Abstract
Gross chromosomal rearrangements (GCRs) have been observed in many cancers. Previously, we have demonstrated many mechanisms for suppression of GCR formation in yeast. However, pathways that promote the formation of GCRs are not as well understood. Here, we present evidence that the Rad1-Rad10 endonuclease, which plays an important role in nucleotide excision and recombination repairs, has a novel role to produce GCRs. A mutation of either the RAD1 or the RAD10 gene reduced GCR rates in many GCR mutator strains. The inactivation of Rad1 or Rad10 in GCR mutator strains also slightly enhanced methyl methanesulfonate sensitivity. Although the GCRs induced by treatment with DNA-damaging agents were not reduced by rad1 or rad10 mutations, the translocation- and deletion-type GCRs created by a single double-strand break are mostly replaced by de novo telomere-addition-type GCR. Results presented here suggest that Rad1-Rad10 functions at different stages of GCR formation and that there is an alternative pathway for the GCR formation that is independent of Rad1-Rad10.
DIFFERENT types of genomic instabilities are observed in many cancers (Lengauer et al. 1998; Vessey et al. 1999; Kolodner et al. 2002). High levels of gross chromosomal rearrangements (GCRs), such as translocations, deletions of chromosome arms, interstitial deletions, inversions, and gene amplification have been reported in many different cancers (Rennstam et al. 2001; Matzke et al. 2003). Such high levels of GCRs in cancer cells could be caused by mutator mutations and as a result facilitate further accumulation of genetic changes (Kolodner et al. 2002; Loeb et al. 2003). It has been documented that many cancer susceptibility syndromes have inherited mutations that cause problems in DNA damage responses or DNA recombination/repair and result in higher frequencies of spontaneous and/or DNA-damage-induced chromosomal aberrations (Khanna and Jackson 2001; Kolodner et al. 2002).
To understand the mechanisms for suppression of GCRs, quantitative assays that can measure different GCR events were developed in Saccharomyces cerevisiae (Chen and Kolodner 1999; Myung et al. 2001c; Kolodner et al. 2002; Huang and Koshland 2003). Currently, at least seven different pathways have been identified for the suppression of GCRs by using the following assays: (1) three different cell cycle checkpoints at S phase (Myung et al. 2001c; Lengronne and Schwob 2002; Myung and Kolodner 2002; Tanaka and Diffley 2002; Huang and Koshland 2003; Banerjee and Myung 2004); (2) a recombination pathway known as break-induced replication (Myung et al. 2001a); (3) pathways that suppress de novo telomere addition (Myung et al. 2001a); (4) two pathways for proper chromatin assembly (Myung et al. 2003); (5) pathways that prevent chromosome ends from being joined to each other (Myung et al. 2001a; Chan and Blackburn 2003; Pennaneach and Kolodner 2004); (6) a mismatch repair pathway that prevents recombination between divergent DNA sequences (Myung et al. 2001b); and (7) pathways that prevent oxidative damage to DNA (Huang et al. 2003; Smith et al. 2004).
More than 50 GCR mutator genes have been identified, and a mutation in each of these GCR mutator genes produces preferentially one or two different types of GCRs (Chen and Kolodner 1999; Myung et al. 2001a,b,c, 2003; Kolodner et al. 2002; Lengronne and Schwob 2002; Tanaka and Diffley 2002; Huang and Koshland 2003; Huang et al. 2003; Myung and Kolodner 2002, 2003; Smith et al. 2004). Strains carrying a mutation in some GCR mutator genes preferentially generate the de novo telomere addition type of GCR (Myung et al. 2001a,c; Smith et al. 2004). GCR mutator genes in this group include PIF1, which encodes a telomerase inhibitor (Zhou et al. 2000; Myung et al. 2001a); MEC1, which is the mammalian ataxia telangiectasia and Rad3-related gene homolog in yeast and is important in all S-phase checkpoints (Foiani et al. 2000; Kolodner et al. 2002; Osborn et al. 2002); RAD5 and RAD18, proteins that function in the postreplication repair presumably through their ubiquitin ligase activity (Broomfield et al. 2001; Matunis 2002); and ELG1, which encodes a protein participating in an alternative replication complex during DNA replication (Bellaoui et al. 2003; Ben-Aroya et al. 2003; Kanellis et al. 2003; Smith et al. 2004).
Mutations in the next group of GCR mutator genes preferentially increase ligase-4-dependent translocations as well as the de novo telomere-addition type of GCRs (Chen and Kolodner 1999; Myung et al. 2001a, 2003). The GCR mutator genes in this group are MRE11, which encodes a protein functioning in DNA recombination, S-phase cell cycle checkpoint and DNA repair (Haber 1998; Symington 2002); RAD52, which participates in almost all known DNA recombination pathways (Symington 2002); RFA1, which encodes a single-strand DNA (ssDNA)-binding protein that is important during DNA replication and recombination (Wold 1997); and CAC1, which functions in chromatin assembly (Krude and Keller 2001).
One additional GCR mutator gene is RAD27, which encodes a flap endonuclease necessary for the processing of Okazaki fragments during DNA replication (Lieber 1997; Chen and Kolodner 1999; Myung et al. 2001a). The inactivation of the RAD27 gene increases both de novo telomere-addition and translocation types of GCRs. However, the translocations observed in rad27 strains are not dependent on ligase 4 (Myung et al. 2001a, 2003).
Although >50 genes whose mutations increase GCR formation have been identified, genes encoding proteins that participate in the formation of GCRs are poorly understood. Currently, there are only a few known proteins that function to generate GCRs. Telomerase (Est2) and other telomere maintenance proteins, such as the yKu70-80 heterodimer, Est1, Est3, and Cdc13, function to add telomeric sequences at the ends of broken chromosomes to form de novo telomere-addition-type GCR (Myung et al. 2001a). Ligase 4 and Lif1 are required for at least one type of translocation, presumably at the ligation step (Myung et al. 2001a).
As in normal DNA repair, GCR formation by misrepair first requires the conversion of damaged DNA to proper substrates for further processing. Such modification of damaged DNA is performed by certain endo- and/or exonucleases to incise unmatched DNA-DNA hybrid intermediates. One endonuclease complex, Rad1-Rad10, has been shown to participate in excising a single-stranded DNA from an unmatched DNA structure during different DNA repair pathways. This suggests that the Rad1-Rad10 could be the enzyme functioning in the GCR formation.
Rad1 and Rad10 were first identified as members of the RAD3 epistasis group required for nucleotide excision repair (NER) of ultraviolet (UV)-damaged DNA in the yeast S. cerevisiae (Friedberg 2001). The human homolog of the RAD1 gene, XPF, is frequently mutated in xeroderma pigmentosum (XP), a cancer-prone syndrome (Bootsma et al. 1998; Friedberg 2001). Mutation of the RAD10 mammalian homolog in murine cells, ERCC1, generates a typical XP phenotype, including extreme sensitivity to UV light and a deficiency in NER (McWhir et al. 1993; Weeda et al. 1997).
RAD1 and RAD10 encode subunits of an endonuclease complex that excises the 5′-end of damaged DNA during NER (Park and Sancar 1994; Aboussekhra et al. 1995; Ivanov and Haber 1995; Prakash and Prakash 2000). In addition, the Rad1-Rad10 endonuclease complex plays a role in processing intermediates in homologous recombination (Schiestl and Prakash 1988, 1990), in resolving DNA interstrand crosslink-induced double-strand breaks (DSBs) (Niedernhofer et al. 2004), in removing 3′-blocked termini from DSBs induced by reactive oxygen species (Guzder et al. 2004), and in the microhomology-mediated end-joining process that requires only a few nucleotide homologies at the joining junction (Ma et al. 2003). Recently, a new role has been discovered for the human Rad1-Rad10 homolog, ERCC1-XPF, in the production of the end-to-end telomere fusion in the absence of TRF2 (Zhu et al. 2003).
The enzymatic activities of the Rad1-Rad10 complex, especially the endonuclease activity for unpaired DNA intermediates in different DNA metabolisms, suggest that this complex could be the enzyme required to process DNA intermediates during GCR formation. Here, we present a novel role for the Rad1-Rad10 complex in the generation of both de novo telomere-addition and translocation types of GCRs. Furthermore, we propose mechanisms for how the Rad1-Rad10 complex functions during GCR formation.
MATERIALS AND METHODS
General genetic methods:
Media for the propagation of strains were as previously described (Myung et al. 2001c; Smith et al. 2004). All S. cerevisiae strains were propagated at 30°. Yeast transformation, yeast chromosomal DNA isolation for use as template in polymerase chain reaction (PCR), and PCRs were performed as previously described (Myung et al. 2001c; Smith et al. 2004).
Strains:
The strains used in this study are all isogenic to RDKY3615 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2-Bgl hom3-10 ade2Δ1 ade8 hxt13::URA3) for general GCR assay and to YKJM941 (MATa ura3::KAN HO::hisG leu2Δ1 trp1Δ63 his3Δ200 lys2-Bgl, hom3-10 ade2Δ1 ade8 sit1::HO-URA3). Both are of the Winston S288c background. All strains were generated using standard PCR-based gene disruption methods and correct gene disruptions were verified by PCR as described previously (Myung et al. 2001c; Smith et al. 2004). The sequences of primers used to generate disruption cassettes and to confirm disruption of indicated genes are available upon request. The detailed genotypes of strains are listed in Table 1.
TABLE 1.
S. cerevisiae strains used in this study
| Strains | Relevant genotype | Plasmid | Reference |
|---|---|---|---|
| RDKY3615 background | |||
| RDKY3615 | Wild type | Chen and Kolodner (1999) | |
| RDKY3617 | rfa1-t33 | Chen and Kolodner (1999) | |
| RDKY3630 | rad27::KAN | Chen and Kolodner (1999) | |
| RDKY3633 | mre11::TRP1 | Chen and Kolodner (1999) | |
| RDKY3735 | sml1::KAN mec1::HIS3 | Myung et al. (2001c) | |
| RDKY4343 | pif1-m2 | Myung et al. (2001a) | |
| RDKY4421 | rad52::HIS3 | Myung et al. (2001a) | |
| RDKY4753 | cac1::TRP1 | Myung et al. (2003) | |
| YKJM219 | mus81::TRP1 | This study | |
| YKJM1385 | rad5::HIS3 | Smith et al. (2004) | |
| YKJM1389 | rad18::HIS3 | Smith et al. (2004) | |
| YKJM1397 | rad1::HIS3 | This study | |
| YKJM1399 | pif1-m2 rad1::HIS3 | This study | |
| YKJM1405 | elg1::HIS3 | Smith et al. (2004) | |
| YKJM1433 | rad10::HIS3 | This study | |
| YKJM1435 | pif1-m2 rad10::HIS3 | This study | |
| YKJM1525 | mms4::TRP1 | Smith et al. (2004) | |
| YKJM1684 | sml1::KAN mec1::HIS3 rad1::TRP1 | This study | |
| YKJM1686 | mre11::TRP1 rad1::HIS3 | This study | |
| YKJM1688 | cac1::TRP1 rad1::HIS3 | This study | |
| YKJM1692 | mre11::TRP1 rad10::HIS3 | This study | |
| YKJM1694 | cac1::TRP1 rad10::HIS3 | This study | |
| YKJM1696 | rad27::KAN rad10::TRP1 | This study | |
| YKJM1698 | rfa1-t33 rad1::TRP1 | This study | |
| YKJM1700 | rfa1-t33 rad10::TRP1 | This study | |
| YKJM1707 | rad27::KAN rad1::TRP1 | This study | |
| YKJM1709 | rad5::TRP1 rad1::HIS3 | This study | |
| YKJM1713 | rad52::TRP1 rad1::HIS3 | This study | |
| YKJM1722 | sml1::KAN mec1::HIS3 rad10::TRP1 | This study | |
| YKJM1724 | elg1::HIS3 rad1::TRP1 | This study | |
| YKJM1726 | elg1::HIS3 rad10::TRP1 | This study | |
| YKJM1755 | rad5::HIS3 rad10::TRP1 | This study | |
| YKJM1833 | rad1::HIS3 rad10::TRP1 | This study | |
| YKJM1897 | rad18::HIS3 rad1::TRP1 | This study | |
| YKJM1899 | rad18::TRP1 rad10::HIS3 | This study | |
| YKJM2341 | mms4::TRP1 rad1::HIS3 | This study | |
| YKJM2343 | mms4::TRP1 rad10::HIS3 | This study | |
| YKJM2345 | mus81::TRP1 rad1::HIS3 | This study | |
| YKJM2347 | mus81::TRP1 rad10::HIS3 | This study | |
| YKJM2440 | Wild type | p42K-TEF, pKJM362 | This study |
| YKJM2442 | Wild type | pKJM358, pKJM373 | This study |
| YKJM2448 | pif1::HYG | p42K-TEF, pKJM362 | This study |
| YKJM2450 | pif1::HYG | pKJM358, pKJM373 | This study |
| YKJM941 background | |||
| YKJM1659 | Wild type | pRS315 | This study |
| YKJM1661 | Wild type | pRDK899 | This study |
| YKJM1885 | rad10::TRP1 | pRS315 | This study |
| YKJM1886 | rad10::TRP1 | pRDK899 | This study |
| YKJM1887 | rad1::TRP1 | pRS315 | This study |
| YKJM1888 | rad1::TRP1 | pRDK899 | This study |
All strains are isogenic to Winston S288w background, RDKY3615 [ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 YEL069::URA3] or YKJM941 [ura3::KAN leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 ΔHO sit1::URA3-HO], for general GCR assay or for an HO-inducible assay, respectively, except for the mutations and plasmids indicated. The pif1-m2 mutation inactivates only the nuclear form of Pif1 since only the second methionine codon for the translation start of the nuclear form of Pif1 is mutated while the first methionine codon for the translation start of the mitochondrial form of Pif1 is intact. pRS315 (LEU2) is a backbone vector containing an ARS1 for a replication origin used to create pRDK899. pRDK899 encodes an HO endonuclease under a galactose-inducible promoter. p42K-TEF (KAN) and pKJM362 (TRP1) are backbone vectors for overexpression. pKJM358 and pKJM373 overexpress Rad1 and Rad10, respectively.
Construction of Rad1 and Rad10 overexpression strains:
The RAD1 and RAD10 genes were amplified from yeast chromosomal DNA by PCR with the primers PRKJM791 (5′-cgcggatccCTTTCCAGATGTCTCAGTTATTTTATCAGGGCG) and PRKJM792 (5′-ccggagctcCTTATAACATATACGGTCGAAGTCACCAAATG) for RAD1 or PRKJM793 (5′-cgcggatccGGTTATCCTAGAAGATGAACAATACTGATCC) and PRKJM794 (5′-ccggagctcCAAGGTTAACAAATTAATCCTTCGAAAAG) for RAD10, respectively. The sequences in lowercase are additional sequences for restriction enzyme digestion for cloning purposes. The amplified RAD1 and RAD10 genes were cloned in the pCR2.1 vector (Invitrogen, San Diego) and labeled pKJM345 (RAD1) and pKJM346 (RAD10). The RAD1 and RAD10 genes were then moved to p42K-TEF (Dualsystems Biotech) or pKJM362, which contains the TEF promoter, multicloning sites, and CYC terminator that are the same as p42K-TEF but in the pRS424 (HIS3) vector backbone. The expressions of RAD1 and RAD10 were then confirmed by UV sensitivity complementation of rad1 or rad10 strains, respectively (data not shown). The overexpression plasmids for RAD1 or RAD10 were then labeled pKJM358 and pKJM373, respectively. pKJM358 and pKJM373 were then transformed into YKJM2366 (pif1::HYG) to create a RAD1 and RAD10 overexpression strain and the resulting strain was labeled YKJM2450. p42K-TEF and pKJM362 were transformed into YKJM2366 to generate a control strain and the resulting strain was labeled YKJM2448.
Characterization of GCR rates and breakpoints:
All GCR rates were determined independently by fluctuation analysis using the method of the median with at least two independent clones two or more times using 5 or 11 cultures for each clone. The average value is reported as previously described (Lea and Coulson 1948; Myung et al. 2001c). The breakpoint spectra from mutants carrying independent rearrangements were determined and classified as described (Myung et al. 2001c; Smith et al. 2004). The significance of the difference among GCR rates or breakpoint spectra was tested by the Fisher's exact probability test using programs available at http://www.leeds.ac.uk/acb/software/capsules/fept.htm.
Induction of GCRs by a single DSB, by an HO endonuclease, or by MMS treatment:
GCR assays after induction of a single DSB by HO endonuclease or treatment with MMS was performed as previously described (Myung and Kolodner 2002, 2003). Briefly, for a HO-inducible GCR assay, S. cerevisiae cells were cultured in synthetic drop-out (SD) media lacking amino acids required for selection of the plasmids that contain a galactose-inducible HO endonuclease gene until a cell density of 1–2 × 107 cells/ml was obtained. Cells were then washed twice with distilled water and incubated further for 5 hr in an equal volume of yeast extract-peptone (YP) media containing 2% (w/v) glycerol and 1% succinic acid. Freshly made 40% galactose was then added to a final concentration of 2% to induce HO-endonuclease expression and cells were incubated for 2 hr. Cells were washed with distilled water twice and suspended in 10 times volume of YP media containing 2% glucose (YPD) and incubated overnight until the culture reached saturation. The cells were then plated onto YPD plates and plates containing both 5-fluoroorotic acid (5-FOA) and canavanine (FC). The frequency of cells resistant to both drugs was determined. Five independent cultures of each strain were used in each experiment and each experiment was performed at least twice. For MMS treatment GCR assays, yeast were cultured as above for HO-inducible GCR assays, to obtain 1–2 × 107 cells/ml densities and incubated with the indicated concentration of MMS for 2 hr. After three washes with distilled water, cells were resuspended in 10 volumes of YPD media and cultured overnight. The next day, GCR frequencies were determined as in HO-inducible GCR assays. Five independent cultures of each strain were used in each experiment and each experiment was performed at least twice. The average fold increases in the GCR frequency of treatment relative to that of each control are described in the results.
Sensitivity to MMS:
Exponential phase S. cerevisiae cells were serially diluted, and 3 μl of cells was spotted onto different plates. The cells were spotted onto a YPD plate and onto YPD plates containing 0.005% MMS for MMS sensitivity. After 2–3 days incubation at 30°, pictures were taken. To test sensitivities of strains for acute exposure to MMS, log-phase yeast cells were treated with various doses of MMS for 2 hr followed by washing two times with distilled water. Serially diluted cells were then plated onto YPD plates and surviving colonies were counted. The percentage of survival was calculated by comparing cell numbers obtained from no treatment controls. For both chronic and acute survival tests, two independent clones for each mutant strain were tested at least twice.
RESULTS
Although GCR suppression mechanisms have been studied extensively (Chen and Kolodner 1999; Myung et al. 2001a,b,c, 2003; Kolodner et al. 2002; Lengronne and Schwob 2002; Myung and Kolodner 2002, 2003; Tanaka and Diffley 2002; Huang and Koshland 2003; Huang et al. 2003; Smith et al. 2004), the mechanism of GCR formation when DNA damage is not properly repaired is poorly characterized. In the present study, we demonstrate that the Rad1-Rad10 endonuclease complex, which normally functions in NER and recombination repair (Schiestl and Prakash 1988, 1990; Friedberg 2001), is also important for the formation of GCRs.
Mutations in the RAD1 or RAD10 genes reduced spontaneous GCR formation:
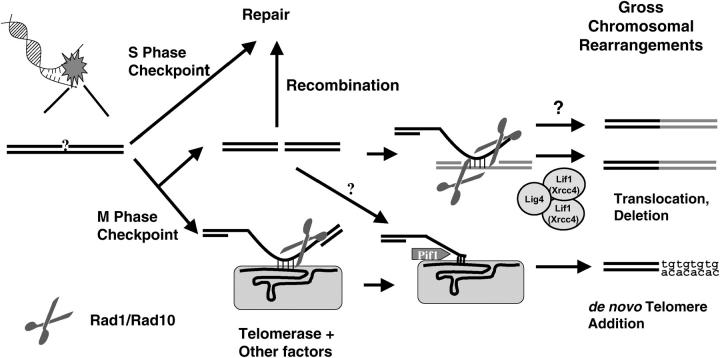
The breakpoint junction structures investigated from both de novo telomere additions and translocations showed minimal homology with 2–10 nucleotide identities (Chen and Kolodner 1999; Myung et al. 2001a,c, 2003; Kolodner et al. 2002; Myung and Kolodner 2002; Pennaneach and Kolodner 2004; Smith et al. 2004). Such minimal homology at the breakpoint junction suggests that intermediate DNA structures during GCR formations might have 3′ overhanging flap structures (see Figure 4). In the case of translocations, an invading single-stranded DNA annealed to the donor strand might cause such 3′ overhanging flap structures; and in the de novo telomere-addition case, a telomerase RNA subunit, Tlc1, hybridized with small TG repeat sequences, might be the cause. These structures need to be removed by endonucleases to continue the GCR formation process. To find such endonucleases, mutations that inactivate different nucleases were tested using the GCR assay.
Figure 4.—
Hypothetical model of how the Rad1-Rad10 endonuclease complex functions for the formation of both de novo telomere-addition and translocation- or deletion-type GCRs. DNA damage, which might be generated during DNA replication or telomere erosion, activates the S-phase checkpoint for proper repair. However, if DNA damage is too high or there is a mutation that allows for GCR formation, DNA damage may escape from proper repair and the mitotic checkpoint signals the generation of GCRs. DNA damage at this point can be processed to generate a DSB, if not properly repaired by recombination, or could invade another DNA region and the imperfectly matched DNA hybrid will be trimmed by the Rad1-Rad10 endonuclease complex. Then, ligase 4/Lif1-dependent or -independent translocation/deletion-type GCRs are produced. However, DNA damage can be directly recognized by the telomerase complex and make a DNA-RNA hybrid for de novo telomere addition. The Rad1-Rad10 endonuclease complex would remove the unhybridized portion of DNA. Then, telomerase starts to add telomeric sequences to cap the end of the chromosome. The Pif1 helicase seems to inhibit at this step. There also appears to be an alternative pathway from a DSB to a de novo telomere addition that does not require the Rad1-Rad10 endonuclease complex.
Inactivation of Mre11 or Rad27, which both have endo/exonuclease activity, increased the GCR rate (Table 2). Mutation of another endonuclease, MUS81, which functions in producing D loops during recombination (Boddy et al. 2001; Osman et al. 2003), and MMS4, which encodes an interacting factor to Mus81, also increased the GCR rate 109- and 169-fold, respectively, compared to wild type (Table 2; Smith et al. 2004). Inactivation of ExoI, which encodes a 5′-3′ exonuclease that preferentially degrades double-stranded DNA in different types of DNA metabolism (Fiorentini et al. 1997; Kolodner and Marsischky 1999), also causes a 10-fold increase in the GCR rate compared to wild type (S. Smith, A. Gupta, R. D. Kolodner and K. Myung, unpublished data). Therefore, it is very unlikely that these endo- or exonucleases are responsible for the removal of 3′ flap overhang structures. However, a mutation in the RAD1 or RAD10 genes or mutations in both RAD1 and RAD10 genes slightly reduced the GCR rate compared to wild type (Table 2).
TABLE 2.
Inactivation ofRAD1 and/orRAD10 genes reduces GCR formation in different GCR mutator strains
| Wild type
|
rad1Δ
|
rad10Δ
|
||||
|---|---|---|---|---|---|---|
| Relevant genotype | Strain | Mutation rate | Strain | Mutation rate | Strain | Mutation rate |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | YKJM1397 | 1.9 × 10−10 (0.5) | YKJM1433 | 1.0 × 10−10 (0.3) |
| pif1-m2 | RDKY4343 | 6.3 × 10−8 (180) | YKJM1399 | 5.2 × 10−8 (149) | YKJM1435 | 5.6 × 10−8 (160) |
| mec1Δ | RDKY3735 | 6.4 × 10−8 (183) | YKJM1684 | 5.4 × 10−9 (15) | YKJM1722 | 2.0 × 10−8 (56) |
| rad5Δ | YKJM1385 | 4.5 × 10−8 (127) | YKJM1709 | 5.3 × 10−9 (15) | YKJM1755 | 2.2 × 10−8 (61) |
| rad18Δ | YKJM1389 | 3.6 × 10−8 (103) | YKJM1897 | 5.2 × 10−9 (15) | YKJM1899 | 6.7 × 10−9 (19) |
| elg1Δ | YKJM1405 | 1.7 × 10−8 (49) | YKJM1724 | <1.1 × 10−9 (3) | YKJM1726 | 1.5 × 10−9 (4) |
| mre11Δ | RDKY3633 | 2.2 × 10−7 (629) | YKJM1686 | 3.9 × 10−8 (111) | YKJM1692 | 6.0 × 10−8 (171) |
| rad52Δ | RDKY4421 | 4.4 × 10−8 (126) | YKJM1713 | 2.4 × 10−9 (7) | NDa | |
| rfa1-t33 | RDKY3617 | 4.7 × 10−7 (1343) | YKJM1698 | 7.4 × 10−9 (21) | YKJM1700 | 5.1 × 10−9 (15) |
| cac1Δ | RDKY4753 | 1.2 × 10−7 (343) | YKJM1688 | <7.7 × 10−10 (2) | YKJM1694 | 4.3 × 10−10 (1) |
| mus81Δ | YKJM219 | 3.8 × 10−8 (109) | YKJM2345 | 4.6 × 10−9 (13) | YKJM2347 | 2.1 × 10−9 (6) |
| mms4Δ | YKJM1525 | 5.9 × 10−8 (169) | YKJM2341 | 1.5 × 10−9 (4) | YKJM2343 | 2.3 × 10−9 (6) |
| rad27Δ | RDKY3630 | 5.0 × 10−7 (1429) | YKJM1707 | 1.7 × 10−7 (471) | YKJM1696 | 2.9 × 10−7 (814) |
All strains are isogenic to the wild-type strain, RDKY3615 [ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 YEL069::URA3], with the exception of the indicated mutations. The numbers in parentheses indicate the fold induction of GCR relative to wild type. The GCR rate of the rad1Δ rad10Δ (YKJM1833) strain was <4.1 × 10−10.
Not determined. The pif1-m2 mutation inactivates only the nuclear form of Pif1, since only the second methionine codon for the translation start of the nuclear form of Pif1 is mutated while the first methionine codon for the translation start of the mitochondrial form of Pif1 is intact. Mutation rates are CanR-5-FOAR per generation.
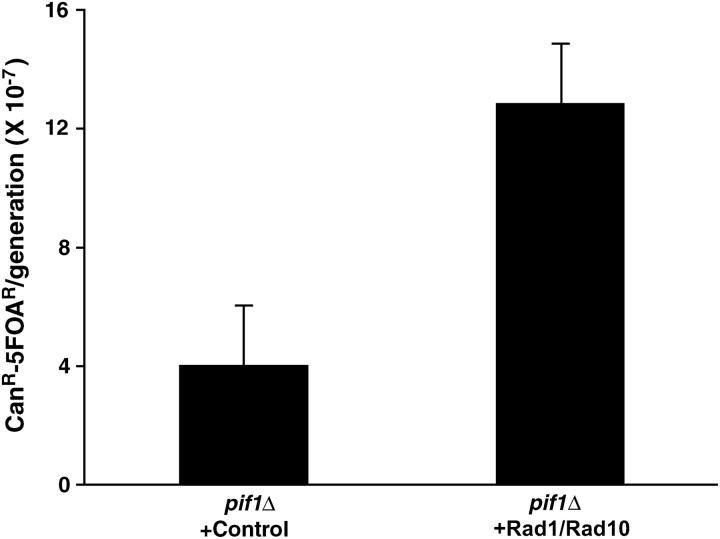
To confirm that the Rad1-Rad10 endonuclease contributes to GCR formation, we determined the GCR rates of strains carrying mutations in a GCR mutator gene along with either rad1 or rad10 (Table 2). When mutations specifically increasing de novo telomere addition types of GCRs, such as mec1, elg1, rad5, or rad18, were combined with either rad1 or rad10, the GCR rates were decreased significantly, by 3- to 50-fold. Strains carrying either rad1 or rad10 and a mutation in a GCR mutator gene that increases both de novo telomere-addition and ligase-4-dependent translocation types of GCRs, such as mre11, rad52, or rfa1-t33, also showed reductions in the GCR formation rate compared to those observed in strains carrying only a GCR mutator gene mutation (Table 2). The GCR rate observed in the rad27 strain was decreased 3- and 2-fold by the additional rad1 or rad10 mutation, respectively. Additional rad1 or rad10 mutations also decreased the GCR rate observed in strains containing a mus81 or mms4 mutation (Table 2). Furthermore, the overexpression of Rad1 and Rad10 proteins in the pif1 strain increased the GCR rate >3-fold, as compared to a pif1 strain with normal Rad1 and Rad10 expression (Figure 1), while overexpression in the wild-type strain did not increase GCR rates (data not shown). Such induction of GCR rate by Rad1-Rad10 overexpression was not observed when we overexpressed several other proteins including Siz1, Ubc9, or Smt3 in the pif1 strain (data not shown). However, the addition of rad1 or rad10 mutations to a pif1-m2 strain, where only the nuclear Pif1 is absent, did not change the GCR rate observed in that strain (Table 2).
Figure 1.—
Overexpression of Rad1 and Rad10 in a pif1Δ strain increases GCR rates. The RAD1 and RAD10 genes were expressed under a strong TEF promoter in high-copy-number plasmids and their effect on the GCR rate was determined. The same high-copy-number plasmids with a TEF promoter without any genes were used as a control. pif1Δ + control, YKJM2448; pif1Δ + Rad1/Rad10,YKJM2450.
The Rad1-Rad10 endonuclease complex promotes both de novo telomere-addition and translocation types of GCRs:
GCR mutator mutations increase different types of GCRs (Table 3; Chen and Kolodner 1999; Myung et al. 2001a,c, 2003; Kolodner et al. 2002; Myung and Kolodner 2002; Pennaneach and Kolodner 2004; Smith et al. 2004). To know whether the Rad1-Rad10 endonuclease complex can promote the formation of a specific GCR or all types of GCR, breakpoint spectra of GCRs generated from strains carrying the rad1 or rad10 mutation with GCR mutator mutations were compared to those observed from strains carrying only a GCR mutator mutation (Table 3). If the Rad1-Rad10 endonuclease complex promotes a specific GCR type, a mutation in the RAD1 or RAD10 genes will reduce only a specific type of GCR.
TABLE 3.
rad1 orrad10 mutations reduce different types of GCR formation in different mutator strains
| Relevant genotype | Strain | Telomere addition | Translocation or deletion |
|---|---|---|---|
| Wild typea | RDKY3615 | 5 (2.9 × 10−10) | 1 (6.0 × 10−11) |
| mec1Δ sml1Δa | RDKY3735 | 9 (6.4 × 10−8) | 0 (<2.9 × 10−9) |
| mec1Δ sml1Δ rad1Δ | YKJM1684 | 8 (4.0 × 10−9) | 2 (1.4 × 10−9) |
| mec1Δ sml1Δ rad10Δ | YKJM1722 | 8 (1.6 × 10−8) | 2 (4.0 × 10−9) |
| rfa1-t33b | RDKY3617 | 5 (2.1 × 10−7) | 6 (2.6 × 10−7) |
| rfa1-t33 rad1Δ | YKJM1698 | 8 (5.9 × 10−9) | 2 (1.5 × 10−9) |
| rfa1-t33 rad10Δ | YKJM1700 | 6 (3.1 × 10−9) | 4 (2.0 × 10−9) |
| mre11Δb | RDKY3633 | 3 (6.6 × 10−8) | 7 (1.5 × 10−7) |
| mre11Δ rad1Δ | YKJM1686 | 4 (1.6 × 10−8) | 6 (2.3 × 10−8) |
| mre11Δ rad10Δ | YKJM1692 | 1 (6.0 × 10−9) | 9 (5.4 × 10−8) |
| rad27Δb | RDKY3630 | 4 (2.0 × 10−7) | 6 (3.0 × 10−7) |
| rad27Δ rad1Δ | YKJM1707 | 3 (5.1 × 10−8) | 7 (1.2 × 10−7) |
| rad27Δ rad10Δ | YKJM1696 | 5 (1.5 × 10−7) | 5 (1.5 × 10−7) |
| MMS treatment | |||
| Wild typec | RDKY3615 | 8 | 3 |
| rad1Δ | YKJM1397 | 11 | 2 |
| rad10Δ | YKJM1433 | 12 | 1 |
| HO induction | |||
| Wild type | YKJM1661 | 5 | 15 |
| rad1Δ | YKJM1888 | 10 | 7 |
| rad10Δ | YKJM1886 | 18 | 4 |
The number of individual GCR structures from different strains is presented. The rates in the parentheses are calculated by multiplying the GCR rates in Table 2 by the proportion of different GCR types observed.
Data from Myung et al. (2001c).
Data from Chen and Kolodner (1999).
Data from Myung and Kolodner (2002).
However, breakpoint spectra analysis in different strains carrying a GCR mutator mutation along with a rad1 or rad10 mutation showed that all types of GCRs detected by our system were reduced by the rad1 or rad10 mutation (Table 3). The high increase of de novo telomere-addition GCRs observed in the mec1 strain was reduced by either rad1 or rad10 mutations. Translocations were observed among the GCRs from the rad1 mec1 or rad10 mec1 strain; these might be due to the decrease of the de novo telomere-addition type of GCR, allowing a low level of translocations to be detected. However, the rate of translocation generated in rad1 mec1 or rad10 mec1 strains was higher than that seen in wild type. Thus, it is also possible that, in mec1 strains, rad1 or rad10 mutations promote translocation GCR formation. Both translocation and de novo telomere-addition GCRs observed in the rfa1-t33, mre11, and rad27 strains were decreased by an additional mutation in the RAD1 or RAD10 gene (Table 3). Therefore, the Rad1-Rad10 endonuclease complex promotes both de novo telomere-addition and translocation GCR formation in most backgrounds.
Incorporation of rad1 or rad10 mutation in GCR mutator strains slightly increases sensitivity to MMS:
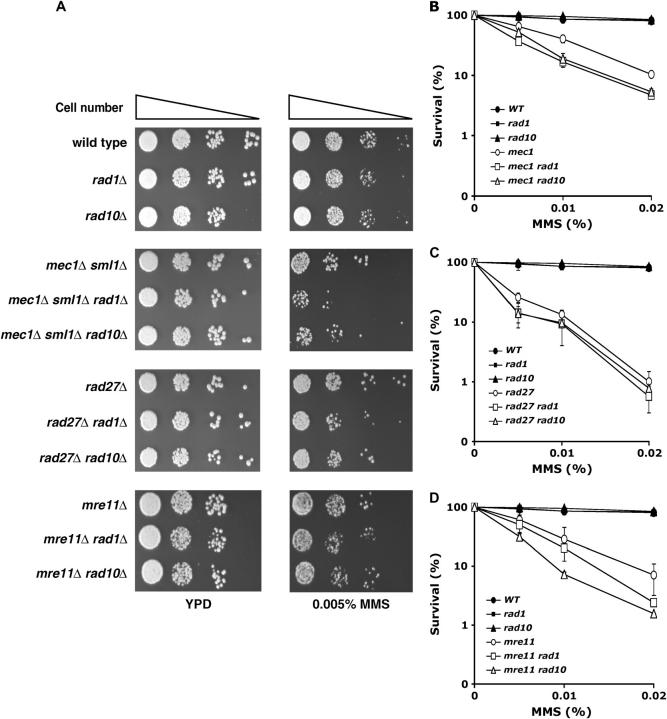
The reduced GCR rates produced by rad1 or rad10 mutations could be due to the inability to process DNA intermediates during GCR formation. If this is the case, strains carrying rad1 or rad10 mutations along with a GCR mutator gene might not tolerate DNA-damaging conditions such as MMS treatment, because at least two different pathways, a proper repair pathway and a GCR formation pathway for survival in MMS treatment, are absent. The rad1 or rad10 strains showed no increased sensitivity compared to wild type when they were grown on a YPD plate containing 0.005% MMS or when they were exposed to various doses of MMS for 2 hr (Figure 2). However, the addition of either rad1 or rad10 mutations to a mec1 strain slightly increased MMS sensitivity, when exposed either chronically (Figure 2A) or acutely (Figure 2B). A similarly modest increase of MMS sensitivity was observed in the rad27 and mre11 strains by an additional rad1 or rad10 mutation (Figure 2, A, C, and D). Recently, we reported that a deficiency in the mitotic checkpoint decreases GCR rates in many GCR mutator strains. Strains carrying mutations in a mitotic checkpoint gene and a GCR mutator gene also increased sensitivity to MMS (Myung et al. 2004). Thus, it is possible that the inactivation of the GCR formation pathway in strains carrying a GCR mutator mutation increases MMS sensitivity in general. However, it should be noted that, although the reduction of GCR formation by either rad1 or rad10 mutations in GCR mutator strains is substantial, the increase of MMS sensitivity is not.
Figure 2.—
An additional rad1 or rad10 mutation in GCR mutator strains increases sensitivity to MMS. (A) Individual strains were serially diluted and spotted onto YPD or YPD containing 0.005% MMS and incubated for 3 days at 30°. (B–D) Individual strains were exposed to the indicated doses of MMS for 2 hr during exponential growth phase and their survivals were compared to untreated controls. (B) mec1Δ sml1Δ; (C) rad27Δ; (D) mre11Δ in wild type, rad1, or rad10 backgrounds. Wild type, RDKY3615; rad1Δ, YKJM1397; rad10Δ, YKJM1433; mec1Δ sml1Δ, RDKY3735; mec1Δ sml1Δ rad1Δ, YKJM1684; mec1Δ sml1Δ rad10Δ, YKJM1722; rad27Δ, RDKY3630; rad27Δ rad1Δ, YKJM1707; rad27Δ rad10Δ, YKJM1696; mre11Δ, RDKY3633; mre11Δ rad1Δ, YKJM1686; mre11Δ rad10Δ, YKJM1692.
The inactivation of Rad1 or Rad10 does not suppress GCR formations induced by either MMS treatment or a single DSB:
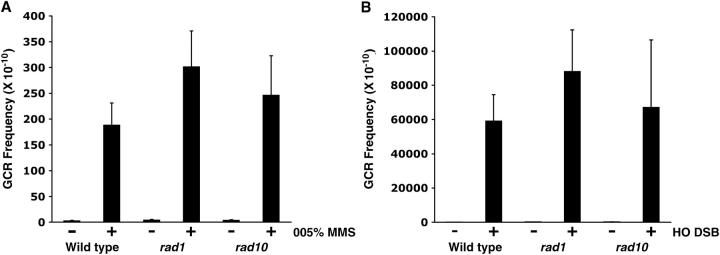
Previously, we demonstrated that GCR frequencies could be increased by MMS treatment or by the introduction of a single DSB by the HO endonuclease (Myung and Kolodner 2002, 2003). GCR formation by MMS treatment or by the introduction of a single DSB was compared in wild type and in strains carrying either the rad1 or the rad10 mutation. A 2-hr treatment with 0.05% MMS in wild type induced GCR frequencies up to 74-fold (Figure 3A). The rad1 and rad10 strains also showed a 77- and 66-fold induction, respectively, of GCR frequencies with 0.05% MMS treatment. The fold induction by MMS treatment of the rad1 and rad10 strains was not significantly different from that in wild type. The introduction of a single DSB by HO endonuclease increased the GCR frequency 462-fold in wild type (Figure 3B). When the same single DSB was introduced in rad1 or rad10 strains, similar levels of GCR induction (611- and 429-fold, respectively) were observed. Therefore, GCR formation induction by two DNA-damaging treatments does not require the Rad1-Rad10 endonuclease complex.
Figure 3.—
GCR induced by 0.05% MMS treatment or the introduction of a single DSB is not markedly affected by the rad1 or rad10 mutations. (A) The indicated yeast strains were treated with 0.05% MMS for 2 hr in log-phase growth condition and released to YPD media to measure the induction of GCR frequency (CanR-5FOAR/total). Wild type, RDKY3615; rad1Δ, YKJM1397; rad10Δ, YKJM1433. The same procedures were performed without MMS treatment for control groups. (B) A single double-strand break was introduced in the indicated yeast strain and the induction of GCR frequency was measured. Wild type, YKJM1661; rad1Δ, YKJM1888; rad10Δ, YKJM1886. As a control, the same strain containing a plasmid without the HO endonuclease was used. Wild type, YKJM1659; rad1Δ, YKJM1887; rad10Δ, YKJM1885. Five different cultures for each experiment were repeated at least twice. Average values with standard deviations are reported.
Breakpoint spectrum analysis revealed that MMS treatment preferentially generated de novo telomere-addition type GCR in wild type (Table 3). The rad1 or rad10 mutations did not change this spectrum. The most common GCRs formed upon the introduction of a single, HO-catalyzed DSB are large-deletion or translocation types of GCRs (Table 3). However, when either Rad1 or Rad10 is inactive, a much higher incidence of de novo telomere addition was observed. The breakpoint spectrum shifts by the rad1 or rad10 mutations are statistically significant (P = 0.04 and 0.0002, respectively). Thus, although the GCR frequency upon formation of a single DSB is not reduced by the rad1 or rad10 mutation, the de novo telomere-addition type GCR becomes preferred.
DISCUSSION
Previously, we demonstrated that there are >50 proteins that function in the suppression of GCRs in S. cerevisiae (Myung et al. 2001a,b,c, 2003; Kolodner et al. 2002; Myung and Kolodner 2002, 2003; Smith et al. 2004). Much less is known about the proteins participating in the formation of GCRs under conditions in which DNA repair is impaired. In this study, we demonstrate that the Rad1-Rad10 endonuclease complex makes major contributions for GCR formation under these conditions. If translocation or de novo telomere addition is mediated through ssDNA invasion of a donor strand using homology consisting of a small number of nucleotides, the unmatched 3′ overhang ssDNA should be removed by a nuclease (Figure 4). Spontaneously generated GCRs in different GCR mutator strains were drastically decreased by an additional mutation in either the RAD1 or the RAD10 gene (Table 2). Therefore, the endonuclease activity of the Rad1-Rad10 complex seems to perform this incision, to allow GCR formation to proceed, although we cannot exclude the possibility that there is a Rad1-Rad10 function other than the nuclease that participates in GCR formation.
This model is supported by the observation that the overexpression of Rad1 and Rad10 in a pif1 strain increased the GCR rate more than threefold (Figure 1). DNA damage, which is normally repaired, could be channeled to GCR formation by the overexpression of the Rad1-Rad10 proteins in the pif1 strain. However, the de novo telomere-addition type of GCR that predominates in the pif1-m2 strain is not reduced by either a rad1 or a rad10 mutation (Table 2). The inactivation of both Rad1 and Rad10 in the pif1-m2 strain caused no further increase in GCR above that observed in strains carrying only the pif1-m2 mutation. The Pif1 protein functions as a telomerase inhibitor in normal telomere maintenance (Figure 4; Zhou et al. 2000; Myung et al. 2001a). The pif1-m2 strain increases de novo telomere addition, due to its deficiency in the inhibition of telomere sequence addition to broken chromosomes by telomerase (Zhou et al. 2000; Myung et al. 2001a). The Pif1 inhibition of de novo telomere addition might happen when telomerase begins to add telomeric sequences after the Rad1-Rad10 complex trims the intermediate DNA hybrid structure. In the absence of the Rad1-Rad10 endonuclease complex, DNA damage that is to become the translocation-type GCR might be channeled to another route to be a substrate for the de novo telomere-addition type GCR by currently unknown endo- or exonucleases (Figure 4). In support of this hypothesis, breakpoint junction structures from strains carrying rad1 or rad10 mutations still showed 2- to 10-nucleotide homology or TG repeat sequences similar to those seen in RAD1 RAD10 strains (data not shown). This explains why the inactivation of Rad1 or Rad10 in a pif1-m2 strain did not alter the increased GCR rate observed in the pif1-m2 strain. Alternately, because the human homolog of Rad1-Rad10, ERCC1-XPF, interacts with TRF2 (a telomere protection protein) (Zhu et al. 2003), it is possible that a mutation in the RAD1 or RAD10 gene causes problems in telomere maintenance. As a result, the intermediate process defect and the telomere maintenance imbalance caused by a rad1 or rad10 mutation may compensate for each other. Since other GCR mutator strains carrying the rad1 or rad10 mutation decreased de novo telomere-addition type GCR, it is more likely that Pif1 inhibits telomerase after the trimming of the DNA intermediate by the Rad1-Rad10 endonuclease complex (Table 2 and Figure 4).
GCR frequencies are increased by either MMS treatment or the introduction of a single DSB (Figure 3; Myung and Kolodner 2002, 2003). Mutations of RAD1 or RAD10 genes did not reduce the GCR frequency induced by these treatments (Figure 3). However, the GCR spectra observed from the rad1 or rad10 strains show a preference for de novo telomere-addition type GCR (Table 3). This effect was most dramatic in the GCR spectra produced by a single DSB. Therefore, in the absence of Rad1-Rad10, the DSB, which is one of the DNA intermediates for GCR formation, is preferentially channeled to de novo telomere addition (Figure 4). However, because some translocations or deletions are still produced, an alternative pathway, which does require the Rad1-Rad10 complex, exists. This can also explain why the inactivation of Rad1 or Rad10 in rfa1-t33, mre11, or rad27 does not eliminate translocation or deletion types of GCRs (Table 3).
A mutation in the RAD1 or RAD10 gene in different GCR mutator strains slightly increased sensitivity to MMS (Figure 2). Similar increases of MMS sensitivity were observed when a mitotic checkpoint gene was mutated in strains carrying a GCR mutator gene (Myung et al. 2004). However, GCR rates were greatly reduced compared to those from strains carrying only a GCR mutator gene mutation. When DNA cannot be repaired properly, GCR formation might be the major pathway for repair. However, GCR can also cause haplo-lethal rearrangements. If most GCR events are haplo-lethal, this might explain why the inactivation of the GCR machinery in GCR mutator strains only slightly increases MMS sensitivity.
One of the interesting findings of this study is that Rad1 and Rad10, proteins that normally promote genome stability through DNA repair, can also be used to produce misrepair products such as GCRs. Similarly, other proteins such as the mitotic checkpoint proteins telomerase and ligase 4, which normally promote genome stability, are also required to produce GCRs (Figure 4). It is still unclear what might cause such drastic changes in the function of these proteins. However, we envision that a clear understanding of both the mechanism of GCR formation and the mechanism of GCR suppression will give insight into such regulation.
Acknowledgments
We thank S. Lee (University of Texas at San Antonio) for helpful discussions. We greatly appreciate L. Brody [National Institutes of Health (NIH)], S. Lee, P. Liu (NIH), J. Swyers (NIH), and members of the Myung laboratory for comments on the manuscript. K. Myung thanks E. Cho and God for great support. This work was supported by the National Human Genome Research Institute (NHGRI) intramural research grant (to K.M.)
References
- Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin et al., 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80: 859–868. [DOI] [PubMed] [Google Scholar]
- Banerjee, S., and K. Myung, 2004. Increased genome instability and telomere length in the elg1-deficient S. cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot. Cell 3: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone et al., 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22: 4304–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya, S., A. Koren, B. Liefshitz, R. Steinlauf and M. Kupiec, 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100: 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates et al., 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Bootsma, D., K. H. Kraemer, J. E. Cleaver and J. H. H. Hoeijmakers, 1998 Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy, pp. 245–274 in The Genetics Basis of Human Cancer, edited by B. Vogelstein and K. W. Kinzler. McGraw-Hill, New York.
- Broomfield, S., T. Hryciw and W. Xiao, 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486: 167–184. [DOI] [PubMed] [Google Scholar]
- Chan, S. W., and E. H. Blackburn, 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11: 1379–1387. [DOI] [PubMed] [Google Scholar]
- Chen, C., and R. D. Kolodner, 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner and L. S. Symington, 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17: 2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., A. Pellicioli, M. Lopes, C. Lucca, M. Ferrari et al., 2000. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res. 451: 187–196. [DOI] [PubMed] [Google Scholar]
- Friedberg, E. C., 2001. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1: 22–33. [DOI] [PubMed] [Google Scholar]
- Guzder, S. N., C. Torees-Ramos, R. E. Johnson, L. Haracska, L. Prakash et al., 2004. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3′-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev. 18: 2283–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J., 1998. The many interfaces of Mre11. Cell 95: 583–586. [DOI] [PubMed] [Google Scholar]
- Huang, D., and D. Koshland, 2003. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 17: 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M. E., A. G. Rio, A. Nicolas and R. D. Kolodner, 2003. A genome wide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, E. L., and J. E. Haber, 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis, P., R. Agyei and D. Durocher, 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13: 1583–1595. [DOI] [PubMed] [Google Scholar]
- Khanna, K. K., and S. P. Jackson, 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254. [DOI] [PubMed] [Google Scholar]
- Kolodner, R. D., and G. T. Marsischky, 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9: 89–96. [DOI] [PubMed] [Google Scholar]
- Kolodner, R. D., C. D. Putnam and K. Myung, 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Krude, T., and C. Keller, 2001. Chromatin assembly during S phase: contributions from histone deposition, DNA replication and the cell division cycle. Cell. Mol. Life Sci. 58: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Lengauer, C., K. W. Kinzler and B. Vogelstein, 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., and E. Schwob, 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9: 1067–1078. [DOI] [PubMed] [Google Scholar]
- Lieber, M. R., 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination, and repair. BioEssays 19: 233–240. [DOI] [PubMed] [Google Scholar]
- Loeb, L. A., K. R. Loeb and J. P. Anderson, 2003. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA 100: 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.-L., E. M. Kim, J. E. Haber and S. E. Lee, 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M. J., 2002. On the road to repair: PCNA encounters SUMO and ubiquitin modifications. Mol. Cell 10: 441–442. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., M. F. Mette, T. Kanno and A. J. Matzke, 2003. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 19: 253–256. [DOI] [PubMed] [Google Scholar]
- McWhir, J., J. Selfridge, D. J. Harrison, S. Squires and D. W. Melton, 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5: 217–224. [DOI] [PubMed] [Google Scholar]
- Myung, K., and R. D. Kolodner, 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., and R. D. Kolodner, 2003. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Rep. 2: 243–258. [DOI] [PubMed] [Google Scholar]
- Myung, K., C. Chen and R. D. Kolodner, 2001. a Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta, C. Chen and R. D. Kolodner, 2001. b SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta and R. D. Kolodner, 2001. c Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408. [DOI] [PubMed] [Google Scholar]
- Myung, K., V. Pennaneach, E. S. Kats and R. D. Kolodner, 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., S. Smith and R. D. Kolodner, 2004. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 15980–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas et al., 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24: 5776–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, A. J., S. J. Elledge and L. Zou, 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12: 509–516. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Park, C. H., and A. Sancar, 1994. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc. Natl. Acad. Sci. USA 91: 5017–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach, V., and R. D. Kolodner, 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36: 612–617. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13–24. [DOI] [PubMed] [Google Scholar]
- Rennstam, K., B. Baldetorp, S. Kytola, M. Tanner and J. Isola, 2001. Chromosomal rearrangements and oncogene amplification precede aneuploidization in the genetic evolution of breast cancer. Cancer Res. 61: 1214–1219. [PubMed] [Google Scholar]
- Schiestl, R. H., and S. Prakash, 1988. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol. 8: 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R. H., and S. Prakash, 1990. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol. 10: 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., J.-Y. Hwang, S. Banerjee, A. Majeed, A. Gupta et al., 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., and J. F. Diffley, 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 16: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey, C. J., C. J. Norbury and I. D. Hickson, 1999. Genetic disorders associated with cancer predisposition and genomic instability. Prog. Nucleic Acid Res. Mol. Biol. 63: 189–221. [DOI] [PubMed] [Google Scholar]
- Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens et al., 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7: 427–439. [DOI] [PubMed] [Google Scholar]
- Wold, M. S., 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66: 61–92. [DOI] [PubMed] [Google Scholar]
- Zhou, J., E. K. Monson, S. Teng, V. P. Schulz and V. A. Zakian, 2000. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289: 771–774. [DOI] [PubMed] [Google Scholar]
- Zhu, X.-D., L. Niedernhofer, B. Kuster, M. Mann, J. H. H. Hoeijmakers et al., 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12: 1489–1498. [DOI] [PubMed] [Google Scholar]