Cell polarity, defined by the asymmetric distribution of cellular components, is a fundamental property of virtually all cell types. The yeast Saccharomyces cerevisiae is an important model system to study the molecular mechanisms of polarity, and lessons learned in this organism have proven applicable to more complex species (1). During cell division, S. cerevisiae undergo a stereotypical form of polarized growth that leads to the formation of a daughter cell. In this process, known as budding, cortical cues deposited during previous cycles of cell division lead to activation of the small GTPase Cdc42, which triggers the formation of actin cables that orient toward the plasma membrane (1). These structures guide the segregation of organelles and act as tracks for secretory vesicles that deliver essential components for bud formation. In close proximity to the exocytic sites, the cells organize sites of endocytosis, which are necessary to counter the potential diffusion of membrane-bound proteins away from the site of polarized growth (2). Endocytic sites are composed of patches of actin interacting with a complex series of endocytic regulatory proteins. Several of the endocytic proteins contain independently folded protein modules, such as the epsin N-terminal homology (ENTH) domain, that interact with phospholipids, including inositol phospholipids, to induce and stabilize membrane curvature (3). Intriguingly, a new study by Aguilar et al. (4) in this issue of PNAS demonstrates that ENTH domains use a surface patch, distinct from their inositol phospholipid-binding pocket, to interact with GTPase activating proteins (GAPs) for Cdc42. Surprisingly, this leads to activation of Cdc42 signaling pathways, perhaps by acting as a sink to sequester GAP activity from Cdc42. The finding that ENTH domains interact with GAPs is exciting not only because it reveals an increased complexity of the ENTH module but also because it provides a new and direct link between endocytosis and events downstream of Cdc42 activation, including actin dynamics and cell polarity.
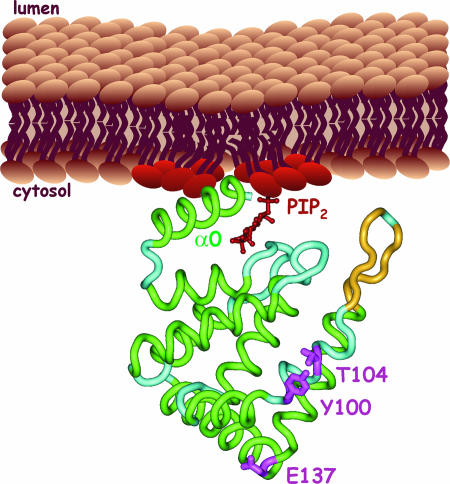
The ENTH domain was originally identified when sequence alignments with the N terminus of epsin 1 revealed an ≈150-residue region that was conserved in proteins from a diverse range of species (5, 6). Members of the epsin family had been implicated in clathrin-mediated endocytosis in mammalian cells because they localize to endocytic sites and contain motifs in their unstructured C-terminal region that allow for interactions with clathrin and other components of the endocytic machinery (7–9). The epsin ENTH domain has a compact α-helical structure (10) (Fig. 1) and binds to phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] (11, 12), which is enriched on the plasma membrane. Intriguingly, when the epsin ENTH domain binds to PtdIns(4,5)P2-rich membranes, unstructured residues at the N terminus form a new α-helix, called α0, that inserts into the inner leaflet of the bilayer, inducing curvature (13, 14) (Fig. 1). The creation of membrane curvature has been considered a major function for the ENTH domain.
Fig. 1.
Backbone trace of the ENTH domain of rat epsin crystallized in complex with inositol (1,4,5)P3 (red), which is used as a mimic of PtdIns(4,5)P2 (PIP2) head groups at the membrane. The structure is from PTB1H0A. α0 is shown to insert into the inner leaflet of the bilayer. Y100, T104, and E137, which form a surface patch for interactions with Cdc42 GAPs, are in pink.
The current study by Aguilar et al. (4) examines the function of ENTH domains in S. cerevisiae, which encode four ENTH domain-bearing proteins termed Ent1p through Ent4p. Ent1/2p are homologues of epsins, whereas Ent3p is a homologue of enthoprotin/epsinR/CLINT, an ENTH domain-bearing protein that functions in clathrin trafficking at the trans-Golgi network and endosomes (3). The role of Ent4p is not known. Similar to their mammalian counterparts, Ent1/2p have an N-terminal ENTH domain followed by a C-terminal region encoding binding motifs for clathrin and endocytic regulatory proteins (15). Individual deletion of ENT1 and ENT2 genes has no functional effect, whereas cells with a double deletion (ent1Δ/ent2Δ or simply ΔΔ) are inviable (15). However, the endocytic function of Ent1/2p could be studied in ΔΔ cells bearing ent1 temperature-sensitive alleles (15). At the nonpermissive temperature, these cells displayed a marked defect in the endocytosis of the lipophilic dye FM4-64, which accumulated in punctate structures at the cell periphery. Also obvious was a defect in the organization of the actin cytoskeleton in which cortical actin patches were enlarged and improperly polarized (15). The role of cortical actin patches, which has been recognized for >20 years, has been elusive, but in fact mutations in many endocytic proteins lead to disruption of these structures, suggesting that their function is intimately linked to endocytosis (16). Recent studies using live cell imaging and electron microscopy have led to a breakthrough in understanding the temporal and spatial relationship between actin and endocytosis (17–19). These studies have revealed that endocytic regulatory proteins, including clathrin and Ent1/2p, assemble on the plasma membrane along with Las17p, the yeast homologue of WASP (Wiskott–Aldrich syndrome protein). In addition to initiating membrane deformation and curvature, these proteins also contribute to actin recruitment, which, through the actions of Las17p and the Arp2/3 complex, forms an actin patch, a conical, branched actin network rising out of the plasma membrane (18, 19). As the endocytic proteins anchor actin filaments to the invaginating membrane with new actin monomers added at the plasma membrane, the growing filaments push the endocytic complex off the membrane, contributing to membrane invagination (19). It is thus compelling that in the new study by Aguilar et al. (4), expression of the Ent1/2p ENTH domain alone is sufficient to rescue the viability phenotype and the perturbation of actin structures seen after epsin disruption. Thus, a single protein module contributes both to membrane curvature during endocytosis and to regulation of actin dynamics.
The phenotypic rescue seen with ENTH domains from Ent1/2p was also observed with the ENTH domain from rat epsin but not with the ENTH domains of Ent3p or Ent4p, demonstrating an evolutionarily conserved function specific to ENTH domains from epsin family members (4). Alignment of Ent1/2p ENTH domains with invertebrate and vertebrate orthologues revealed several invariant residues (Y100, T104, and E137) that contribute to a surface-exposed patch that is distinct from the binding site for PtdIns(4,5)P2 and is oriented toward the cytosol (Fig. 1). In a carefully controlled series of experiments, Aguilar et al. (4) demonstrated that the Ent1p ENTH domain (ENTH1) with Y100R and T104D mutations, when expressed off the endogenous promoter, was not able to complement the viability phenotype (4). Moreover, cortical actin patches were not polarized at buds, and actin cables were reduced or absent, despite the fact the ENTH1 mutants were properly expressed and folded and were able to bind inositols with similar affinity as ENTH1 WT (4). In contrast, ENTH1 with mutations that disrupt lipid-binding activity were able to partially complement the ΔΔ cells. Thus, the membrane binding and curvature activities of yeast epsins, which are linked to endocytosis, may contribute to cell viability, but the Y100/T104-based surface patch is absolutely critical for cell viability, actin organization, and polarity.
To identify the mechanisms linking the ENTH domain to cell polarity, Aguilar et al. (4) performed a two-hybrid screen with WT ENTH1 and identified all three known yeast Cdc42 GAPs: Rga1, Rga2, and Bem3 (4). Their interaction with the ENTH domain is direct, dependent on the Y100/T104-based surface patch, and necessary for viability and normal actin organization. Down-regulation of ENTH1 interactions with the GAPs in an ENTH1Y100R mutant strain leads to a striking decrease in Cdc42·GTP levels. Because activation of Cdc42 is a key signal for establishing an axis of polarity (1), the decrease in Cdc42·GTP likely forms the basis for the polarity phenotype. Thus, sequestration of Cdc42 GAPs into ENTH·GAP complexes provides the mechanism by which ENTH domains regulate cell polarity. Although it has been established that sites of endocytosis and exocytosis localize in close proximity during bud formation, epsins represent the first common regulatory link. Interestingly, in a search for Cdc42 effectors whose overexpression could bypass the ENTH1Y100R phenotype, the authors identified Gic1, Gic2, and Bem1, and a gic1Δ/gic2Δ double knockout strain phenocopies the cell polarity defects observed for ENTH1Y100R mutants (4). These effector proteins have previously been linked to cell polarity and are thought to recruit the formin Bni1 to the tip of the forming bud (1). Once localized and activated through its interaction with Cdc42, Bni1 controls nucleation and elongation of actin cables. The epsin ENTH domain thus clearly regulates establishment and maintenance of cell polarity.
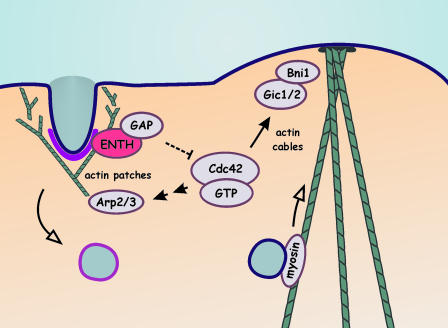
A model of the role of Ent1/2p is shown in Fig. 2. Under normal conditions of polarized growth, endocytic sites are established near to sites of secretion/bud formation. Ent1/2p is recruited to nascent endocytic sites, likely through interactions with PtdIns(4,5)P2, clathrin, and other components of the endocytic machinery (15). Through interactions with PtdIns(4,5)P2, the ENTH domain contributes to membrane destabilization and invagination (13). Binding of Cdc42 GAPs to the ENTH domain could prevent their access to Cdc42 or could down-regulate their catalytic activity, leading to localized activation of Cdc42 at bud sites. Activation of Cdc42 would influence polarized growth through the formation of actin cables. Cdc42·GTP can also contribute to activation of the Arp2/3 complex (20), thereby facilitating the formation of actin patches necessary for endocytosis. In both yeast and mammalian cells, sites of polarized cell growth often coincide with regions of secretion and endocytosis. Thus, epsins may be master regulators bringing together these key cellular functions.
Fig. 2.
Model of the potential roles of the ENTH domain of Ent1/2p. Clathrin and endocytic regulatory proteins (purple), including Ent1/2p (the ENTH domain alone is shown), localize to sites of endocytosis where they contribute to membrane deformation and curvature. By means of the actions of Arp2/3, actin (green) assembles into an actin patch that contributes to membrane invagination. The ENTH domains bind and sequester/inhibit Cdc42 GAPs, allowing for local activation of Cdc42. Through a Gic1/2/Bni1 pathway, actin cables are assembled that direct secretory vesicle in a myosin-dependent manner. Cdc42·GTP can also contribute to actin patch formation by signaling to activation of Arp2/3.
Conflict of interest statement: No conflicts declared.
See companion article on page 4116.
References
- 1.Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 2.Valdez-Taubas J., Pelham H. R. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Legendre-Guillemin V., Wasiak S., Hussain N. K., Angers A., McPherson P. S. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 4.Aguilar R. C., Longhi S. A., Shaw J. D., Yeh L.-Y., Kim S., Schön A., Freire E., Hsu A., McCormick W. K., Watson H. A., Wendland B. Proc. Natl. Acad. Sci. USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kay B. K., Yamabhai M., Wendland B., Emr S. D. Protein Sci. 1999;8:435–438. doi: 10.1110/ps.8.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenthal J. A., Chen H., Slepnev V. I., Pellegrini L., Salcini A. E., Di Fiore P. P., De Camilli P. J. Biol. Chem. 1999;274:33959–33965. doi: 10.1074/jbc.274.48.33959. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Fre S., Slepnev V. I., Capua M. R., Takei K., Butler M. H., Di Fiore P. P., De Camilli P. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 8.Yamabhai M., Hoffman N. G., Hardison N. L., McPherson P. S., Castagnoli L., Cesareni G., Kay B. K. J. Biol. Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 9.Hussain N. K., Yamabhai M., Ramjaun A. R., Guy A. M., Baranes D., O’Bryan J. P., Der C. J., Kay B. K., McPherson P. S. J. Biol. Chem. 1999;274:15671–15677. doi: 10.1074/jbc.274.22.15671. [DOI] [PubMed] [Google Scholar]
- 10.Hyman J., Chen H., Di Fiore P. P., De Camilli P., Brunger A. T. J. Cell Biol. 2000;149:537–546. doi: 10.1083/jcb.149.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford M. G., Pearse B. M., Higgins M. K., Vallis Y., Owen D. J., Gibson A., Hopkins C. R., Evans P. R., McMahon H. T. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 12.Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 13.Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 14.Stahelin R. V., Long F., Peter B. J., Murray D., De Camilli P., McMahon H. T., Cho W. J. Biol. Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 15.Wendland B., Steece K. E., Emr S. D. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engqvist-Goldstein A. E., Drubin D. G. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 17.Huckaba T. M., Gay A. C., Pantalena L. F., Yang H. C., Pon L. A. J. Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodal A. A., Kozubowski L., Goode B. L., Drubin D. G., Hartwig J. H. Mol. Biol. Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaksonen M., Toret C. P., Drubin D. G. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Lechler T., Jonsdottir G. A., Klee S. K., Pellman D., Li R. J. Cell Biol. 2001;155:261–270. doi: 10.1083/jcb.200104094. [DOI] [PMC free article] [PubMed] [Google Scholar]