until no additional marker exhibited a statistically significant LRS value (Manly et al. 2001). For each CIM analysis, a marker (for QTL near a chromosome end) or markers (for QTL not near a chromosome end) flanking the test interval as well as the peak marker associated with the five other significant QTL were added to the model as cofactors. The permutation-derived threshold was then calculated by performing 1000 permutations of the phenotypic data and CIM was conducted to determine the effect of the test interval with these cofactors included in the model. This process was performed for each significant QTL. To determine the effect of the significant QTL on the suggestive QTL, CIM was performed with the peak marker associated with the six significant QTL added to the model as cofactors. The confidence interval for each significant QTL identified by CIM analysis was estimated using bootstrapping analysis (Visscher et al. 1996), which is part of the interval-mapping function of MapManager QTX. For each significant QTL, the percentage of trait variance explained and the additive effect was determined by CIM analysis (Manly et al. 2001). Degree of dominance was calculated using CIM analysis and the method of Falconer and Mackay (1996).
Evaluation of genetic interactions:
Potential interaction between the 58 markers resident on the six chromosomes that yielded suggestive or significant evidence of a QTL was evaluated pairwise using MapManager QTX. For this analysis, the threshold for significance was obtained by performing 1000 permutations of the phenotypic data using the interaction model with the level of significance set at P = 0.01 (Manly et al. 2001). Interaction testing was performed with the probability of a type I error set at ≤10−5 (Manly et al. 2001).
Statistical analyses:
Differences in pituitary mass between experimental groups were assessed using the Wilcoxon rank sum test (GB-STAT, version 6.5; Dynamic Microsystems, Silver Spring, MD). P-values ≤0.05 were considered to be indicative of statistical significance. The distribution of pituitary mass and log10-transformed pituitary mass values within the combined F2 population was tested for normality using SPSS version 12.0 (SPSS, Chicago). The contribution of environmental factors to phenotypic variance was estimated using the method of Wright (1968).
RESULTS
Phenotypic characterization of progeny from a COP × ACI cross:
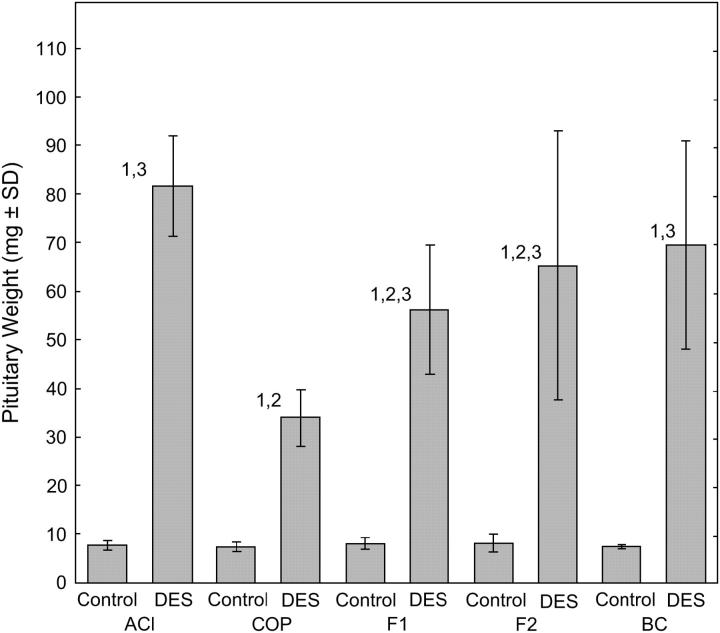
Treatment with the synthetic estrogen DES for 12 weeks resulted in significant increases in pituitary mass in each of the experimental groups evaluated in the context of the COP × ACI (strain of the female is indicated first) cross, relative to that observed in untreated male rats (Figure 1). However, the magnitude of the pituitary growth response to DES was rat strain specific and genetically determined. DES increased pituitary mass in male ACI rats 10.6-fold, from 7.7 mg [standard deviation (SD) = 0.9] in untreated male ACI rats to 81.7 mg (SD = 10.3). By contrast, DES increased pituitary mass only 4.4-fold in male COP rats, from 7.6 mg (SD = 1.0) to 34.2 mg (SD = 5.9). The pituitary growth response of the DES-treated (COP × ACI)F1 progeny was intermediate to that exhibited by the parental ACI and COP strains, indicating that this phenotype behaves as an incompletely dominant trait. In these F1 progeny, DES increased pituitary mass 6.9-fold, from 8.2 mg (SD = 1.2) to 56.6 mg (SD = 13.2). Male (COP × ACI)F2 rats exhibited a 7.8-fold increase in pituitary mass when treated with DES, whereas DES treatment increased pituitary mass 9.2-fold in male (COP × ACI)BC rats. Pituitary mass in untreated rats did not differ significantly between groups (Figure 1).
Figure 1.—
Sensitivity to DES-induced pituitary growth in progeny from a COP × ACI cross is genetically determined. Male ACI, COP, F1, F2, and BC rats from a cross originating with COP females and ACI males were generated and treated with DES for 12 weeks beginning at 9 weeks of age as described in materials and methods. Each data bar represents the mean mass of the anterior pituitary gland in milligrams ± the standard deviation of the mean. Each of the untreated control groups consisted of 5–6 rats. The numbers of DES-treated rats were: ACI, 14; COP, 14; F1, 18; F2, 163; and BC, 49. The Wilcoxon rank sum test was performed to assess differences in pituitary mass between experimental groups. P-values ≤0.05 were considered indicative of statistical significance. Numeral 1 indicates a statistically significant difference between a DES-treated population and its corresponding sham-treated control population. Numeral 2 indicates a significant difference between the indicated population and the treatment-matched ACI population. Numeral 3 indicates a significant difference between the indicated population and the treatment-matched COP population.
The pituitary growth responses exhibited by the DES-treated male F1, F2, and BC populations evaluated in this COP × ACI cross were compared to the responses exhibited by DES-treated F1, F2, and BC populations evaluated in an ACI × COP cross that was described by us previously (Spady et al. 1999d). DES-induced pituitary growth was similar for each of the F1, F2, and BC populations from the two crosses (Table 1), indicating that the parental orientation of the crosses generating the F1 generation did not exert a discernible effect on the phenotypes exhibited by the DES-treated F1, F2, or BC progeny from these reciprocal crosses between the ACI and COP strains. Table 2 summarizes the combined data from the two crosses.
TABLE 1.
Pituitary weights in DES-treated male rats from reciprocal intercrosses between the ACI and COP rat strains
| Progeny type |
COP × ACIa | ACI × COPa | P-valueb |
|---|---|---|---|
| F1 | 56.6 ± 13.6 (18) | 58.8 ± 7.5 (30) | 0.81 |
| F2 | 65.3 ± 26.5 (163) | 60.9 ± 23.9 (103) | 0.15 |
| BC | 71.8 ± 25.1 (49) | 68.2 ± 12.8 (19) | 1.00 |
Mean pituitary weight and standard deviation are presented. The number of animals in each group is in parentheses.
P-values were calculated using the Wilcoxon rank sum test.
TABLE 2.
Combined data from reciprocal crosses between the ACI and COP rat strains
| Group | Untreateda | DES treateda |
P-value vs. ACIb |
P-value vs. COPb |
|---|---|---|---|---|
| ACI | 8.1 ± 1.1 (7) | 72.7 ± 14.6 (28) | — | <0.0001 |
| COP | 9.5 ± 2.7 (8) | 36.2 ± 7.4 (28) | <0.0001 | — |
| F1 | 9.2 ± 1.5 (12) | 58.0 ± 10.0 (48) | 0.0003 | <0.0001 |
| F2 | 8.9 ± 1.7 (8) | 63.6 ± 25.6 (266) | 0.0024 | <0.0001 |
| BC | 7.8 ± 0.4 (5) | 70.7 ± 22.2 (68) | 0.2819 | <0.0001 |
Mean pituitary weight and standard deviation are shown for each of the pooled populations from the COP × ACI and ACI × COP crosses. The number of animals in each group is in parentheses.
Pituitary mass in each group of DES-treated animals was compared to that observed in the parental ACI and COP strains using the Wilcoxon rank sum test.
Mapping of QTL that control estrogen-induced pituitary growth:
Genotypes were initially determined at 178 SSLP markers distributed across the 20 autosomes for 45 DES-treated (COP × ACI)F2 and 44 DES-treated (ACI × COP)F2 progeny that exhibited the extreme phenotypes, i.e., the largest or smallest pituitary masses, within their respective populations. Interval-mapping analyses of each of these F2 subpopulations provided suggestive evidence for the presence of QTL affecting pituitary mass on RNO3, RNO6, and RNO10 (data not shown). Because the (COP × ACI)F2 and (ACI × COP)F2 populations did not differ from one another phenotypically (Table 1) and the preliminary interval-mapping analyses of the two subpopulations of phenotypically extreme F2 progeny were suggestive of similar QTL, the two subpopulations were combined to increase statistical power. Interval-mapping analysis of this combined population of 89 phenotypically extreme F2 progeny revealed a total of eight regions on RNO1, RNO3, RNO4, RNO5, RNO6, and RNO10 where LRS values exceeded 10.6, the permutation-derived threshold indicative of suggestive evidence for the presence of a QTL affecting pituitary mass (data not shown). Genotypes were subsequently determined at 58 SSLP markers spanning these six chromosomes for the remaining 117 DES-treated (COP × ACI)F2 progeny and the remaining 58 DES-treated (ACI × COP)F2 progeny. Interval-mapping analyses of this combined population of 264 DES-treated F2 progeny revealed one locus on RNO3 where the peak LRS value exceeded 20.9, the permutation-derived threshold indicative of highly significant evidence of a QTL; a total of four loci on RNO1, RNO3, RNO6, and RNO10 where the peak LRS values exceeded 14.0, the permutation-derived threshold indicative of significant evidence of a QTL; and three additional loci on RNO1, RNO4, and RNO5 where the peak LRS values exceeded 7.9, the permutation-derived threshold for suggestive evidence of a QTL (data not shown).
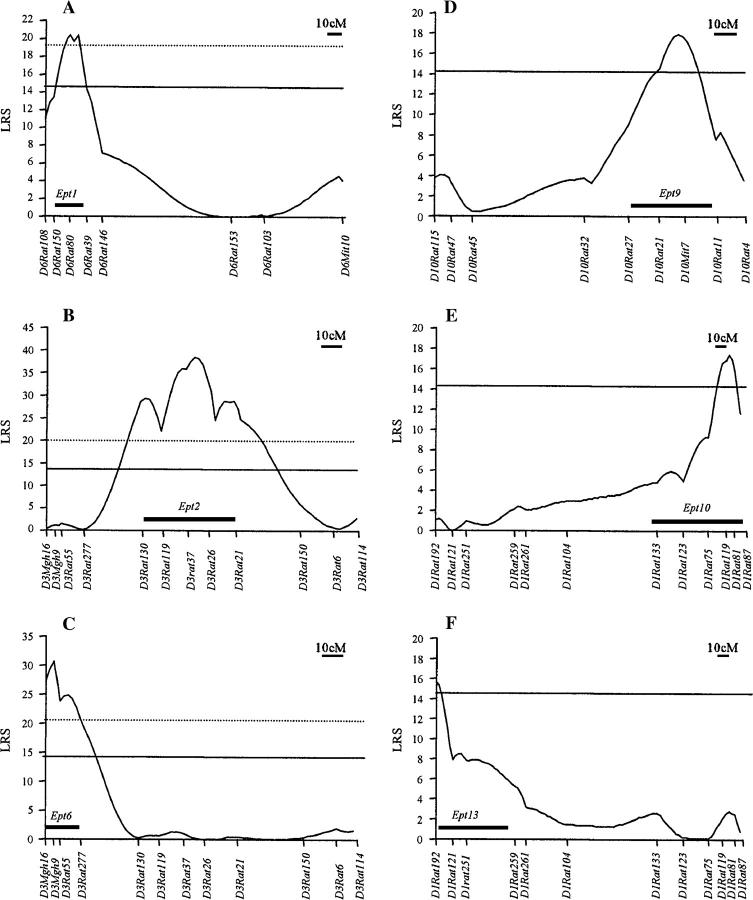
CIM was performed to evaluate these QTL further. The CIM analyses revealed six statistically significant QTL residing on RNO1, RNO3, RNO6, and RNO10 (Figure 2). These loci have been designated as Estrogen-induced pituitary tumor (Ept) loci: Ept1, Ept2, Ept6, Ept9, Ept10, and Ept13. The numbering of these QTL is not continuous because other Ept loci not described here have been mapped in other crosses between the ACI and Brown Norway strains (T. E. Strecker and J. D. Shull, unpublished data). Ept1 is defined by a peak LRS value of 20.4 at D6Rat80 and a confidence interval extending from D6Rat150 to D6Rat39 (Figure 2A). This locus is estimated to account for 8% of the phenotypic variance exhibited by the combined F2 population (Table 3). Ept2 is defined by a peak LRS value of 38.4 near D3Rat26, with the confidence interval defined by D3Rat130 and D3Rat21 (Figure 2B). Ept2 is estimated to account for 14% of the phenotypic variance and exerts the strongest effect on DES-induced pituitary mass of the six QTL (Table 3). Ept6 also maps to RNO3 (Figure 2C). This QTL is defined by a peak LRS value of 30.6 at D3Mgh9, a confidence interval extending from D3Mgh16 to D3Rat277, and accounts for ∼11% of the phenotypic variance (Table 3). Ept9 is defined by a peak LRS value of 17.9 near D10Mit7 and a confidence interval extending from D10Rat27 to D10Rat11 (Figure 2D). This QTL is estimated to account for 7% of the phenotypic variance (Table 3). Both Ept10 and Ept13 map to RNO1. Ept10 is defined by an LRS peak of 17.4 near D1Rat119 and a confidence interval extending from D1Rat133 to D1Rat81 (Figure 2E). Ept13 is defined by a peak LRS value of 15.6 near D1Rat192 and a confidence interval extending from D1Rat192 to D1Rat259 (Figure 2F). Ept10 and Ept13 are estimated to account for 7 and 6% of the phenotypic variance, respectively (Table 3). When CIM analysis was performed for the suggestive QTL on RNO4, a peak LRS value of 11.1 was observed at D4Mgh7. Although this LRS value exceeded the permutation-derived threshold suggestive of a QTL (7.6), it did not exceed the threshold indicative of a significant QTL (14.0). By contrast, CIM-derived LRS values on RNO5 did not exceed the permutation-derived threshold suggestive of a QTL (7.6). The six significant Ept loci and the suggestive QTL on RNO4 together are estimated to account for 40% of the phenotypic variance, whereas environmental factors are estimated to account for 34% of the variance.
Figure 2.—
Six quantitative trait loci determine sensitivity to DES-induced pituitary growth in F2 progeny from reciprocal crosses between the ACI and COP rat stains. Genotypes were determined at the indicated polymorphic SSLP markers for a total of 264 phenotypically defined F2 progeny from reciprocal crosses between the ACI and COP strains. Each horizontal axis represents the genetic map of the indicated rat chromosome in Haldane centimorgans and the markers at which genotypes were determined. Each vertical axis represents the LRS value for the correlation between log10-transformed pituitary mass and genotype along each chromosomal interval as determined by CIM. Each solid box represents the bootstrapping-derived confidence interval. (A) Ept1 is defined by an LRS peak of 20.4 at D6Rat80 and a confidence interval of ∼15 cM extending from D6Rat150 to D6Rat39. Permutation-derived thresholds for significant (14.3; solid line) and highly significant (19.6; dotted line) evidence of a QTL are shown. (B) Ept2 is defined by an LRS peak of 38.4 between D3Rat37 and D3Rat26 and a confidence interval of ∼48 cM from D3Rat130 to D3Rat21. Permutation-derived thresholds for significant (13.7; solid line) and highly significant (20.0; dotted line) evidence of a QTL are shown. (C) Ept6 is defined by an LRS peak of 30.6 at D3Mgh9 and a confidence interval of ∼17 cM from D3Mgh16 to D3Rat277. Permutation-derived thresholds for significant (14.0; solid line) and highly significant (20.5; dotted line) evidence of a QTL are shown. (D) Ept9 is defined by an LRS peak of 17.9 between D10Rat21 and D10Mit7 and a confidence interval of ∼35 cM from D10Rat27 to D10Rat11. The permutation-derived threshold (14.1) for significant evidence of a QTL is shown. (E) Ept10 is defined by an LRS peak of 17.4 between D1Rat119 and D1Rat81 and a confidence interval of ∼60 cM from D1Rat133 to D1Rat81. The permutation-derived threshold (14.2) for significant evidence of a QTL is shown. (F) Ept13 is defined by an LRS peak of 15.6 at D1Rat192 and a confidence interval of ∼57 cM from D1Rat192 to D1Rat259. The permutation-derived threshold for significant (14.5) evidence of a QTL is shown.
TABLE 3.
Actions ofEpt loci on DES-induced pituitary mass
| QTL | Markera | Position (cM)b |
Peak LRS valuec |
% varianced |
Additive effecte |
Degree of dominancef |
Phenotype A/A − C/Cg |
|---|---|---|---|---|---|---|---|
| Ept1 | D6Rat80 | 11 | 20.4 | 8 | 0.05 | −0.41 | 16.3 |
| Ept2 | D3Rat26 | 73 | 38.4 | 14 | 0.07 | 0.00 | 24.2 |
| Ept6 | D3Mgh9 | 4 | 30.6 | 11 | −0.07 | −0.28 | −19.7 |
| Ept9 | D10Mit7 | 104 | 17.9 | 7 | 0.05 | 0.00 | 13.8 |
| Ept10 | D1Rat119 | 212 | 17.4 | 7 | −0.04 | 0.73 | −15.8 |
| Ept13 | D1Rat192 | 0 | 15.6 | 6 | 0.04 | −0.50 | 14.6 |
SSLP marker nearest the LRS peak.
Position of peak LRS score in Haldane units relative to position of first marker on the chromosome.
Peak LRS value from CIM analyses.
Percentage of phenotypic variance attributed to the QTL as determined by CIM analyses.
Additive effect (Falconer and Mackay 1996) was generated using MapManager QTX and log10-transformed pituitary mass data. Positive values indicate the ACI allele is associated with increased pituitary mass. Negative values indicate the COP allele is associated with increased pituitary mass.
Degree of dominance (Falconer and Mackay 1996). 0 indicates additive affects of the ACI and COP alleles; 1 indicates full dominance of the ACI allele; −1 indicates full dominance of the COP allele.
Mean pituitary mass (milligrams) in the F2 subpopulation that is homozygous for the ACI allele at the indicated marker minus mean pituitary mass in the F2 subpopulation that is homozygous for the COP allele at that marker.
Impact of Ept loci on pituitary mass:
Data presented above indicate that growth response of the pituitary gland to DES is determined by a minimum of six QTL. CIM analyses indicate that ACI alleles of Ept1, Ept2, Ept9, and Ept13 confer increased pituitary mass in response to DES, whereas COP alleles for Ept6 and Ept10 confer increased mass (Table 3). Whereas the ACI alleles for Ept2 and Ept9 appeared to act additively, the ACI alleles for the remaining four Ept loci deviated from an additive mode of action (Table 3). When the combined F2 population was classified into subpopulations according to genotype at the marker closest to the LRS peak for each Ept locus, homozygosity for the ACI allele at Ept1, Ept2, Ept9, and Ept13 was associated with 16.3, 24.2, 13.8, and 14.6 mg, respectively, of additional pituitary mass relative to rats homozygous for the COP allele at these loci (Table 3). By contrast, homozygosity for the COP allele at Ept6 and Ept10 was associated with 19.7 and 15.8 mg, respectively, of additional pituitary mass relative to rats homozygous for the ACI allele. For each of the six Ept loci, the average pituitary mass in F2 rats homozygous for the ACI allele differed significantly from that observed in the F2 rats that were homozygous for the COP allele.
Epistatic interaction between Ept9 and Ept10:
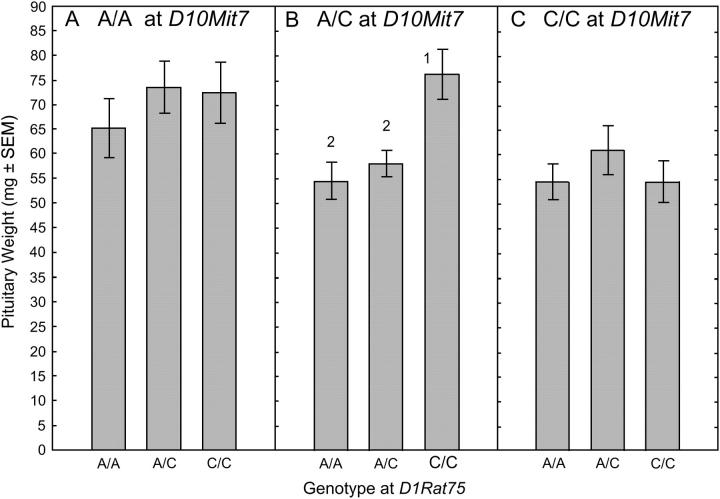
Map Manager QTX was used to evaluate potential pairwise interactions between the 58 markers residing on the six chromosomes demonstrated to harbor the six Ept loci or to have yielded suggestive evidence of a QTL during interval mapping. A single, statistically significant interaction was detected between D10Mit7, which resides within Ept9, and D1Rat75, which resides within Ept10. This interaction yielded an LRS value of 44.1, which exceeded the permutation-derived threshold, 42.0, indicative of highly significant evidence of interaction. To illustrate the nature of this interaction, the F2 population was initially subdivided on the basis of genotype at D10Mit7 and then further subdivided by genotype at D1Rat75 (Figure 3). In F2 rats homozygous for either the ACI allele (Figure 3A) or the COP allele (Figure 3C) at the Ept9 marker D10Mit7, genotype at the Ept10 marker D1Rat75 had no significant effect on pituitary mass. By contrast, among the F2 rats that were heterozygous at the Ept9 marker D10Mit7, rats homozygous for the COP allele at the Ept10 marker D1Rat75 exhibited significantly increased pituitary mass, relative to that exhibited by F2 rats that were either homozygous for the ACI allele or heterozygous at D1Rat75 (Figure 3B). The biological significance of this interaction is not known at this time.
Figure 3.—
Characterization of the interaction between Ept9 and Ept10. Each of the 264 phenotypically defined F2 rats from the reciprocal crosses between the ACI and COP rat strains was classified by genotype at the Ept9 marker D10Mit7 and then further classified by genotype at the Ept10 marker D1Rat75. Each data bar represents the mean mass of the anterior pituitary gland in milligrams ± the standard error of the mean. The Wilcoxon rank sum test was performed to assess differences in pituitary mass between the genotypically defined subpopulations. P-values ≤0.05 were considered indicative of statistical significance. Numeral 1 indicates that pituitary mass differs significantly from that observed in the subpopulation homozygous for the ACI allele at D1Rat75. Numeral 2 indicates that pituitary mass differs significantly from that observed in the subpopulation homozygous for the COP allele at D1Rat75.
DISCUSSION
It has been known for >60 years that chronic treatment with estrogens, either naturally occurring or synthetic, leads to development of pituitary tumors in rats (Mceuen et al. 1936; Segaloff and Dunning 1945; Clifton and Meyer 1956) and mice (Gardner and Strong 1940; Gardner 1941; Spady et al. 1999b). Moreover, substantial evidence suggests that estrogens may contribute to development of pituitary tumors in humans (Landolt et al. 1984; Holmgren et al. 1986; Miyai et al. 1986; Gooren et al. 1988; Panteon et al. 1988; Bevan et al. 1989; Kovacs et al. 1994). However, the mechanisms through which estrogens induce pituitary tumor development remain poorly defined. Elucidation of the genetic bases of the differing sensitivities of the genetically related ACI and COP rat strains to DES-induced pituitary growth will likely provide novel insights into the mechanisms through which estrogens regulate pituitary lactotroph homeostasis and induce pituitary tumor development.
The data presented in this study indicate that estrogen-induced pituitary growth behaves as a quantitative trait in reciprocal crosses between the ACI and COP rat strains. Six QTL that determine sensitivity to DES-induced pituitary growth have been mapped through CIM analyses of the F2 populations generated in these crosses. Thus, the genetic bases of estrogen-induced pituitary tumor development in these crosses are more complex than proposed by us previously (Spady et al. 1999d). We have estimated that these six Ept loci, together with one suggestive locus, account for 40% of the phenotypic variance exhibited by the combined F2 population from the two crosses. Environmental factors were estimated to account for an additional 34% of phenotypic variance. These data suggest that one or more unmapped Ept loci account for the estimated 26% of the phenotypic variance that remains unexplained. It is possible that an unmapped Ept locus may reside within a region of the rat genome too distant from the nearest polymorphic marker to allow the locus to be detected. Seven segments of the rat genome exist where an additional Ept locus, were it to reside there, would be 20–28 cM from the nearest polymorphic marker. These segments are located on RNO1, -2, -5, -6, -7, -10, and -17. Moreover, the X and Y chromosomes were not fully evaluated in this study. Therefore, we cannot exclude these chromosomes as determinants of the pituitary growth response to DES. Alternatively, the remaining 26% of the phenotypic variance could result from the actions of multiple unmapped Ept loci, each of which would exert an effect on pituitary mass that was too small to be detected in our analyses (Van Ooijen 1992; Muranty and Goffinet 1997).
The six Ept loci mapped in this study segregate and appear, for the most part, to function independently of one another. ACI alleles at Ept1, Ept2, Ept9, and Ept13 are associated with increased DES-induced pituitary growth, relative to COP alleles at these loci. In contrast, COP alleles at Ept6 and Ept10 are associated with increased pituitary growth. Thus, the sensitivity of the COP strain to DES-induced pituitary growth is not simply due to the action of growth-conferring alleles shared with the genetically related ACI strain. Rather, the sensitivity of the COP strain to DES-induced pituitary growth is due, at least in part, to the action of growth-conferring alleles not carried by the ACI strain. Interestingly, we have previously demonstrated that a 40% restriction of dietary energy consumption inhibits estrogen-induced pituitary growth in COP, but not ACI, rats (Spady et al. 1999a,c; Harvell et al. 2001, 2002, 2003). Together these data suggest that the mechanism underlying estrogen-induced pituitary growth in the ACI rat strain differs, at least in part, from that in the COP strain.
Wendell et al. have mapped to RNO2, RNO3, RNO5, and RNO9 a total of six QTL, referred to as Estrogen-dependent pituitary mass (Edpm) loci, that affect DES-induced pituitary growth in female F2 and BC progeny derived from crosses between the F344 and BN rat strains (Wendell and Gorski 1997; Wendell et al. 2000). Data from our study of male F2 progeny from crosses between the ACI and COP rat strains specifically exclude pituitary growth-controlling loci from the regions of RNO2, RNO5, and RNO9 to which Wendell et al. mapped five of the six loci that control DES-induced pituitary growth in the F344 × BN crosses. The Ept2 locus mapped by us to RNO3 overlaps with the Edpm3 locus mapped by Wendell et al. Thus, five of the six Ept loci mapped in the ACI × COP crosses are clearly distinct from those mapped by Wendell et al. in crosses between the F344 and BN strains. We hypothesize that the dissimilarities in the QTL mapped in our study compared to those of Wendell et al. result from differences in the inbred strains being evaluated. However, we cannot exclude the possibility that the differences in the QTL identified in the two studies are due to variation in experimental design, such as the gender of rats used for linkage analysis.
We observed highly significant evidence of an epistatic interaction between Ept9 and Ept10. The potential interaction between Ept9 and Ept10 is particularly intriguing because Jak2 resides within the Ept10 interval, tightly linked to D1Rat75, and Stat5a and Stat5b, which are phosphorylated by Jak2, reside within the Ept9 interval, linked to D10Mit7. Together, Jak2 and Stat5a/5b mediate PRL signaling through the PRL receptor to regulate lactotroph function and number (Bole-Feysot et al. 1998; Schuff et al. 2002). The known role of Jak2 and both Stat5a and Stat5b in mediating the effects of PRL signaling makes these genes attractive candidates for Ept10 and Ept9, respectively.
In studies performed in parallel to those presented herein, analysis of female F2 progeny from reciprocal crosses between the ACI and COP strains localized to RNO5 and RNO18 two loci, referred to as Emca1 and Emca2, respectively, that determine susceptibility to 17β-estradiol-induced mammary cancer (Gould et al. 2004). Although the interval-mapping analyses performed in our study provided suggestive evidence of a QTL modifying the pituitary growth response to DES on RNO5, this putative QTL was not confirmed during CIM analyses. Similarly, the genetic analyses described herein indicate that RNO18 does not harbor a genetic determinant of DES-induced pituitary growth. It is also interesting that the regions of the genome in which the six Ept loci reside do not exert discernible effects on susceptibility to 17β-estradiol-induced mammary cancer in the female F2 progeny (Gould et al. 2004). Thus, the QTL that determine sensitivity to DES-induced pituitary growth in male F2 progeny are distinct from those that determine susceptibility to 17β-estradiol-induced mammary cancer in female F2 progeny evaluated in these reciprocal crosses between the ACI and COP rat strains. Together, these data indicate that the genes that determine the tumorigenic response to continuous estrogen stimulation act, in large part, in a rat strain- and tissue-specific manner.
In summary, the data presented herein indicate that a minimum of six QTL control sensitivity to DES-induced pituitary growth in F2 progeny produced in crosses between the ACI and COP rat strains. Congenic rat lines are under development that will allow the impact of each Ept locus on estrogen-induced pituitary growth to be assessed, both qualitatively and quantitatively, independently of the other loci. These congenic lines will also allow each Ept locus to be mapped with greater precision. The recent assembly of the draft sequence of the rat genome will facilitate the identification of those genes residing within each Ept locus that determine sensitivity to DES-induced pituitary growth, and this, in turn, should significantly enhance our understanding of how estrogens regulate lactotroph homeostasis and contribute to development of PRL-producing pituitary tumors.
Acknowledgments
We thank the staff of the Eppley Institute Animal Care Facility for their excellent care of the research animals. We thank Douglas Wendell for many helpful suggestions on data analysis. Finally, we thank Cindy Lachel for her technical assistance and critical evaluation of this manuscript. This work was supported by National Institutes of Health (NIH) grants R01-CA68529 and R01-CA77876. NIH Cancer Center Support Grant P30-CA36727 supported shared resources within the University of Nebraska Medical Center Eppley Cancer Center. T.E.S. was supported in part by NIH training grant T32-CA09476. T.E.S. and B.S.S. were supported in part by training grant DAMD17-00-1-0361 from the U.S. Army Breast Cancer Training Program. B.S.S. was supported in part by grant DAMD17-03-1-0477 from the U.S. Army Breast Cancer Training Program. T.J.S. was supported in part by a Bukey Presidential Fellowship from the Graduate College of the University of Nebraska.
References
- Bevan, J. S., J. Sussman, A. Roberts, M. Hourihan and J. R. Peters, 1989. Development of an invasive macroprolactinoma: a possible consequence of prolonged oestrogen replacement. Case report. Br. J. Obstet. Gynecol. 96: 1440–1444. [DOI] [PubMed] [Google Scholar]
- Bole-Feysot, C., V. Goffin, M. Edery, N. Binart and P. A. Kelly, 1998. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocrinol. Rev. 19: 225–268. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton, K. H., and R. K. Meyer, 1956. Mechanism of anterior pituitary tumor induction by estrogen. Anat. Rec. 125: 65–81. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., 2002. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3: 43–52. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996 Introduction to Quantitative Genetics, Ed. 4. Longman Group, Harlow, UK.
- Furth, J., G. Ueda and K. H. Clifton, 1973. The pathophysiology of pituitaries and their tumors: methodological advances. Methods Cancer Res. 10: 201–277. [Google Scholar]
- Gardner, W. U., 1941. The effect of estrogen on the incidence of mammary and pituitary tumors in hybrid mice. Cancer Res. 1: 345–358. [Google Scholar]
- Gardner, W. U., and L. C. Strong, 1940. Strain-limited development of tumors of the pituitary gland in mice receiving estrogens. Yale J. Biol. Med. 12: 543–548. [PMC free article] [PubMed] [Google Scholar]
- Gooren, L. J. G., J. Assies, H. Asscheman, R. De Slegte and H. Van Kessel, 1988. Estrogen-induced prolactinoma in a man. J. Clin. Endocrinol. Metab. 66: 444–446. [DOI] [PubMed] [Google Scholar]
- Gould, K. A., M. Tochacek, B. S. Schaffer, T. M. Reindl, C. R. Murrin et al., 2004. Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics 168: 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell, D. M. E., T. E. Strecker, M. Tochacek, B. Xie, K. L. Pennington et al., 2000. Rat strain specific actions of 17β-estradiol in the mammary gland: correlation between estrogen-induced lobuloalveolar hyperplasia and susceptibility to estrogen-induced mammary cancers. Proc. Natl. Acad. Sci. USA 97: 2779–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell, D. M. E., T. J. Spady, T. E. Strecker, A. M. Lemus-Wilson, K. L. Pennington et al., 2001 Dietary energy restriction inhibits estrogen induced pituitary tumorigenesis in a rat strain specific manner, pp. 496–501 in Hormonal Carcinogenesis III, edited by J. J. Li, S. A. Li and J. R. Daling. Springer, New York.
- Harvell, D. M. E., T. E. Strecker, B. Xie, K. L. Pennington, R. D. McComb et al., 2002. Dietary energy restriction inhibits estrogen-induced mammary, but not pituitary, tumorigenesis in the ACI rat. Carcinogenesis 23: 161–169. [DOI] [PubMed] [Google Scholar]
- Harvell, D. M. E., L. K. Buckles, K. A. Gould, K. L. Pennington, R. D. McComb et al., 2003. Rat strain specific attenuation of estrogen action in the anterior pituitary gland by dietary energy restriction. Endocrine 21: 175–183. [DOI] [PubMed] [Google Scholar]
- Holmgren, U., G. Bergstrand, K. Hagenfeldt and S. Werner, 1986. Women with prolactinoma—effect of pregnancy and lactation on serum prolactin and on tumour growth. Acta Endocrinol. 111: 452–459. [DOI] [PubMed] [Google Scholar]
- Holtzman, S., J. P. Stone and C. J. Shellabarger, 1979. Influence of diethylstilbestrol treatment on prolactin cells of female ACI and Sprague-Dawley rats. Cancer Res. 39: 779–784. [PubMed] [Google Scholar]
- Kovacs, K., L. Stefaneanu, S. Ezzat and H. S. Smyth, 1994. Prolactin-producing pituitary adenoma in a male-to-female transsexual patient with protracted estrogen administration: a morphologic study. Arch. Pathol. Lab. Med. 118: 562–565. [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt, A. M., E. Del Pozo and J. Hayek, 1984. Injectable bromocriptine to treat acute, oestrogen-induced swelling of invasive prolactinoma. Lancet 2: 111. [DOI] [PubMed] [Google Scholar]
- Manly, K. F., R. H. Cudmore, Jr. and J. M. Meer, 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- Mceuen, C. S., H. Selye and J. B. Collip, 1936. Prolonged administration of oestrin in rats. Lancet 1: 775–776. [Google Scholar]
- Miyai, K., K. Ichihara, K. Kondo and S. Mori, 1986. Asymptomatic hyperprolactinaemia and prolactinoma in the general population-mass screening by paired assays of serum prolactin. Clin. Endocrinol. 25: 549–554. [DOI] [PubMed] [Google Scholar]
- Muranty, H., and B. Goffinet, 1997. Selective genotyping for location and estimation of the effect of a quantitative trait locus. Biometrics 53: 629–643. [Google Scholar]
- Noble, R. L., C. S. Mceuen and J. B. Collip, 1940. Mammary tumours produced in rats by the action of oestrone pellets. Can. Med. Assoc. J. 42: 413–417. [PMC free article] [PubMed] [Google Scholar]
- Pandey, J., A. Bannout and D. L. Wendell, 2004. The Edpm5 locus prevents the ‘angiogenic switch’ in an estrogen-induced rat pituitary tumor. Carcinogenesis 25: 1829–1838. [DOI] [PubMed] [Google Scholar]
- Panteon, E., E. Loumaye, M. Maes and P. Malvaux, 1988. Occurrence of prolactinoma after estrogen treatment in a girl with constitutional tall stature. J. Pediatr. 113: 337–339. [DOI] [PubMed] [Google Scholar]
- Schuff, K. G., S. T. Hentges, M. A. Kelly, N. Binart, P. A. Kelly et al., 2002. Lack of prolactin receptor signaling in mice results in lactotroph proliferation and prolactinomas by dopamine-dependent and -independent mechanisms. J. Clin. Invest. 110: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segaloff, A., and W. F. Dunning, 1945. The effect of strain, estrogen, and dosage on the reaction of the rat's pituitary and adrenal to estrogenic stimulation. Endocrinology 36: 238–240. [Google Scholar]
- Shull, J. D., 2002 Hormonal carcinogenesis, pp. 417–428 in Encyclopedia of Cancer, Vol. 2, Ed. 2, edited by J. R. Bertino. Academic Press, San Diego.
- Shull, J. D., T. J. Spady, M. C. Snyder, S. L. Johansson and K. L. Pennington, 1997. Ovary intact, but not ovariectomized female ACI rats treated with 17β-estradiol rapidly develop mammary carcinoma. Carcinogenesis 18: 1595–1601. [DOI] [PubMed] [Google Scholar]
- Shull, J. D., K. L. Pennington, T. M. Reindl, M. C. Snyder, T. E. Strecker et al., 2001. Susceptibility to estrogen-induced mammary cancer segregates as an incompletely dominant phenotype in reciprocal crosses between the ACI and Copenhagen rat strains. Endocrinology 142: 5124–5130. [DOI] [PubMed] [Google Scholar]
- Spady, T. J., D. M. E. Harvell, M. C. Snyder, K. L. Pennington, R. D. Mccomb et al., 1998. a Estrogen-induced tumorigenesis in the Copenhagen rat: disparate susceptibilities to development of prolactin-producing pituitary tumors and mammary carcinomas. Can. Lett. 124: 95–103. [DOI] [PubMed] [Google Scholar]
- Spady, T. J., A. M. Lemus-Wilson, K. L. Pennington, D. J. Blackwood, T. M. Paschall et al., 1998. b Dietary energy restriction abolishes development of prolactin-producing pituitary tumors in Fischer 344 rats treated with 17β-estradiol. Mol. Carcinogen. 23: 86–95. [DOI] [PubMed] [Google Scholar]
- Spady, T. J., D. M. E. Harvell, A. Lemus-Wilson, T. E. Strecker, K. L. Pennington et al., 1999. a Modulation of estrogen action in the pituitary and mammary glands by dietary energy consumption. J. Nutr. 129: 587S–590S. [DOI] [PubMed] [Google Scholar]
- Spady, T. J., R. D. Mccomb and J. D. Shull, 1999. b Estrogen action in the regulation of cell proliferation, cell survival, and tumorigenesis in the rat anterior pituitary gland. Endocrine 11: 217–233. [DOI] [PubMed] [Google Scholar]
- Spady, T. J., K. L. Pennington, R. D. Mccomb, D. F. Birt and J. D. Shull, 1999. c Estrogen-induced pituitary tumor development in the ACI rat is not inhibited by dietary energy restriction. Mol. Carcinogen. 26: 239–253. [PubMed] [Google Scholar]
- Spady, T. J., K. L. Pennington, R. D. Mccomb and J. D. Shull, 1999. d Genetic bases of estrogen-induced pituitary growth in an intercross between the ACI and Copenhagen rat strains: dominant Mendelian inheritance of the ACI phenotype. Endocrinology 140: 2828–2835. [DOI] [PubMed] [Google Scholar]
- Teuscher, C., R. J. Butterfield, R. Z. Ma, J. F. Zachary, R. W. Doerge et al., 1999. Sequence polymorphisms in the chemokines Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5) are candidates for eae7, a locus controlling susceptibility to monophasic remitting/nonrelapsing experimental allergic encephalomyelitis. J. Immunol. 163: 2262–2266. [PubMed] [Google Scholar]
- Van Ooijen, J. W., 1992. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 84: 803–811. [DOI] [PubMed] [Google Scholar]
- Visscher, P. M., R. Thompson and C. S. Haley, 1996. Confidence intervals in QTL mapping by bootstrapping. Genetics 143: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell, D. L., and J. Gorski, 1997. Quantitative trait loci for estrogen-dependent pituitary tumor growth in the rat. Mamm. Genome 8: 823–829. [DOI] [PubMed] [Google Scholar]
- Wendell, D. L., A. Herman and J. Gorski, 1996. Genetic separation of tumor growth and hemorrhagic phenotypes in an estrogen-induced tumor. Proc. Natl. Acad. Sci. USA 93: 8112–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell, D. L., S. B. Daun, M. B. Stratton and J. Gorski, 2000. Different functions of QTL for estrogen-dependent tumor growth of the rat pituitary. Mamm. Genome 11: 855–861. [DOI] [PubMed] [Google Scholar]
- Wendell, D. L., J. Pandey and P. Kelley, 2002. A congenic strain of rat for investigation of control of estrogen-induced growth. Mamm. Genome 13: 664–666. [DOI] [PubMed] [Google Scholar]
- Wiklund, J., J. Rutledge and J. Gorski, 1981. a A genetic model for the inheritance of pituitary tumor susceptibility in F344 rats. Endocrinology 109: 1708–1714. [DOI] [PubMed] [Google Scholar]
- Wiklund, J., N. Wertz and J. Gorski, 1981. b A comparison of estrogen effects on uterine and pituitary growth and prolactin synthesis in F344 and Holtzman rats. Endocrinology 109: 1700–1707. [DOI] [PubMed] [Google Scholar]
- Wiklund, J. A., and J. Gorski, 1982. Genetic differences in estrogen-induced deoxyribonucleic acid synthesis in the rat pituitary: correlations with pituitary tumor susceptibility. Endocrinology 111: 1140–1149. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1968 Evolution and the Genetics of Populations: A Treatise in Three Volumes, Vol. 1, pp. 373–420. University of Chicago Press, Chicago.
- Zeng, Z-B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]